Abstract
Background
α-adrenergic receptor 2A (ADRA2A) and angiotensin-converting enzyme (ACE) genes have been variably associated with obesity and its related phenotypes in different populations worldwide.
Objectives
This cross-sectional study aims to investigate the association of adrenergic receptor α2A (ADRA2A) rs553668 and angiotensin-converting enzyme (ACE) I/D single nucleotide polymorphisms (SNPs) with obesity traits (body mass index-BMI; waist-hip ratio-WHR; total body fat percentage - TBF) in a Malaysian population.
Materials and Methods
Demographic and clinical variables were initially collected from 230 subjects via convenience sampling among residents and workers in Setapak, Malaysia, but in the end only 214 multi-ethnic Malaysians (99 males; 45 Malays, 116 ethnic Chinese, and 53 ethnic Indians) were available for statistical analysis. Genotyping was performed by polymerase chain reaction using DNA extracted from mouthwash samples.
Results
The overall minor allele frequencies (MAFs) for ADRA2A rs553668 and ACE I/D were 0.55 and 0.56, respectively. Allele distribution of ACE I/D was significantly associated with ethnicity and WHR class. Logistic regression analysis showed that subjects with the ACE II genotype and I allele were, respectively, 2.15 and 1.55 times more likely to be centrally obese, but when adjusted for age and ethnicity, this association was abolished. Covariate analysis controlling for age, gender, and ethnicity also showed similar results, where subjects carrying the II genotype or I allele did not have significantly higher WHR. Combinatory genotype and allele analysis for ADRA2A rs553668 and ACE I/D showed that subjects with both ADRA2A rs553668 GG and ACE I/D II genotypes had significant lowest WHR compared to other genotype combinations.
Conclusions
The ACE II genotype might be a protective factor against central adiposity risk among the Malaysian population when in combination with the ADRA2A rs553668 GG genotype.
Keywords: Receptors, Adrenergic, Alpha-2, ACE Protein, Polymorphism, Single Nucleotide, Obesity, Malaysia, Human
1. Background
Obesity, a multi-factorial disorder, is rising at an alarming rate worldwide, including Malaysia, where the 4th national health and morbidity survey (NHMS IV) in 2011 reported its prevalence at 15.1% (1). There are more than 120 candidate genes for obesity-related phenotypes (2), and α-adrenergic receptor 2A (ADRA2A) and angiotensin-converting enzyme (ACE) genes are two of them.
ADRA2A, a cell-surface G-protein-coupled receptor (GPCR) for catecholamines, controls various physiological functions such as modulation of neuron transmission, smooth muscle contraction, thermoregulation energy homeostasis, and metabolism of glucose and lipids (3). The ADRA2A gene has a common single nucleotide polymorphism (SNP) at the 3’ untranslated region (UTR) (HGVS NM_000681.3:c.*427A > G; dbSNP rs553668), and this SNP has been associated with obesity traits, elevated glucose levels, type 2 diabetes, and increased cardiovascular disease risks (4, 5).
Angiotensin-converting enzyme (ACE) is a zinc metallopeptidase involved in the conversion of angiotensin I into angiotensin II, which in turn increases lipolysis and inhibits adipocyte differentiation (6). A polymorphism identified in ACE gene intron 16, in which a 287-bp Alu sequence was found to be present (insertion, or I) or absent (deletion, or D) (dbSNP rs4646994) in the population, has been inconsistently associated with obesity (7-9).
The two SNPs mentioned above have been previously associated with obesity in different populations, but with inconclusive findings. To date, data about the association of the abovementioned SNPs with the prevalence of obesity in the Malaysian population have not been reported. The genetic and environmental backgrounds of the Malaysian population are distinct from the populations studied previously. Hence, the data from previous association studies cannot be extrapolated for the Malaysian population. Therefore, studying the association of ADRA2A rs553668 and ACE I/D SNPs with obesity is important because it could serve as a potential genetic marker for obesity that could aid in the early detection of obesity among the Malaysian population.
2. Objectives
This study aimed to determine the prevalence of the ADRA2A rs553668 and ACE I/D SNPs and their possible association with obesity traits (body mass index-BMI; waist-hip ratio-WHR; total body fat percentage-TBF) in a sample of a multi-ethnic Malaysian population.
3. Materials and Methods
3.1. Subjects
A convenience sampling method was adopted for this cross-sectional study. Questionnaires and sample collections were conducted from 1 October-31 December 2008, and from 1 October-31 December 2009, among unrelated subjects comprised of staff and students from Tunku Abdul Rahman university (Kuala Lumpur campus), a private institute of higher learning, as well as residents of Setapak, Kuala Lumpur, Malaysia. These data extract centres were non-referral. Based on the population size of Setapak (274,639, according to the 2010 Malaysia population and housing census (10)), in order to detect a margin of error of 5%, with a confidence level of 90% and a response distribution of at least 30%, a minimum sample size of 228 subjects had to be recruited (Raosoft® sample size calculator software, Raosoft Inc., US). The exclusion criteria were pregnancy, patients undergoing dialysis, bodybuilders or highly trained athletes, people with fever or swelling, and patients with osteoporosis who have very low bone density. Due to these exclusion criteria, 16 people were excluded from the study, so the available sample size for statistical analysis was 214 subjects (99 males, 115 females; 45 Malays, 116 ethnic Chinese, and 53 ethnic Indians; mean age of 26.27 ± 11.93 years). The ethnicities of the subjects were self-identified. This study received ethical approval from the UTAR scientific and ethical review committee (UTAR/SERC/08-09-SAYYH/01). Patients’ privacy was respected, and all subjects signed informed consent forms. The study was conducted in accordance with the declaration of Helsinki (amended in Seoul, 2008).
3.2. Clinical and Anthropometric Measurements
The systolic and diastolic blood pressures (SBP; DBP) and pulse rates of the subjects were measured using the HEM-712C Omron blood pressure monitor, and duplicate readings were obtained after the subjects were in resting condition for at least 10 minutes. Height, waist, and hip circumferences were determined using a measuring tape. Waist-hip ratio (WHR) was calculated by dividing the waist circumference (WC) by the hip circumference. The HBF-362 Omron Karada scan bio-impedance scale was used to analyze body compositions: weight, body mass index (BMI), total body fat (TBF). All equipment was calibrated according to the manufacturer’s protocol at the start of the day before measurements were performed by the same observer throughout the study. The cut-off points for obesity, overall adiposity (TBF), and central adiposity (WHR) were ≥ 25 kg/m2 (11), 20% (males) or 30% (females) (12), and 0.85 (13), respectively.
3.3. DNA Extraction and Genotyping
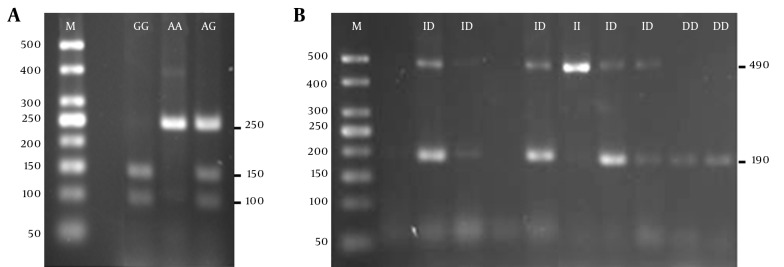
The DNA extraction protocol from mouthwash samples was conducted based on the methods outlined previously (14). The ADRA2A rs553668 and ACE I/D SNPs were analyzed by Polymerase Chain Reaction (PCR) using the primers, reagents, and conditions adopted from Lima et al. (4) for ADRA2A and Rigat et al. (15) for ACE. The sequences of forward primers and reverse primers used were 5’CAGAGCAGCACTGGACTAC-3’ and 5’ TGGAAGGATCTCCCAAG-3’, respectively, for ADRA2A; and 5’CTGGAGACCACTCCCATCCTTTCT-3’ and 5’GATGTGGCCATCACATTCGTCAGAT-3’, respectively, for ACE. PCR was performed in a 20 µL total reaction mixture containing 1 × Taq buffer without MgCl2 (PhileKorea, Korea), 1.0 mM MgCl2, 10 μM dNTPs (PhileKorea, Korea), 10 pmol of forward primer, 10 pmol of reverse primer, 100 ng of DNA template, 0.5 U of Taq polymerase (PhileKorea, Korea), and an appropriate volume of sterile deionized water to top up. The PCR amplification protocol was carried out using the Eppendorf Mastercycler Gradient PCR machine, which began with initial denaturation at 94°C for 3 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 63°C for 30 seconds (for ADRA2A), or 58°C for 1 minute (for ACE), extension at 72°C for 45 seconds, and final elongation at 72°C for 10 minutes. The genotyping for ADRA2A rs553668 SNP was performed by digesting PCR products using Dra1 restriction enzyme (Thermo Scientific, MA, USA) at 37°C for at least 5 hours. ACE PCR products were electrophoresed, then stained with ethidium bromide and visualized under UV light after electrophoresis. Figure 1 shows the sizes of the cleaved or uncleaved PCR products of ADRA2A and ACE after agarose gel electrophoresis.
Figure 1. ADRA2A rs553668 and ACE I/D SNPs Genotype Analysis on 2.5% Agarose Gel.
A, ADRA2A rs553668 SNP; M-50 bp DNA ladder (PhileKorea, Korea), GG-cleaved homozygous mutant GG (150 and 100 bp fragments), AA-uncleaved homozygous wild-type AA (250 bp), AG-heterozygous (250, 150 and 100 bp fragments); B, ACE I/D SNP; M-50 bp DNA Ladder (PhileKorea, Korea), DD-homozygous DD (190 bp fragment), ID-heterozygous ID (490 and 190 bp fragments), II-homozygous II (490 bp fragment). The smears at around 50 bp indicate primer-dimers.
3.4. Statistical Analysis
Statistical power calculation was performed using Quanto version 1.2.4 (http://biostats.usc.edu/Quanto.html) with the following parameters: outcome: disease; design: unmatched case control (1: 1.97); hypothesis: gene only; significance: 0.05, 2-sided; mode of inheritance: log-additive; allele frequency: ADRA2A/ACE - 0.55; population risk [based on the prevalence of obesity in 2011 NMHS IV (1)]: 0.151; Genetic effect: 2. With the case-control pairs’ sample size of 214, a statistical power of more than 90% was achieved for all the gene association studies.
IBM SPSS Statistics software was used to analyze the data from the study. Allelic frequencies were estimated by gene counting, and the distribution of genotypes was tested for the Hardy-Weinberg equilibrium using the Chi-square (χ2) test. Data for continuous variables were presented as means ± standard deviations (SD) or adjusted means ± standard error of the mean (SEM), and as frequency for categorical variables. The normality of the distributions of continuous variables was tested using the Kolmogorov-Smirnov test, and variables that were not distributed normally were log-transformed prior to statistical analysis. Genotype and allele frequencies of the polymorphism were assessed for associations with demographic and anthropometric classes using Pearson’s χ2 test. Logistic regression analysis (enter method) was performed with adjustments for age, gender, and ethnicity to evaluate if the polymorphism could predict the risk of increased central adiposity. Analysis of covariance using the univariate general linear model with adjustment for covariates (age, gender, and ethnicity) was performed for anthropometric and clinical measurements. A P value of less than 0.05 was considered statistically significant.
4. Results
Table 1 shows the baseline characteristics of the subjects. More than half of the subjects were females, though there were more obese males than obese females. Except for the pulse rate, all the clinical and anthropometric measurements were significantly higher in obese subjects, regardless of gender.
Table 1. Baseline Characteristics of the Subjectsa.
| Variables | Males (n = 99) | P Valueb | Females (n = 115) | P Valueb | ||
|---|---|---|---|---|---|---|
| Non-Obese | Obese | Non-Obese | Obese | |||
| Numbers | 60 (60.6) | 39 (39.4) | 82 (71.3) | 33 (28.7) | ||
| SBP, mmHg | 122.39 ± 13.78 | 132.09 ± 14.58 | < 0.001c | 111.28 ± 13.20 | 124.62 ± 15.70 | < 0.001c |
| DBP, mmHg | 74.66 ± 7.84 | 81.46 ± 9.27 | < 0.001c | 68.65 ± 8.42 | 76.32 ± 9.12 | < 0.001c |
| Pulse rate, bpm | 81.46 ± 11.17 | 82.64 ± 14.82 | 0.95 | 83.66 ± 15.09 | 82.30 ± 11.83 | 0.826 |
| BMI, kg/m 2 | 21.30 ± 2.23 | 30.23 ± 4.39 | < 0.001c | 20.42 ±2.35 | 30.15 ± 3.87 | < 0.001c |
| WHR | 0.84 ± 0.05 | 0.92 ± 0.07 | < 0.001c | 0.79 ± 0.06 | 0.87 ± 0.06 | < 0.001c |
| TBF, % | 14.98 ± 5.37 | 34.34 ± 10.08 | < 0.001c | 20.29 ± 5.62 | 40.74 ± 9.46 | < 0.001c |
Abbreviations: BMI, body mass index; DBP, diastolic blood pressure; PR, pulse rate; SBP, systolic blood pressure; TBF, total body fat; WHR, waist-to-hip ratio.
aValues are presented as mean ± SD or No. (%).
bP values from Mann-Whitney U test.
cSignificant at < 0.05.
Table 2 shows the genotype and allele distribution of subjects who did not deviate from the Hardy-Weinberg equilibrium, and are categorized under different demographic and anthropometric classes. The overall minor allele frequencies (MAFs) for ADRA2A rs553668 and ACE I/D SNPs were quite similar at 0.55 and 0.56, respectively, while, according to Malay/Chinese/Indian ethnicities, their MAFs were 0.49/0.55/0.59 and 0.69/0.55/0.48, respectively. Therefore, the allele distribution of ACE I/D was significantly associated with ethnicity, with the MAF of Malays being significantly higher. Meanwhile, the MAFs for non-obese/obese for ADRA2A rs553668 and ACE I/D SNPs were 0.56/0.53 and 0.53/0.63, respectively. The allele distribution of the ACE I/D SNP was significantly associated with WHR class, but not gender, BMI class, or TBF class.
Table 2. Association of ADRA2A rs553668 and ACEI/D Genotype and Allele Distribution With Demographic and Anthropometric Classesa,b.
| Variables | SNP | Genotype | Allele | |||||
|---|---|---|---|---|---|---|---|---|
| 1/1 | 1/2 | 2/2 | χ2, P Value | 1 | 2 | χ2, P Value | ||
| Gender | 2.85, 0.24; 1.62, 0.45 | 2.28, 0.13; 0.94, 0.33 | ||||||
| Male | ||||||||
| ADRA2A rs553668 | 12 (12.1) | 58 (58.6) | 29 (29.3) | 82 (41.4) | 116 (58.6) | |||
| ACE I/D | 22 (22.2) | 38 (38.4) | 39 (39.4) | 82 (41.4) | 116 (58.6) | |||
| Female | ||||||||
| ADRA2A rs553668 | 22 (19.1) | 68 (59.1) | 25 (21.7) | 112 (48.7) | 118 (51.3) | |||
| ACE I/D | 27 (23.5) | 52 (45.2) | 36 (31.3) | 106 (46.1) | 124 (53.9) | |||
| Ethnicity | 2.39, 0.67; 8.23, 0.08 | 1.86, 0.39; 8.90, 0.01c | ||||||
| Malay | ||||||||
| ADRA2A rs553668 | 9 (20.0) | 28 (62.2) | 8 (17.8) | 46 (51.1) | 44 (48.9) | |||
| ACE I/D | 5 (11.1) | 18 (40.0) | 22 (48.9) | 28 (31.1) | 62 (68.9) | |||
| Chinese | ||||||||
| ADRA2A rs553668 | 18 (15.5) | 68 (58.6) | 30 (25.9) | 104 (44.8) | 128 (55.2) | |||
| ACE I/D | 27 (23.3) | 51 (44.0) | 38 (32.8) | 105 (45.3) | 127 (54.7) | |||
| Indian | ||||||||
| ADRA2A rs553668 | 7 (13.2) | 30 (56.6) | 16 (30.2) | 44 (41.5) | 62 (58.5) | |||
| ACE I/D | 17 (32.1) | 21 (39.6) | 15 (28.3) | 55 (51.9) | 51 (48.1) | |||
| BMI class | ||||||||
| Non-obese | 2.83, 0.24; 3.75, 0.15 | 0.32, 0.58; 3.64, 0.06 | ||||||
| ADRA2A rs553668 | 24 (16.9) | 78 (54.9) | 40 (28.2) | 126 (44.4) | 158 (55.6) | |||
| ACE I/D | 38 (26.8) | 58 (40.8) | 46 (32.4) | 134 (47.2) | 150 (52.8) | |||
| Obese | ||||||||
| ADRA2A rs553668 | 10 (13.9) | 48 (66.7) | 14 (19.4) | 68 (47.2) | 76 (52.8) | |||
| ACE I/D | 11 (15.3) | 32 (44.4) | 29 (40.3) | 54 (37.5) | 90 (62.5) | |||
| TBF class | ||||||||
| Normal | 1.66, 0.44; 2.46, 0.29 | 0.93, 0.33; 1.70, 0.19 | ||||||
| ADRA2A rs553668 | 24 (15.4) | 89 (57.1) | 43 (24.6) | 137 (43.9) | 175 (56.1) | |||
| ACE I/D | 40 (25.6) | 63 (40.4) | 53 (34.0) | 143 (45.8) | 169 (54.2) | |||
| High | ||||||||
| ADRA2A rs553668 | 10 (17.2) | 37 (63.8) | 11 (19.0) | 57 (49.1) | 59 (50.9) | |||
| ACE I/D | 9 (15.5) | 27 (46.6) | 22 (37.9) | 45 (38.8) | 71 (61.2) | |||
| WHR class | 3.14, 0.21; 4.31, 0.12 | 2.42, 0.12; 4.93, 0.03c | ||||||
| Normal | ||||||||
| ADRA2A rs553668 | 16 (13.6) | 67 (56.8) | 35 (29.7) | 99 (41.9) | 137 (58.1) | |||
| ACE I/D | 32 (27.1) | 51 (43.2) | 35 (29.7) | 115 (48.7) | 121 (51.3) | |||
| High | ||||||||
| ADRA2A rs553668 | 18 (18.8) | 59 (61.5) | 19 (19.8) | 95 (49.5) | 97 (50.5) | |||
| ACE I/D | 17 (17.7) | 39 (40.6) | 40 (41.7) | 73 (38.0) | 119 (62.0) | |||
Abbreviations: BMI, body mass index; WHR, TBF, total body fat; waist-to-hip ratio; 1/1, homozygous wild-type AA (ADRA2A rs553668) or II (ACE), 1/2, heterozygous genotypes, 2/2, variant GG (ADRA2A rs553668) or DD (ACE).
aValues are expressed as No. (%), which the percentages indicate the percentage within the same demographic/anthropometric class.
bP values from Pearson’s chi-square test.
csignificant at < 0.05.
Since the ACE I/D allele was associated with WHR, a logistic regression analysis to estimate the risk of being centrally obese (high WHR) by having different ACE I/D genotypes and alleles was performed as shown in Table 3. Subjects with the II genotype or I allele were 2.15 times and 1.55 times, respectively, more likely to be centrally obese. However, when adjusted for age, gender, and ethnicity, this association of ACE I/D SNP with central obesity was abolished for both II genotype and I allele (Table 3). Indeed, covariate analysis of variance after controlling for age, gender, and ethnicity also showed similar results, where subjects carrying the II genotype or I allele did not have significantly higher WHR (Table 4). Also, all other clinical and anthropometric measurements were not significantly different among the ACE I/D genotypes and alleles after controlling for age, gender, and ethnicity (Table 4).
Table 3. Logistic Regression Analysis for the Association of ACEI/D SNP With Central Adipositya.
| Genotypes/Alleles | Unadjusted | Adjustedb | ||
|---|---|---|---|---|
| Central Adiposity (Based on WHR) | Odds Ratio (95% CI) | P Value | Odds Ratio (95% CI) | P Value |
| DD | 1.00 | NA | 1.00 | NA |
| ID | 1.44 (0.70, 2.96) | 0.32 | 1.57 (0.70, 3.52) | 0.28 |
| II | 2.15 (1.02, 4.52) | 0.04c | 2.02 (0.87, 4.70) | 0.10 |
| D | 1.00 | NA | 1.00 | NA |
| I | 1.55 (1.05, 2.28) | 0.03c | 1.46 (0.95, 2.26) | 0.09 |
Abbreviation: NA, not available.
aValues are from logistic regression enter method.
bAdjusted for co-variates: gender, age, and ethnicity.
cP value significant at < 0.05.
Table 4. Adjusted Means of Clinical and Anthropometric Measurements for Different Genotypes and Alleles of ACEI/D SNPa,b.
| Variables | Genotype | Allele | P Value (Genotype) | P Value (Allele) | |||
|---|---|---|---|---|---|---|---|
| DD | ID | II | D | I | |||
| SBP, mmHg | 118.34 ± 2.10 | 120.49 ± 1.53 | 121.20 ± 1.69 | 119.38 ± 1.06 | 120.92 ± 0.93 | 0.53 | 0.24 |
| DBP, mmHg | 73.53± 1.29 | 73.77 ± 0.94 | 74.11 ± 1.04 | 73.65 ± 0.65 | 73.98 ± 0.57 | 0.91 | 0.66 |
| Pulse rate, bpm | 81.51 ± 1.95 | 84.24 ± 1.42 | 81.47 ± 1.57 | 82.82 ± 0.99 | 82.51 ± 0.87 | 0.44 | 0.92 |
| BMI, kg/m 2 | 23.71 ± 0.74 | 24.27 ± 0.54 | 23.73 ± 0.60 | 23.98 ± 0.38 | 23.93 ± 0.33 | 0.81 | 0.90 |
| WHR | 0.84 ± 0.01 | 0.84 ± 0.01 | 0.84 ± 0.01 | 0.84 ± 0.01 | 0.84 ± 0.00 | 0.90 | 0.66 |
| TBF, % | 24.10 ± 1.61 | 25.46 ± 1.17 | 23.67 ± 1.30 | 24.74 ± 0.81 | 24.34 ± 0.72 | 0.66 | 0.98 |
Abbreviations: BMI, body mass index; DBP, diastolic blood pressure; PR, pulse rate; SBP, systolic blood pressure; WHR, waist-to-hip ratio; TBF, Total body fat.
aAll values (except WHR) were log-transformed before analysis using univariate analysis of variance (general linear model), adjusted for co-variates: age, gender, and ethnicity.
bValues are presented as adjusted mean ± SEM (estimated marginal means ± standard error of the mean).
When the subjects were grouped based on wild-type/mutant combinations of ADRA2A rs553668 and ACE I/D genotypes and alleles and anthropometric measurements, WHR was significantly different among the genotype combinations (Table 5). Subjects with the GG/II genotype combination had significantly lower WHR compared with other genotype combinations, while, although statistically insignificant, they also had the lowest BMI and TBF. Meanwhile, WHR, BMI, and TBF were not significantly different among the different allele combinations (Table 5).
Table 5. Adjusted Means of Anthropometric Measurements for the Genotype and Allele Combinations of ADRA2A rs553668 and ACEI/D SNPsa,b.
| Variables | Number | WHR | BMI, kg/m2 | TBF, % |
|---|---|---|---|---|
| Genotype combinations | ||||
| AA/DD | 8 | 0.86 ± 0.02 | 26.98 ± 1.65 | 32.10 ± 3.60 |
| AA/II | 11 | 0.85 ± 0.02 | 22.71 ± 1.42 | 21.88 ± 3.11 |
| GG/DD | 16 | 0.85 ± 0.01 | 22.87 ± 1.19 | 22.79 ± 2.60 |
| GG/II | 12 | 0.79 ± 0.02 | 21.66 ± 1.37 | 19.67 ± 3.00 |
| P value | 0.01c | 0.19 | 0.36 | |
| Allele combinations | ||||
| A/D | 105 | 0.84 ± 0.01 | 24.66 ± 0.50 | 26.12 ± 1.08 |
| A/I | 89 | 0.86 ± 0.01 | 24.30 ± 0.54 | 24.71 ± 1.17 |
| G/D | 83 | 0.84 ± 0.01 | 23.09 ± 0.56 | 23.00 ± 1.22 |
| G/I | 151 | 0.84 ± 0.01 | 23.73 ± 0.42 | 24.15 ± 0.90 |
| P value | 0.105 | 0.26 | 0.51 |
Abbreviations: BMI, body mass index; TBF, total body fat; WHR, waist-to-hip ratio.
aBMI and TBF were log-transformed before analysis using univariate analysis of variance (general linear model), adjusted for co-variates: age, gender, and ethnicity.
bValues are presented as adjusted mean ± SEM (estimated marginal means ± standard error of the mean).
cP value significant at < 0.05.
5. Discussion
For ADRA2A rs553668, we found that the majority of the Malaysian subjects had the AG genotypes, followed by AA and GG genotypes, while the overall MAF was 0.55, similar to the Chinese Han population (16). In contrast, the MAF for this SNP was lower in Caucasian and African-American populations, ranging from 0.15 - 0.21 in the former (4, 17, 18) and 0.25 in the latter (4). Therefore, the similar MAF found between the Chinese ethnic group in Malaysia and those in China reflect the genetic ancestral origins based on their migration history (19). Regarding Malays and Indians, a comparison could not be made because no published literature is available for comparison. As for ACE I/D, a recent meta-analysis concluded that the average MAF was 0.43/0.45 for obese/non-obese subjects among Caucasians, 0.41/0.53 among Asians, and 0.35/0.42 among Africans (20). However, our study found the MAF to be 0.53/0.63, even higher than the average MAFs among Asians (20). Nevertheless, our findings are consistent with the trend that the MAF for ACE I/D is higher among Asians in comparison with other ethnic populations.
Our study is consistent with previous studies that found that the ADRA2A rs553668 SNP was not associated with BMI or obesity in Chinese Han (16) and British Caucasians (17). However, among African Americans, there was a significant gene dose-response relationship for the GA genotype of this SNP with both BMI and TBF (4). The abovementioned previous studies found that this SNP was associated with fasting glucose and type 2 diabetes risks instead, which were not assessed in the current study. While for ACE I/D, numerous studies concerning the association between ACE I/D gene polymorphism and obesity risks have generated mixed results (7, 20-25). For example, Strazzullo et al. (7) reported that Italian men with the D/D genotype had 82% higher odds of being overweight compared to those with the I/D or I/I genotype, while associations between this SNP and obesity were not found in African populations (25). However, a recent meta-analysis concluded that the DD genotype/D allele was associated with the obesity risk factor in Africans, but not Caucasians or Asians, while II genotype might not be a protective factor against obesity risks in overall populations, such as Africans, Caucasians, and Asians (20). In this study, an association was not found between this SNP and BMI/obesity, but it was with WHR. However, after controlling for gender, age, and ethnicity, WHR was no longer associated with this SNP.
Aside from obesity, there have been a number of meta-analyses and studies investigating the association between the ACE I/D gene SNP and the risk of obesity-related diseases, where the D allele was positively associated with metabolic syndrome (26), type 2 diabetes (27), essential hypertension (28), and reduced high-density lipoprotein cholesterol levels (29). In this study, we found that this SNP was not associated with blood pressure, while the association with other obesity-related parameters such as glucose level and lipid profile remains to be determined.
In a combinatory genotype-and-allele analysis involving ADRA2A rs553668 and ACE I/D SNPs, we found that subjects with both ADRA2A rs553668 GG and ACE I/D II genotypes had the significantly lowest WHR compared to other genotype combinations. Those having the two genotypes alone did not report a lower WHR. Therefore, the II genotype, but not the I allele, might be a protective factor against central adiposity (as indicated by WHR) risks in the Malaysian population only when it is in combination with the GG genotype of ADRA2A rs553668. This is perhaps the first study reporting on the existence of a gene interaction between ADRA2A rs553668 and ACE I/D SNPs in influencing central adiposity. Gene interactions between ACE I/D and PPAR-γ2 SNPs influencing BMI and fat mass have been reported previously (30). Because ADRA2A and ACE genes play a common role in blood pressure regulation (31, 32), they might also play another common role in regulating adipogenesis in the abdominal area.
To the best of our knowledge, this is the first study investigating the prevalence of the ADRA2A rs553668 and ACE I/D SNPs and their association with obesity traits within a sample of the multi-ethnic Malaysian population in Setapak, Kuala Lumpur. However, we are aware of several limitations in our study; these include the small sample size, which limits statistical power and generalization to the overall Malaysian population. The case-control design in this study also does not allow for a causality conclusion to be made. Also, as only two SNPs of ADRA2A and ACE were evaluated in this study, it is unclear whether another common ADRA2A SNP (rs1800544), or SNPs in other adrenergic receptor families (4) and ACE (26) that are in tight linkage disequilibrium with these two SNPs, might have associations with obesity instead. Therefore, a larger and more ethnically diverse sample size are needed in the future. The direct effect of these SNPs on physiological phenotypes, such as α-adrenergic receptor activity and ACE plasma level, could also be investigated in future studies.
In conclusion, the angiotensin-converting enzyme gene (ACE) II genotype might be a protective factor against central adiposity risks within the Malaysian population only when it is in combination with α-adrenergic receptor 2A gene (ADRA2A) rs553668 GG genotype, suggesting the existence of a gene interaction between ADRA2A and ACE in influencing central adiposity.
Acknowledgments
We gratefully acknowledge all the volunteers who participated in this study.
Footnotes
Authors’ Contribution:Vicneswari Shunmugam collected the data and performed the experiments. Yee-How Say developed the original idea and the protocol, abstracted and analyzed the data, wrote the manuscript, and is the guarantor.
Funding/Support:This project was funded by the department of biomedical Sciences, faculty of sciences, Tunku Abdul Rahman university.
References
- 1.Institute for Public Health Malaysia. Kuala Lumpur. Kuala Lumpur: Ministry of Health; 2011. The Fourth National Health and Morbidity Survey (NHMS III) 2011. [Google Scholar]
- 2.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, et al. The human obesity gene map: the 2005 update. Obesity. 2006;14(4):529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 3.Yasuda K, Matsunaga T, Adachi T, Aoki N, Tsujimoto G, Tsuda K. Adrenergic receptor polymorphisms and autonomic nervous system function in human obesity. Trends Endocrinol Metab. 2006;17(7):269–75. doi: 10.1016/j.tem.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Lima JJ, Feng H, Duckworth L, Wang J, Sylvester JE, Kissoon N, et al. Association analyses of adrenergic receptor polymorphisms with obesity and metabolic alterations. Metabolism. 2007;56(6):757–65. doi: 10.1016/j.metabol.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langberg EC, Seed Ahmed M, Efendic S, Gu HF, Ostenson CG. Genetic association of adrenergic receptor alpha 2A with obesity and type 2 diabetes. Obesity (Silver Spring). 2013;21(8):1720–5. doi: 10.1002/oby.20162. [DOI] [PubMed] [Google Scholar]
- 6.Jones BH, Standridge MK, Moustaid N. Angiotensin II increases lipogenesis in 3T3-L1 and human adipose cells. Endocrinology. 1997;138(4):1512–9. doi: 10.1210/endo.138.4.5038. [DOI] [PubMed] [Google Scholar]
- 7.Strazzullo P, Iacone R, Iacoviello L, Russo O, Barba G, Russo P, et al. Genetic variation in the renin-angiotensin system and abdominal adiposity in men: the Olivetti Prospective Heart Study. Ann Intern Med. 2003;138(1):17–23. doi: 10.7326/0003-4819-138-1-200301070-00007. [DOI] [PubMed] [Google Scholar]
- 8.Ryan AS, Nicklas BJ, Berman DM, Ferrell RE. The insertion/deletion polymorphism of the ACE gene is related to insulin sensitivity in overweight women. Diabetes Care. 2001;24(9):1646–52. doi: 10.2337/diacare.24.9.1646. [DOI] [PubMed] [Google Scholar]
- 9.Um JY, Mun KS, An NH, Kim PG, Kim SD, Song YS, et al. Polymorphism of angiotensin-converting enzyme gene and BMI in obese Korean women. Clin Chim Acta. 2003;328(1-2):173–8. doi: 10.1016/s0009-8981(02)00428-x. [DOI] [PubMed] [Google Scholar]
- 10.Department of Statistics, Malaysia. Population distribution by local authority areas and mukims, 2010. 2015 Available from: http://www.statistics.gov.my/portal/download_Population/files/population/05Jadual_Mukim_negeri/Mukim_WPKL.pdf.
- 11.International Association for the Study of Obesity. Hong Kong: World Health Organization; 2000. The Asia-Pacific perspective: redefining obesity and its treatment; International Obesity Task Force. [Google Scholar]
- 12.Omron Healthcare Co. Japan: 2010. Omron KARADA Scan Body Composition & Scale (HBF-362). [Google Scholar]
- 13.WHO. Geneva: WHO Press; 2011. Waist circumference and waist-hip ratio: report of a WHO expert consultation. [Google Scholar]
- 14.Aidar M, Line SR. A simple and cost-effective protocol for DNA isolation from buccal epithelial cells. Braz Dent J. 2007;18(2):148–52. doi: 10.1590/s0103-64402007000200012. [DOI] [PubMed] [Google Scholar]
- 15.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86(4):1343–6. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T, Zhu X, Wu X, Li J, Pan L, Li P, et al. Evaluation of the association between the ADRA2A genetic polymorphisms and type 2 diabetes in a Chinese Han population. Genet Test Mol Biomarkers. 2012;16(12):1424–7. doi: 10.1089/gtmb.2012.0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talmud PJ, Cooper JA, Gaunt T, Holmes MV, Shah S, Palmen J, et al. Variants of ADRA2A are associated with fasting glucose, blood pressure, body mass index and type 2 diabetes risk: meta-analysis of four prospective studies. Diabetologia. 2011;54(7):1710–9. doi: 10.1007/s00125-011-2108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Cerqueira CC, Polina ER, Contini V, Marques FZ, Grevet EH, Salgado CA, et al. ADRA2A polymorphisms and ADHD in adults: possible mediating effect of personality. Psychiatry Res. 2011;186(2-3):345–50. doi: 10.1016/j.psychres.2010.08.032. [DOI] [PubMed] [Google Scholar]
- 19.Teo YY, Sim X, Ong RT, Tan AK, Chen J, Tantoso E, et al. Singapore Genome Variation Project: a haplotype map of three Southeast Asian populations. Genome Res. 2009;19(11):2154–62. doi: 10.1101/gr.095000.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao S, Huang S. A meta-analysis of the association between angiotensin-converting enzyme insertion/ deletion gene polymorphism and the risk of overweight/obesity. J Renin Angiotensin Aldosterone Syst. 2015;16(3):687–94. doi: 10.1177/1470320313501218. [DOI] [PubMed] [Google Scholar]
- 21.Yang SJ, Kim S, Park H, Kim SM, Choi KM, Lim Y, et al. Sex-dependent association between angiotensin-converting enzyme insertion/deletion polymorphism and obesity in relation to sodium intake in children. Nutrition. 2013;29(3):525–30. doi: 10.1016/j.nut.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Mehri S, Mahjoub S, Hammami S, Zaroui A, Frih A, Betbout F, et al. Renin-angiotensin system polymorphisms in relation to hypertension status and obesity in a Tunisian population. Mol Biol Rep. 2012;39(4):4059–65. doi: 10.1007/s11033-011-1187-2. [DOI] [PubMed] [Google Scholar]
- 23.Akin F, Turgut S, Bastemir M, Turgut G, Kursunluoglu R, Karasu U, et al. Angiotensin-converting enzyme gene polymorphism in overweight and obese Turkish patients with insulin resistance. DNA Cell Biol. 2010;29(4):207–12. doi: 10.1089/dna.2009.0934. [DOI] [PubMed] [Google Scholar]
- 24.Kim K. Association of angiotensin-converting enzyme insertion/deletion polymorphism with obesity, cardiovascular risk factors and exercise-mediated changes in Korean women. Eur J Appl Physiol. 2009;105(6):879–87. doi: 10.1007/s00421-008-0973-6. [DOI] [PubMed] [Google Scholar]
- 25.Kramer H, Wu X, Kan D, Luke A, Zhu X, Adeyemo A, et al. Angiotensin-converting enzyme gene polymorphisms and obesity: an examination of three black populations. Obes Res. 2005;13(5):823–8. doi: 10.1038/oby.2005.94. [DOI] [PubMed] [Google Scholar]
- 26.Xi B, Ruiter R, Chen J, Pan H, Wang Y, Mi J. The ACE insertion/deletion polymorphism and its association with metabolic syndrome. Metabolism. 2012;61(6):891–7. doi: 10.1016/j.metabol.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 27.Niu W, Qi Y, Gao P, Zhu D. Angiotensin converting enzyme D allele is associated with an increased risk of type 2 diabetes: evidence from a meta-analysis. Endocr J. 2010;57(5):431–8. doi: 10.1507/endocrj.k09e-360. [DOI] [PubMed] [Google Scholar]
- 28.Li Y. Angiotensin-converting enzyme gene insertion/deletion polymorphism and essential hypertension in the Chinese population: a meta-analysis including 21,058 participants. Intern Med J. 2012;42(4):439–44. doi: 10.1111/j.1445-5994.2011.02584.x. [DOI] [PubMed] [Google Scholar]
- 29.You CH, Hong YS, Kwak JY, Kim NY, Roh MS, Jung KY, et al. [The relationship between ACE I/D polymorphism and HDL cholesterol]. J Prev Med Public Health. 2006;39(6):505–10. [PubMed] [Google Scholar]
- 30.Passaro A, Dalla Nora E, Marcello C, Di Vece F, Morieri ML, Sanz JM, et al. PPARgamma Pro12Ala and ACE ID polymorphisms are associated with BMI and fat distribution, but not metabolic syndrome. Cardiovasc Diabetol. 2011;10:112. doi: 10.1186/1475-2840-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Civantos Calzada B, Aleixandre de Artinano A. Alpha-adrenoceptor subtypes. Pharmacol Res. 2001;44(3):195–208. doi: 10.1006/phrs.2001.0857. [DOI] [PubMed] [Google Scholar]
- 32.Kalupahana NS, Moustaid-Moussa N. The adipose tissue renin-angiotensin system and metabolic disorders: a review of molecular mechanisms. Crit Rev Biochem Mol Biol. 2012;47(4):379–90. doi: 10.3109/10409238.2012.694843. [DOI] [PubMed] [Google Scholar]