Abstract
Objective
To evaluate HPV16 CpG methylation and methyl-haplotypes and their association with cervix precancer and cancer utilizing massively parallel single molecule next-generation sequencing (NGS).
Methods
A nested case-control study of HPV16 positive women was performed in a prospective cohort from Guanacaste, Costa Rica designed to study the natural history of HPV and cervical neoplasia. Controls encompassed 31 women with transient infections; there were 44 cases, including 31 women with CIN3 and 13 with cervical cancer. DNA samples from exfoliated cervical cells were treated with bisulfite and four regions (E6, E2, L2 and L1) were amplified with barcoded primers and tested by NGS. CpG methylation was quantified using a bioinformatics pipeline.
Results
Median methylation levels were significantly different between the CIN3+ cases versus controls in the E2, L2, and L1 regions. Methyl-haplotypes, specifically in 5 CpG sites included in the targeted L2 region, with the pattern “−−+−+” had the highest Area Under the Curve value (AUC = 88.40%) observed for CIN3 vs. controls. The most significant CpG site, L2 4277, determined by bisulfite NGS had an AUC = 78.62%.
Conclusions
This study demonstrates that NGS of bisulfite treated HPV DNA is a useful and efficient technique to survey methylation patterns in HPV16. This procedure provides quantitative information on both individual CpG sites and methyl-haplotypes that identify women with elevated present or subsequent risk for HPV16 CIN3 and cancer.
Introduction
Cervical cancer is the third most commonly diagnosed cancer and the fourth leading cause of cancer deaths in women worldwide [1]. There are approximately 530,000 new cases and 275,000 associated deaths annually [2]. Nearly all cases of cervical cancer are caused by a persistent infection with a high-risk (HR) type of human papillomavirus (HPV), which includes approximately 12 genotypes within the genus Alphapapillomavirus [3]. Of these, HPV16 and HPV18 are the two most important carcinogenic HPV types and together cause 70% of cervical cancer and 50% of the precancerous lesions, specifically cervical intraepithelial neoplasia (CIN) grade 3 (CIN3) [4-6]. Cervical cancer develops over decades from an acute infection with a carcinogenic HPV type that is maintained as a persistent infection [7], whereas the majority of infections clear spontaneously [8].
Prevention of cervix cancer has been accomplished through Pap test screening and more recently co-testing that also includes Pap and HPV testing. Most recently, primary stand-alone HPV testing is also an option [9]. Management options for abnormal screening tests include colposcopy versus close-surveillance [10]. A positive HPV result, however, has insufficient specificity since it does not discriminate between HPV-associated cancer-relevant lesions (CIN2+) and transient, clinically benign infections [11, 12]. HPV persistence represents a critical distinction between infections with substantial risk of progression to CIN3 or cancer and those that are benign or transient [13]. Referral rates to colposcopy are high in screening settings and diagnostic tests, which can readily distinguish between women with cervical precancer from those transiently infected are necessary to improve the utility of HR-HPV testing [11, 12]. With the FDA approval of multiple HR-HPV tests and recent data indicating the advantages of HPV testing as part of the secondary prevention of cervical cancer, differentiation between benign cervical HR-HPV infections and those at risk to progress represents an urgent and significant clinical challenge [10].
The molecular mechanisms underlying persistence and progression to precancer and cancer are largely unknown [10, 14]. As a result, there is a clinical need for additional biomarkers, particularly amongst HPV+/cytology- women; a substantial proportion of women tested for HPV in cervix cancer screening programs [10]. An area of promising study is the epigenetic modifications, i.e., DNA methylation of the viral double-stranded DNA genome as a biomarker for women at risk for cancer [15]. Moreover, these epigenetic changes may also provide insights into the molecular mechanisms of progression [16, 17]. In recent years, multiple studies have focused on evaluating the association of HPV16 genome methylation with cervical precancer and cancer [18-25]. These studies provide a consistent model whereby the HPV16 genome shows increased DNA CpG methylation in women with precancerous lesions and cervical cancer, compared to women who are able to clear their infection [18-20, 26].
In this study, we use an emerging technology for detection of HPV methylation by sequencing bisulfite-treated HPV16 DNA using massively parallel single molecule sequencing. Specifically, we evaluated the HPV16 methylome for methyl-haplotypes, the combination of methylated CpG sites on a single molecule as previously described [24], combined with quantifying CpG methylation sites. The methyl-haplotype is the combination of the methylation statuses of CpG sites (+, the C is methylated; −, the C is unmethylated) in cis determined from a single read. For example, a DNA amplicon with 3 CpG sites has the following eight possible methyl-haplotypes: −−−, +−−, −+−, −−+, ++−, +−+, −++, +++. With the anticipated widespread implementation of HPV DNA testing for cervix cancer prevention, the clinical objective of this study was to determine the significance of HPV16 methyl-haplotypes to identify HPV16-positive women with or at risk for CIN3+ in a prospective study previously evaluated for single site methylation [23].
Methods
Study population
Cervical specimens were selected from a random sample, population-based longitudinal study of the natural history of HPV infection and cervical neoplasia amongst 10,049 women in Costa Rica [27]. Women over the age of 18 were recruited for screening between June 1993 and December 1994 and were followed for up to 7 years. At each clinical visit, a cervical sample was taken from the cervix for HPV testing [27, 28]. The study protocol was reviewed and reapproved annually by the National Cancer Institute and Costa Rican Institutional Review Boards [27].
Cervical samples, DNA isolation, and HPV16 detection
For the current nested case-control study, 75 women with single HPV16 type infections were selected for 3 main infection outcomes: (1) transient or cleared, 31 women; (2) CIN3, 31 women; and (3) cervical cancer, 13 women. In addition, there were 13 women who had multiple specimens prior to the diagnosis of CIN3 or cancer. DNA was isolated and previously tested for the presence of HPV16 by MY09/MY11 PCR and type specific oligonucleotide (dot blot) hybridization [29].
Bisulfite treatment
Cervical DNA samples were treated with bisulfite using the EZ DNA methylation kit (Zymo Research, Orange, CA), as recommended by the manufacturer. In brief, 25 μl of extracted DNA from cervical samples were mixed with dilution buffer and denatured at 37° C for 15 minutes. Following denaturation, 100 μl of freshly prepared sodium bisulfite was added and the samples were incubated at 50° C for 16 hours. Post-treatment, samples were desulphonated, washed with 70% ethanol, eluted in 30 μl of elution buffer and stored at −20°C. Following bisulfite conversion and PCR amplification, unmethylated C’s are converted to T’s, whereas methylated C’s are retained as C’s [30]. The ratio of C/C+T indicates the proportion of methylated cytosines at each CpG site in the assayed sample.
Primer design, PCR amplification and NG sequencing
Four regions within the HPV16 genome were selected for testing using NG sequencing assays for quantitating CpG methylation based on our previous data using pyrosequencing [20]. Methylation sensitive primers based on converted C to T changes at non-CpG sites after bisulfite treatment were designed using MethPrimer (http://www.urogene.org/methprimer/index1.html) [31]. The primers were designed to amplify fragments containing- 3 CpG sites in the E6 Open Reading Frame (ORF) region (494, 502, 506); 9 CpG sites in the E2 ORF region (3412, 3415, 3417, 3433, 3436, 3448, 3462, 3473, 3496); 5 CpG sites in the L2 ORF (4240, 4249, 4261, 4270, 4277); and 4 CpG sites in the L1 ORF (7034, 7091, 7136, 7145). In total, 21 CpG sites across four open reading frames were surveyed. Each forward primer contained a specific 8 base pair barcode to identify each sample (see Bioinformatics below). Oligonucleotide primers were synthesized by Integrated DNA Technologies (IDT, Coralville, IA) (see TableS1).
Each reaction mixture contained 1 μl of bisulfite-treated DNA, 400 nM dNTP, 4.0 uM MgCl2, 2.5 μl 10× Buffer, 0.2 uM of the forward and reverse primers, and 0.08 units of HotStart-IT polymerase (United States Biochemicals, Cleveland, OH) in a total reaction volume of 25 μl. The PCR conditions and annealing temperatures for each assay are provided in Table S1. After confirming the signal intensity of each PCR product on a 3% agarose gel, the amplicons were pooled at similar molar quantities. The PCR products from each of the four assays were pooled separately and isolated by electro-elution, precipitated with isopropanol and 3M NaOAc (Sodium Acetate) and resuspended in 50 μl of Elution Buffer as previously described [25]. The isolated DNA’s were submitted for library construction and sequenced using paired-end reads on an Illumina HiSeq 2000 at the Albert Einstein Epigenomics Core Facility.
Bioinformatics and determination of CpG methylation status
Illumina sequencing data files were first filtered for low quality reads using a minimum average read PHRED score of 25 and minimum nucleotide PHRED score of 25 in a 5 bp sliding window [32, 33]. The data was then demultiplexed based on the 8 bp DNA Hamming barcodes to assign sequencing reads to their sample of origin [34]. Quality control and demultiplexing were performed using the mubiomics package [35]. Following demultiplexing, we employed Bismark (The Babraham Institute, Cambridge, UK) [36]. Bismark first builds a bisulfite-converted reference library from DNA sequences (e.g., HPV16 genomic sequences). It then maps paired-end reads to the bisulfite converted library to determine the methylation state of CpG, and non-CpG cytidines [36]. Three Python scripts (Python is a programming language with many open source commands or scripts), which operate in a pipeline, were developed in our laboratory to process the Bismarck output. The methtable.py script produced a pattern of methylation for every sequenced molecule, indicating whether cytidines in the assay were methylated (+), unmethylated (−), missing (o) or in disagreement for reads from each direction (x). The methcount.py script then generated counts of each unique pattern for each sample. Finally, the methsummarize.py script produced site-specific methylation percentages for each assayed cytidine in each sample by comparing the ratio of methylated cytidines (C) to the total number of methylated and unmethylated cytidines (T + C) at each cytidine in the assayed fragment. All scripts are available from the authors upon request.
Statistical Methods
All observations with a total number of reads less than 500 were removed from analysis. To evaluate this cut point, data were randomly sampled at increasing read counts and median methylation was calculated and stratified by infectious outcome. In each instance, the median was stable after 500 reads. Based on these criteria the number of samples varies for each PCR assay (see Table 1).
Table 1. Association of HPV16 methylation levels at specific CpG sites and infection outcomes.
| Median (Range) | FDR Adjusted | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 3 groupsa |
3 groupsb |
Controls | Controls | CIN 3 | |||||
| Gene | CpG Site | Controls | CIN 3 | Cancer | KW | KW | vs. CIN3c | vs. Cancerc | vs. Cancerc |
| N=21 | N=29 | N=13 | |||||||
| E6 | 494 | 1.16(0.60, 3.06) | 1.54(0.32, 82.98) | 1.76(0.62, 83.62) | 0.26 | 0.35 | 0.35 | 0.35 | 0.75 |
| 502 | 1.45(0.68, 4.63) | 2.24(0.49, 86.32) | 2.52(0.64, 84.86) | 0.24 | 0.32 | 0.32 | 0.32 | 0.71 | |
| 506 | 1.05(0.48, 1.92) | 1.24(0.43, 84.27) | 1.12(0.53, 84.02) | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | |
| N=18 | N=28 | N=13 | |||||||
| E2 | 3412 | 0.43(0.18, 5.36) | 1.68(0.18, 97.03) | 5.40(0.22, 91.66) | 0.01 | 0.02 | 0.02 | 0.02 | 0.34 |
| 3415 | 0.42(0.25, 4.38) | 1.09(0.12, 90.25) | 4.56(0.26, 84.60) | 0.03 | 0.05 | 0.16 | 0.02 | 0.16 | |
| 3417 | 0.28(0.19, 1.90) | 0.62(0.12, 95.48) | 2.07(0.22, 84.91) | 0.01 | 0.01 | 0.07 | 0.01 | 0.12 | |
| 3433 | 0.57(0.32, 8.64) | 3.19(0.21, 93.04) | 5.82(0.23, 89.15) | 0.05 | 0.07 | 0.07 | 0.07 | 0.56 | |
| 3436 | 0.42(0.23, 7.23) | 1.94(0.22, 95.96) | 4.91(0.26, 90.15) | 0.00 | 0.01 | 0.01 | 0.01 | 0.26 | |
| 3448 | 1.12(0.28, 7.53) | 1.94(0.11, 90.73) | 5.72(0.21, 91.77) | 0.45 | 0.60 | 0.60 | 0.60 | 0.60 | |
| 3462 | 0.39(0.22, 4.98) | 1.23(0.11, 83.41) | 1.45(0.21, 75.67) | 0.11 | 0.19 | 0.19 | 0.18 | 0.40 | |
| 3473 | 0.48(0.25, 3.78) | 0.88(0.21, 77.72) | 2.13(0.28, 74.86) | 0.05 | 0.10 | 0.34 | 0.06 | 0.11 | |
| 3496 | 0.74(0.32, 11.11) | 2.89(0.11, 92.41) | 12.80(0.51, 83.27) | 0.00 | 0.00 | 0.05 | 0.00 | 0.05 | |
| N=25 | N=29 | N=11 | |||||||
| L2 | 4240 | 5.55(0.34, 96.31) | 20.71(1.32, 88.35) | 34.64(2.01, 87.83) | 0.00 | 0.01 | 0.02 | 0.01 | 0.12 |
| 4249 | 1.17(0.34, 95.34) | 3.98(0.49, 69.56) | 4.64(0.52, 67.18) | 0.01 | 0.02 | 0.02 | 0.03 | 0.50 | |
| 4261 | 1.24(0.10, 95.66) | 8.81(0.75, 87.82) | 17.20(1.41, 84.90) | 0.00 | 0.00 | 0.00 | 0.00 | 0.21 | |
| 4270 | 0.87(0.30, 32.12) | 7.11(0.59, 75.48) | 6.24(0.69, 67.13) | 0.00 | 0.00 | 0.00 | 0.00 | 0.42 | |
| 4277 | 1.67(0.25, 95.55) | 7.78(0.84, 82.03) | 12.63(0.52, 83.74) | 0.00 | 0.00 | 0.00 | 0.00 | 0.27 | |
| N=17 | N=28 | N=13 | |||||||
| L1 | 7034 | 0.76(0.11, 12.09) | 3.52(0.37, 45.65) | 9.27(1.22, 86.42) | 0.01 | 0.01 | 0.04 | 0.01 | 0.06 |
| 7091 | 13.06(0.76, 41.66) | 15.66(0.59, 82.04) | 32.49(1.78, 93.89) | 0.09 | 0.12 | 0.91 | 0.10 | 0.10 | |
| 7136 | 1.46(0.26, 28.20) | 3.37(0.48, 75.76) | 9.27(0.89, 92.19) | 0.02 | 0.05 | 0.10 | 0.04 | 0.10 | |
| 7145 | 1.03(0.18, 14.79) | 6.02(0.31, 76.45) | 12.10(1.01, 91.50) | 0.01 | 0.01 | 0.01 | 0.01 | 0.20 | |
P value <0.05, or significant at the accepted rate of FDR
N, number of samples; KW, Kruskal-Wallis test; FDR, False Discovery Rate
p-value corresponding to association between methylation at specific CpG sites with the 3 infection outcomes
p-value corresponding to association between methylation at specific CpG sites with the 3 infection outcomes
p-value corresponding to association between methylation at specific CpG sites with Controls versus CIN3 or Cancer; and CIN3 versus Cancer, FDR Adjusted (Benjamini-Hochberg)
Single site CpG methylation was summarized by median and range within each of the samples for controls, CIN3, or cancer. In order to assess differences amongst the three outcome groups, Kruskal-Wallis tests were performed and where significant, the corresponding Mann-Whitney test was further utilized. The p-values were adjusted to account for false discovery rates (FDR) that were below a nominal 0.05 [37].
To determine the magnitude of association of single CpG sites and methyl-haplotype patterns with clinical outcome, odds ratios (ORs) and 95% confidence intervals were computed by dichotomizing results using the median methylation levels of a single site or pattern. ROC analyses were performed to assess the discrimination between CIN3 and transient infection of using either methylation at single sites or methyl-haplotype patterns. The area under the curve (AUC) and 95% CI of single sites and the best 5 patterns are reported. ROC curves of the single sites and patterns with highest AUC were overlaid to compare the discrimination achieved with methyl-haplotypes compared to a single CpG site.
The longitudinal serial samples from subjects who developed CIN3+ were grouped in time periods based on years to diagnosis. For each time period, the median methylation percentages were calculated. This procedure was performed for single sites (data not shown) in each segment and for the methyl-haplotype patterns within each region (top 5 patterns in this analysis) and are presented. All inference based tests were held at an α level of 0.05 and all statistical computing was performed in the R environment version 2.12.2 [38].
NG Sequencing Validation
A subset of CpG methylation data from NG assays were compared with pyrosequencing methylation data that was previously determined [15]. Agreement with available pyrosequencing data was evaluated using linear regression and a Pearson’s correlation coefficient (r). Linear regression curves indicated excellent agreement between NG sequencing and pyrosequencing results for single CpG site methylation with overall r = 0.99 for the E6 region (n=14), r = 0.91 for E2 (n=16), r = 0.97 for L2 (n=16), and r = 0.97 for the L1 region (n=11). This agreement is similar to that detected in our previous studies comparing HPV16 pyrosequencing and NGS methylation data [24, 25].
Results
Analyses of Single CpG Site Quantitative Methylation determined by bisulfite NGS
The association of HPV16 methylation levels at specific CpG sites among 3 infectious outcomes (transient infections no disease, CIN3, and cancer) was evaluated using NGS (see Table 1). Prior to accounting for multiple testing (unadjusted), methylation levels were significantly different among the 3 groups; 6 of 9 CpG sites in the E2 region, all 5 CpG sites in the L2 region, and 3 of 4 CpG sites in the L1 region. Methylation levels were significantly higher in patients with CIN3 and cancer. After accounting for multiple testing using the Benjamini-Hochberg method (FDR Adjustment), median methylation levels were significantly different among the 3 groups in 4 of the 9 CpG sites in E2, all 5 CpG sites in L2, and 3 of 4 CpG sites in L1. Overall, the level of CpG methylation was low in the E6 region and no significant differences were noted among the 3 groups.
We examined the differential methylation among patients in the control group versus patients with CIN3 and cancer. Comparing the control group to patients with CIN3, we observed that 3 of 9 CpG sites in the E2 region, all 5 CpG sites in the L2 region, and 2 of 4 CpG sites in the L1 region had significantly higher median methylation percent levels in patients with CIN3. As expected, there were also significantly higher median methylation levels noted when comparing patients with cancer vs. controls in 5 of 9 CpG sites in the E2 region, all 5 CpG sites in the L2 region, and 3 of 4 CpG sites in the L1 region (see Table 1). Of the analyzed sites, one CpG site in the E2 region (site 3496) revealed significant differences in median methylation levels in cervical cells from patients in the CIN3 group compared to those with cancer.
Magnitude of association of CIN3 and cancer with NG determined methyl-haplotype and single CpG site methylation levels
Next we compared association of methylation levels of single CpG sites versus methyl-haplotypes for CIN3. Table 2 shows the Odds Ratios (OR) and the mean Area Under the Curve (AUC) values at single HPV16 CpG sites. The association of single CpG sites in patients with CIN3 vs. patients in whom the HPV16 infection cleared was statistically significant in 5 of 9 CpG sites in the E2 region, 5 of 5 CpG sites in the L2 region, and 3 of 4 CpG sites in the L1 region. The OR was highest (30.3) at sites 4270 and 4277 within the L2 region (p<0.0001). The highest AUC for single sites were noted at the CpG sites 3436 in the E2 region (72.70%), 7145 in the L1 region (73.32%), and 3 sites in the L2 region – 4261 (75.45%), 4270 (77.93%), and 4277 (78.62%), the last having the highest mean AUC (p<0.0001).
Table 2. Odds Ratios and ROC Analyses of CIN3 vs. Controls based on NGS determined HPV16 Methylation Levels.
| Gene | CpG Site | Odds Ratio (95% Confidence Interval) |
p-value | Mean AUC % (95% Confidence Interval) |
p-value |
|---|---|---|---|---|---|
| E6 | 494 | 1.8 (0.59, 5.49) | 0.15 | 62.89% (58.61, 67.40) | 0.06 |
| 502 | 2.09 (0.68, 6.43) | 0.10 | 63.22% (58.97, 67.05) | 0.06 | |
| 506 | 2.09 (0.68, 6.43) | 0.10 | 53.04% (47.51, 57.51) | 0.36 | |
| E2 | 3412 | 3.61 (1.12, 11.68) | 0.02 | 69.84% (65.00, 71.85) | 0.01 |
| 3415 | 2.20 (0.72, 6.74) | 0.08 | 61.59% (58.20, 65.22) | 0.08 | |
| 3417 | 3.61 (1.12, 11.68) | 0.02 | 66.19% (61.45, 69.61) | 0.03 | |
| 3433 | 3.61 (1.12, 11.68) | 0.02 | 67.94% (66.21, 71.67) | 0.02 | |
| 3436 | 5.50 (1.58, 19.09) | 0.00 | 72.70% (66.89, 76.67) | 0.00 | |
| 3448 | 1.65 (0.55, 4.97) | 0.19 | 59.05% (56.94, 62.22) | 0.14 | |
| 3462 | 2.20 (0.72, 6.74) | 0.08 | 62.06% (57.37, 63.84) | 0.07 | |
| 3473 | 1.65 (0.55, 4.97) | 0.19 | 57.94% (49.17, 66.67) | 0.17 | |
| 3496 | 3.03 (0.96, 9.55) | 0.03 | 66.51% (64.40, 71.94) | 0.02 | |
| L2 | 4240 | 3.40 (1.10, 10.53) | 0.02 | 69.93% (67.00, 74.75) | 0.01 |
| 4249 | 5.20 (1.56, 17.32) | 0.00 | 71.72% (66.38, 75.42) | 0.00 | |
| 4261 | 14.63 (3.24, 65.94) | 0.00 | 75.45% (72.07, 78.93) | 0.00 | |
| 4270 | 30.33 (4.95, 185.94) | 0.00 | 77.93% (74.83, 81.44) | 0.00 | |
| 4277 | 30.33 (4.95, 185.94) | 0.00 | 78.63% (75.00, 81.77) | 0.00 | |
| L1 | 7034 | 6.75 (1.72, 26.47) | 0.00 | 69.33% (65.38, 74.75 | 0.02 |
| 7091 | 1.30 (0.40, 4.22) | 0.33 | 51.05% (49.26, 56.59) | 0.46 | |
| 7136 | 5.18 (1.40, 19.19) | 0.01 | 65.13% (60.16, 70.75) | 0.05 | |
| 7145 | 6.75 (1.72, 26.47) | 0.00 | 73.32% (69.50, 80.00) | 0.00 |
P value <0.05 is significant
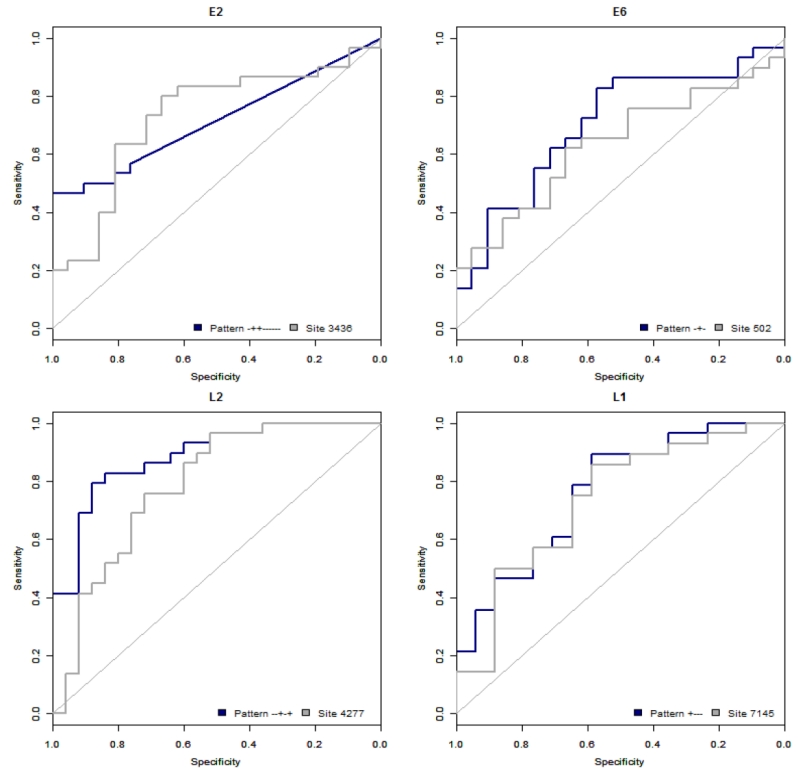
We examined the utility of using methyl-haplotypes that are generated from single molecule reads that reveal CpG methylation states “in phase”. As previously described, a pattern is comprised of “+’s” which represent methylated sites at a given position and “−’s”, which represent sites that were not methylated [24, 25]. For example, a pattern “+−+−“ with a count of 1,400 represents 1400 single molecules sequenced by Illumina in which site 1 and site 3 were methylated, while sites 2 and 4 were unmethylated in each read. We hypothesized that methy-haplotypes may be more informative of the methylation status in a specific region of the HPV16 genome than single CpG sites and thus analyzed the ORs and AUCs for methyl-haplotypes calculated from each assay comparing cases and controls (Table 3). Table 3 displays the 5 methyl-haplotypes with the highest mean AUCs and corresponding OR’s per segment of HPV16. The highest ORs and mean AUCs observed were in the L2 region, with the pattern “−−+−+” being most strongly associated with CIN3 compared to patients that cleared HPV, having an OR of 25.85 and an AUC of 88.40%. When comparing single CpG sites and methyl-haplotypes, the L2 methyl-haplotypes had the highest AUCs (AUC 88.40% vs. 78.62%) compared to the single CpG site L2 4277 that had the highest AUC for single sites (see Table 2 and Figure 1)
Table 3. HPV16 Methyl-haplotypes associated with CIN3 vs. Controls.
| Gene | Pattern | Odds Ratio (95% Confidence Interval) |
p-value | AUC % (95% Confidence Interval) |
p-value |
|---|---|---|---|---|---|
| E6 | −+− | 5.68 (1.54, 20.96) | 0.00 | 69.90% (68.79, 72.93) | 0.01 |
| − − − | 0.41 (0.13, 1.27) | 0.06 | 67.72% (66.38, 70.24) | 0.98 | |
| +−− | 1.73 (0.56, 5.31) | 0.17 | 64.97% (63.62, 67.59) | 0.04 | |
| −−+ | 0.97 (0.32, 2.93) | 0.48 | 60.78% (58.62, 63.28) | 0.11 | |
| ++− | 1.73 (0.56, 5.31) | 0.17 | 60.10% (58.36, 62.41) | 0.11 | |
| E2 | −++−−−−−− | 1.42 (0.03, 74.31) | 0.43 | 71.08% (70.44, 72.67) | 0.00 |
| +−−−+−−−− | 1.42 (0.03, 74.31) | 0.43 | 70.25% (69.79, 74.17) | 0.00 | |
| +−−+−−−−− | 1.42 (0.03, 74.31) | 0.43 | 69.5% (68.97, 72.50) | 0.00 | |
| −−−−+−−−+ | 1.42 (0.03, 74.31) | 0.43 | 68.56% (68.06, 71.25) | 0.01 | |
| −−++−−−−− | 1.42 (0.03, 74.31) | 0.43 | 68.47% (67.82, 71.00) | 0.01 | |
| L2 | −−+−+ | 25.85 (4.22, 158.43) | 0.00 | 88.40% (88.07, 90.52) | 0.00 |
| ++−+− | 1.16 (0.02, 60.42) | 0.47 | 82.40% (82.14, 85.49) | 0.00 | |
| +−−+− | 5.77 (1.63, 20.41) | 0.00 | 82.29% (81.97, 86.06) | 0.00 | |
| −−−−+ | 12.46 (2.76, 56.19) | 0.00 | 80.89% (80.57, 84.14) | 0.00 | |
| −−−++ | 8.00 (2.06 31.00) | 0.00 | 78.94% (78.59, 82.00) | 0.00 | |
| L1 | +−−− | 7.41 (1.74, 31.60) | 0.00 | 75.82% (74.78, 79.69) | 0.00 |
| −−+− | 5.33 (1.36, 20.91) | 0.01 | 73.44% (72.10, 78.35) | 0.00 | |
| −−−+ | 4.09 (1.10, 15.17) | 0.02 | 72.99% (71.88, 77.90) | 0.00 | |
| −+−+ | 3.26 (0.91, 11.63) | 0.03 | 70.98% (69.98, 75.22) | 0.01 | |
| ++−+ | 1.63 (0.03, 85.85) | 0.40 | 70.15% (69.72, 74.55) | 0.01 |
P value <0.05 is significant
Figure 1. Receiver operating characteristic curves for highest AUC of methyl-haplotype and single-site per region.
Receiver operating characteristic (ROC) curves with highest AUCs for methyl-haplotypes and single-sites in HPV16 regions (A) Region E2, (B) Region E6, (C) Region L2, (D) Region L1. The % sensitivity, the true positive rate, is given along the y-axis versus 1-specificity along the x-axis, with a diagonal reference line.
Longitudinal analyses
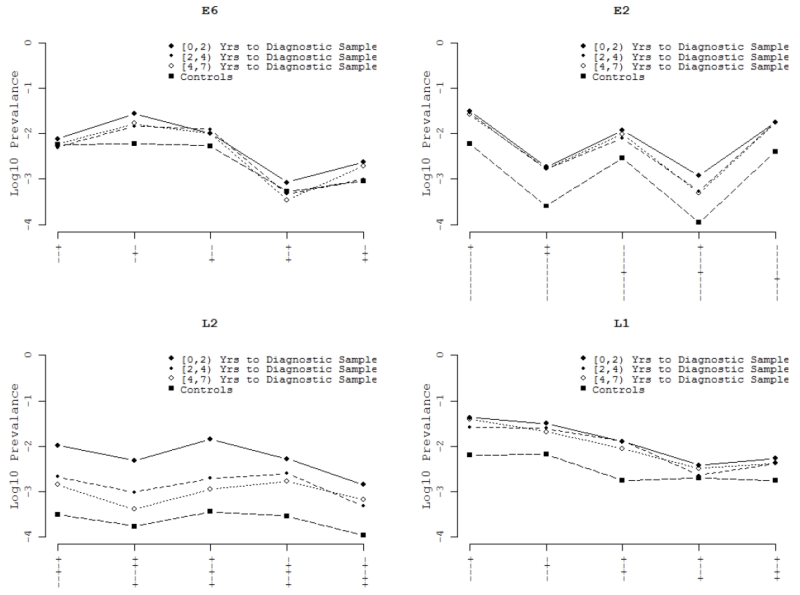
To explore the predictive value of CpG methylation, we analyzed the data on 13 women with persistent HPV16 infection, who had multiple cervical samples (ranging from 2-6 samples per patient) collected at successive screening visits prior to their CIN3 or cancer diagnosis. In this sub-analysis, we evaluated HPV16 methyl-haplotypes in each successive specimen up to and including the diagnostic specimen. Two distinct patterns were observed, one where the median percent methyl-haplotype increased as the interval to diagnosis decreased (see L2 patterns), and a second pattern where the median percent methyl-haplotype remained relatively constant over the 7 year period (see E2 and L1 patterns, Figure 2).
Figure 2. Longitudinal analysis of methyl-haplotypes with highest AUC per region.
Mean % methylation by methyl-haplotypes for 30 serial samples from 13 women collected 0-7 years before diagnosis of CIN3+ in HPV16 regions (A) Region E6, (B) Region E2, (C) Region L2, (D) Region L1. The legend within the figure indicates the symbol and time interval to diagnostic samples. The x-axis indicates each individual methyl-haplotype by gene region. The y-axis indicates mean log10 prevalence or the % of pattern over the total patterns.
Discussion
This study tested the use of next-gen sequencing of bisulfite treated DNA from exfoliated cervical cells to quantitate HPV16 CpG methylation sites and methyl-haplotypes and their association with cervix precancer/cancer. This study builds upon the previous validation of the bisulfite-NG method and utilizes this technology to evaluate HPV16 CpG methylation and methyl-haplotypes as a marker for cervix precancer and cancer. Moreover, the analysis on methyl-haplotypes is only beginning to emerge as a method of evaluating cervix cancer risk and disease detection. We analyzed HPV16 DNA methylation at CpG sites in 4 regions (E6, E2, L1, and L2) amplified as single fragments of the HPV16 genome utilizing barcoded primers to identify individual samples and specific fragments. This is the third study by our group to utilize NG high-throughput sequencing for epigenomic analysis of HPV16 and supports the laboratory efficiency compared to other methods [24, 25]. The current study also reports analyses of new CpG sites in the E2 ORF that were not previously analyzed by pyrosequencing, in addition to applying this technique to a cohort of women from a population-based longitudinal study. The main conclusion is that this method is simpler, cheaper and provides additional information than the current “gold standard” of pyrosequencing [15].
Our findings further strengthen HPV methylation studies by the additive ability to analyze HPV16 methyl-haplotypes using NG sequencing technology. Next-generation (NG) sequencing can be utilized to evaluate the methylation status of bisulfite-treated DNA by sequencing a particular amplicon at the single molecule level evaluating multiple CpG sites in a gene region [15, 39]. This contrasts the laborious pyrosequencing method that provides individual site quantitative methylation data by sampling a heterogeneous DNA population of DNA molecules [40]. The use of NG sequencing also provides additional information about methylation at multiple CpG sites in cis across a DNA fragment, allowing reconstruction of methyl-haplotypes or patterns of methylation by evaluating a string of CpG sites on single molecules. By analyzing methyl-haplotypes or patterns of methylation, we were able to conclude that specific patterns in the E2, L2, and L1 regions were as strong or showed stronger association with CIN3, the true precancer lesion needing treatment, than single CpG sites alone. Specifically, comparing cases and controls, the ORs and mean AUC’s were highest in the L2 region, reaching 88.4% for the pattern “−−+−+” compared to highest mean AUC of 78.62% for single CpG site 4277 in the L2 region (Figure 1). In the same cohort, Mirabello et al. [23], reported the highest AUC (82%) at position L1 6457. This region was not evaluated in the current report. We have also developed a nomenclature to organize the massive amounts of data, naming each assay by the ORF or region of the genome, giving an assay number, and listing the CpG sites within each assay (see [25] for a detailed description). This study focused on a single fragment from 4 different regions of the HPV16 genome (assays: E6.1, E2.1, L2.1 and L1.2) based on previous data of significant CpG sites in the HPV16 genome [20]. Nevertheless, larger studies will need to be performed to establish the most significant regions for CpG analyses for clinical risk stratification.
The analyses comparing women with CIN3 and cancer identified a single CpG site E2 3496, not previously analyzed, that showed significant differences between samples from women with precancer and cancer (see Table 1). In fact, in most cases median methylation levels increased in cross-sectional diagnostic samples from women with precancer and cancer suggesting an ongoing process of CpG methylation. Similar differences were observed between samples from women with CIN2/CIN3 and cancer in a previous report, but whether this was related to combining CIN2 and CIN3 was not discerned [23]. Understanding whether CpG methylation is mechanistically involved in precancer development and progression will require additional multidisciplinary research. This study provides a basis for further investigation in this area.
Our study has several strengths and limitations. We used a well-established cohort for which we had prior knowledge on site-specific methylation and CIN3+, allowing analytical evaluation of NGS of bisulfite DNA. In addition, we were able to apply our recent published technology to studying methyl-haplotypes in longitudinal samples over time. This study had limitations in sample size, the focus on a few regions of the HPV16 genome and the inability to analyze possible etiologic cofactors in small subsets of women. There were also a limited number of longitudinal samples, allowing only preliminary evaluation of the long-term outcome of women over time with different levels of methyl-haplotypes. This study also used median methylation levels to categorize methylation states for odds ratio estimations and cannot be directly compared to previous work comparing tertiles. The best analytical techniques for evaluation of quantitative HPV CpG methylation data with precancer and cancer risk still remain to be defined.
In conclusion, this work continues to support the use of NG sequencing technology as a robust method to identify and interpret HPV16 methylation. It has advantages over pyrosequencing with its ability to provide methylation data at multiple sites within a region now referred to as a methyl-haplotype. Using these methods, we demonstrate that methylation is significantly higher in women with CIN3 and cancer and we suggest that methyl-haplotypes deserve further consideration for risk assessment and disease association. Given that most HPV16 infections are transient and regress over time, it is increasingly important to identify the women that require colposcopy and treatment to prevent and/or treat cervical cancer. The advantages of bisulfite NG sequencing of HPV16 genomes indicate that this a technology that requires further investigation in additional populations, development of high-volume methods and establishment of best regions of CpG analyses to evaluate whether this promising marker of precancer/cancer can be moved into the clinical arena.
Supplementary Material
Research Highlights.
Next Gen sequencing is an efficient method to quantitate HPV16 CpG methylation
Methyl-haplotypes of HPV16 methylation are associated with cervical precancer
L2 Methylation increases over time in HPV16 infections leading to precancer
Acknowledgments
This work was supported in part by the National Cancer Institute (CA78527), the Einstein Cancer Research Center (P30CA013330) from the National Cancer Institute, and the Einstein-Montefiore Center for AIDS funded by the NIH (AI-51519). The authors acknowledge the women who participated in this study and the Guanacaste Project staff.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors have no conflicts of interest to report.
References
- [1].Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- [2].Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, et al. Global burden of human papillomavirus and related diseases. Vaccine. 2012;30(Suppl 5):F12–23. doi: 10.1016/j.vaccine.2012.07.055. [DOI] [PubMed] [Google Scholar]
- [3].Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, et al. A review of human carcinogens--Part B: biological agents. Lancet Oncol. 2009;10:321–2. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- [4].Guan P, Howell-Jones R, Li N, Bruni L, de Sanjose S, Franceschi S, et al. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer. 2012;131:2349–59. doi: 10.1002/ijc.27485. [DOI] [PubMed] [Google Scholar]
- [5].Li N, Franceschi S, Howell-Jones R, Snijders PJ, Clifford GM. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: Variation by geographical region, histological type and year of publication. Int J Cancer. 2011;128:927–35. doi: 10.1002/ijc.25396. [DOI] [PubMed] [Google Scholar]
- [6].Smith JS, Lindsay L, Hoots B, Keys J, Franceschi S, Winer R, et al. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. Int J Cancer. 2007;121:621–32. doi: 10.1002/ijc.22527. [DOI] [PubMed] [Google Scholar]
- [7].Schiffman M, Wentzensen N. Human papillomavirus infection and the multistage carcinogenesis of cervical cancer. Cancer Epidemiol Biomarkers Prev. 2013;22:553–60. doi: 10.1158/1055-9965.EPI-12-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ho GY, Bierman R, Beardsley L, Chang CJ, Burk RD. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med. 1998;338:423–8. doi: 10.1056/NEJM199802123380703. [DOI] [PubMed] [Google Scholar]
- [9].Wright TC, Stoler MH, Behrens CM, Sharma A, Zhang G, Wright TL. Primary cervical cancer screening with human papillomavirus: End of study results from the ATHENA study using HPV as the first-line screening test. Gynecol Oncol. 2015;136:189–97. doi: 10.1016/j.ygyno.2014.11.076. [DOI] [PubMed] [Google Scholar]
- [10].Schiffman M, Solomon D. Clinical practice. Cervical-cancer screening with human papillomavirus and cytologic cotesting. N Engl J Med. 2013;369:2324–31. doi: 10.1056/NEJMcp1210379. [DOI] [PubMed] [Google Scholar]
- [11].Hansel A, Steinbach D, Greinke C, Schmitz M, Eiselt J, Scheungraber C, et al. A Promising DNA Methylation Signature for the Triage of High-Risk Human Papillomavirus DNA-Positive Women. PLoS One. 2014;9:e91905. doi: 10.1371/journal.pone.0091905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cuzick J, Clavel C, Petry KU, Meijer CJ, Hoyer H, Ratnam S, et al. Overview of the European and North American studies on HPV testing in primary cervical cancer screening. Int J Cancer. 2006;119:1095–101. doi: 10.1002/ijc.21955. [DOI] [PubMed] [Google Scholar]
- [13].Schiffman M, Wentzensen N, Wacholder S, Kinney W, Gage JC, Castle PE. Human papillomavirus testing in the prevention of cervical cancer. J Natl Cancer Inst. 2011;103:368–83. doi: 10.1093/jnci/djq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hebner CM, Laimins LA. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev Med Virol. 2006;16:83–97. doi: 10.1002/rmv.488. [DOI] [PubMed] [Google Scholar]
- [15].Clarke MA, Wentzensen N, Mirabello L, Ghosh A, Wacholder S, Harari A, et al. Human papillomavirus DNA methylation as a potential biomarker for cervical cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:2125–37. doi: 10.1158/1055-9965.EPI-12-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20:274–81. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- [18].Sun C, Reimers LL, Burk RD. Methylation of HPV16 genome CpG sites is associated with cervix precancer and cancer. Gynecol Oncol. 2011;121:59–63. doi: 10.1016/j.ygyno.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fernandez AF, Rosales C, Lopez-Nieva P, Grana O, Ballestar E, Ropero S, et al. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 2009;19:438–51. doi: 10.1101/gr.083550.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mirabello L, Sun C, Ghosh A, Rodriguez AC, Schiffman M, Wentzensen N, et al. Methylation of human papillomavirus type 16 genome and risk of cervical precancer in a Costa Rican population. J Natl Cancer Inst. 2012;104:556–65. doi: 10.1093/jnci/djs135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kalantari M, Osann K, Calleja-Macias IE, Kim S, Yan B, Jordan S, et al. Methylation of human papillomavirus 16, 18, 31, and 45 L2 and L1 genes and the cellular DAPK gene: Considerations for use as biomarkers of the progression of cervical neoplasia. Virology. 2014;448C:314–21. doi: 10.1016/j.virol.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lorincz AT, Brentnall AR, Vasiljevic N, Scibior-Bentkowska D, Castanon A, Fiander A, et al. HPV16 L1 and L2 DNA methylation predicts high-grade cervical intraepithelial neoplasia in women with mildly abnormal cervical cytology. Int J Cancer. 2013;133:637–44. doi: 10.1002/ijc.28050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mirabello L, Schiffman M, Ghosh A, Rodriguez AC, Vasiljevic N, Wentzensen N, et al. Elevated methylation of HPV16 DNA is associated with the development of high grade cervical intraepithelial neoplasia. Int J Cancer. 2013;132:1412–22. doi: 10.1002/ijc.27750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mirabello L, Frimer M, Harari A, McAndrew T, Smith B, Chen Z, et al. HPV16 methyl-haplotypes determined by a novel next-generation sequencing method are associated with cervical precancer. Int J Cancer. 2015;136:E146–53. doi: 10.1002/ijc.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sun C, McAndrew T, Smith BC, Chen Z, Frimer M, Burk RD. Characterization of HPV DNA methylation of contiguous CpG sites by bisulfite treatment and massively parallel sequencing-the FRAGMENT approach. Front Genet. 2014;5:150. doi: 10.3389/fgene.2014.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brandsma JL, Sun Y, Lizardi PM, Tuck DP, Zelterman D, Haines GK, 3rd, et al. Distinct human papillomavirus type 16 methylomes in cervical cells at different stages of premalignancy. Virology. 2009;389:100–7. doi: 10.1016/j.virol.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bratti MC, Rodriguez AC, Schiffman M, Hildesheim A, Morales J, Alfaro M, et al. Description of a seven-year prospective study of human papillomavirus infection and cervical neoplasia among 10000 women in Guanacaste, Costa Rica. Rev Panam Salud Publica. 2004;15:75–89. doi: 10.1590/s1020-49892004000200002. [DOI] [PubMed] [Google Scholar]
- [28].Herrero R, Castle PE, Schiffman M, Bratti MC, Hildesheim A, Morales J, et al. Epidemiologic profile of type-specific human papillomavirus infection and cervical neoplasia in Guanacaste, Costa Rica. J Infect Dis. 2005;191:1796–807. doi: 10.1086/428850. [DOI] [PubMed] [Google Scholar]
- [29].Castle PE, Porras C, Quint WG, Rodriguez AC, Schiffman M, Gravitt PE, et al. Comparison of two PCR-based human papillomavirus genotyping methods. J Clin Microbiol. 2008;46:3437–45. doi: 10.1128/JCM.00620-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hayatsu H. Discovery of bisulfite-mediated cytosine conversion to uracil, the key reaction for DNA methylation analysis--a personal account. Proc Jpn Acad Ser B Phys Biol Sci. 2008;84:321–30. doi: 10.2183/pjab/84.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- [32].Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–4. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38:1767–71. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat methods. 2008;5:235–7. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Smith BC, McAndrew T, Chen Z, Harari A, Barris DM, Viswanathan S, et al. The cervical microbiome over 7 years and a comparison of methodologies for its characterization. PLoS One. 2012;7:e40425. doi: 10.1371/journal.pone.0040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–2. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics. 2001;29:1165–88. [Google Scholar]
- [38].RDCT . R: A language and environment for statistical computing. 2.12.2 ed. RDCT; Vienna, Austria: 2011. [Google Scholar]
- [39].Krueger F, Kreck B, Franke A, Andrews SR. DNA methylome analysis using short bisulfite sequencing data. Nat methods. 2012;9:145–51. doi: 10.1038/nmeth.1828. [DOI] [PubMed] [Google Scholar]
- [40].Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–5. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.