Abstract
Background
MicroRNAs (miRNAs) act as post-transcriptional regulators of gene expression. Genetic variation in miRNA-encoding sequences or their corresponding binding sites may affect the fidelity of the miRNA-messenger RNA interaction and subsequently alter risk of cancer development.
Methods
This study expanded the search for miRNA-related polymorphisms contributing to the etiology of colorectal cancer (CRC) across the genome using a novel platform, the Axiom® miRNA Target Site Genotyping Array (237,858 markers). After quality control, the study included 596 cases and 429 controls from the Molecular Epidemiology of Colorectal Cancer study, a population-based case-control study of CRC in northern Israel. The association between each marker and CRC status was examined assuming a log-additive genetic model using logistic regression adjusted for sex, age, and two principal components.
Results
Twenty-three markers had p-values less than 5.0E-04, and the most statistically significant association involved rs2985 (chr6:34845648; intronic of UHRF1BP1; OR=0.66; p-value=3.7E-05). Further, this study replicated a previously published locus, rs1051690 in the 3’-untranslated region of the insulin receptor gene INSR (OR = 1.38; p = 0.03), with strong evidence of differences in INSR gene expression by genotype.
Conclusions
This study is the first to examine associations between genetic variation in miRNA target sites and CRC using a genome-wide approach. Functional studies to identify allele-specific effects on miRNA binding are needed to confirm the regulatory capacity of genetic variation to influence risk of CRC.
Impact
This study demonstrates the potential for a miRNA-targeted genome-wide association study to identify candidate susceptibility loci and prioritize them for functional characterization.
Keywords: miRNA, genome-wide association study, CRC, GWAS, susceptibility
Introduction
In addition to protein-coding messenger RNAs (mRNAs), other classes of small RNA molecules exist with specialized regulatory and processing functions. Among these types of regulatory RNAs are microRNAs (miRNAs), short (18–24 nucleotide) non-protein-coding molecules that act as post-transcriptional regulators of gene expression (1). The biogenesis of a miRNA begins with transcription from a small, stand-alone gene or an intron or exon of a known protein-coding gene and transitions through a series of conversion steps from hairpin precursors to duplexed pre-miRNA intermediates to mature, single-stranded miRNAs (2, 3). MiRNAs exert their regulatory effects via binding to complementary ~6–8 nucleotide target seed sites in the 3’ untranslated regions (3’-UTRs) of one or more mRNAs. Depending on the fidelity and context of the interaction, this binding acts to repress translation of the messenger into protein or to signal for degradation of the targeted mRNA (1, 4, 5). Each miRNA typically binds multiple, even thousands, of messenger targets, offering the potential for widespread downstream effects (6, 7).
Deregulated miRNA profiles have been described across a range of cancers including colorectal cancer (CRC) (8, 9). Further, some suggest that miRNA biology can be integrated into the molecular sub-typing of colorectal tumors and into the traditional model of genetic alterations accompanying progression from normal mucosa to carcinoma, particularly among tumors that develop through the chromosomal instability pathway (7, 10–13). As an extension of this work, several miRNAs have been proposed as biomarkers for CRC early detection, prognosis, and progression (14–16).
Despite extensive miRNA profiling in colorectal tumors, the factors driving aberrations in miRNA expression and their impact on CRC development and progression are not yet well defined. One hypothesis proposes that single nucleotide polymorphisms (SNPs) in genes encoding the miRNA sequence or 3’-UTR regions of the corresponding binding sites affect miRNA transcription, miRNA processing, and/or the fidelity of the miRNA-mRNA interaction. Any of these alterations could plausibly impact target mRNA translation into proteins critical for cellular differentiation and proliferation. Evidence from studies of candidate miRNA-related genetic alterations supports this hypothesis and suggests that such SNPs may alter expression of some miRNAs in CRC(17) and increase or decrease the risk of tumor development (18). Target site polymorphisms that confer risk in specific populations have been identified in INSR (18), CD86(18), IL16(19), RPA2(20), and GTF2H1(20); however, replication of these findings has been limited with the exception of rs1051690 in INSR and rs17281995 in CD86 (21). To date, published studies have not comprehensively investigated polymorphisms implicated in the miRNA regulatory pathway across the genome.
In this study, we expanded the search for miRNA-related genetic variants important in the etiology of CRC across the genome and investigated the association between thousands of genetic variants in miRNA target sites in 3’-UTR regions and miRNA-encoding genes and CRC risk using a novel genotyping platform. In contrast to a classical genome-wide association study (GWAS) approach that relies on haplotype-tagging SNPs, we leveraged genotyping of SNPs bioinformatically predicted to have functional implications specific to the miRNA regulatory pathway. We then characterized the predicted miRNA binding consequences of our most significantly associated SNPs and further explored these associations with expression quantitative trait loci (eQTL) analyses. This study was designed to evaluate the feasibility of a targeted GWAS approach for identifying lead candidates and prioritizing them for functional characterization based on biologically relevant hypotheses. The genetically homogeneous founder population of Ashkenazi Jewish individuals experiences a high burden of CRC and served as the focus of this study (22).
Materials and Methods
Study population: Molecular Epidemiology of Colorectal Cancer (MECC) Study
MECC is a population-based, case-control study of pathologically-confirmed, incident cases of CRC recruited from a geographically-defined region of northern Israel (23). Subject recruitment began in 1998 and remains on-going. Individually-matched controls with no prior history of CRC are selected from the same source population that gave rise to cases based on the Clalit Health Services database. Matching factors include age, sex, Jewish ethnicity (Jewish versus non-Jewish), and primary clinic site. Subjects are interviewed to obtain demographic data, clinical information, family history, and dietary habits. Biospecimens including blood, paraffin blocks, and snap frozen tumors are collected. Based on resource limitations, 1,266 cases and controls (approximately 15% of all MECC participants) were initially selected for genotyping. Following sample quality control (see Supplementary Figure 1 for details), genome-wide analysis was performed on 596 cases and 429 controls enriched for Ashkenazi Jewish ancestry (Table 1). Informed consent was obtained according to Institutional Review Board-approved protocols at Carmel Medical Center (Haifa) and the University of Southern California.
Table 1.
Demographic and clinical characteristics of MECC participants (n=1,025) genotyped on the Axiom® miRNA Target Site Genotyping Array platform.
| Cases (n=596) | Controls (n=429) | |
|---|---|---|
| Age [mean(sd)] | 70.9 (10.7) | 74.3 (10.6) |
| Sex (%) | ||
| Male | 291 (48.8) | 221 (51.5) |
| Female | 305 (51.2) | 208 (48.5) |
| Self-reported race/ethnicity (%) | ||
| Ashkenazi | 595 (99.8) | 413 (96.3) |
| Ashkenazi/Sephardi | 1 (0.2) | 4 (0.9) |
| Sephardi | 0 | 7 (1.6) |
| Ashkenazi/non-Jewish | 0 | 2 (0.5) |
| Missing | 0 | 3 (0.7) |
Genotyping and Quality Control
Germline DNA was extracted from peripheral blood samples, purified, quantified by nanodrop and Qubit fluorometric quantitation, and genotyped by Affymetrix on a novel Axiom® miRNA Target Site Genotyping array with 237,858 SNPs and indels (Supplementary Table 1). Markers were selected for the microarray from four online bioinformatic databases: PolymiRTS (86,340), dPORE (10,400), Patrocles (1,200), and microRNA.org (158,400). These databases were leveraged to select polymorphic loci for the array that overlap genes encoding miRNAs, miRNA gene regulatory regions, proteins important for miRNA processing, and/or target seed sites (24–28). For microRNA.org, Affymetrix used the database’s high quality predictions of miRNA binding sites (both conserved and non-conserved) and intersected microRNA.org’s predicted sites with the 1000 Genomes Phase 1 (March 2012) release to identify markers. In addition, the array included a panel of n =4,470 ancestry informative markers (AIMs) and loci with known complex trait associations from the August 16, 2011 National Human Genome Research Institute (NHGRI) GWAS Catalog (29). In an ancillary study, MECC samples were also genotyped on a custom Affymetrix Axiom® platform with ~1.3 million SNPs and indels as part of the ColoRectal Transdisciplinary (CORECT) Study, and concordance was compared across the genotyping platforms. IMPUTE2 v2.2.2 was used to impute missing genotypes based on reference haplotypes from Phase I of the 1000 Genomes Project (March 2012 release; n = 1092) (30, 31).
MECC genotype data was filtered based on quality control metrics at the individual subject and SNP levels (Supplementary Figure 1). Samples with >5% missing genotypes, sex mismatches (between self-reported and genotypic predicted sex), and duplicate samples were identified and subsequently removed. Monomorphic markers and markers with <95% call rate were excluded. Further, SNPs that were not consistent with Hardy Weinberg Equilibrium (HWE) in controls were excluded. Principal components analysis (PCA) was conducted using a panel of AIMs and the pcaMethods Bioconductor package (32) in R to identify ethnic outliers for removal and to later adjust for population stratification. Principal component (PC) definitions of ancestry were used to exclude ethnic outliers and non-Ashkenazi Jewish individuals from the analysis. Thus, all individuals in the final analysis dataset (described in Table 1) were genetically of Ashkenazi Jewish descent, regardless of their self-reported ethnicity.
Gene expression quantification
Gene expression levels from 419,473 probe sets derived from two Affymetrix expression arrays were quantified on RNA isolated from snap frozen tumors of 331 MECC CRC cases. Of these 331 cases, 135 also had high-throughput genotype data available (63 on the Affymetrix Axiom® CORECT custom array and 72 on the Illumina HumanOmni 2.5S-v1 BeadChip). Methods for gene expression quantification via hybridization to GeneChip® Human Genome U133A 2.0 and Human Genome U133 Plus 2.0 Arrays have been described elsewhere (33). Briefly, expression was measured in two batches (one for each array) followed by quantile normalization and log2 transformation of MAS 5.0-calculated signal intensities. Data from the two batches were aligned after individual batch preprocessing and quality control. These microarray data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE26682) to comply with Minimum Information About a Microarray Gene Experiment (MIAME) guidelines.
Statistical Analysis
Logistic regression was employed to examine the marginal association between each marker on the miRNA target site array with MAF>=1% (nmarker = 55,208) and CRC risk assuming a log-additive genetic model. Here, each additional copy of the minor allele was assumed to confer the same magnitude of risk or protection. Each model was run both unadjusted and adjusted for sex, age, and the first two PCs. We calculated beta coefficients, standard errors, odds ratios (OR) with associated 95% confidence intervals, and p-values from unconditional logistic regression. The Bonferroni-corrected alpha level was set at 9.0×10−7 (0.05/55,406 SNPs). After taking this genome-wide approach, we then examined previously published SNPs from three studies in the candidate miRNA-related polymorphism literature to assess our ability to replicate purported risk loci (18–20).
To begin the bioinformatic characterization of functional consequences of our most significantly associated SNPs, we investigated predicted changes in miRNA binding using a combination of databases: microrna.org, miRBase, PolymiRTS, and dPORE (24, 25, 28, 34–37). MicroSNiPer was also used to identify the potential disruption or creation of miRNA binding sites for the following 3’-UTR SNPs in Table 2: rs3180466, rs1972820, and rs2985 (38). A seed site of a minimum of either 7 or 8 bases was specified for each of these SNPs. In addition, we conducted analysis of variance (ANOVA) to compare differences in gene expression by genotype for all SNPs with association p-values less than 5 × 10−4 as well as for a previously published risk locus, where expression and genotype data permitted. Expression of the gene nearest to each SNP was considered.
Table 2.
Summary of variants from the Axiom® miRNA Target Site Genotyping Array with the most statistically significant associations from age, sex, and PC 1-2 adjusted logistic regression models. A1 miRNA and A2 miRNA refer to miRNAs predicted to bind in the presence of each alternative allele. AIM = ancestry informative marker. GWAS = GWAS compatibility marker. NA = no prediction found. Grey indicates the availability of high-throughput genotype data and gene expression data for the nearest gene.
| rsID | CHR | BP | A1a | MAF | OR | SEb | P | Gene | A1 miRNA | A2 miRNA |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2985 | 6 | 34845648 | C | 0.30 | 0.66 | 0.10 | 3.7E-05 | UHRF1BP1 (intron) | miR-4529-5p | miR-885-5p |
| rs1139139 | 10 | 5020625 | T | 0.27 | 1.54 | 0.11 | 6.0E-05 | AKR1C1 (downstream) | NA | miR-451b miR-556-5p |
| rs6827968 | 4 | 161399652 | A | 0.34 | 1.48 | 0.10 | 1.0E-04 |
RAPGEF2 (downstream) FSTL5 (downstream) |
AIM | AIM |
| rs12130051 | 1 | 19545053 | T | 0.06 | 2.27 | 0.21 | 1.2E-04 | KIAA0090 (3'-UTR) | NA | miR-222 miR-1244 miR-3129 |
| rs80350662 | 1 | 178819064 | A | 0.04 | 2.63 | 0.26 | 2.1E-04 | RALGPS2 (intron) | miR-32-3p miR-4775 |
miR-1277-5p miR-889 |
| rs1834481 | 11 | 112023827 | G | 0.12 | 0.60 | 0.14 | 2.4E-04 | IL18 (intron) | NA | miR-637 miR-5009-5p miR-541-3p |
| rs1044724 | 6 | 125412231 | C | 0.08 | 0.54 | 0.17 | 2.5E-04 |
RNF217 (downstream) TPD52L1 (upstream) |
miR-3978 | miR-3978 |
| rs4766991 | 12 | 113137384 | T | 0.16 | 1.58 | 0.13 | 2.7E-04 |
PTPN11 (downstream) RPH3A (upstream) |
In promoter for miR-1302-1 | In promoter for miR-1302-1 |
| rs7746892 | 6 | 125408263 | G | 0.08 | 0.54 | 0.17 | 2.9E-04 |
RNF217 (downstream) TPD52L1 (upstream) |
miR-545 | miR-1252 miR-4476 miR-4533 miR-873 |
| rs7746860 | 6 | 125408221 | G | 0.08 | 0.54 | 0.17 | 2.9E-04 |
RNF217 (downstream) TPD52L1 (upstream) |
miR-2681 | miR-1295 miR-4747-3p |
| rs2489495 | 10 | 38502333 | T | 0.20 | 0.66 | 0.12 | 3.6E-04 | LOC1001290 55 (exon) | miR-635 | NA |
| rs853158 | 5 | 142605172 | C | 0.36 | 0.71 | 0.10 | 3.7E-04 | ARHGAP26 (3'-UTR) | miR-3926 | miR-4480 |
| rs9374072 | 6 | 109591586 | G | 0.31 | 0.71 | 0.10 | 3.7E-04 |
CEP57L1 (downstream) CCDC162 (upstream) |
miR-605 | NA |
| rs471429 | 6 | 125409031 | G | 0.09 | 0.57 | 0.16 | 3.9E-04 |
RNF217 (downstream) TPD52L1 (upstream) |
miR-3126-5p miR-3174 miR-3591-5p miR-3606 miR-4419a miR-4510 miR-921 |
miR-4270 miR-4441 |
| rs12268559 | 10 | 32856746 | C | 0.07 | 0.54 | 0.18 | 4.3E-04 | CCDC7 (coding) | miR-578 | NA |
| rs142004998 | 23 | 119760042 | C | 0.08 | 0.51 | 0.19 | 4.4E-04 | C1GALT1C1 (3'-UTR) | miR-1284 miR-337-3p miR-520d-5p |
NA |
| rs3180466 | 2 | 129023866 | G | 0.03 | 0.38 | 0.28 | 4.5E-04 | HS6ST1 (3'-UTR) | miR-4758-5p miR-574-5p miR-615-3p miR-1238-5p miR-4745-5p miR-3677-5p |
miR-4669 miR-3659 |
| rs79029362 | 1 | 178516408 | G | 0.03 | 2.91 | 0.30 | 4.6E-04 | C1orf220 (exon) | miR-455-3p | NA |
| rs4766992 | 12 | 113137531 | A | 0.26 | 1.45 | 0.11 | 4.7E-04 |
PTPN11 (downstream) RPH3A (upstream) |
Upstream of miR-1302- encoding gene | Upstream of miR-1302-encoding gene |
| rs56391924 | 10 | 32745248 | C | 0.07 | 0.54 | 0.18 | 4.7E-04 | CCDC7 (missense) | miR-4273 | NA |
| rs117299563 | 19 | 52095600 | C | 0.02 | 0.28 | 0.37 | 4.9E-04 | FLJ30403 (exon,3'-UTR) | NA | miR-3684 |
| rs6072275 | 20 | 39743905 | A | 0.15 | 1.59 | 0.13 | 4.9E-04 | TOP1 (intron) | GWAS | NA |
| rs107321 | 22 | 18512282 | T | 0.41 | 0.72 | 0.09 | 4.9E-04 | FLJ41941 (exon) | miR-1284 miR-337-3p miR-374a-5p |
NA |
| rs1972820 | 2 | 212243422 | G | 0.36 | 1.41 | 0.10 | 4.9E-04 | ERBB4 (3'-UTR) | miR-4633-5p miR-532-5p |
miR-3144-3p miR-875-5p |
| rs12247495 | 10 | 32802829 | C | 0.07 | 0.54 | 0.18 | 4.9E-04 | CCDC7 (intron) | miR-539-5p | NA |
A1 = effect allele for the corresponding odds ratios;
SE = standard error of the beta estimate
Results
Targeted genome-wide association analysis
Plots of the first 3 eigenvalues from MECC PCA indicated that the original samples selected for analysis included some non-Ashkenazi Jewish individuals (almost exclusively among subjects without CRC) that inhibited our ability to control for confounding due to population stratification through PC adjustment (data not shown). However, following the removal of 5 ethnic outliers and 211 non-Ashkenazis identified based on genetic definitions, the first 2 PCs were sufficient to control for the remaining population stratification, as indicated by genomic control lambda (GC λ) values shown below. The distributions of demographic and clinical characteristics of the final analysis dataset were comparable across case and control groups (Table 1).
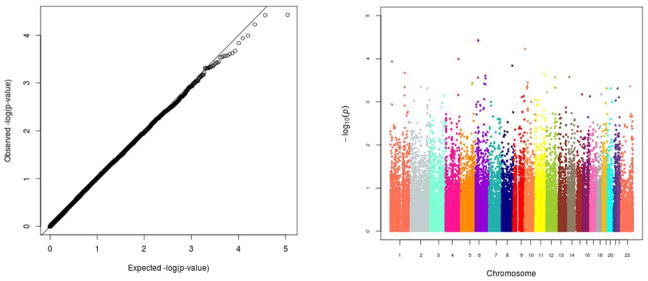
Quantile-quantile (Q-Q) and Manhattan plots visually display –log10(p-values) resulting from the logistic regression models adjusted for age, sex, and 2 PCs (Figure 1). The Q-Q plot in the left panel plots the rank-ordered observed –log10(p-value) against the rank-ordered expected –log10(p-value). It demonstrates that, on average, we did not observe SNPs with associations more statistically significant than expected under a uniform distribution of p-values. The GC λ value of 1 suggests that PCs 1 and 2 were sufficient to control for population stratification in our ethnically homogenous study sample. The Manhattan plot displays the summary results by ordered chromosomal position and shows that our lowest p-values are in the 10−5 range with none reaching genome-wide statistical significance after correction for multiple testing.
Figure 1.
Quantile-quantile and Manhattan plots of p-values for association between each genetic marker and colorectal cancer (CRC) from logistic regression adjusted for sex, age, and 2 PCs. Minor allele frequency>=1%; nmarker=55,406. Genomic control λ = 1.
Although none of the individual SNPs achieved genome-wide significance, our top findings are detailed in Table 2. Interestingly, seven out of our nine most statistically significant SNPs yield a predicted change in miRNA binding in an allele-specific manner. Each of these seven significant variants predict either a change from no miRNA binding to one or more miRNAs binding or from one set of miRNAs to a different set. None of the most significant miRNA SNPs has been previously reported as significantly associated with risk of CRC.
Gene expression analysis for top association findings
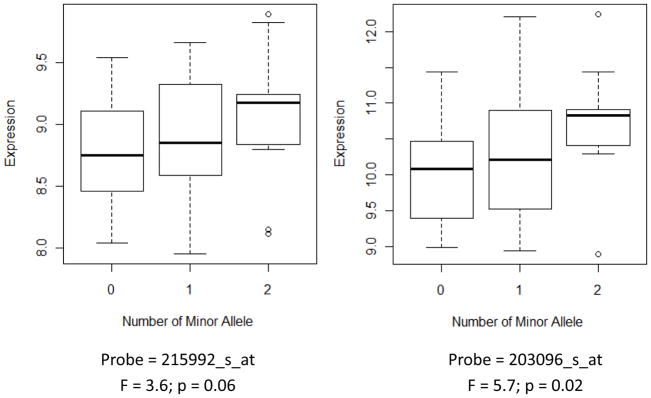
Of the top 25 SNPs that met our p-value threshold of 5 × 10−4 from the association analysis, 11 corresponding nearest genes had at least 1 matching probe in our gene expression dataset. Among the 21 total probes quantifying gene expression from these 11 genes (some genes had multiple probes), 13 probes for 6 genes had a corresponding genotype measured in MECC cases from the custom Affymetrix CORECT Axiom and/or Illumina Omni platforms. ANOVA results for gene expression [log2(normalized intensity)] by genotype for the 6 represented nearest genes with appropriate data availability revealed only one statistically significant SNP, intergenic rs6827968 that falls downstream of the RAP guanine nucleotide exchange factor 2 (RAPGEF2) gene (F=5.71; p-value=0.02). RAPGEF2 expression levels for two probes plotted against the number of copies of the minor allele at this SNP locus in our study sample can be visualized in Figure 2 , which provides evidence of an eQTL (probe 215992_s_at: F = 3.6, P = 0.06; probe 203096_s_at: F = 5.7, P = 0.02).
Figure 2.
RAPGEF2 gene expression [log2(normalized intensity)] in colorectal cancers measured by two separate probes in 72 MECC CRC cases by rs6827968 genotype.
Replication of previously published risk loci
We also examined the CRC association with 19 candidate miRNA SNPs previously presented in the literature (8 from Landi et al(18), 5 from Azimzadeh et al(19), and 6 from Naccarati et al(20)), of which 6 were statistically significant in the original report. In our dataset, we replicated only one of the previously reported findings (Table 3). The replicated variant (rs1051690), originally reported in Landi et al (18), is located in the 3’-UTR region of the insulin receptor gene INSR (Table 3; OR = 1.38; p = 0.03) and has predicted miRNA binding consequences. Our eQTL analysis demonstrated a statistically significant association between the rs1051690 variant and expression of the INSR gene, with increasing expression tracking with each additional copy of the minor allele (Supplementary Figure 2; F = 21.3; p = 8.98×10−6).
Table 3.
MECC association results from logistic regression adjusted for age, sex, and PC 1-2 for Axiom® miRNA Target Site Genotyping Array markers previously studied as miRNA-related variants. Data row 1 shows replication of a variant in INSR that was suggestively associated with CRC risk in prior publications by Landi et al in 2008 and 2011 (OR (95% CI) = 1.86 (0.99,3.50); P=0.052).
| SNP | CHR | Position | Gene | A1d | Published OR | Published P | MECC OR | MECC SEe | MECC P |
|---|---|---|---|---|---|---|---|---|---|
| rs1051690a | 19 | 7116963 | INSR | T | 1.86 | 0.05 | 1.38 | 0.15 | 0.03 |
| rs1368439a | 5 | 158742014 | IL12B | G | 1.17 | 0.65 | 0.80 | 0.12 | 0.06 |
| rs11515b | 9 | 21968199 | CDKN2A | G | 1.16 | 0.71 | 1.14 | 0.12 | 0.30 |
| rs1126547c | 3 | 14186757 | XPC | C | 1.13 | 0.73 | 1.14 | 0.13 | 0.32 |
| rs3135500a | 16 | 50766886 | NOD2 | A | 1.22 | 0.07 | 1.09 | 0.09 | 0.37 |
| rs1131445a | 15 | 81601782 | IL16 | C | 0.99 | 1.00 | 0.93 | 0.09 | 0.41 |
| rs1131445b | 15 | 81601782 | IL16 | C | 2.21 | 0.004 | 0.93 | 0.09 | 0.41 |
| rs1051208b | 3 | 12625747 | RAF1 | T | 1.11 | 0.85 | 0.90 | 0.14 | 0.44 |
| rs4596c | 11 | 18388128 | GTF2H1 | C | 0.79 | 0.03 | 0.93 | 0.10 | 0.44 |
| rs2229090c | 3 | 14187345 | XPC | C | 0.91 | 0.38 | 1.08 | 0.12 | 0.54 |
| rs17281995a | 3 | 121839641 | CD86 | C | 2.93 | 0.01 | 1.05 | 0.12 | 0.66 |
| rs11677a | 1 | 20301964 | PLA2G2A | T | 1.02 | 0.97 | 1.06 | 0.14 | 0.70 |
| rs7356c | 1 | 28218100 | RPA2 | C | 1.33 | 0.04 | 1.02 | 0.09 | 0.81 |
| rs1803541c | 2 | 128014913 | ERCC3 | T | 0.96 | 0.70 | 0.97 | 0.14 | 0.86 |
| rs16870224a | 5 | 40692940 | PTGER4 | A | 2.31 | 0.14 | 0.99 | 0.12 | 0.94 |
| rs16870224b | 5 | 40692940 | PTGER4 | A | 0.29 | 0.11 | 0.99 | 0.12 | 0.94 |
| rs4781563c | 16 | 14045399 | ERCC4 | A | 0.68 | 0.09 | 1.00 | 0.10 | 0.97 |
| rs916055a | 17 | 4534834 | ALOX15 | C | 0.98 | 0.91 | 1.00 | 0.10 | 0.98 |
| rs743554b | 17 | 73754248 | ITGB4 | A | 0.76 | 0.36 | NA | NA | NA |
Genotype concordance: miRNA targeted array vs. custom GWAS array
Only 14,436 markers were directly measured on both the Affymetrix miRNA Target Site Genotyping Array and the CORECT Axiom 1.3M custom array in the same set of samples. For the 14,436 overlapping markers, there was a 99.89% overall genotype concordance across arrays. However, because the number of directly measured markers shared by both arrays was low, we then compared genotypes for directly measured markers on the miRNA targeted array with 1000 Genomes imputed genotypes from the Axiom 1.3M custom array. Of the 88,205 directly genotyped, post-quality control markers on the miRNA targeted array, 63,407 were imputed with high quality from the Axiom 1.3M custom array. A comparison of the 63,407 miRNA targeted array genotypes and the corresponding best call genotypes derived from the Axiom 1.3M custom array imputation showed that heterozygote genotype concordance was severely depressed for SNPs with MAF<=5%. That is, consistent with prior GWAS studies using imputed genotypes, imputation did not perform well for the less common alleles important in the miRNA pathway and did not accurately reflect the directly measured genotypes.
Discussion
To our knowledge, this is the first study to examine associations between genetic variations in miRNA genes or target seed sites and CRC risk using a genome-wide approach informed by bioinformatic miRNA prediction algorithms. While we did not identify any genome-wide significant associations meeting the traditional threshold of 5×10−8, this study did highlight suggestive variants with predicted miRNA binding implications. These findings led us to replicate a previously reported association between rs1051690 in INSR and CRC risk and to demonstrate variability in INSR gene expression by genotype at this locus. While limited with respect to power, our initial study of only 596 cases and 429 controls demonstrated the potential for a targeted miRNA GWAS approach to identify candidate susceptibility loci and to prioritize them based on biological insights for further functional characterization.
Alterations of expression from miRNA targets may be mediated by seed site polymorphisms that strengthen or weaken the miRNA-mRNA interaction. We illustrate a relevant example in this study from an association finding through eQTL analysis, and more generally, demonstrate our novel approach (Figure 3). The INSR 3’-UTR variant (rs1051690) association with CRC had previously been detected in both Czech Republic and Spanish case-control studies assuming a co-dominant model (18, 21). We were able to replicate this risk locus based on a log-additive genetic model assumption. To date, few studies have examined the functional consequences miRNA-related SNPs. However, INSR is a notable exception. The same group that originally identified the INSR association later conducted in vitro luciferase reporter assays to show that the minor allele differentially regulates reporter gene expression (21). Evidence from our eQTL analysis corroborates this finding and provides an example of how such a target site polymorphism could influence that same gene’s expression in a dose-response manner (Supplementary Figure 2). A link between insulin resistance and CRC has long been recognized (39). It is possible that each additional copy of the minor/risk allele reduces miRNA-mRNA binding to the point of inhibiting mRNA degradation, which is what may lead to the increased INSR gene expression observation. It is also possible that the SNP exerts an effect analogous to haploinsufficiency, such that one copy of the major allele is not sufficient to appropriately repress INSR protein expression. Further functional work is necessary to elucidate this particular SNP’s mechanism of action.
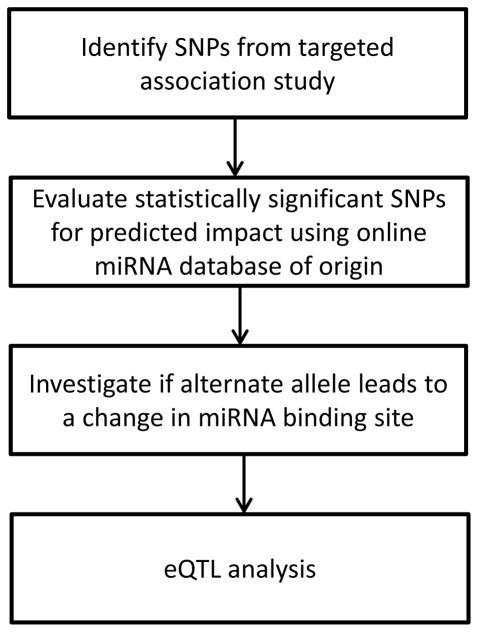
Figure 3.
Bioinformatic pipeline for predicted impact of statistically significant SNPs on miRNA target seed sites and cis gene expression. SNPs identified from the targeted association study were evaluated using PolymiRTS, Patrocles, and microRNA.org (in conjunction with markers identified by the 1000 Genomes Project) for indications of target seed sites that would be disrupted by the presence of the alternate allele for each genetic variant. Markers from the PolymiRTS database were further annotated for potential creation of new seed sites. Statistically significant markers were further investigated with MicroSNiPER to identify additional target seed sites both disrupted and potentially created by the presence of the alternate allele. The final step was cis-eQTL analysis where gene expression data was available.
Another illustrative example for the success of our approach lies with rs6827968, our third most statistically significant finding from the targeted GWAS that also showed evidence as a cis eQTL. Although rs6827968 is highly unlikely to exert a direct regulatory influence via the miRNA pathway on the nearest gene since it is an AIM, RAPGEF2 encodes a protein that could plausibly be linked to CRC etiology. RAPGEF2 activates RAS through promotion of the active GTP-bound state in a GTP/GDP-regulated signal transduction switch (40).
Sethupathy and Collins suggested in 2008 that studies elucidating the role of miRNA-related polymorphisms in complex diseases such as CRC should focus on three domains: genetic, functional (testing altered miRNA targeting mediated by genetic variation), and mechanistic (testing the mechanism by which altered miRNA leads to tumor development) (41). We provide evidence with respect to genetic and functional studies. The next step is to expand our genotyped dataset to increase power for detecting novel risk loci. With respect to genotyping platform for future studies, this study highlighted the advantage of this novel genotyping array over a traditional GWAS array with imputation based on the 1000 Genomes Project haplotypes, particularly when rare variants are of interest. A comparison of concordance between the Affymetrix miRNA Target Site Genotyping Array genotypes and imputed best call genotypes from the CORECT Axiom 1.3M custom array showed that the targeted miRNA array has added value over the GWAS array for the thousands of markers from this regulatory pathway with MAF<=5%. Also, functional studies are underway to identify SNP effects on miRNA binding fidelity (for rs1051690 as well as other top association findings) and to find the best in vitro model for allele-specific effects. Further, replication and fine-mapping will strengthen our confidence in both novel and previously published findings. Finally, this study suggests the benefit of reexamining previously published CRC susceptibility regions identified through GWAS for potential functional SNPs in miRNA binding sites or other miRNA pathway-related sequences. Given that most GWAS risk loci identified to date have MAF>=5%, reanalysis of existing, imputed GWAS datasets using the bioinformatic approaches described here has a high probability of yielding insights into the functional relevance of these regions.
This study has limitations with respect to power and modeling assumptions. Our sample size is limited to 1,025 samples. However, this analysis, which was able to replicate a previously identified miRNA risk locus and characterize preliminary functionality, provides justification for study in a larger sample. Our lack of genome-wide significant findings is likely attributable to a lack of power, and our sample size did not permit the investigation of effects for rare variants with MAF<1%. Also, not all SNPs exert their effects according to the assumed log-additive genetic model, and this choice made to restrict multiple testing could inhibit our ability to identify risk loci that are consistent with a recessive, dominant, or co-dominant model. Further, we did not consider interactions between these potentially risk-conferring variants or variant effects in the context of environmental risk factors. Finally, our ability to examine gene expression was limited by data availability and restriction to studying the SNP’s nearest gene.
Despite these limitations, we provide evidence that a targeted genome-wide approach for studying germline susceptibility can be extended beyond known or purported cancer biology pathways to the exploration of a regulatory pathway with widespread post-transcriptional effects. A better understanding of the mechanisms by which aberrations in miRNA expression and binding impact CRC development and progression may offer insights for prevention and targeted therapeutics. Specifically, the INSR variant warrants further investigation in a functional setting to elucidate its role in the alteration of CRC risk.
Supplementary Material
Acknowledgments
Financial Support
This work was supported by the National Cancer Institute at the National Institutes of Health (U19 CA148107 to S.B. Gruber, P30 CA014089 to S.B. Gruber, and R01 CA81488 to S.B. Gruber); the National Human Genome Research Institute at the National Institutes of Health (T32 HG000040 to S.L. Schmit); the National Institute of Environmental Health Sciences at the National Institutes of Health (T32 ES013678 to S.L. Schmit); and the Rackham Graduate School at the University of Michigan (Rackham Predoctoral Fellowship to S.L. Schmit). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We acknowledge the ColoRectal Transdisciplinary (CORECT) Study and its investigators for contributing genotypes for a comparative analysis of the miRNA targeted array versus a custom GWAS array.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–60. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 6.Fearon ER. Molecular genetics of colorectal cancer. Annual review of pathology. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 7.Vilar E, Tabernero J, Gruber SB. Micromanaging the classification of colon cancer: the role of the microRNAome. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:7207–9. doi: 10.1158/1078-0432.CCR-11-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, et al. The colorectal microRNAome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3687–92. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 11.Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, et al. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes & development. 2009;23:2152–65. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balaguer F, Moreira L, Lozano JJ, Link A, Ramirez G, Shen Y, et al. Colorectal cancers with microsatellite instability display unique miRNA profiles. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:6239–49. doi: 10.1158/1078-0432.CCR-11-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartley AN, Yao H, Barkoh BA, Ivan C, Mishra BM, Rashid A, et al. Complex patterns of altered MicroRNA expression during the adenoma-adenocarcinoma sequence for microsatellite-stable colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:7283–93. doi: 10.1158/1078-0432.CCR-11-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schee K, Fodstad O, Flatmark K. MicroRNAs as biomarkers in colorectal cancer. The American journal of pathology. 2010;177:1592–9. doi: 10.2353/ajpath.2010.100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu M, Chen H. The role of microRNAs in colorectal cancer. Journal of genetics and genomics = Yi chuan xue bao. 2010;37:347–58. doi: 10.1016/S1673-8527(09)60053-9. [DOI] [PubMed] [Google Scholar]
- 16.Ju J. miRNAs as biomarkers in colorectal cancer diagnosis and prognosis. Bioanalysis. 2010;2:901–6. doi: 10.4155/bio.10.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinci S, Gelmini S, Mancini I, Malentacchi F, Pazzagli M, Beltrami C, et al. Genetic and epigenetic factors in regulation of microRNA in colorectal cancers. Methods. 2013;59:138–46. doi: 10.1016/j.ymeth.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, Vodickova L, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–84. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 19.Azimzadeh P, Romani S, Mohebbi SR, Mahmoudi T, Vahedi M, Fatemi SR, et al. Association of polymorphisms in microRNA-binding sites and colorectal cancer in an Iranian population. Cancer genetics. 2012;205:501–7. doi: 10.1016/j.cancergen.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 20.Naccarati A, Pardini B, Stefano L, Landi D, Slyskova J, Novotny J, et al. Polymorphisms in miRNA-binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis. 2012;33:1346–51. doi: 10.1093/carcin/bgs172. [DOI] [PubMed] [Google Scholar]
- 21.Landi D, Moreno V, Guino E, Vodicka P, Pardini B, Naccarati A, et al. Polymorphisms affecting micro-RNA regulation and associated with the risk of dietary-related cancers: a review from the literature and new evidence for a functional role of rs17281995 (CD86) and rs1051690 (INSR), previously associated with colorectal cancer. Mutation research. 2011;717:109–15. doi: 10.1016/j.mrfmmm.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Fireman Z, Sandler E, Kopelman Y, Segal A, Sternberg A. Ethnic differences in colorectal cancer among Arab and Jewish neighbors in Israel. The American journal of gastroenterology. 2001;96:204–7. doi: 10.1111/j.1572-0241.2001.03476.x. [DOI] [PubMed] [Google Scholar]
- 23.Poynter JN, Gruber SB, Higgins PD, Almog R, Bonner JD, Rennert HS, et al. Statins and the risk of colorectal cancer. The New England journal of medicine. 2005;352:2184–92. doi: 10.1056/NEJMoa043792. [DOI] [PubMed] [Google Scholar]
- 24.Ziebarth JD, Bhattacharya A, Chen A, Cui Y. PolymiRTS Database 2.0: linking polymorphisms in microRNA target sites with human diseases and complex traits. Nucleic acids research. 2012;40:D216–21. doi: 10.1093/nar/gkr1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmeier S, Schaefer U, MacPherson CR, Bajic VB. dPORE-miRNA: polymorphic regulation of microRNA genes. PloS one. 2011;6:e16657. doi: 10.1371/journal.pone.0016657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiard S, Charlier C, Coppieters W, Georges M, Baurain D. Patrocles: a database of polymorphic miRNA-mediated gene regulation in vertebrates. Nucleic acids research. 2010;38:D640–51. doi: 10.1093/nar/gkp926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome biology. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic acids research. 2008;36:D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stacklies W, Redestig H, Scholz M, Walther D, Selbig J. pcaMethods--a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 2007;23:1164–7. doi: 10.1093/bioinformatics/btm069. [DOI] [PubMed] [Google Scholar]
- 33.Vilar E, Bartnik CM, Stenzel SL, Raskin L, Ahn J, Moreno V, et al. MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer research. 2011;71:2632–42. doi: 10.1158/0008-5472.CAN-10-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic acids research. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic acids research. 2008;36:D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic acids research. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffiths-Jones S. The microRNA Registry. Nucleic acids research. 2004;32:D109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barenboim M, Zoltick BJ, Guo Y, Weinberger DR. MicroSNiPer: a web tool for prediction of SNP effects on putative microRNA targets. Human mutation. 2010;31:1223–32. doi: 10.1002/humu.21349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giovannucci E. Insulin and colon cancer. Cancer causes & control : CCC. 1995;6:164–79. doi: 10.1007/BF00052777. [DOI] [PubMed] [Google Scholar]
- 40.Rebhun JF, Castro AF, Quilliam LA. Identification of guanine nucleotide exchange factors (GEFs) for the Rap1 GTPase. Regulation of MR-GEF by M-Ras-GTP interaction. The Journal of biological chemistry. 2000;275:34901–8. doi: 10.1074/jbc.M005327200. [DOI] [PubMed] [Google Scholar]
- 41.Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends in genetics : TIG. 2008;24:489–97. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.