Abstract
Human embryonic stem cell (hESC) neural differentiation models have tremendous potential for evaluating environmental compounds in terms of their ability to induce neurodevelopmental toxicity. Genomic based-approaches are being applied to identify changes underlying normal human development (in vitro and in vivo) and the effects of environmental exposures. Here, we investigated whether mechanisms that are shared between hESC neural differentiation model systems and human embryos are candidate biomarkers of developmental toxicities for neurogenesis. We conducted a meta-analysis of transcriptomic datasets with the goal of identifying differentially expressed genes that were common to the hESC-model and human embryos. The overlapping NeuroDevelopmental Biomarker (NDB) gene set contained 304 genes which were enriched for their roles in neurogenesis. These genes were investigated for their utility as candidate biomarkers in the context of toxicogenomic studies focused on the effects of retinoic acid, valproic acid, or carbamazepine in hESC models of neurodifferentiation. The results revealed genes, including 13 common targets of the 3 compounds, that were candidate biomarkers of neurotoxicity in hESC-based studies of environmental toxicants.
Keywords: human embryonic stem cells, human development, neuron, neurotoxicity, genomics, meta-analyses, environmental exposures, neurodevelopment
1. Introduction
Environmental factors may underlie a variety of developmental anomalies, the majority of which arise during the early stages of pregnancy. Driven by the desire to reduce costs and animal usage in developmental toxicology studies, the transition from in vivo to in vitro testing is gaining momentum [1, 2]. Cell culture models are an attractive alternative that enable high throughput evaluation of environmental factors that may contribute to developmental toxicity. Of note, less than 1% of the 80,000 Environmental Protection Agency (EPA)-registered compounds have been properly tested for their potential to cause neuro (developmental) toxicity [3]. Therefore, established in vitro models of neurodevelopment are needed to study the multitude of prevalent compounds with unknown toxicities.
Human embryonic stem cells (hESCs), which can be differentiated into neurons, have been proposed as screening tools for neurodevelopmental toxicity testing [4, 5]. There are well-recognized differences in embryonic development [6], ESC properties [7] and chemical-sensitivity [8] between rodents and humans. Therefore, hESC models offer advantages over rodent ESCs for human hazard assessment, enabling extrapolation of data to our species. Several multi-step protocols have been established to induce hESC neural differentiation. For example, in a commonly used approach, pluripotent hESCs are suspended as aggregates (embryoid bodies (EBs)) in serum-containing medium, which initiates spontaneous differentiation of the three germ layers. These changes model the initial steps of differentiation that occur during gastrulation [9].
Added during the EB stage, defined media and growth factor cocktails are used to specify ectodermal, mesodermal or endodermal fates. As to neuronal development, growth factors/cytokines and small molecules that work in specific pathways (e.g., BMP2 and/or SMAD [10] inhibitors, LIF [11] and retinoic acid [RA] [12]) are added to direct an ectodermal fate. EBs are placed on select substrates [13], which promote neural rosette (NR) formation. Patterned NRs contain neural precursor cells (NPCs) that express markers that are upregulated in the neural tube during human embryogenesis [14, 15]. Then multipotent NPCs are differentiated into cells of the neural lineage, and additional factors added to influence patterning and the emergence of neuronal subtypes [16]. The addition of environmental compounds at one or more stages of the hESC neural differentiation protocol enables testing of their effects on the major developmental transitions [17].
Genomics-based approaches are used to investigate the underlying molecular changes during human embryogenesis at cellular and organismic levels. For example, investigators have used hESC models and global transcriptomic approaches to profile neuronal progenitor states [18]. These studies suggest multiple genes and related-pathways are modulated during this process. Along these lines, using the UCSF4 hESC line, our group profiled pluripotent and NP cells with the goal of identifying key genes/pathways that are expressed in this model of human development [19]. At an organismal level, transcriptomic technologies have been employed at specific time periods during human embryonic development, ranging from the first cell divisions to the later stages of organogenesis [20–22]. The data provide valuable insights into the dynamic molecular transformations occurring at select stages. These investigations also enable cross-species analyses of shared mechanisms [23].
For assessing developmental toxicity of environmental chemicals, genomic responses, i.e., toxicogenomic approaches, are more sensitive and compound-specific than classical morphological endpoints [24]. Furthermore, toxicogenomic effects can be evaluated across models. Specifically, responses in vitro and in vivo can be directly compared, providing context for using cell, organ and embryo culture models for risk assessment [25, 26]. Therefore, genomics-based approaches may add tremendous value to standardized in vitro methodologies in screening for neurodevelopmental toxicity.
Recent toxicogenomic studies used a standardized (mouse) ESC differentiation model to study the developmental toxicity of various compounds [27]. The results identified genes that are differentially expressed as a function of differentiation (neural or cardiac) in culture and highlighted the effects of the test compounds on these patterns [28–31]. These studies support the notion that the altered expression of molecules that are coordinated with normal differentiation in vitro can successfully predict the potential developmental toxicity of environmental compounds. While these reports point to molecular correlates for the assessment of developmental toxicity, uncertainty remains over their relevance and specificity as applied to human development.
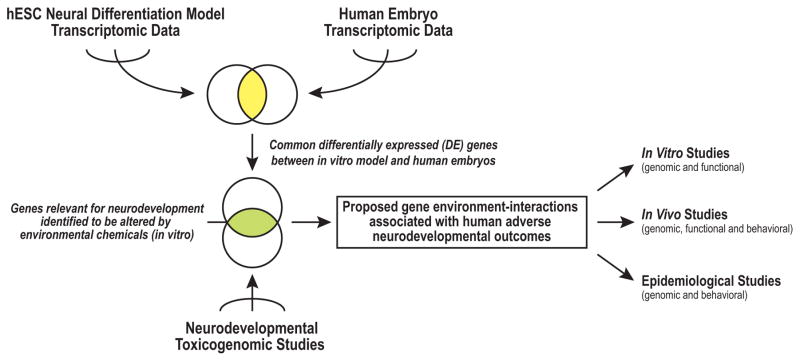
Over the past decade, researchers have produced volumes of genomic data under diverse conditions using human and non-human, in vivo and in vitro models. Submitted to repositories, such as NCBI (Gene Expression Omnibus (GEO)) [32] or EBI (ArrayExpress) [33], these publicly available datasets are a valuable resource for post hoc analyses. Taking advantage of this resource, recent comparison studies provide potential strategies for determining: 1) commonalities between in vitro and in vivo models [23]; and 2) the cross-model consistency of molecular responses to environmental exposures [34, 35]. In this context, we devised an analysis framework that we used to integrate human transcriptomic datasets for identifying biomarkers of neurodevelopmental toxicity (Figure 1). We conducted a meta-analysis of in vitro differentiation data, specifically for the hESC neural differentiation model. These transcriptomic datasets included results of experiments completed in our laboratory [19] and previously published studies. We compared the results to transcriptomic datasets that were acquired during the initial stages of human embryo development, including the period of neurogenesis. Thus, we identified a core set of differentially expressed (DE) genes which change during the early stages of human neurodevelopment, common to both in vitro and human embryos. In preliminary verification experiments, we demonstrated the utility of this gene set for studying the effects of environmental chemicals in hESC models of this process.
Figure 1. An analysis framework for integrating human transcriptomic datasets to identify biomarkers of neurodevelopmental toxicity.
Meta-analyses of transcriptomic data generated in human embryonic stem cell (hESC) neural differentiation models revealed signatures at the various stages, which were compared to datasets that interrogated gene expression at specific days of human embryonic development, including neurogenesis. Differentially expressed (DE) genes that were shared between the two models were chosen as candidate biomarkers of neural development. They were applied to toxicogenomic studies that employed hESCs to identify potential gene-environment interactions associated with adverse neurodevelopmental outcomes. These results could inform the design of future studies that use in vitro and in vivo models or investigate effects at a population level.
2. Material and Methods
2.1 Transcriptomic Data Collection
Datasets were acquired via public NCBI (GEO) and EBI (ArrayExpress) repositories. The criteria for selection of transcriptomic datasets relevant to hESC neural differentiation were as follows. Search terms were defined as “human”, “embryonic stem cell”, “differentiation”, and “neural” or “neuron”. A minimum of 2 biological replicates per group and 6 microarray assays (Affymetrix or Illumina array platforms) was required. Fifty-two GEO and 8 ArrayExpress studies met these criteria. Secondary filters eliminated studies that used genetically modified lines, non-neural-related differentiation routes, and non-human cell lines. In total, we identified four unique hESC neural differentiation datasets [36–39]. We added a transcriptomic dataset that was generated in our laboratory that compared the UCSF4 hESC line in a pluripotent state and their NPC derivatives [19]. Using a similar approach, we searched the two repositories to obtain human embryo transcriptomic datasets. We selected studies that analyzed at least 6 stages and a minimum of 3 biological replicates per group. In total, 3 datasets from different stages of human embryonic development met these criteria [20–22].
2.2 Data Processing and Gene Expression Comparisons
Each transcriptomic dataset was downloaded and individually processed using the Affymetrix Expression Console [40] and Transcriptome Analysis Console (TAC) [41] software packages or BRB Arraytools (for Illumina arrays) [42]. Raw values were normalized via the Robust Multi-array Average (RMA) algorithm [43]. One-way-ANOVA (OWA) was independently applied within each of the five in vitro and three human embryo datasets to determine significance of differentially expressed (DE) genes across time. Average fold change (FC) values between hESCs and their derivatives (in vitro) or the earliest embryonic stage vs. later periods (human embryos) were determined. Datasets were annotated using the Affymetrix TAC or Illumina databases (10/1/14). For comparisons across studies, we used the Official Gene Symbol (OGS). In the case of multiple probes per gene, the one with the lowest p-value, i.e., most significant changes over time, was used for comparison purposes. Datasets were merged using the R statistical package [44] via the OGS identifier. Significantly DE genes were defined as p ≤ 0.01 OWA and FC ≥ 1.5 (absolute). To describe regulation over time across groups, Pearson correlation (PC) coefficients were calculated for each gene within each dataset across time using time in culture or human embryo stage as a categorical variable. The comparability of DE genes for the five in vitro studies was determined by making iterative comparisons of each dataset with the other four. Hierarchical clustering of FC values was computed using average linkage and Euclidean distance (TIGR MEV [45]). Finally, the DE genes that emerged from the five in vitro studies were used to identify similar expression patterns over time in the three human embryo datasets. Similar patterning of common DE genes between the two models was determined by comparing PC coefficients [in vitro (median of the five datasets) vs. human embryo]. We determined the degree of enrichment of DE-IVND genes as compared to the DE genes identified within each independent dataset by conducting a Z-test for two population proportions [ ]. The results enabled identification of the timed series that most closely mirrored the stages modeled in culture i.e., early neurogenesis, which was termed the NeuroDevelopmental Biomarker (NDB) set.
2.3 Characterization of the NeuroDevelopmental Biomarker Set
Functional enrichment of Gene Ontology (GO) Biological Processes (Level 4) was evaluated using DAVID [46]. For the entire dataset, GO terms containing ≥7 DE genes, p ≤ 0.0005 and fold enrichment (FE) ≥ 1.5 were considered significant. Additionally, corresponding p-values and FE scores were determined for upregulated or downregulated genes within individual GO terms. To describe relative overrepresentation across GO terms, an enrichment index was calculated [− log (p – value) * FE] as described previously [25]. Terms were grouped based on GO classifications (http://geneontology.org) into related themes. Clustering of FC values associated with genes involved in CNS development were investigated by using TIGR MEV as described above. Using OPOSSUM [47], we identified enriched motifs within promoter regions, defined as 1000 base pairs upstream of our gene of interest. Overrepresented motifs (number of gene targets with enriched motif ≥2 and Fisher score ≥4) were determined for up or downregulated NDB genes.
2.4 Conservation of the NeuroDevelopmental Biomarker Set in Human, Rat, and Zebrafish
We assessed patterns of expression of the NDB set in two models commonly used for neurodevelopmental toxicological studies. We obtained whole embryo rat [23] and zebrafish datasets [48] (Supplemental Table 1). As described above, datasets were processed, annotated, and merged with the NDB set (and related human data) using the OGS identifier. Significantly DE genes in these studies were also defined as p ≤ 0.01 OWA and (absolute) FC ≥ 1.5. To assure human and zebrafish homology of common DE-NDB genes, we added an additional filter—identifiers designated as homologs via NCBI and ≥ 60% protein similarity (BLAST, [49]). Common trends in regulation between human, rat, and zebrafish systems were evaluated by comparing PC coefficients.
2.5 Applying the NDB Set as a Functional Correlate for Toxicogenomic Studies
Next, we evaluated the expression of the NDB set in two hESC-toxicogenomic studies (Supplemental Table 2). These datasets were selected [50, 51] from the GEO or ArrayExpress databases according to the following criteria: 1) use of a hESC model; 2) exposure during hESC neural differentiation; 3) n ≥ 3 concentrations tested; and 4) n ≥ 2 replicates for each concentration group. The data were generated by using Affymetrix platforms (HG-U133_Plus_2 or HG-U133_PM+). In one of the studies [50], which used a neural rosette hESC differentiation model, retinoic acid (RA) effects on the transcriptome were investigated after eight days exposure (0.002– 2 μM). In the other study [51], which applied a “rapid, standardized” hESC neural differentiation model system, the effects of two anticonvulsants and known developmental toxicants [52, 53], valproic acid (VPA, 100–1000 μM) and carbamazepine (CBZ, 33–333 μM), on the transcriptome were investigated following 1 or 7 days exposure. As described above, these datasets were independently normalized (RMA) and annotated. OWA was applied across all exposure groups (i.e,, compound and vehicle control) and duplicate probes were managed by using the probe demonstrating the most significant response to RA, VPA, or CBZ as compared to the vehicle control. We examined only the 7 day exposure group for VPA and CBZ due to similarity in exposure duration with the RA study. Average FC values were determined by calculating the log2 ratio between the average intensities of each exposure group vs. the average intensities of the vehicle control. In this context, DE genes were defined as having a significant chemical response of p ≤ 0.01 (OWA) and an absolute FC ≥ 1.5. We conducted a Z-test for two population proportions to determine DE-NDB gene enrichment as compared to all DE genes following exposure to each compound. We calculated the average FC in gene expression response of the NDB set as well as two subsets of the NDB set, DE genes and non-DE genes, by using the absolute mean of log2 FC values. To determine the overall significance of the NDB set as a functional correlate, we conducted a Fisher’s combined probability test for the complete NDB set across all concentrations of each compound tested. Cellular and molecular phenotypes associated with each concentration of the three compounds were described as previously reported [50, 51, 54]
3. Results
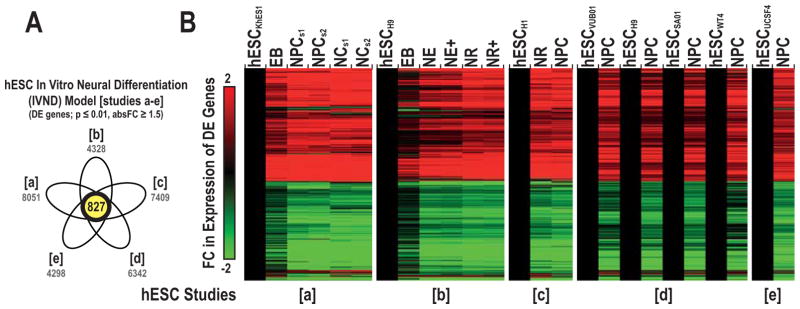
Based on our established criteria (see methods), we acquired data from five neural differentiation transcriptomic studies (a–e) that employed eight hESC lines (Table 1). These datasets were from the GEO or ArrayExpress repositories, which included a transcriptomic analysis completed in our laboratory that compared hESCs and NPCs. Each had a unique experimental design and focused on a different hESC line with the exception of H9, which was used twice (b and d). Alignment via the OGS enabled evaluation of 19,026 genes across the five studies. Each study was individually normalized and processed before OWA was applied to determine the number of significant DE genes at various time points in each study. Iterative comparisons of each dataset with the other 4 revealed an average overlap of 78% (range of overlap: 66% – 94%) in terms of common DE genes (p ≤ 0.01, absolute FC ≥ 1.5). In total, we identified 827 DE genes that were shared among the five studies (Figure 2), of which 89% were similarly regulated (up or down) across time.
Table 1. hESC neural differentiation model: published transcriptomic studies.
We compared a dataset generated in our lab using the hESC neural differentiation model with additional datasets obtained from GEO or ArrayExpress repositories. Within each dataset, significantly DE genes (DEstudy) were defined using a cutoff of p ≤ 0.01 and absolute FC ≥ 1.5. The total number of DE genes and the percentage of DE genes vs. unique targets evaluated are shown for each study. The comparability of DE genes for the five in vitro studies was determined by making iterative comparisons of each dataset with the other four. The % Overlap indicated the percentage of common DE genes between each dataset versus the common DE genes of the other four datasets. Abbreviations: hESC (human embryonic stem cell), EB (embryoid body), NE (primitive neural ectoderm), NR (neural rosette), NPC (neural precursor cell), NC (neuron cell), s (stage), and + (with FGF2).
| ID | Study # | hESC | Samples (n) | Platform | DEstudy genes (#, %) | % Overlap | Ref |
|---|---|---|---|---|---|---|---|
| a | GSE39144 | KhES1 | ESC, EB, NPCs1, NPCs2, NCs1, NCs2 (n=3/group, n=18 arrays) | Affymetrix HG- U133_Plus_2 | 8051 (36%) | 85% (827/978) | [38] |
| b | GSE9940 | H9 | ESC, EB, NE, NE+, NR, NR+ (n=3/group, n=18 arrays) | Affymetrix HG-U133_Plus_2 | 4328 (19%) | 71% (827/1161) | [39] |
| c | GSE56906 | H1 | ESC, NR, NPC (n=2/group, n=6 arrays) | Affymetrix HG-U133_Plus_2 | 7409 (33%) | 94% (827/879) | [37] |
| d | GSE34201 | VUB01, H9, SA01, WT4 | ESC, NPC (n=3/group, n=24 arrays) | Illumina HumanWG-6 v3.0 expression beadchip | 6342 (17%) | 72% (827/1154) | [36] |
| e | GSE74378 | UCSF4 | ESC, NPC (n=3/group, n=6 arrays) | Affymetrix Human Gene 2.0 ST Array | 4298 (17%) | 66% (827/1253) | [19] |
Figure 2. Common DE genes underlying hESC neural differentiation.
The shared overlap in DE genes over time across five independent hESC neural differentiation datasets (Venn diagram, studies a–e) (A). Four of the hESC datasets were obtained via the GEO or ArrayExpress repositories and one was generated in our laboratory. The total number of DE genes identified in each study is noted in grey. Hierarchical clustering of the 827 common DE genes (B). This subset was termed the in vitro neural differentiation (IVND) gene set. Average FC log2 ratios were the difference in expression between each neural derivative and their respective hESC group. Abbreviations: hESC (human embryonic stem cell), EB (embryoid body), NE (primitive neural ectoderm), NR (neural rosette), NPC (neural precursor cell), NC (neuron cell), s (stage) and + (with FGF2).
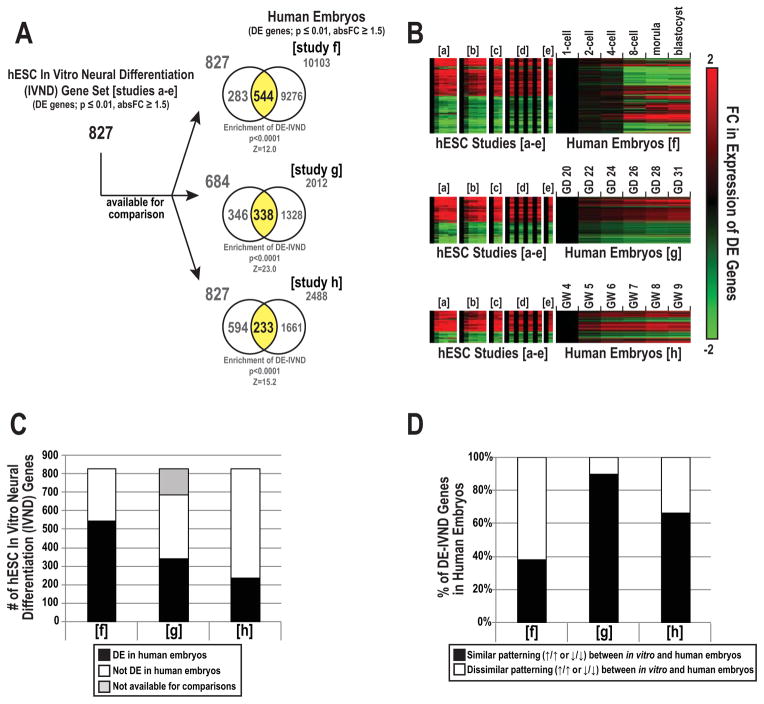
We used the in vitro neural differentiation (IVND) common gene set to interrogate the early stages of human embryonic development. We acquired published datasets that investigated the transcriptome during three critical periods: 1-cell to blastocyst stage (study f), early neurogenesis (study g), and late organogenesis (study h; Table 2). Pairwise comparisons with IVND genes showed an overlap of 66% (544 genes, study f), 49% (338 genes, study g), and 28% (233 genes, study h) (p ≤ 0.01, FC ≥ absolute 1.5) (Figure 3A–C). Based on expected distributions of DE genes, the three datasets were enriched for DE-IVND genes (p ≤ 0.0001). As to the expression patterns of DE-IVND genes over time (e.g., up vs. down) between models, hierarchical clustering analysis revealed significant variations in similar patterning (black bar) between in vitro and human embryos: 38% (208 genes, study f), 90% (304 genes, study g), and 67% (155 genes, study h) (Figure 3D). Thus, the results of this analysis suggested that gene expression patterns during hESC neural differentiation most closely modeled the early stages of neurogenesis in the embryo as a whole (study g).
Table 2. Early human embryos: published transcriptomic studies.
We compared DE genes in the hESC neural differentiation model with human embryo transcriptomic studies obtained from GEO or ArrayExpress repositories. Within each dataset, significantly DE genes (DEstudy) were defined using a cutoff of p ≤ 0.01 and absolute FC ≥ 1.5. The total number of DE genes and the percentage of DE genes vs. unique targets evaluated are shown for each study. Abbreviations: CNS (central nervous system), GD (gestational day), GW (gestational week).
| ID | Study ID | Time | Samples (n) | CNS Development Stage | Platform | DEstudy genes (#, %) | Ref |
|---|---|---|---|---|---|---|---|
| f | GSE18290 | <GW 1 | human embryo: 1-cell, 2-cell, 4-cell, 8-cell, morula, blastocyst (n=3/group, n=18 arrays) | Pre-CNS development | Affymetrix HG- U133_Plus_2 | 10103 (45%) | [22] |
| g | GSE18887 | GW 3–4 | human embryo: (GD 20–32) (n=3/group, n=18 arrays) | Early neurogenesis | Affymetrix Human Genome U133A Array | 2012 (15%) | [21] |
| h | GSE15744 | GW 4–9 | human embryo: (GW 4–9) (n=3/group, n=18 arrays) | Late organogenesis | Affymetrix HG- U133_Plus_2 | 2488 (11%) | [20] |
Figure 3. Overlap of DE genes between the in vitro neural differentiation set and human embryonic transcriptomes at various time points.
We used the in vitro neural differentiation (IVND) common gene set to interrogate the human embryo transcriptome during the 1-cell to blastocyst (study f), early neurogenesis (study g), and late organogenesis (study h) stages. The shared overlap in DE genes between the IVND set and each of the three human embryo studies (A). Z-scores and p-values corresponding to enrichment of DE-IVND genes within each human embryo study are listed below each respective Venn. Expression of common DE genes over time (B). FC ratios (log2) were expressed as comparisons between hESCs and their derivatives or the earliest embryonic stage vs. later periods (human embryos). The distribution of DE and non-DE-IVND genes in each human embryo study (C). The shared overlap in the number of DE-IVND genes with similar (or dissimilar) gene expression patterns, i.e., up or downregulated over time, between the IVND set and human embryo studies (D). Abbreviations: gestational day (GD) and gestational week (GW).
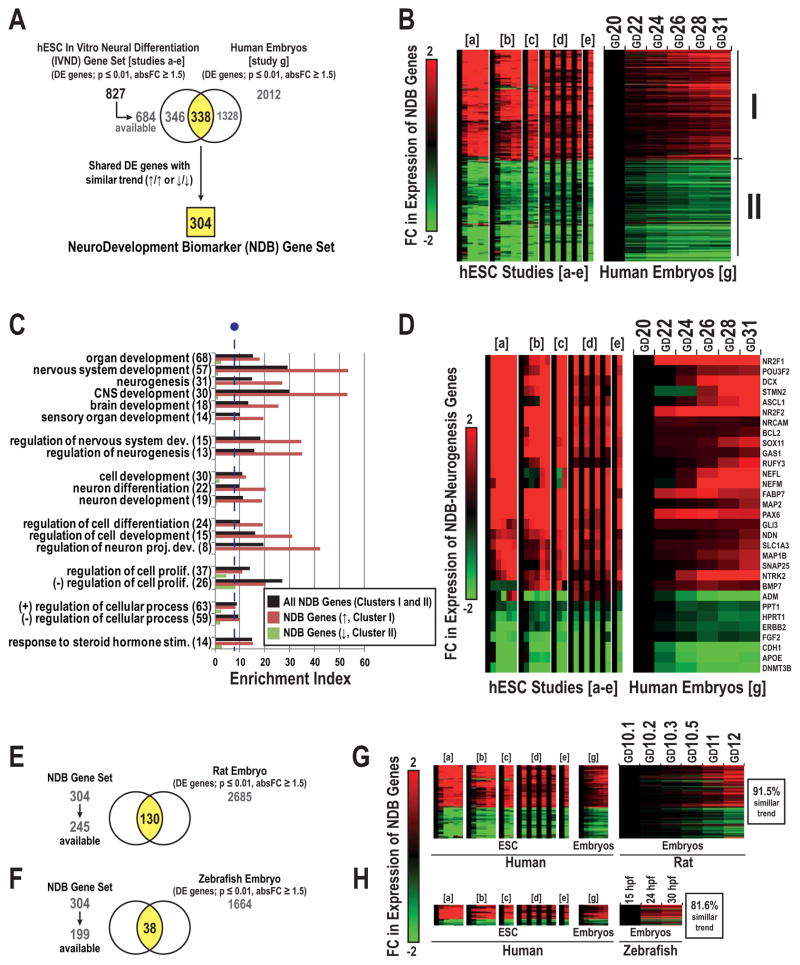
Additional analyses focused on the 304 DE genes that were similarly regulated in the IVND dataset and study g (Figure 4A), which we termed the “NeuroDevelopmental Biomarker” (NDB) set. In general, gene expression over time was either up- (Cluster I) or downregulated (Cluster II) rather than showing more complex patterns (Figure 4B). Within the NDB set, we observed significant enrichment of genes involved in central nervous system (CNS), neurogenesis, and neuron development related terms (Figure 4C), which were confined to Cluster I. In total, 31 DE genes were associated with the term “neurogenesis”; 23 were upregulated (e.g., NR2F1, POU3F2, DCX, STMN2, ASCL1, NR2F2, NRCAM, NEFL, NEFM, MAP2, PAX6, GLI3, and NTRK2) and 8 were downregulated (e.g., PPT1, HPRT1, CDH1, DNMT3B) over time (Figure 4D). Motif enrichment analysis revealed transcription factor binding sites that were overrepresented in the promoter regions of NDB genes, including PAX6 and LHX3 which were unique to Cluster I and POU5F1 which was unique to Cluster II (not shown).
Figure 4. Characterization of the NeuroDevelopmental Biomarker set.
We identified 304 DE genes that were similarly regulated in the IVND dataset and the human embryo transcriptome (early neurogenesis, study g) (A). We defined this gene subset as the NeuroDevelopmental Biomarker (NDB) set. Hierarchical clustering of the expression of the NDB set across developmental time in hESC neural differentiation models and human embryos (B). Cluster I and II correspond to subsets of up or downregulated genes over time. Enrichment analysis of GO Biological Processes for the complete NDB set and Clusters I or II (C). The enrichment index was calculated via -log (p-value) * FE. The line break (purple) indicates the cutoff for significance (p ≤ 0.0005, fold enrichment ≥ 1.5). Hierarchical clustering of the expression of NDB genes associated with the term “neurogenesis” (31 total) over time (D). NDB-genes that were DE in rat (130 total, E) or zebrafish embyros (38 total, F). Expression of NDB genes during stages of rat neural tube development (G) and zebrafish neurogenesis (H). For all heatmaps, FC ratios were expressed as comparisons between hESCs and their derivatives (in vitro) or the earliest embryonic stage vs. later stages (embryos). Abbreviations: GD (gestational day) and (hpf) hours post-fertilization.
Using published datasets, we compared the expression of the NDB set in human (in vitro and embryos) with the rat or zebrafish during embryonic neural development (Supplemental Table 1). In total, 130 genes of the NDB set were also DE in the rat (Figure 4E). Fewer of the NDB genes (38) were DE in the zebrafish dataset (Figure 4F). With regard to the DE-NDB gene expression patterns, 91.5% and 81.6% were shared between human with rat or zebrafish, respectively (Figure 4G, H). These analyses suggested that the NDB gene set included multiple conserved master regulators of programs that promote early CNS and neuron development through established transcription factor networks.
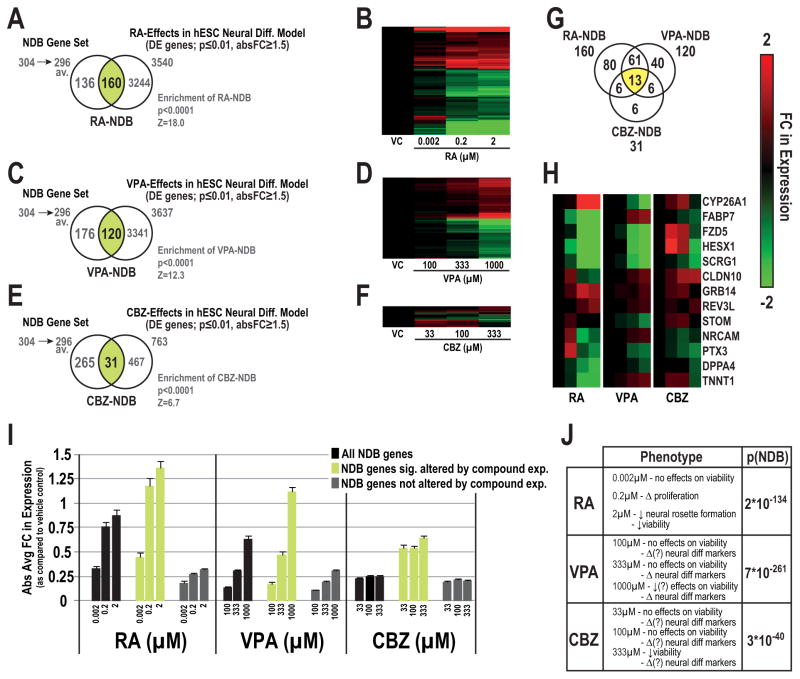
As a proof-of-principle, we applied our NDB genes to toxicogenomic datasets, which investigated the concentration-dependent effects of RA [50], VPA or CBZ [51] exposures for ~1 week on neurodevelopment using various hESC neural differentiation models (Supplemental Table 2). Overall, we observed significant concentration-dependent effects in the NDB gene set for each of the three compounds (enrichment score, p ≤ 0.0001; Figure 5). Over 50% of the NDB set (160 genes) were altered upon RA treatment (p ≤ 0.01, absolute FC ≥ 1.5) (Venn, Figure 5A, heat map 5B). In similar comparisons, VPA and CBZ significantly altered NDB-genes, but to a lesser degree than RA. In total, we observed 120 and 31 NDB genes to be significantly altered by VPA or CBZ, respectively (Figure 5C–F). In general, the majority of NDB genes altered by RA or VPA displayed monotonic dose-dependent relationships. Thirteen NDB genes were identified as significantly altered by the three exposures (Figure 5G), which included CYP26A1, FABP7, FZD5, HESX1, SCRG1, CLDN10, NRCAM, and PTX3 (Figure 5H), suggesting particular targets that could be especially vulnerable to neurotoxicants. Absolute average FC values of chemical effects on all NDB genes, (DE and non-DE genes) were concentration-dependent (Figure 5I). Maximum effects, peaking at a log2 FC response of 1.4 (~2.6 fold), occurred at 2 μM with RA as compared to VPA, which peaked at a log2 FC response of 1.1 (2.2 fold) with 1000 μM VPA. As compared to RA and VPA, CBZ concentration-dependent effects on NDB genes, in terms of absolute FC (Figure 5I) and overall significance (p(NDB); Figure 5J), were less pronounced. These observations were associated with differences in cellular or molecular phenotypic changes that were associated with the three compounds (Figure 5J).
Figure 5.
Application of the NDB set to toxicogenomic studies investigating the effects of developmental toxicants in hESC neural differentiation models. Concentration-dependent effects on NDB genes whose expression was significantly altered by retinoic acid (RA-NDB; Venn, A, heatmap, B), valproic acid (VPA-NDB set; C, D) or carbmazepine (CBZ-NDB set, E, F) in hESC models of neural differentiation. Z-scores and p-values corresponding to enrichment of DE-NDB genes for each compound are listed below each respective Venn diagram. Distribution of shared and compound-specific NDB genes for the three compounds (G). Clustering analysis of shared NDB genes significantly altered by the three compounds (H). Absolute average FC values of all NDB genes together with DE- and non DE-NDB genes by compound (I). FC values are displayed for each concentration in relation to their respective vehicle control with standard error bars (log2 scale). Cellular and/or molecular phenotypes described by Colleoni et al.[50] or Schulpen et al. [51, 54] and Fisher’s combined probability test p-values (p[NDB]) for the complete NDB set across all concentrations (Table, J). Abbreviations: available for comparisons (av.).
4. Discussion
hESC models are proposed as screening tools to assess environmental chemicals for neurodevelopmental toxicity. Defining in vitro functional correlates for in vivo processes can enable data extrapolation, hazard identification, and risk assessment. Thus, in this study, following our devised framework (Figure 1), we integrated human transcriptomic datasets to identify biomarkers of neurodevelopmental toxicity for application in the hESC neural differentiation model. In a four-step process, we: 1) identified common DE genes associated with hESC neural differentiation; 2) examined the expression of these genes during the initial stages of human development; 3) characterized a subset of these genes postulated as essential for hESC neuronal differentiation and/or the early stages of human neurogenesis; and 4) demonstrated the utility of this gene set for studying the effects of environmental chemicals on neurodevelopment in vitro. Although we used this data analysis pipeline for a very specific purpose, the general experimental strategy could be broadly applied to study the effects of chemicals on many other developmental processes.
In independent studies using diverse growth conditions and cell lines, transcriptomic-based approaches have been applied to profile the underlying transitions that comprise hESC-neural differentiation. In this study, we identified DE genes associated with this process that were common to five studies, including one that was performed in our laboratory. Overall, our results suggested that despite differences in experimental conditions there were conserved patterns of gene expression. Using this list of DE genes (the IVND set), we investigated their expression patterns in the human embryo at three critical intervals beginning at the 1-cell stage. In general, genes that were regulated during hESC neural differentiation were also modulated during human embryogenesis. However, the extent of overlap was stage-specific. Our results suggested that transcriptional changes underlying hESC neural differentiation mirrored most closely early neurogenesis—the intended period of representation of this in vitro model. We termed the shared genes as the NDB set. These findings agree with previous studies demonstrating that hESC neural differentiation models capture aspects of neural induction and patterning on molecular and cellular levels. For example, hESC-derived neural rosettes resemble cell arrangements observed in the neural tube and developing neocortex. This is consistent with the fact that these structures contain gradients of differentiated cell types, with less differentiated neural cells (NPCs) in the center and more mature cells at the basal surface [55].
The NDB set contained many molecules that are involved in human neurodevelopment, including well-documented master regulators of ectoderm cell fate (e.g., NANOG, PAX6, POU5F1), neuronal differentiation (e.g., NTRK2, POU3F2, STMN2), microtubule and filament organization (e.g., NEFL, TUBA1A, DCX, MAP1B, MAP2), neural adhesion molecules (e.g., CDH1, CDH3, CDH7, NCAM1, NRCAM) and genes implicated in complex neurobehavioral functions (e.g., AMMECR1, ASCL1, IQGAP1, SNAP25) (Supplemental Table 5). In all, ~20% of the NDB set were linked with GO terms related to “Nervous System Development” and 10% with “Neurogenesis”. Our analysis also identified molecules whose functions during CNS development are not yet understood. Specifically, for ~30% of our NDB set, we identified, via PubMed [56] searches, ≤ 3 articles associated with a particular gene as related to CNS development or neural differentiation. For example, it is possible that transcriptional regulators, SCML1 [57] and ZFHX4 [58] may be involved in these processes based on their expression in the developing mammalian brain and suspected roles in other biological processes. Thus, it could be interesting to explore their actions in this context. Conversely, this study highlighted differences in gene expression between the two models, which are expected due to the lack of complexity of the in vitro model, which has other advantages in terms of tractability. While our results suggested a high level of correlation between the hESC-neural differentiation model and the earliest stages of neurodevelopment in human embryos, molecular changes in the in vitro model also correspond to other transition periods in embryonic development. For example, 233 IVND genes were differentially regulated across late organogenesis; 155 displayed similar expression patterns in the hESC model (Figure 3). This subset included genes with roles in nervous system development (e.g., ELAVL3, ST8SIA2, DCX, MAP2, STMN2), suggesting in vitro molecular correlates for later stages of human neural development.
Over the past decade, toxicogenomic signatures have been explored as potential indicators of developmental toxicity. However, an all-defining “developmental toxicity” signature has yet to be discovered due to the complexities of cell type-specific responses to environmental chemicals. Therefore, recent attempts have been made using models of individual processes with an emphasis on identifying genes that change during normal development, and subsequently, the compounds that directly impact their expression in relationship to toxicity. For example, using a set of 29 genes associated with (mouse) ESC differentiation, known neurodevelopmental toxicants were classified with a success rate of 84% [31]. These studies and others provide evidence that genes involved in ESC differentiation are useful biomarkers for testing the effects of compounds that are potentially toxic at critical developmental stages. Moving forward, we anticipate that our strategy, which coalesces hESC and human embryo data, may improve this general approach by identifying common elements between models, thereby establishing relevant targets for in vitro modeling.
As a proof of principle, we analyzed expression of the NDB set in two toxicogenomic studies. One determined the effects of RA, a classic neurodevelopmental toxicant and morphogen, in a hESC neural rosette model, and the other assayed the effects of VPA and CBZ, anticonvulsants and known teratogens, in a hESC neural differentiation model. Our results suggested that the NDB set is useful for detecting neurodevelopmental toxicity for the three compounds. RA and VPA exposures altered the expression of ≥ 40% of the genes in the NDB set. Effects were dose-dependent and observed at concentrations that failed to significantly elicit cellular phenotypic changes. In general, CBZ effects were less pronounced, but still significant. Differences in the response among compounds may be due to differing modes-of-action or other experimental factors. These observations are supported by evaluation of individual NDB targets previously altered by environmental exposures at levels that perturb hESC neural differentiation. For example, VPA or lead both reduce PAX6 expression [17, 59], and methylmercury decreases expression of NCAM1 and MAP2 [60]. Other genes, such as ASCL1, have also been investigated as biomarkers of neurodevelopment in environmental studies using NPCs [61]. Our initial validation of the NDB set by applying this metric to pre-existing toxicogenomic studies suggested its potential promise as a correlate of neurodevelopmental toxicity in hESC neural differentiation models.
5. Conclusions
While our initial results are promising, the application of the NDB set to future toxicogenomic studies will be key to defining the specificity and sensitivity of these biomarkers and their potential to predict adverse neurodevelopmental outcomes. As additional genomic datasets are generated and incorporated, our framework will become more useful for identifying the gene targets in hESC models that are relevant to human developmental exposures. Specifically, we anticipate the acquisition of more quantitative (e.g., RNA-seq) and complementary data (e.g., bisulfite-seq) in the context of detailed developmental time-course studies. Many in vitro models of developmental transitions have been proposed as alternatives for in vivo analysis of developmental toxicity [62]. As we begin to incorporate data from these experimental systems into the testing paradigm, endpoints that unite in vitro and in vivo models—shared anchors—will be particularly useful for predicting adverse neurodevelopmental outcomes.
Supplementary Material
Research Highlights.
Identified differentially expressed (DE) genes in hESC neurogenesis (n=5 studies, in vitro)
Described regulation of in vitro DE genes during human embryo development
Shared patterns (in vitro vs. human embryos) were enriched for neurogenesis genes
Carried out a preliminary verification of the candidate biomarkers
Initial results showed utility as sensitive markers of neurodevelopmental toxicity
Acknowledgments
The authors would like to thank the scientists who contributed to the experimental research studies and related genomic datasets used in our meta-analysis. Support for this project was generously provided by a grant from the National Institute of Environmental Health Sciences, K99 ES023846.
Abbreviations
- hESC
Human embryonic stem cell
- NDB
Neurodevelopmental biomarker
- DE
Differentially expressed gene
- RA
Retinoic Acid
- VPA
Valproic Acid
- CBZ
Carbamazepine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Judson R, et al. Perspectives on validation of high-throughput assays supporting 21st century toxicity testing. ALTEX. 2013;30(1):51–6. doi: 10.14573/altex.2013.1.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knudsen TB, et al. Developmental toxicity testing for safety assessment: new approaches and technologies. Birth defects research Part B, Developmental and reproductive toxicology. 2011;92(5):413–20. doi: 10.1002/bdrb.20315. [DOI] [PubMed] [Google Scholar]
- 3.Miodovnik A. Environmental neurotoxicants and developing brain. The Mount Sinai journal of medicine New York. 2011;78(1):58–77. doi: 10.1002/msj.20237. [DOI] [PubMed] [Google Scholar]
- 4.Colleoni S, et al. Characterisation of a neural teratogenicity assay based on human ESCs differentiation following exposure to valproic acid. Current medicinal chemistry. 2012;19(35):6065–71. [PubMed] [Google Scholar]
- 5.Talens-Visconti R, et al. Neural differentiation from human embryonic stem cells as a tool to study early brain development and the neuroteratogenic effects of ethanol. Stem cells and development. 2011;20(2):327–39. doi: 10.1089/scd.2010.0037. [DOI] [PubMed] [Google Scholar]
- 6.Gerstein MB, et al. Comparative analysis of the transcriptome across distant species. Nature. 2014;512(7515):445–8. doi: 10.1038/nature13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginis I, et al. Differences between human and mouse embryonic stem cells. Developmental biology. 2004;269(2):360–80. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 8.Burgess-Herbert SL, Euling SY. Use of comparative genomics approaches to characterize interspecies differences in response to environmental chemicals: challenges, opportunities, and research needs. Toxicology and applied pharmacology. 2013;271(3):372–85. doi: 10.1016/j.taap.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Sharon N, et al. Molecular and functional characterizations of gastrula organizer cells derived from human embryonic stem cells. Stem cells. 2011;29(4):600–8. doi: 10.1002/stem.621. [DOI] [PubMed] [Google Scholar]
- 10.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature biotechnology. 2009;27(3):275–80. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majumder A, et al. Neurotrophic effects of leukemia inhibitory factor on neural cells derived from human embryonic stem cells. Stem cells. 2012;30(11):2387–99. doi: 10.1002/stem.1201. [DOI] [PubMed] [Google Scholar]
- 12.Schuldiner M, et al. Induced neuronal differentiation of human embryonic stem cells. Brain research. 2001;913(2):201–5. doi: 10.1016/s0006-8993(01)02776-7. [DOI] [PubMed] [Google Scholar]
- 13.Hall PE, et al. Laminin enhances the growth of human neural stem cells in defined culture media. BMC neuroscience. 2008;9:71. doi: 10.1186/1471-2202-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koch P, et al. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(9):3225–30. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Germain N, Banda E, Grabel L. Embryonic stem cell neurogenesis and neural specification. Journal of cellular biochemistry. 2010;111(3):535–42. doi: 10.1002/jcb.22747. [DOI] [PubMed] [Google Scholar]
- 16.Denham M, Dottori M. Signals involved in neural differentiation of human embryonic stem cells. Neuro-Signals. 2009;17(4):234–41. doi: 10.1159/000231890. [DOI] [PubMed] [Google Scholar]
- 17.Senut MC, et al. Lead exposure disrupts global DNA methylation in human embryonic stem cells and alters their neuronal differentiation. Toxicological sciences : an official journal of the Society of Toxicology. 2014;139(1):142–61. doi: 10.1093/toxsci/kfu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fathi A, et al. Comprehensive gene expression analysis of human embryonic stem cells during differentiation into neural cells. PloS one. 2011;6(7):e22856. doi: 10.1371/journal.pone.0022856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson JF, et al. Transcriptomic data of UCSF4 human embryonic stem cells (hESCs) and their neural derivatives (GSE74378) 2015 [Google Scholar]
- 20.Yi H, et al. Gene expression atlas for human embryogenesis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24(9):3341–50. doi: 10.1096/fj.10-158782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang H, et al. Transcriptome analysis of early organogenesis in human embryos. Developmental cell. 2010;19(1):174–84. doi: 10.1016/j.devcel.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Xie D, et al. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome research. 2010;20(6):804–15. doi: 10.1101/gr.100594.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson JF, Verhoef A, Piersma AH. Transcriptomic analysis of neurulation and early organogenesis in rat embryos: an in vivo and ex vivo comparison. Toxicological sciences : an official journal of the Society of Toxicology. 2012;126(1):255–66. doi: 10.1093/toxsci/kfr343. [DOI] [PubMed] [Google Scholar]
- 24.Robinson JF, Piersma AH. Toxicogenomic approaches in developmental toxicology testing. Methods in molecular biology. 2013;947:451–73. doi: 10.1007/978-1-62703-131-8_31. [DOI] [PubMed] [Google Scholar]
- 25.Robinson JF, et al. Dose-response analysis of phthalate effects on gene expression in rat whole embryo culture. Toxicology and applied pharmacology. 2012;264(1):32–41. doi: 10.1016/j.taap.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Robinson JF, et al. A comparison of gene expression responses in rat whole embryo culture and in vivo: time-dependent retinoic acid-induced teratogenic response. Toxicological sciences : an official journal of the Society of Toxicology. 2012;126(1):242–54. doi: 10.1093/toxsci/kfr342. [DOI] [PubMed] [Google Scholar]
- 27.Seiler AE, Spielmann H. The validated embryonic stem cell test to predict embryotoxicity in vitro. Nature protocols. 2011;6(7):961–78. doi: 10.1038/nprot.2011.348. [DOI] [PubMed] [Google Scholar]
- 28.Pennings JL, et al. Gene set assembly for quantitative prediction of developmental toxicity in the embryonic stem cell test. Toxicology. 2011;284(1–3):63–71. doi: 10.1016/j.tox.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 29.Theunissen PT, et al. Complementary detection of embryotoxic properties of substances in the neural and cardiac embryonic stem cell tests. Toxicological sciences : an official journal of the Society of Toxicology. 2013;132(1):118–30. doi: 10.1093/toxsci/kfs333. [DOI] [PubMed] [Google Scholar]
- 30.van Dartel DA, et al. Early gene expression changes during embryonic stem cell differentiation into cardiomyocytes and their modulation by monobutyl phthalate. Reproductive toxicology. 2009;27(2):93–102. doi: 10.1016/j.reprotox.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Pennings JL, Theunissen PT, Piersma AH. An optimized gene set for transcriptomics based neurodevelopmental toxicity prediction in the neural embryonic stem cell test. Toxicology. 2012;300(3):158–67. doi: 10.1016/j.tox.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 32.Barrett T, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic acids research. 2013;41(Database issue):D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolesnikov N, et al. ArrayExpress update--simplifying data submissions. Nucleic acids research. 2015;43(Database issue):D1113–6. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson JF, et al. Comparison of MeHg-induced toxicogenomic responses across in vivo and in vitro models used in developmental toxicology. Reproductive toxicology. 2011;32(2):180–8. doi: 10.1016/j.reprotox.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 35.Euling SY, et al. Use of genomic data in risk assessment case study: II. Evaluation of the dibutyl phthalate toxicogenomic data set. Toxicology and applied pharmacology. 2013;271(3):349–62. doi: 10.1016/j.taap.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Feyeux M, et al. Early transcriptional changes linked to naturally occurring Huntington’s disease mutations in neural derivatives of human embryonic stem cells. Human molecular genetics. 2012;21(17):3883–95. doi: 10.1093/hmg/dds216. [DOI] [PubMed] [Google Scholar]
- 37.Kim JJ, et al. Molecular effect of ethanol during neural differentiation of human embryonic stem cells. Genomics data. 2014;2:139–143. doi: 10.1016/j.gdata.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miya F, et al. GSE39144: Expression data of human induced pluripotent stem cells (hiPSCs), human embryonic stem cells (hESCs) and those differentiated cells. 2013 [Google Scholar]
- 39.Zhang S. GSE9940: ES cells, EBs grown in suspension (d6), PEL (d10) stage and neural rosettes (d17) (zhang-affy-human-346640) 2007 [Google Scholar]
- 40.Affymetrix. Expression Console™ Software 1.4. 2015 http://media.affymetrix.com/support/downloads/manuals/expression_console_userguide.pdf.
- 41.Affymetrix. Transcriptome Analysis Console (TAC) 3.0. 2015 http://media.affymetrix.com/support/downloads/manuals/tac_user_manual.pdf.
- 42.Simon R, et al. Analysis of gene expression data using BRB-ArrayTools. Cancer informatics. 2007;3:11–7. [PMC free article] [PubMed] [Google Scholar]
- 43.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 44.R Development Core Team. R: A Language and Environment for Statistical Computing. 2015 https://www.R-project.org.
- 45.Saeed AI, et al. TM4 microarray software suite. Methods in enzymology. 2006;411:134–93. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 46.Huang DW, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic acids research. 2007;35(Web Server issue):W169–75. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon AT, et al. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3. 2012;2(9):987–1002. doi: 10.1534/g3.112.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonventre JA, et al. Manipulation of the HIF-Vegf pathway rescues methyl tert-butyl ether (MTBE)-induced vascular lesions. Toxicology and applied pharmacology. 2013;273(3):623–34. doi: 10.1016/j.taap.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altschul SF, et al. Basic local alignment search tool. Journal of molecular biology. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 50.Colleoni S, et al. Development of a neural teratogenicity test based on human embryonic stem cells: response to retinoic acid exposure. Toxicological sciences : an official journal of the Society of Toxicology. 2011;124(2):370–7. doi: 10.1093/toxsci/kfr245. [DOI] [PubMed] [Google Scholar]
- 51.Schulpen SH, Pennings JL, Piersma AH. Gene Expression Regulation and Pathway Analysis After Valproic Acid and Carbamazepine Exposure in a Human Embryonic Stem Cell-Based Neurodevelopmental Toxicity Assay. Toxicological sciences : an official journal of the Society of Toxicology. 2015;146(2):311–20. doi: 10.1093/toxsci/kfv094. [DOI] [PubMed] [Google Scholar]
- 52.Matalon S, et al. The teratogenic effect of carbamazepine: a meta-analysis of 1255 exposures. Reproductive toxicology. 2002;16(1):9–17. doi: 10.1016/s0890-6238(01)00199-x. [DOI] [PubMed] [Google Scholar]
- 53.Ornoy A. Valproic acid in pregnancy: how much are we endangering the embryo and fetus? Reproductive toxicology. 2009;28(1):1–10. doi: 10.1016/j.reprotox.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 54.Schulpen SH, et al. Distinct gene expression responses of two anticonvulsant drugs in a novel human embryonic stem cell based neural differentiation assay protocol. Toxicology in vitro : an international journal published in association with BIBRA. 2015;29(3):449–57. doi: 10.1016/j.tiv.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Banda E, Grabel L. Directed Differentiation of Human Embryonic Stem Cells into Neural Progenitors. Methods in molecular biology. 2014 doi: 10.1007/7651_2014_67. [DOI] [PubMed] [Google Scholar]
- 56.National Library of Medicine (US) 2011 doi: 10.1080/15360280801989377. PubMed Health [Internet] Available from: http://www.ncbi.nlm.nih.gov/pubmedhealth/ [DOI] [PubMed]
- 57.van de Vosse E, et al. Characterization of SCML1, a new gene in Xp22, with homology to developmental polycomb genes. Genomics. 1998;49(1):96–102. doi: 10.1006/geno.1998.5224. [DOI] [PubMed] [Google Scholar]
- 58.Kostich WA, Sanes JR. Expression of zfh-4, a new member of the zinc finger-homeodomain family, in developing brain and muscle. Developmental dynamics : an official publication of the American Association of Anatomists. 1995;202(2):145–52. doi: 10.1002/aja.1002020206. [DOI] [PubMed] [Google Scholar]
- 59.Balmer NV, et al. Epigenetic changes and disturbed neural development in a human embryonic stem cell-based model relating to the fetal valproate syndrome. Human molecular genetics. 2012;21(18):4104–14. doi: 10.1093/hmg/dds239. [DOI] [PubMed] [Google Scholar]
- 60.Stummann TC, Hareng L, Bremer S. Hazard assessment of methylmercury toxicity to neuronal induction in embryogenesis using human embryonic stem cells. Toxicology. 2009;257(3):117–26. doi: 10.1016/j.tox.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 61.Hoelting L, et al. A 3-dimensional human embryonic stem cell (hESC)-derived model to detect developmental neurotoxicity of nanoparticles. Archives of toxicology. 2013;87(4):721–33. doi: 10.1007/s00204-012-0984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schenk B, et al. The ReProTect Feasibility Study, a novel comprehensive in vitro approach to detect reproductive toxicants. Reproductive toxicology. 2010;30(1):200–18. doi: 10.1016/j.reprotox.2010.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.