Abstract
Weight loss is diagnostic of cachexia, a debilitating syndrome contributing mightily to morbidity and mortality in cancer. Most research has probed mechanisms leading to muscle atrophy and adipose wasting in cachexia; however cachexia is a truly systemic phenomenon. Presence of the tumor elicits an inflammatory response and profound metabolic derangements involving not only muscle and fat, but also the hypothalamus, liver, heart, blood, spleen and likely other organs. This global response is orchestrated in part through circulating cytokines that rise in conditions of cachexia. Exogenous Interleukin-6 (IL6) and related cytokines can induce most cachexia symptomatology, including muscle and fat wasting, the acute phase response and anemia, while IL-6 inhibition reduces muscle loss in cancer. Although mechanistic studies are ongoing, certain of these cachexia phenotypes have been causally linked to the cytokine-activated transcription factor, STAT3, including skeletal muscle wasting, cardiac dysfunction and hypothalamic inflammation. Correlative studies implicate STAT3 in fat wasting and the acute phase response in cancer cachexia. Parallel data in non-cancer models and disease states suggest both contributory and protective functions for STAT3 in other organs during cachexia. Finally, STAT3 contributes to cancer cachexia through enhancing tumorigenesis, metastasis and immune suppression, particularly in tumors associated with high prevalence of cachexia. This review examines the evidence linking STAT3 to multi-organ manifestations of cachexia in cancer and evidence for targeting STAT3 for anti-cachexia therapies.
1. INTRODUCTION
Cachexia is a devastating complication of cancer and other chronic diseases, including burn injury, organ failure, trauma, sepsis and HIV/AIDS[1,2]. In all cachexia etiologies, weight loss and rate of weight loss are directly correlated with mortality [3]. Although malnutrition through anorexia and gastrointestinal dysfunction are common, cachexia is more than mere starvation. Catabolic pathways prevail in cachexia, and over-feeding is insufficient to prevent weight loss [4]. The most overt manifestation of cachexia is loss of muscle and fat mass. This progressive muscle wasting impairs function and activities of daily living, increases toxicity and complications of anti-cancer treatments, and left unabated results in decreased mobility, impaired respiration and eventually demise[5]. While the only definitive cure for cachexia is cure of the primary disease, recent work in mice indicates that blocking muscle loss prolongs life and function even in the absence of effects on tumor growth [6,7]. Thus cachexia represents not an unrelenting progression to death but a tractable problem within our ability to treat. Currently there is a single approved drug for cachexia therapy—a ghrelin mimetic showing modest benefit in patients [8,9]. Understanding the mechanisms underlying cachexia and systemic dysmetabolism is a prerequisite for finding additional and more effective therapeutic options.
The IL-6/GP130/Janus Kinase (JAK)/STAT3 pathway has been studied for the past 25 years for its roles in cachexia due to the association of circulating IL-6 and muscle wasting, the biological activities of IL-6 that mimic cancer cachexia, and the availability of powerful molecular and genetic tools [10,11]. Most effort has centered on IL-6, which in normal biology is necessary for a proper immune response and muscle growth and regeneration and which at higher levels is convincingly and causally linked with the systemic inflammation and wasting phenomena of cachexia [12]. Less effort has been expended on parallel activators and downstream mediators of the pathway, although genetic ablation studies of muscle GP130 [13] and clinical pharmacological inhibition of JAKs [14] support their pro-catabolic roles in cancer cachexia. Recently a central role for STAT3 in muscle wasting of cancer has been described [15,16], although the associated mechanisms are incompletely known. Furthermore, STAT3’s contribution to non-muscle organs in cachexia is even less clear. Here we will review briefly the basic biology of STAT3, the systemic dysmetabolism of cancer cachexia, the known and suspected roles for STAT3 in that process, and the potential for targeting STAT3 for therapeutic benefit.
2. STAT3
STAT3 was initially identified as the downstream effector of IL-6 and other pro-inflammatory mediators, the primary output of which is to modulate gene expression. It is now clear that multiple pathways activate STAT3, which has been shown to possess not only transcription factor activity, but also the ability to alter epigenetic regulation of gene expression and mitochondrial function [17].
2.1 STAT3 structure/function
STAT3 belongs to a family of seven proteins (STATs 1, 2, 3, 4, 5a, 5b, and 6) that generally transduce signals from activated receptors or intracellular kinases to the nucleus, thus activating and regulating gene transcription [18,19]. STAT3 function is essential for development and not redundant with other STAT proteins given STAT3 knockout mice are early embryonic lethal, dying at day 7.5 [20]. Subsequently it has become clear that STAT3 modulates transcription of a variety of genes involved in regulation of critical functions in multiple tissues through both cell autonomous and non-autonomous mechanisms [21–23].
Like the other STAT proteins, STAT3 possesses an N-terminal coiled-coiled domain mediating protein-coactivator interactions, a DNA-binding domain, a SH2 domain required for docking to receptor phophotyrosine (pY) sites, and a C-terminal transactivation domain [24]. Binding of STAT3 at a receptor (such as GP130) leads to its activation [as through Janus kinases (JAKs)], consisting of phosphorylation of a specific tyrosine residue (Y705) in the C-terminal domain. The phosphorylation of STAT3, promotes homodimerization and subsequently nuclear localization, where STAT3 interacts with coactivators and binds to specific response elements in the promoter regions of target genes, regulating transcription both positively and negatively. Phosphorylation on a serine (S727) in the C-terminal domain promotes association of STAT3 with transcription co-activators, including p300/CBP, providing for maximal activation of particular target genes [25–27]. Reversible acetylation on lysine residues in the SH2 domain (K685) and in the NH2 terminus (K49, K87) also promotes STAT3 dimer stabilization, DNA binding, interaction with transcriptional coactivators, and target gene expression [28–31]. Recently, a novel phospho-Threonine (pT714)/pS727 form of STAT3 has been implicated in renal cell carcinoma [32]. Moreover, distinct roles for unphosphorylated STAT3 in oncogenesis and gene transcription have also been described [33]. K685 acetylation is required for expression of most unphosphorylatedSTAT3-dependent genes[31]. Which of these modifications of STAT3, if any, are necessary for cancer cachexia is as yet unknown, although increased pY705-STAT3 has been documented in muscle [13,15,16,34–42], and pS727-STAT in liver [39] and fat [40] in murine cancer cachexia.
2.2 Activators of STAT3
Originally identified as a mediator of IL-6 and related cytokines, multiple other upstream inputs initiate STAT3 activation. IL-6-type cytokines (IL-6, IL-10, IL-11, LIF, CT-1, OSM, CNTF), bind GP130 and activate JAKs, which in turn phosphorylate STAT3, among other signaling mediators [43]. Leptin [44] and G-CSF [45], both with homologous receptors to GP130 also activate STAT3, as do several receptor tyrosine kinases including the epidermal growth factor (EGF) [46,47], platelet-derived growth factor (PDGF) [48], and Colony Stimulating Factor-1 (CSF-1) [49] receptors, and receptors for the class 2 alpha-helical cytokines IL-10, Interferon (IFN)-γ and IFN-α [50], and the IL-2 family [51]. MicroRNAs [52–54], Src Family Kinases (SRKs) including Src, Lck, Fyn, Hck and Fgr, and RhoGTPases also are reported to activate STAT3 [55–59].
The upstream mediators of STAT3 best characterized in cancer cachexia are the IL-6 family of cytokines, predominantly IL-6 itself [10], but also LIF [37,60,61], OSM [62] and CNTF [63], each of which has been shown either necessary or sufficient to induce weight loss and cachexia in mice. While IL-10 activates STAT3, it has been shown to reduce cachexia, consistent with its antagonistic role versus IL-6 in other tissues and systems [64]. A host-derived factor implicated in cancer cachexia and found in the urine of patients with cachexia, Proteolysis-Inducing Factor (PIF), regulates NF-κB and STAT3 activation and gene expression in hepatocytes [65]. Intriguingly, genetic data demonstrate that STAT3 expression and up-regulation of the IL-6 pathway in locomotor and diaphragm muscles in C26 cachexia requires intact function of FoxO transcription factors [66], while FoxO3 has been identified as a STAT3 target gene [67]. Like STAT3, FoxO1/FoxO3 is tightly linked to muscle wasting in general, including cancer cachexia, and is necessary and sufficient for myofiber atrophy [68–71]. Whether STAT3 is necessary for FoxO-induced wasting or vice versa has not yet been tested. Overall, given its position downstream of this variety of cachexia-promoting factors, STAT3 would seem to present a key node for targeting.
2.3 STAT3 functions and outputs
STAT3 was initially identified as a transcription factor modulating gene expression. Identification and categorization of STAT3 target genes and functions is complicated by the cell-type specific, highly divergent binding sites and pleiotrophic effects of STAT3 in diverse tissues, as well as STAT3’s ability to heterodimerize with other STAT proteins. Genome wide STAT3 binding data obtained from ChIP-seq studies in four different cell types (embryonic stem cells, CD4+ T cells, macrophages, and AtT-20 mouse pituitary tumor cells) reveals two modes of STAT3 binding, general and cell-type specific. In the first mode, STAT3 binds a set of conserved elements regulating expression of core proteins important for a STAT3 self-regulatory loop in all cell types. Out of the 35 universally regulated genes, 15 are transcription factors thatseem to coincide with the regulation of STAT3 signaling in general. The second mode of STAT3 binding is cell-type specific and proposed to be controlled by discrete transcriptional regulatory modules and cell-type specific proteins that assemble prior to and after STAT3 binding, ensuring that the specificity of signaling required for that cell type remains intact.
Studies using manipulation of STAT3 in combination with expression profiling and chromatin profiling and DNA binding assays have identified several hundred STAT3 target genes that vary considerably across different cell types and tissues [67,72–77]. These are not collected in a single repository, however generally genes increased or decreased by STAT3 include those modulating proliferation/survival, pluripotency, angiogenesis, wound healing or invasion/metastasis, and inflammation/immunity In this last class, STAT3 is essential for normal induction of acute phase response (APR) genes in the liver [78,79]. The APR is a massive shift in the liver transcriptome and proteome in response to infection, inflammation, LPS, IL-6, or TNF, in which dozens to thousands of genes are up-regulated as part of the innate immune response [80]. Among these are secreted pathogen recognition receptors, components of the complement system, proteinase inhibitors, regulators of iron homeostasis and others of unknown function. These protein changes are detectable in the circulation as increased Fibrinogen, C-reactive Protein (CRP), hepcidin, alpha2-microglobulin, etc., and are frequently increased in cancer cachexia. STAT3 represses gene expression as well, including some interferon signaling genes, which is consistent with the opposing roles of STAT3 and STAT1 in many biological contexts [81–83]. Finally, genes induced by STAT3 encode proteins both expected to promote signaling through this pathway (including IL6 [84], other cytokines, IL6R and STAT3 itself [85]) as well as genes known to terminate signaling, including Suppressors of Cytokine Signaling 3 (SOCS3). Thus, normal physiological regulation of this pathway is achieved by a burst of activity followed by feedback inhibition and eventual termination of signaling.
In addition to its effects on expression of specific target genes, STAT3 has also been implicated in epigenetic switches involving metabolic reprogramming, inflammation, and transformation [17,73,86–92]. In addition, accumulating data demonstrate a role for mitochondrial STAT3 in altering mitochondrial DNA copy number [93], mitochondrial gene expression [94], and maintenance of the electron transport chain [95–97], thereby influencing cell survival [98] [69], transformation [97] and metabolism [99]. To date no studies have directly addressed these other highly relevant functions of STAT3 in cachexia.
3. SYSTEMIC INFLAMMATION IN CANCER CACHEXIA
Many forms of cachexia, including burn/trauma [100–103], sepsis [104–106], organ failure [107–112], HIV/AIDS [112,113], and cancer [114] are associated with elevated serum inflammatory cytokines and activation of the acute phase response. After burn injury, for example, patients can lose up to 25% of body mass [115], accompanied by elevations in circulating cytokines orders of magnitude greater than normal [101,103]. The connection between cytokines and muscle wasting is more variable in cancer, however, with some patients exhibiting profound wasting with low evidence of inflammation (low CRP, normal albumin) and others with little wasting but high CRP and low albumin [114,116]. Thus, inflammation is not included in the 2011 consensus definition of cancer cachexia [117].
Circulating mediators of inflammation implicated in cancer-induced muscle wasting include members of the TNF, IL-6, IFN, and IL-2 families of cytokines, although the only factors thus far consistently correlated with cachexia overall are IL-6, Activin A and Growth Differentiation Factor (GDF)-15, the latter two members of the Transforming Growth Factor-β superfamily [118–121]. The relationship is a general one, however, with the concentrations of these factors highly heterogeneous across patients. Such variability in presentation hints at differences in underlying mechanisms that might be cancer-type, cancer-stage, treatment-type, genotype [122–124] or organ-site specific, although this is largely unknown.
Manifestations of systemic inflammation in cancer cachexia include effects on multiple tissues. The most obvious of these and the most functionally consequential is wasting of skeletal muscle through activation of protein catabolism [125] and impaired myogenesis [126]. Adipose tissue inflammation is also observed, with wasting mediated through lipolytic and thermogenic processes [127]. Inflammation of the brain and hypothalamus leads to anorexia, anosmia and results in lowered food intake [128]. Liver inflammation takes the form of hypertrophy and activation of the acute phase response [39]. Dysregulation of hematopoiesis including anemia, thrombocytosis, and immunosuppression are observed [129–132]. Cardiac wasting leads to impaired function [133–135]. Impairment of gut absorptive and barrier functions as well as hypogonadism is common.
Recently germline activating mutations in STAT3 have been described and linked to systemic auto-immune disease [136]. Simplistically, systemic activation of STAT3 in cancer could lead to similar multi-organ inflammation. Here we first review the evidence for STAT3 functions in muscle during cachexia, followed by its functions in other organs.
4.0 STAT3 IN SKELETAL MUSCLE OF CANCER CACHEXIA
Most research on STAT3 in cachexia has focused upon skeletal muscle. Here we review the substantial correlative and existing functional evidence for STAT3 involvement in muscle wasting. Current data support a pro-atrophic role for STAT3 in myofibers through transcriptional regulation of known “atrogenes”. Less clear is any role for STAT3 in mitochondrial and metabolic dysfunction and impaired myogenesis of cancer cachexia.
4.1 STAT3 activation in muscle of cancer cachexia
Correlative data strongly implicate a causal role for STAT3 in skeletal muscle wasting of cancer cachexia. Increased STAT3 RNA, protein and pY705-STAT3 have been observed in skeletal muscle in diverse models of cancer cachexia, including IL-6 administration, C26 adenocarcinoma, Lewis lung carcinoma (LLC), B16 melanoma, and ApcMin intestinal cancer [13,15,16,34–42]. A STAT3 gene signature and elevated expression of STAT3 interacting genes are also evident in C26 muscles [34,37,66,137,138] and in muscles from a genetically engineered mouse model of pancreatic cancer [42]. The extent of STAT3 phosphorylation tends to increase with severity of cachexia and serum concentrations of IL-6, such that the highest pY705-STAT3 levels are observed in ApcMin and the lowest in B16 melanoma [15]. Conversely, mice with muscle-specific deletion of GP130 and LLC cachexia exhibit reduced muscle loss with reduced pSTAT3 [13], with similar results after treatment of ApcMin cachexia with IL6R neutralizing antibodies [139].
A single study to date has examined STAT3 activation in patient cancer cachexia samples, where no increased pY705-STAT3 was observed in surgical rectus muscle biopsies of patients with cachexia and high CRP levels versus those with cachexia and low CRP levels [116]. No non-cancer controls were tested. Such negative evidence is difficult to interpret, however. Given the diversity of factors contributing to cachexia, variances upstream of STAT3 might explain the lack of differential pSTAT3. Alternatively, pY705-STAT3 might not be the relevant form in human muscle wasting. Furthermore, analysis of STAT3 pathway activity might be technically challenging, given that general anesthesia and surgery strongly activate gene expression of IL-6, JAK1, STAT3, SOCS1, SOCS2 and SOCS3 in diaphragm and thus potentially in other skeletal muscles [140]. Such dynamic changes during sample collection could obscure evidence of more long-standing pathway activation. Well-controlled studies in other human muscle wasting conditions, including COPD [141], chronic kidney disease [142], aging [143] and pediatric burn injury [144] (but not in a study of adult burns [145]), do demonstrate increased JAK/STAT3 pathway activation and target gene expression in muscle, particularly SOCS3, strongly suggestive that increased STAT3 transcriptional activity could be a common mediator of human cachexia.
4.2 STAT3 functions in myofibers in cancer cachexia
Functional genetic data in adult mice also support a pro-catabolic role for STAT3 in muscle wasting of cancer. A constitutively activated mutant form of STAT3 (cSTAT3) introduced via plasmid electroporation induces muscle fiber atrophy and enhances C26- and IL-6-induced cachexia in mice [15]. Conversely, gene transfer of a dominant negative STAT3 or shSTAT3 is protective of muscle in cancer cachexia [15]. Thus local manipulation of STAT3 modulates fiber size in conditions of inflammation.
Two separate reports of mice with skeletal muscle-specific STAT3 deletion (through Cre-mediated recombination driven by the muscle creatine kinase (MCK) promoter and hence deleted for STAT3 from early muscle differentiation) reveal no significant difference in body composition or baseline muscle mass, suggesting redundant, compensated or insignificant roles for STAT3 in the normal regulation of muscle mass [142,146]. Moreover, STAT3 deletion did not alter resting energy expenditure in normal or diet-induced obesity (a low inflammation condition), nor did it affect glucose tolerance or in vivo insulin action [146]. In contrast, when muscle-specific STAT3 knockout mice were challenged with cachexia either through chronic kidney disease or LLC cachexia, muscle mass and grip strength were preserved and myofiber wasting was reduced [16,142]. These results demonstrate a necessity for myofiber STAT3 in inflammation-associated muscle wasting.
These loss-of-function studies in which STAT3 is deleted specifically from the myofiber have not yet been complemented by STAT3 knockout in other resident muscle cell types, including satellite cells, or by STAT3 gain of function studies in muscle at all.
4.3 Mechanisms for STAT3-induced muscle atrophy in cancer cachexia
The strongest mechanistic data to date are from cachexia studies in myofiber-specific STAT3 knockout mice [16,142]. Those experiments demonstrate elevated myostatin expression in cachexia of renal failure, streptozotocin-induced diabetes, and LLC cachexia, all conditions of high IL-6 and skeletal muscle STAT3 activation. Myostatin is a TGF-β family member that tonically inhibits muscle growth and promotes cachexia through effects on myofiber protein homeostasis and myogenesis. STAT3 deletion reduces expression of CCAAT/enhancer-binding protein δ (C/EBPδ), which in turn reduces myostatin expression in cachexia of renal failure or streptozotocin-induced diabetes. Caspase-3 was also identified as a STAT3 target in cachexia. Whether this is the only pathway associated with JAK/STAT3-dependent muscle wasting remains to be determined. Other pathways were not evaluated.
An alternative mechanism for STAT3-induced muscle wasting could be through re-prioritization of the muscle transcriptome and proteome. As detailed above, STAT3 is important for normal APR expression in liver. Studies in C26 cachexia demonstrate a robust skeletal muscle APR transcriptomic response [34]. The levels of fibrinogen expressed in liver versus muscle in this model suggest that muscle might be a greater source of certain APR proteins than liver. If other APR proteins are similarly increased in cachectic muscle, it is reasonable to speculate that a large portion of amino acids freed from skeletal muscle structural proteins through proteolysis would be re-synthesized into these secreted proteins and exported from the cell. Calculations by others suggest that catabolism of 2.6 grams of muscle protein is required to produce 1 gram of fibrinogen [114,147]. Thus, such a diversion of amino acids to serum proteins would be metabolically draining and energetically expensive.
Skeletal muscle in cachexia is also characterized by metabolic shifts, with loss of oxidative phosphorylation in favor of glycolysis. STAT3 deletion in other cell types impairs mitochondrial function through apparently non-transcriptional mechanisms, suggesting that investigation of STAT3 localization and functions in muscle mitochondria are warranted.
4.4 Potential roles for STAT3 in impaired myogenesis of cancer cachexia
While much work has focused on protein homeostasis in cachexia, recent studies demonstrate accumulation of muscle progenitor cells, including satellite cells in murine and human cancer cachexia [126,148,149]. It is posited these cells are activated in response to cancer or cytokine-induced myocyte damage, but are unable to differentiate thereby contributing to muscle wasting by impaired nuclear accretion and loss of subsequent hypertrophic stimulus. While its role in this pathology has not yet been addressed directly, STAT3 clearly influences proliferation and differentiation of muscle progenitor cells. In vitro, STAT3 knockdown reduced LIF-stimulated proliferation of C2C12 myoblasts [150]; furthermore, STAT3 knockdown reduced markers of myogenesis in C2C12 cells cultured in differentiation conditions [151]. In mice, ablation of STAT3 in Pax7+ satellite cells leads to enhanced proliferation during the regeneration process, but compromises differentiation of these cells into new myofibers [152]. STAT3 deletion or pharmacological inhibition enhances symmetric renewal of satellite cells and engraftment and enhances muscle repair in both adult and aged animals, as well as in dystrophic mice [153]. These studies suggest STAT3 likely plays a role in the expanded muscle progenitor pool in cachexia.
5.0 STAT3 OUTSIDE MUSCLE IN CACHEXIA
Although most work has focused on STAT3 in muscle of cachexia, virtually all cells express GP130 and are capable of activating STAT3. Moreover, essential STAT3 functions have been described in most cell types in normal and disease states. In the hyperinflammatory conditions of cachexia then, STAT3 is potentially globally activated. Next we review organ-specific effects of cancer cachexia and the largely correlative evidence for STAT3 involvement in organ-specific manifestations of cancer cachexia.
5.1 STAT3 in the tumor
STAT3 contributes to cancer development and progression [154–156] and is activated in many cancer types including glioblastoma, breast, prostate, liver, and pancreas [157–160]. STAT3 signaling contributes to tumor cell proliferation, survival, angiogenesis, metastatic potential, immunosuppression, and chemoresistance [17,161]. Most human cancers show elevated STAT3 phosphorylation, while their transcriptional profiles are consistent with constitutively activated STAT3-regulated gene expression [27]. It has been suggested that normal cells can bypass STAT3 pharmacological inhibition for survival (given normal basal phenotypes with several organ-specific STAT3 mutants), thus drugs targeting STAT3 signaling would potentially be well tolerated by the rest of the organism. Intensive effort has identified STAT3 as a promising target for antineoplastic therapies, with particular promise in cancers with high rates of cachexia and systemic inflammation, including pancreas [162–164], non-small cell lung cancer [165], hepatocelluar carcinoma [166], biliary cancers [167], renal clear cell carcinoma [168], and ovarian cancer [169].
In pancreatic cancer in particular, with its highest rates of cachexia, STAT3 contributes to the establishment of early pancreatic lesions in genetically engineered mouse (GEM) models, [155,156] and promotes tumor and stromal cell proliferation and viability along with autocrine and paracrine signals [161,170]. Recent studies also demonstrate STAT3 in tumor stromal cells as contributory to tumor progression and immunosuppression. Pancreatic cancer-associated stellate cells secrete factors including IL-6 that promote differentiation of myeloid-derived suppressor cells (MDSCs) leading to immunosupression and enhanced tumor growth. The STAT3 inhibitor FLLL32 reduces production of IL-6 and blocks expansion of MDSCs [171]. MDSCs, in turn, have been implicated in cachexia [172]. STAT3 is also implicated in pancreas tumor-to-endothelial cell crosstalk. The JAK inhibitor Ruxolitinib suppresses stimulatory signals from endothelial cells and prolongs survival in mice [173]. As well, inhibition of STAT3 via JAK inhibitor AZD1480 in combination with gemcitabine results in increased drug delivery and improved survival in a pancreas cancer GEM model via remodeling of the stroma and downregulation of gemcitabine metabolizing enzyme cytidine deaminase, Cda [174]. Thus, targeting STAT3 might have the additional benefit of slowing muscle wasting indirectly through inhibiting tumor-stroma interaction or reducing tumor burden.
5.2 STAT3 in neuroinflammation
Hypothalamic inflammation is reviewed extensively elsewhere in this issue [128]. Briefly, however, the hypothalamus regulates certain metabolic processes and other activities of the autonomic nervous system, including appetite, thermoregulation, circadian rhythms, and fatigue. All of these are abnormal in patients and mice with cancer cachexia, including persistent stimulation of anorexigenic pathways and inhibition of orexigenic pathways [175]. The effect is to reduce food intake and food seeking behaviors, adding malnutrition to the persistent catabolic state and leading to negative nitrogen balance. Hypothalamic STAT3 activation is observed with increased GP130 cytokine and leptin signaling [176]. Brain-specific deletion of the STAT3 inhibitor SOCS3 results in increased hypothalamic STAT3 phosphorylation, body weight loss and suppression of food intake [177]. In related studies, mice with STAT3 deletion in arcuate neurons exhibit weight gain and mild hyperphagia [178]. Finally, hypothalamic double knockout of the STAT3 phosphatases PTP1B and TCPTP/PTPN2 additively antagonize obesity [179], which alternatively might be described as promoting cachexia or a pre-cachectic state. These studies are consistent with a causal role for CNS and hypothalamic STAT3 in anorexia and related symptoms in inflammatory cancer conditions, although as far as we can determine, no definitive genetic studies specifically targeting brain-derived STAT3 in cancer cachexia have been described.
5.3 STAT3 in adipose wasting and browning
Adipose tissue is lost more rapidly than skeletal muscle in cancer cachexia, although the obesity epidemic tends to obscure this symptom. Adipose wasting and browning of white adipose tissue is reviewed elsewhere in this issue [127]. However, it is important to note that elevated STAT3 and pY705-STAT3 have been noted in white adipose tissue in C26 cachexia [40] and pancreatic cancer GEM models [42]. Moreover, STAT3 is a critical determinant of brown fat differentiation and consistutive activation of STAT3 was sufficient to reverse a TYK2 knockout model of obesity [180]. Given that leptin activates STAT3 and controls energy homeostasis, glucose and lipid metabolism and immune function [181], and that IL-6 is both necessary for fat wasting in C26 cachexia [182] and sufficient to cause fat wasting, STAT3 might integrate multiple cytokine signals mediating adipose tissue loss in cachexia.
5.4 STAT3 in the liver
Effects of cancer cachexia on the liver are incompletely described in animal models [39,183–186] and mostly undescribed in human cancer cachexia with the notable exception of the frequently observed and previously described APR. This massive shift in the liver proteome requires additional protein synthetic capacity. Enhanced ribosomal biogenesis and ER/golgi expansion are observed, with consequent hepatocyte hypertrophy and overall growth in liver mass. Hepatomegaly likely also represents hepatocyte proliferation (as distinct from organ size increase due to hepatic tumors or metastatases). In mice, increased liver mass is observed in C26 and LLC cachexia [34]. In rats with Yoshida AH-130 tumors, hepatomegaly is observed in early cachexia, followed by hepatocyte apoptosis and reduced liver mass towards death [187] [102]. In patients with myeloproliferative neoplasms, cancers associated with profound muscle loss, hepatomegaly and splenomegaly are reduced by JAK inhibition [103]. As well, children with burn injury exhibit liver growth from 2–5 times normal liver mass, concomitant with muscle wasting, while mice with burn injury show lesser hepatomegly and muscle wasting [100,101,103]. The extent to which liver growth occurs in human cancer cachexia remains to be determined. However, all these conditions share high levels of IL-6, and IL-6 administration alone is sufficient to drive hepatocyte proliferation, massive liver hypertrophy and the acute phase response [188].
Speculatively, both the APR and organ growth (either hepatomegaly or splenomegaly) could enhance muscle wasting indirectly by competing for amino acids, fatty acids and other substrates, thereby starving muscle and preventing hypertrophy. Indeed, studies in drosophila demonstrate that organ proliferation (in this case gut or tumor) leads to systemic wasting [189,190]. Finally, cancer cachexia depletes liver glycogen content and alters glucose metabolism and protein synthesis. Ultrastructurally, C26 cancer cachexia is associated with mitochondrial abnormalities and increased endoplasmic reticulum, the last consistent with synthesis and secretion of APR proteins [191]. Given that IL-6 and STAT3 are hepatoprotective in other contexts, promoting proliferation and preventing oxidative damage, inflammation and fibrosis [192–195], functional studies are essential for determining whether STAT3 targeting in cachexia would be protective or counter-productive.
5.4 STAT3 in the spleen
Little direct study of the splenic response to cancer cachexia has been undertaken. Splenomegaly is noted in several cachexia animal models [196,197] and is a notable feature of myelofibrosis. In general, splenomegaly is a manifestation of hyperfunction, including immune hyperplasia, extramedullary hematopoiesis and removal of defective red blood cells. However enlargement of the spleen could also reflect portal hypertension secondary to liver failure or venous obstruction and benign or malignant infiltration as in myeloproliferative diseases and metastatic cancer. The spleen was noted to express certain cachexia-associated GP130 cytokines in LLC cachexia [62], although that study did not rule out expression from invading tumor cells.
5.5 STAT3 in the heart
Cancer cachexia manifests in mice and rats with concomitant cardiac cachexia, with progressive loss of cardiac mass and function [112–114]. Cardiac dysfunction and wasting are also present in patients with colorectal cancer, who exhibit impaired exercise capacity, left ventricular ejection fraction, lean mass, and heart rate variability [115]. Heart failure in the context of cancer is particularly significant because cachexia will predispose to heart failure and heart failure will result in cachexia and muscle wasting [116]. Defining the mechanisms leading to cardiac wasting per se is thus essential. However, to date STAT3 activation in cardiac muscle in cachexia has not been described and increased STAT3, IL6R or SOCS3 were not noted in a comparative analysis of gene expression in C26 cachexia, although elevated STAT3 was observed in skeletal muscle, consistent with prior studies [117].
Careful and numerous genetic studies demonstrate that endogenous STAT3 plays essential, protective roles for cardiac size and function [118]. Male mice with cardiomyocte-specific deletion of STAT3 develop heart failure, while female mice develop post-partum cardiac myopathy. Cardiomyocyte STAT3 deficiency leads to dysregulation of the ubiquitin-proteasome system, loss of microtubule stability, and mitochondrial respiratory deficits. Cardiomyocytes deficient in STAT3 also demonstrate that STAT3 directly regulates β-adrenergic receptor (βAR) signaling. The STAT3 knockout cardiomyocytes displayed a decrease in cardiac contractile response to acute βAR stimulation (Zhang et. al, in press) Moreover, constitutive global STAT3 activation leads to inflammatory myocarditis in mice 24069556, along with systemic auto-immune disease in humans. These data suggest a contrasting role for STAT3 in cardiac versus skeletal muscle. Indeed, whereas IL6/GP130/JAK/STAT3 pathway activity promotes skeletal muscle wasting, scores of studies link this same pathway to cardioprotective events in ischemic injury and other models of heart failure. Genetic studies to determine the precise, non-redundant roles of STAT3 in cardiac muscle during cancer cachexia are essential for resolving this conundrum, preferably prior to clinical trials of STAT3 inhibitors.
5.7 STAT3 in blood
STAT3 plays a central role in hematopoiesis, including in development of T helper cell and B cell subsets, dendritic cell development and maturation, inhibition of neutrophil numbers and regulation of stem cell self-renewal. As well, STAT3 inhibits TLR signaling in macrophages [119]. Which of these functions is important in cancer cachexia has not been tested directly, however data in parallel studies are suggestive. A clinical trial of the JAK1/2 inhibitor INCB018424 in patients with myelofibrosis showed reduced hepatosplenomegaly, increased walking time and reduced levels of blood (cell type or just peripheral blood?) pSTAT3 by western blotting. This suggests that pSTAT3 activation in blood might be a readily available proxy marker for STAT3 action in other organs (e.g. muscle) and that STAT3 inhibition in blood cells might be protective in cachexia. Conversely, deletion of STAT3 in hematopoietic cells enhanced thrombocytosis and shortened survival in a JAK2-V617F mouse model of myeloproliferative neoplasms [120].
Surprisingly, STAT3 has also been shown to regulate collagen-induced platelet aggregation independently of its transcription factor activity [121], as well as mitochondrial gene expression in platelets. Given pre-clinical and clinical data linking muscle wasting, IL-6, thrombocytosis, platelet aggregation and mortality in renal disease and cancer cachexia, these studies suggest yet another role for platelet STAT3 in cancer cachexia [122–124].
5.8 STAT3 elsewhere
Gut function is compromised in cancer cachexia. Gut absorptive function and barrier integrity are abnormal in mouse models, permitting translocation of gut microbiota eliciting systemic inflammation and likely distant organ injury (REF). In patients with cancer cachexia, malabsorption, decreased gut motility and systemic inflammation are observed (REF). Given abundant evidence for pro-survival and anti-apoptotic roles in the gut, some amount of IL-6/STAT3 activation is likely protective in cachexia, although sustained IL-6/STAT3 activation has been linked to gut dysfunction PMC3819310. However, no studies directly examining STAT3 function in the cachetic gut were identified.
Finally, STAT3 is also enhanced in the testes of cachexic ApcMin mice, as STAT3 increases with increasing cachexia severity and circulating IL-6, and concomitant with declining testis mass and testosterone levels [32]. Hypogonadism is frequently observed in cachectic states, particularly burn injury. Whether STAT3 activation in the testis and hypogonadism are functionally related, however, is currently not known.
5.9 Summary of STAT3 function in cachexia
In summary, STAT3 is activated in tumor and its associated microenvironment, muscle, liver, fat, brain, blood, and testis in cancer cachexia. Pro-cachectic functions have been definitively identified in tumor, myofibers, fat, liver and brain. Other studies suggest essential, protective functions of STAT3 in the heart, gut and blood. Clearly, further study is required to clarify organ-specific roles of STAT3 and the clinical potential and perils inherent in general STAT3 inhibition for treatment of cachexia.
6.0 STAT3 INHIBITION
There are various inhibitors of STAT3 activity reported in the literature; however many of these have limitations with respect to mechanisms of action. Several inhibit STAT3 without a clearly defined mechanism and/or have other mechanism of actions, which could contribute to their cellular and in vivo effects. For the purpose of this review then, we will highlight some of the most recent IL-6/IL6R/GP130/JAK/STAT3 pathway inhibitors that show in vivo efficacy for relevant endpoints alongside pharmacodynamic markers of STAT3 inhibition [198]. Although treatment with these inhibitors results in reduced STAT3 activity, some of these inhibitors act upstream of STAT3 and some act directly on STAT3 itself.
6.1 Blocking upstream ligand-receptor interactions
Blockade of ligand binding to receptor would prevent downstream STAT3 activation. Indeed, neutralizing antibodies against IL-6 (Siltuximab) or its receptor (Tocilizumab) are effective at reducing muscle wasting and cachexia in murine cancer models. Antibodies to IL-6 are FDA-approved for rheumatoid arthritis and have shown early promise in case reports (REF) and clinical trials in cancer cachexia (REF). Similarly, an antibody directed against IL6R, Tocilizumab, disrupts IL6/IL6R binding to GP130 and lead to reduced JAK/STAT3 pathway activity and rapid improvement of rheumatoid arthritis, with reduction in B cell hyperactivity and a dramatic normalization of the acute phase response (reviewed in [[79]]). Given the abundance of evidence linking IL-6 to both cancer cachexia and tumor progression, well-designed randomized trials probing body composition, evidence of systemic inflammation and muscle, fat and tumor endpoints are clearly indicated. There are limitations to such studies, including variability in circulating IL-6 levels in patients with cancer cachexia. Levels are elevated overall, but are sufficiently variable between patients to complicate trial design. The trial in lung cancer used CRP levels for inclusion criteria, which is reasonable given that IL-6 promotes CRP expression in hepatocytes, however other cytokines also induce the APR. Moreover, it is unclear whether serum IL-6 levels reflect levels in the relevant target tissues given that IL6 expression in muscle can change >100-fold without corresponding changes in the blood.
Indeed, a soluble GP130 (sGP130) has been tested in other models of hyper-inflammation, but not in cancer cachexia to date. This biologic selectively inhibits IL-6 trans-signaling by binding the IL-6/sIL-6R complex in serum, without affecting classical signaling mediated through IL-6 binding to membrane-bound IL-6R. Due to the ubiquitous expression of GP130, sGP130 could inhibit systemic IL-6 trans-signaling in cachexia [198]. sGP130 shows efficacy in animal models of arthritis, peritonitis, inflammatory bowel disease and colon cancer [127–129], and it is currently in preclinical development for Crohns disease (reviewed in [125]). However, a limitation for the use of sGP130Fc is that, when the IL-6 trans-signaling is counteracted by sGP130Fc binding, free IL-6 is not neutralized, whichcan directly activate STAT3 through classical membrane-bound IL-6R-mediated pathway.
The greatest limitation of IL6-focused therapies, however, is that IL-6 is likely not the sole cause of muscle wasting and STAT3 activation in cachexia. A broader strategy to inhibit various other GP130 ligands might be employed using antibodies directed against GP130 itself.
6.2 Inhibiting STAT3 kinases
To date only tyrosine phosphorylation of STAT3 has been linked to cancer cachexia. Inhibition of Janus kinases might reduce STAT3 activation and slow cancer cachexia. Various small molecule JAK inhibitors are described, many with substantial pre-clinical data showing STAT3 inhibition in vitro and in vivo. The JAK2 inhibitor AG490 reduces pSTAT3 and STAT3 DNA-binding activity in different forms of leukemia [130]. An AG490 analog, LS-104, is now into phase II clinical trials for the therapy of acute lymphoblastic leukemia [131]. Similarly, the FDA-approved JAK1/2 inhibitor INCB018424/ruxolitinib has been shown to reduce pSTAT3 in peripheral blood of patients with myelofibrosis [12]. INCB018424/ruxolitinib might preserve muscle through on-target effects because it markedly reduces IL-6-induced STAT3 activation and myotube atrophy in vitro [13]. The JAK inhibitor CEP701 also shows reduction in STAT3 phosphorylation and efficacy in myelofibrosis [132].
Beyond JAK inhibitors, patients with cholangiocarcinoma (Prado 22510747) treated with the MEK inhibitor selumetinib experienced muscle gain and reduced IL-6 levels versus controls. Similarly, the tyrosine kinase inhibitor Sunitinib was shown to prevent cachexia in mice orthotopically implanted with renal cell carcinoma and in C26 hosts by antagonizing STAT3 activation and inhibiting MuRF-1-associated muscle catabolism [133].
6.3 Inhibiting STAT3 itself
Direct STAT3 inhibition may be more efficacious in cancer cachexia as other mechanisms beyond IL6/GP130/JAK stimulation can activate STAT3 signaling [134]. One strategy for inhibiting STAT3 activity is to prevent its recruitment to activated receptors, phosphorylation, and homodimerization. Along this line, a 29-amino acid cell-permeable peptide derived from the STAT3 SH2 domain is able to replicate STAT3 biochemical properties, binding with high affinity to known STAT3-binding motifs, and thus preventing activation of endogenous STAT3 [135]. This STAT3 inhibitory peptide reduced IL-6-induced myotube wasting in vitro [13]. STATTIC, a non-peptide STAT3 inhibitor, was first identified during a large in vitro high-throughput screening of over 17,000 compounds [136]. STATTIC was shown to antagonize STAT3 phosphorylation, as well as the dimerization and nuclear translocation of active STAT3. STATTIC has demonstrated radiation-sensitizing activity with reduction of pSTAT3 in xenografted colorectal cancers (Spitzner) 23934972. However, the best evidence for direct pharmacological inhibition of STAT3 for preserving muscle comes from studies with C188-9, a cell-permeable binaphthol-sulfonamide that targets the pY binding site in the SH2 domain, blocking recruitment to tyrosine kinase complexes and dimerization. C188-9 reduced pY705-STAT3 in the muscle of chronic kidney disease and C26 mice, concomitant with sparing of muscle mass, grip strength and myofiber size [14, 91].
Derivatives of salicylic acid have also been found to inhibit STAT3 [137–139]. These are unique compared with most STAT3 blockers, in that they do not directly block protein phosphorylation, and for their greater potency. These are the first STAT3 antagonists known to reach IC50 in the nM range, both in vitro and in vivo. These dimerization disruptors emerged from SF-1-066 [140], have affinity (KD) of 300–504 nM for STAT3, and do not inhibit STAT1. Analogs, BP-1-102 and SH-4-54 have potent anti-tumor efficacy in breast, lung, brain and acute lymphoblastic leukemia tumor xenografts [137–139].
Natural products such as withacnistin and curcumin possess inhibitory action on the STAT3 pathway. However curcumin affects many pathways in cells, therefore efforts in drug discovery were initiated to find analogs that were more selective for STAT3. The resulting FLLL compounds, including FLLL31, 32, and 62, are curcumin analogs that inhibit STAT3 signaling and bind to both Jak2 and STAT3 [141, 142]. These compounds demonstrate inhibition of STAT3 signaling via inhibition of phosphorylation at Y705, blockade of DNA binding and reduction of downstream gene expression. Blockade of STAT3 with FLLL32 reduced the production of IL-6 from the cells within the stroma of pancreatic cancer [143]. The effects of IL-6 secretion are far-reaching in that they can stimulate STAT3 signaling within the tumor cell as well as lead to a suppression of the immune system, and increase loss of muscle mass [13, 143]. This reduction in IL-6 secretion by FLLL32 may also provide a reduction in the effects of IL-6 signaling in muscle thereby decreasing the cancer-induced loss of muscle mass.
Another approach to blockade of the STAT3 transcription factor would be to block its DNA binding and thereby abrogate expression of genes that contribute to muscle wasting. One recent publication demonstrated a virtual screening platform to identify small molecules that could target the DNA binding domain of STAT3 and found in S3-54 [144]. Cardoso et al demonstrated that Redox factor-1 (Ref-1) reduces STAT3 protein enabling it to bind to DNA more effectively. Furthermore, the Ref-1 inhibitor E3330 blocks IL-6 induced STAT3 DNA binding without affecting total STAT3 protein levels or the phosphorylation of Y705 [145]. The Ref-1 protein also regulates DNA binding of NF-κB in addition to STAT3 (REF). Both of these transcription factors play a role in cancer cachexia and thus E3330 might have greater efficacy than agents targeting STAT3 alone. E3330 can be tested in animal models and potentially in humans as E3330 shows low toxicity in human subjects. Another compound recently described as a STAT3 inhibitor, Galeillalactone, covalently modify cysteines in the DNA binding domain of STAT3, thereby blocking STAT3 action without affecting phosphorylation [146]. Galeillalactone inhibits growth of prostate cancer cells in vitro and in vivo.
Finally, preventing the transit of activated STAT3 dimers through the nuclear pore would inhibit STAT3 transcriptional activity. However, how exactly STAT3 shuttling through the nuclear membrane occurs has yet to be clearly defined.
6.4 Activating the endogenous STAT3 inhibitors
Physiologic antagonists inhibit STAT3 activity at the level of the receptors, JAKs and STAT3. These include STAT3-activated SOCS proteins, particularly SOCS3 [13, 15, 27]. The SOCS family members (SOCS 1-7 and CIS) are characterized by a central SH2 domain mediating binding to p-Y residues on JAKs, gp130 and other cytokine receptor chains [20, 147, 148]. SOCS3 can inhibit STAT3 by three main mechanisms: direct inhibition of JAK proteins, competition with STAT3 for binding to pY sites on activated receptors, and recruitment and ubiquitylation of signaling proteins and their consequent degradation by the proteasome system [20, 147, 148]. SOCS3 expression is elevated in C26 cachexia, although the vast increase in SOCS3 mRNA was not mirrored by a correspondingly large increase in protein. This raises the potential that augmenting SOCS3 expression or stabilizing SOCS3 protein levels might be protective in cachexia. An alternative mechanism through which the STAT3 signaling is terminated is the dephosphorylation of STAT3, mediated by TC-PTP/PTPN2 in the nucleus and cytoplasm [149]. Enhancement of this phosphatase activity through increasing TC-PTP/PTPN2 expression or activity could be a novel approach to blocking STAT3 in cachexia.
6.5 Multi-modal inhibition
Thus efficient and enduring blockade of STAT3 might be achieved at several different sites along its pathway of activation. By utilizing such different inhibitors alone and in combination, we might be able to arrive at therapeutic regimens to slow or prevent cachexia, while also reducing tumor growth.
7.0 CONCLUDING REMARKS
Overall, strong correlative data and parallel studies in other systems suggest important roles for STAT3 in directly mediating muscle wasting in cancer cachexia through or in addition to effects on other non-muscle tissues. Robust genetic studies have only just begun, however, and while these recent results support a causal role for STAT3 in myofiber atrophy, knowledge of its roles in other processes remain elusive. Muscle specific knockout mice are unlikely to be spared from cancer death for long, given that fat wasting and other organ effects were likely unabated. Further study is required to determine whether cardioprotective and other potential salutary activities of STAT3 in cachexia might render targeting of STAT3 for muscle preservation impossible, or whether organ-specific STAT3 pathways or interactors might be found.
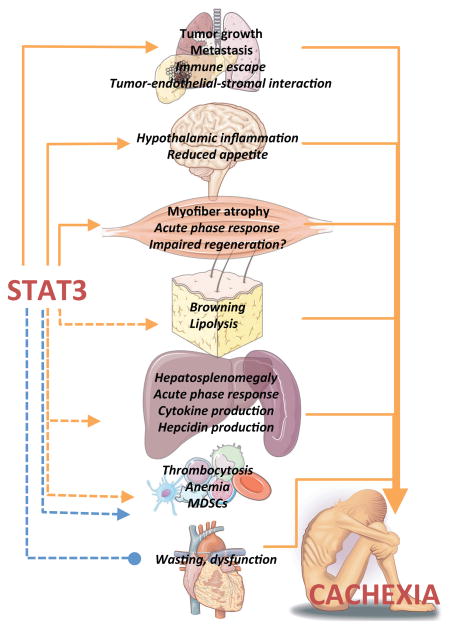
Figure 1. Activities of STAT3 that contribute to the multi-organ, systemic response to tumor.
Activating roles of STAT3 are shown in orange, with the inhibitory role of STAT3 on cardiac hypotrophy shown in blue. Cachexia-promoting effects of organ dysfunction are shown in orange. Genetically confirmed activities of STAT3 in cachexia models are shown in boldface; activities implicated, but not functionally confirmed in cancer cachexia are shown in italics.
Table 1.
| Inhibitor | Target | Stage | Reference |
|---|---|---|---|
| Siltuximab | IL-6 | Phase | |
| Tocilizumab | IL-6 Receptor | FDA approved | Teresa can you add this ref? |
| AZD1480 | Jak1/2/3 | Terminated due to neurotoxicity | (7, 10, 14) |
| Ruxolitinib (INCB018424) | Jak1/2 | Phase II/III | (11, 12) |
| CEP701 | Jak | Phase II | [132] |
| FLLL31, FLLL32 | Jak2/STAT3 | Preclinical | (13) |
| SH-4-54 | STAT3 | Preclinical | (18) |
| E3330 | Ref-1 | Preclinical | (25, 27) |
| C188-9 | STAT3 | Preclinical | (21, 22) |
| Selumetinib | MEK | Phase II/III | |
| Sunitinib | Tyrosine kinase | Phase II/III | [133] |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Teresa A. Zimmers, Email: zimmerst@iu.edu.
Melissa L. Fishel, Email: mfishel@iu.edu.
Andrea Bonetto, Email: abonetto@iupui.edu.
Bibliography
- 1.Cohen S, Nathan JA, Goldberg AL. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat Rev Drug Discov. 2015;14:58–74. doi: 10.1038/nrd4467. [DOI] [PubMed] [Google Scholar]
- 2.Argilés JM, Busquets S, Stemmler B, López-Soriano FJ. Cachexia and sarcopenia: mechanisms and potential targets for intervention. Curr Opin Pharmacol. 2015;22:100–6. doi: 10.1016/j.coph.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Evans WJ, Morley JE, Argilés J, Bales C, Baracos V, Guttridge D, et al. Cachexia: a new definition. Clin Nutr. 2008;27:793–9. doi: 10.1016/j.clnu.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Ströhle A, Zänker K, Hahn A. Nutrition in oncology: the case of micronutrients (review) Oncol Rep. 2010;24:815–28. doi: 10.3892/or.2010.815. [DOI] [PubMed] [Google Scholar]
- 5.Kazemi-Bajestani SM, Mazurak VC, Baracos V. Computed tomography-defined muscle and fat wasting are associated with cancer clinical outcomes. Semin Cell Dev Biol. 2015 doi: 10.1016/j.semcdb.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Benny Klimek ME, Aydogdu T, Link MJ, Pons M, Koniaris LG, Zimmers TA. Acute inhibition of myostatin-family proteins preserves skeletal muscle in mouse models of cancer cachexia. Biochem Biophys Res Commun. 2010;391:1548–54. doi: 10.1016/j.bbrc.2009.12.123. [DOI] [PubMed] [Google Scholar]
- 7.Zhou X, Wang JL, Lu J, Song Y, Kwak KS, Jiao Q, et al. Reversal of cancer cachexia and muscle wasting by ActRIIB antagonism leads to prolonged survival. Cell. 2010;142:531–43. doi: 10.1016/j.cell.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Von Haehling S, Anker SD. Treatment of cachexia: An overview of recent developments. Int J Cardiol. 2015;184:736–42. doi: 10.1016/j.ijcard.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Garcia JM. Anamorelin hydrochloride for the treatment of cancer-anorexia-cachexia in NSCLC. Expert Opin Pharmacother. 2015;16:1245–53. doi: 10.1517/14656566.2015.1041500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narsale AA, Carson JA. Role of interleukin-6 in cachexia: therapeutic implications. Curr Opin Support Palliat Care. 2014;8:321–7. doi: 10.1097/SPC.0000000000000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strassmann G, Fong M, Kenney JS, Jacob CO. Evidence for the involvement of interleukin 6 in experimental cancer cachexia. J Clin Invest. 1992;89:1681–4. doi: 10.1172/JCI115767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16:448–57. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 13.Puppa MJ, Gao S, Narsale AA, Carson JA. Skeletal muscle glycoprotein 130’s role in Lewis lung carcinoma-induced cachexia. FASEB J. 2014;28:998–1009. doi: 10.1096/fj.13-240580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med. 2010;363:1117–27. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonetto A, Aydogdu T, Jin X, Zhang Z, Zhan R, Puzis L, et al. JAK/STAT3 pathway inhibition blocks skeletal muscle wasting downstream of IL-6 and in experimental cancer cachexia. Am J Physiol Endocrinol Metab. 2012;303:E410–E421. doi: 10.1152/ajpendo.00039.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva KA, Dong J, Dong Y, Dong Y, Schor N, Tweardy DJ, et al. Inhibition of Stat3 activation suppresses caspase-3 and the ubiquitin-proteasome system, leading to preservation of muscle mass in cancer cachexia. J Biol Chem. 2015;290:11177–87. doi: 10.1074/jbc.M115.641514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–46. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 18.Heinrich PC, Behrmann I, Müller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998;334(Pt 2):297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiu H, Nicholson SE. Biology and significance of the JAK/STAT signalling pathways. Growth Factors. 2012;30:88–106. doi: 10.3109/08977194.2012.660936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94:3801–4. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hankey PA. Regulation of hematopoietic cell development and function by Stat3. Front Biosci (Landmark Ed) 2009;14:5273–90. doi: 10.2741/3597. [DOI] [PubMed] [Google Scholar]
- 22.Haghikia A, Ricke-Hoch M, Stapel B, Gorst I, Hilfiker-Kleiner D. STAT3, a key regulator of cell-to-cell communication in the heart. Cardiovasc Res. 2014;102:281–9. doi: 10.1093/cvr/cvu034. [DOI] [PubMed] [Google Scholar]
- 23.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19:2548–56. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- 24.Germain D, Frank D. Targeting the cytoplasmic and nuclear functions of signal tranducers and activators of transcription 3 for cancer therapy. Clin Cancer Res. 2007;13:5665–9. doi: 10.1158/1078-0432.CCR-06-2491. [DOI] [PubMed] [Google Scholar]
- 25.Schuringa JJ, Schepers H, Vellenga E, Kruijer W. Ser727-dependent transcriptional activation by association of p300 with STAT3 upon IL-6 stimulation. FEBS Lett. 2001;495:71–6. doi: 10.1016/s0014-5793(01)02354-7. [DOI] [PubMed] [Google Scholar]
- 26.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Müller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank D. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 28.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–73. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 29.Wang R, Cherukuri P, Luo J. Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J Biol Chem. 2005;280:11528–34. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- 30.Ray S, Boldogh I, Brasier AR. STAT3 NH2-terminal acetylation is activated by the hepatic acute-phase response and required for IL-6 induction of angiotensinogen. Gastroenterology. 2005;129:1616–32. doi: 10.1053/j.gastro.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 31.Dasgupta M, Unal H, Willard B, Yang J, Karnik SS, Stark GR. A Critical Role for Lysine 685 in Gene Expression Mediated by Unphosphorylated STAT3. J Biol Chem. 2014 doi: 10.1074/jbc.M114.603894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waitkus MS, Chandrasekharan UM, Willard B, Tee TL, Hsieh JK, Przybycin CG, et al. Signal integration and gene induction by a functionally distinct STAT3 phosphoform. Mol Cell Biol. 2014;34:1800–11. doi: 10.1128/MCB.00034-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, et al. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–47. [PubMed] [Google Scholar]
- 34.Bonetto A, Aydogdu T, Kunzevitzky N, Guttridge DC, Khuri S, Koniaris LG, et al. STAT3 activation in skeletal muscle links muscle wasting and the acute phase response in cancer cachexia. PLoS ONE. 2011;6:e22538. doi: 10.1371/journal.pone.0022538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pretto F, Ghilardi C, Moschetta M, Bassi A, Rovida A, Scarlato V, et al. Sunitinib prevents cachexia and prolongs survival of mice bearing renal cancer by restraining STAT3 and MuRF-1 activation in muscle. Oncotarget. 2015;6:3043–54. doi: 10.18632/oncotarget.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hetzler KL, Hardee JP, Puppa MJ, Narsale AA, Sato S, Davis JM, et al. Sex differences in the relationship of IL-6 signaling to cancer cachexia progression. Biochim Biophys Acta. 2015;1852:816–25. doi: 10.1016/j.bbadis.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seto DN, Kandarian SC, Jackman RW. A Key Role for Leukemia Inhibitory Factor in C26 Cancer Cachexia. J Biol Chem. 2015;290:19976–86. doi: 10.1074/jbc.M115.638411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White JP, Puppa MJ, Narsale A, Carson JA. Characterization of the male ApcMin/+ mouse as a hypogonadism model related to cancer cachexia. Biol Open. 2013;2:1346–53. doi: 10.1242/bio.20136544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narsale AA, Enos RT, Puppa MJ, Chatterjee S, Murphy EA, Fayad R, et al. Liver inflammation and metabolic signaling in ApcMin/+ mice: the role of cachexia progression. PLoS ONE. 2015;10:e0119888. doi: 10.1371/journal.pone.0119888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsoli M, Schweiger M, Vanniasinghe AS, Painter A, Zechner R, Clarke S, et al. Depletion of white adipose tissue in cancer cachexia syndrome is associated with inflammatory signaling and disrupted circadian regulation. PLoS ONE. 2014;9:e92966. doi: 10.1371/journal.pone.0092966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baltgalvis KA, Berger FG, Pena MM, Davis JM, Muga SJ, Carson JA. Interleukin-6 and cachexia in ApcMin/+ mice. Am J Physiol Regul Integr Comp Physiol. 2008;294:R393–R401. doi: 10.1152/ajpregu.00716.2007. [DOI] [PubMed] [Google Scholar]
- 42.Gilabert M, Calvo E, Airoldi A, Hamidi T, Moutardier V, Turrini O, et al. Pancreatic cancer-induced cachexia is Jak2-dependent in mice. J Cell Physiol. 2014;229:1437–43. doi: 10.1002/jcp.24580. [DOI] [PubMed] [Google Scholar]
- 43.Xu S, Neamati N. gp130: a promising drug target for cancer therapy. Expert Opin Ther Targets. 2013;17:1303–28. doi: 10.1517/14728222.2013.830105. [DOI] [PubMed] [Google Scholar]
- 44.Vaisse C, Halaas JL, Horvath CM, Darnell JE, Stoffel M, Friedman JM. Leptin activation of Stat3 in the hypothalamus of wild-type and ob/ob mice but not db/db mice. Nat Genet. 1996;14:95–7. doi: 10.1038/ng0996-95. [DOI] [PubMed] [Google Scholar]
- 45.Tian SS, Lamb P, Seidel HM, Stein RB, Rosen J. Rapid activation of the STAT3 transcription factor by granulocyte colony-stimulating factor. Blood. 1994;84:1760–4. [PubMed] [Google Scholar]
- 46.Zhong Z, Wen Z, Darnell JE. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–8. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, van Boxel-Dezaire AH, Cheon H, Yang J, Stark GR. STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc Natl Acad Sci U S A. 2013;110:16975–80. doi: 10.1073/pnas.1315862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vignais ML, Sadowski HB, Watling D, Rogers NC, Gilman M. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol Cell Biol. 1996;16:1759–69. doi: 10.1128/mcb.16.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novak U, Harpur AG, Paradiso L, Kanagasundaram V, Jaworowski A, Wilks AF, et al. Colony-stimulating factor 1-induced STAT1 and STAT3 activation is accompanied by phosphorylation of Tyk2 in macrophages and Tyk2 and JAK1 in fibroblasts. Blood. 1995;86:2948–56. [PubMed] [Google Scholar]
- 50.Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol. 2004;22:929–79. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- 51.Fung MM, Rohwer F, McGuire KL. IL-2 activation of a PI3K-dependent STAT3 serine phosphorylation pathway in primary human T cells. Cell Signal. 2003;15:625–36. doi: 10.1016/s0898-6568(03)00003-2. [DOI] [PubMed] [Google Scholar]
- 52.Walker SR, Xiang M, Frank DA. STAT3 Activity and Function in Cancer: Modulation by STAT5 and miR-146b. Cancers (Basel) 2014;6:958–68. doi: 10.3390/cancers6020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiang M, Birkbak NJ, Vafaizadeh V, Walker SR, Yeh JE, Liu S, et al. STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-κB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Sci Signal. 2014;7:ra11. doi: 10.1126/scisignal.2004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okamoto M, Nasu K, Abe W, Aoyagi Y, Kawano Y, Kai K, et al. Enhanced miR-210 expression promotes the pathogenesis of endometriosis through activation of signal transducer and activator of transcription 3. Hum Reprod. 2015;30:632–41. doi: 10.1093/humrep/deu332. [DOI] [PubMed] [Google Scholar]
- 55.Cao X, Tay A, Guy GR, Tan YH. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol Cell Biol. 1996;16:1595–603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ram P, Iyengar R. G protein coupled receptor signaling through the src and Stat3 pathway: roe in proliferation and transformation. Oncogene. 2001;20:1601–6. doi: 10.1038/sj.onc.1204186. [DOI] [PubMed] [Google Scholar]
- 57.Silva CM. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene. 2004;23:8017–23. doi: 10.1038/sj.onc.1208159. [DOI] [PubMed] [Google Scholar]
- 58.Faruqi TR, Gomez D, Bustelo XR, Bar-Sagi D, Reich NC. Rac1 mediates STAT3 activation by autocrine IL-6. Proc Natl Acad Sci U S A. 2001;98:9014–9. doi: 10.1073/pnas.161281298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aznar S, Valerón PF, del Rincon SV, Pérez LF, Perona R, Lacal JC. Simultaneous tyrosine and serine phosphorylation of STAT3 transcription factor is involved in Rho A GTPase oncogenic transformation. Mol Biol Cell. 2001;12:3282–94. doi: 10.1091/mbc.12.10.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Metcalf D, Gearing DP. Fatal syndrome in mice engrafted with cells producing high levels of the leukemia inhibitory factor. Proc Natl Acad Sci U S A. 1989;86:5948–52. doi: 10.1073/pnas.86.15.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iseki H, Kajimura N, Ohue C, Tanaka R, Akiyama Y, Yamaguchi K. 1995 doi: 10.1111/j.1349-7006.1995.tb02435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barton BE, Murphy TF. Cancer cachexia is mediated in part by the induction of IL-6-like cytokines from the spleen. Cytokine. 2001;16:251–7. doi: 10.1006/cyto.2001.0968. [DOI] [PubMed] [Google Scholar]
- 63.Henderson JT, Seniuk NA, Richardson PM, Gauldie J, Roder JC. 1994 [Google Scholar]
- 64.Fujiki F, Mukaida N, Hirose K, Ishida H, Harada A, Ohno S, et al. Prevention of adenocarcinoma colon 26-induced cachexia by interleukin 10 gene transfer. Cancer Res. 1997;57:94–9. [PubMed] [Google Scholar]
- 65.Watchorn TM, Waddell I, Dowidar N, Ross JA. Proteolysis-inducing factor regulates hepatic gene expression via the transcription factors NF-(kappa)B and STAT3. FASEB J. 2001;15:562–4. doi: 10.1096/fj.00-0534fje. [DOI] [PubMed] [Google Scholar]
- 66.Judge SM, Wu CL, Beharry AW, Roberts BM, Ferreira LF, Kandarian SC, et al. Genome-wide identification of FoxO-dependent gene networks in skeletal muscle during C26 cancer cachexia. BMC Cancer. 2014;14:997. doi: 10.1186/1471-2407-14-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hutchins AP, Diez D, Takahashi Y, Ahmad S, Jauch R, Tremblay ML, et al. Distinct transcriptional regulatory modules underlie STAT3’s cell type-independent and cell type-specific functions. Nucleic Acids Res. 2013;41:2155–70. doi: 10.1093/nar/gks1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reed SA, Sandesara PB, Senf SM, Judge AR. Inhibition of FoxO transcriptional activity prevents muscle fiber atrophy during cachexia and induces hypertrophy. FASEB J. 2012;26:987–1000. doi: 10.1096/fj.11-189977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem. 2004;279:41114–23. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 70.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/S1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 71.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kruczyk M, Przanowski P, Dabrowski M, Swiatek-Machado K, Mieczkowski J, Wallerman O, et al. Integration of genome-wide of Stat3 binding and epigenetic modification mapping with transcriptome reveals novel Stat3 target genes in glioma cells. Biochim Biophys Acta. 2014;1839:1341–50. doi: 10.1016/j.bbagrm.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 73.Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–15. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Snyder M, Huang XY, Zhang JJ. Identification of novel direct Stat3 target genes for control of growth and differentiation. J Biol Chem. 2008;283:3791–8. doi: 10.1074/jbc.M706976200. [DOI] [PubMed] [Google Scholar]
- 75.Oh YM, Kim JK, Choi Y, Choi S, Yoo JY. Prediction and experimental validation of novel STAT3 target genes in human cancer cells. PLoS ONE. 2009;4:e6911. doi: 10.1371/journal.pone.0006911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alvarez JV, Frank DA. Genome-wide analysis of STAT target genes: Elucidating the mechanism of STAT-mediated oncogenesis. Cancer Biol Ther. 2004;3:1045–50. doi: 10.4161/cbt.3.11.1172. [DOI] [PubMed] [Google Scholar]
- 77.Dauer DJ, Ferraro B, Song L, Yu B, Mora L, Buettner R, et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24:3397–408. doi: 10.1038/sj.onc.1208469. [DOI] [PubMed] [Google Scholar]
- 78.Quinton LJ, Blahna MT, Jones MR, Allen E, Ferrari JD, Hilliard KL, et al. Hepatocyte-specific mutation of both NF-κB RelA and STAT3 abrogates the acute phase response in mice. J Clin Invest. 2012;122:1758–63. doi: 10.1172/JCI59408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sakamori R, Takehara T, Ohnishi C, Tatsumi T, Ohkawa K, Takeda K, et al. Signal transducer and activator of transcription 3 signaling within hepatocytes attenuates systemic inflammatory response and lethality in septic mice. Hepatology. 2007;46:1564–73. doi: 10.1002/hep.21837. [DOI] [PubMed] [Google Scholar]
- 80.Bode JG, Albrecht U, Häussinger D, Heinrich PC, Schaper F. Hepatic acute phase proteins--regulation by IL-6- and IL-1-type cytokines involving STAT3 and its crosstalk with NF-κB-dependent signaling. Eur J Cell Biol. 2012;91:496–505. doi: 10.1016/j.ejcb.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 81.Wan CK, Andraski AB, Spolski R, Li P, Kazemian M, Oh J, et al. Opposing roles of STAT1 and STAT3 in IL-21 function in CD4+ T cells. Proc Natl Acad Sci U S A. 2015;112:9394–9. doi: 10.1073/pnas.1511711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kirchmer MN, Franco A, Albasanz-Puig A, Murray J, Yagi M, Gao L, et al. Modulation of vascular smooth muscle cell phenotype by STAT-1 and STAT-3. Atherosclerosis. 2014;234:169–75. doi: 10.1016/j.atherosclerosis.2014.02.029. [DOI] [PubMed] [Google Scholar]
- 83.Zheng J, van de Veerdonk FL, Crossland KL, Smeekens SP, Chan CM, Al Shehri T, et al. Gain-of-function STAT1 mutations impair STAT3 activity in patients with Chronic Mucocutaneous Candidiasis (CMC) Eur J Immunol. 2015 doi: 10.1002/eji.201445344. [DOI] [PubMed] [Google Scholar]
- 84.Shabo Y, Lotem J, Sachs L. Autoregulation of interleukin 6 and granulocyte-macrophage colony-stimulating factor in the differentiation of myeloid leukemic cells. Mol Cell Biol. 1989;9:4109–12. doi: 10.1128/mcb.9.9.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Narimatsu M, Maeda H, Itoh S, Atsumi T, Ohtani T, Nishida K, et al. Tissue-specific autoregulation of the stat3 gene and its role in interleukin-6-induced survival signals in T cells. Mol Cell Biol. 2001;21:6615–25. doi: 10.1128/MCB.21.19.6615-6625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Q, Wang HY, Woetmann A, Raghunath PN, Odum N, Wasik MA. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood. 2006;108:1058–64. doi: 10.1182/blood-2005-08-007377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Q, Wang HY, Marzec M, Raghunath PN, Nagasawa T, Wasik MA. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci U S A. 2005;102:6948–53. doi: 10.1073/pnas.0501959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fleming JD, Giresi PG, Lindahl-Allen M, Krall EB, Lieb JD, Struhl K. STAT3 acts through pre-existing nucleosome-depleted regions bound by FOS during an epigenetic switch linking inflammation to cancer. Epigenetics Chromatin. 2015;8:7. doi: 10.1186/1756-8935-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hedrich CM, Rauen T, Apostolidis SA, Grammatikos AP, Rodriguez Rodriguez N, Ioannidis C, et al. Stat3 promotes IL-10 expression in lupus T cells through trans-activation and chromatin remodeling. Proc Natl Acad Sci U S A. 2014;111:13457–62. doi: 10.1073/pnas.1408023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tang Y, Luo Y, Jiang Z, Ma Y, Lin CJ, Kim C, et al. Jak/Stat3 signaling promotes somatic cell reprogramming by epigenetic regulation. Stem Cells. 2012;30:2645–56. doi: 10.1002/stem.1225. [DOI] [PubMed] [Google Scholar]
- 92.Gasche JA, Hoffmann J, Boland CR, Goel A. Interleukin-6 promotes tumorigenesis by altering DNA methylation in oral cancer cells. Int J Cancer. 2011;129:1053–63. doi: 10.1002/ijc.25764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gianotti TF, Castaño G, Gemma C, Burgueño AL, Rosselli MS, Pirola CJ, et al. Mitochondrial DNA copy number is modulated by genetic variation in the signal transducer and activator of transcription 3 (STAT3) Metab Clin Exp. 2011;60:1142–9. doi: 10.1016/j.metabol.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 94.Vassilev AO, Lorenz DR, Tibbles HE, Uckun FM. Role of the leukemia-associated transcription factor STAT3 in platelet physiology. Leuk Lymphoma. 2002;43:1461–7. doi: 10.1080/1042819022386716. [DOI] [PubMed] [Google Scholar]
- 95.Phillips D, Reilley MJ, Aponte AM, Wang G, Boja E, Gucek M, et al. Stoichiometry of STAT3 and mitochondrial proteins: Implications for the regulation of oxidative phosphorylation by protein-protein interactions. J Biol Chem. 2010;285:23532–6. doi: 10.1074/jbc.C110.152652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–7. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–6. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boengler K, Hilfiker-Kleiner D, Heusch G, Schulz R. Inhibition of permeability transition pore opening by mitochondrial STAT3 and its role in myocardial ischemia/reperfusion. Basic Res Cardiol. 2010;105:771–85. doi: 10.1007/s00395-010-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shaw PE. Could STAT3 provide a link between respiration and cell cycle progression? Cell Cycle. 2010;9:4294–6. doi: 10.4161/cc.9.21.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pedroso FE, Spalding PB, Cheung MC, Yang R, Gutierrez JC, Bonetto A, et al. Inflammation, organomegaly, and muscle wasting despite hyperphagia in a mouse model of burn cachexia. J Cachexia Sarcopenia Muscle. 2012;3:199–211. doi: 10.1007/s13539-012-0062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jeschke MG, Gauglitz GG, Kulp GA, Finnerty CC, Williams FN, Kraft R, et al. Long-term persistance of the pathophysiologic response to severe burn injury. PLoS ONE. 2011;6:e21245. doi: 10.1371/journal.pone.0021245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Finnerty CC, Przkora R, Herndon DN, Jeschke MG. Cytokine expression profile over time in burned mice. Cytokine. 2009;45:20–5. doi: 10.1016/j.cyto.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jeschke MG, Chinkes DL, Finnerty CC, Kulp G, Suman OE, Norbury WB, et al. Pathophysiologic response to severe burn injury. Ann Surg. 2008;248:387–401. doi: 10.1097/SLA.0b013e3181856241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Callahan LA, Supinski GS. Sepsis-induced myopathy. Crit Care Med. 2009;37:S354–S367. doi: 10.1097/CCM.0b013e3181b6e439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Van Hees HW, Schellekens WJ, Linkels M, Leenders F, Zoll J, Donders R, et al. Plasma from septic shock patients induces loss of muscle protein. Crit Care. 2011;15:R233. doi: 10.1186/cc10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Friedrich O, Reid MB, Van den Berghe G, Vanhorebeek I, Hermans G, Rich MM, et al. The Sick and the Weak: Neuropathies/Myopathies in the Critically Ill. Physiol Rev. 2015;95:1025–109. doi: 10.1152/physrev.00028.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Remels AH, Gosker HR, Langen RC, Schols AM. The mechanisms of cachexia underlying muscle dysfunction in COPD. J Appl Physiol. 2013;114:1253–62. doi: 10.1152/japplphysiol.00790.2012. [DOI] [PubMed] [Google Scholar]
- 108.Londhe P, Guttridge DC. Inflammation induced loss of skeletal muscle. Bone. 2015;80:131–42. doi: 10.1016/j.bone.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hoepers AT, Menezes MM, Fröde TS. Systematic review of anaemia and inflammatory markers in chronic obstructive pulmonary disease. Clin Exp Pharmacol Physiol. 2015;42:231–9. doi: 10.1111/1440-1681.12357. [DOI] [PubMed] [Google Scholar]
- 110.Doehner W, Frenneaux M, Anker SD. Metabolic Impairment in Heart Failure: The Myocardial and Systemic Perspective. J Am Coll Cardiol. 2014;64:1388–400. doi: 10.1016/j.jacc.2014.04.083. [DOI] [PubMed] [Google Scholar]
- 111.Ebner N, Elsner S, Springer J, von Haehling S. Molecular mechanisms and treatment targets of muscle wasting and cachexia in heart failure: an overview. Curr Opin Support Palliat Care. 2014;8:15–24. doi: 10.1097/SPC.0000000000000030. [DOI] [PubMed] [Google Scholar]
- 112.Reid J, Noble HR, Porter S, Shields JS, Maxwell AP. A literature review of end-stage renal disease and cachexia: understanding experience to inform evidence-based healthcare. J Ren Care. 2013;39:47–51. doi: 10.1111/j.1755-6686.2013.00341.x. [DOI] [PubMed] [Google Scholar]
- 113.Badowski M, Pandit NS. Pharmacologic Management of Human Immunodeficiency Virus Wasting Syndrome. Pharmacotherapy. 2014 doi: 10.1002/phar.1431. [DOI] [PubMed] [Google Scholar]
- 114.Stephens NA, Skipworth RJ, Fearon KC. Cachexia, survival and the acute phase response. Curr Opin Support Palliat Care. 2008;2:267–74. doi: 10.1097/SPC.0b013e3283186be2. [DOI] [PubMed] [Google Scholar]
- 115.Newsome TW, Mason AD, Pruitt BA. Weight loss following thermal injury. Ann Surg. 1973;178:215–7. doi: 10.1097/00000658-197308000-00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Johns N, Hatakeyama S, Stephens NA, Degen M, Degen S, Frieauff W, et al. Clinical classification of cancer cachexia: phenotypic correlates in human skeletal muscle. PLoS ONE. 2014;9:e83618. doi: 10.1371/journal.pone.0083618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fearon K, Strasser F, Anker SD, Bosaeus I, Bruera E, Fainsinger RL, et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol. 2011;12:489–95. doi: 10.1016/S1470-2045(10)70218-7. [DOI] [PubMed] [Google Scholar]
- 118.Lerner L, Tao J, Liu Q, Nicoletti R, Feng B, Krieger B, et al. MAP3K11/GDF15 axis is a critical driver of cancer cachexia. J Cachexia Sarcopenia Muscle. 2015:n/a–n/a. doi: 10.1002/jcsm.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lerner L, Hayes TG, Tao N, Krieger B, Feng B, Wu Z, et al. Plasma growth differentiation factor 15 is associated with weight loss and mortality in cancer patients. J Cachexia Sarcopenia Muscle. 2015:n/a–n/a. doi: 10.1002/jcsm.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Loumaye A, de Barsy M, Nachit M, Lause P, Frateur L, van Maanen A, et al. Role of Activin A and Myostatin in human cancer cachexia. J Clin Endocrinol Metab. 2015;100:2030–8. doi: 10.1210/jc.2014-4318. [DOI] [PubMed] [Google Scholar]
- 121.Holmer R, Goumas FA, Waetzig GH, Rose-John S, Kalthoff H. Interleukin-6: a villain in the drama of pancreatic cancer development and progression. HBPD INT. 2014;13:371–80. doi: 10.1016/s1499-3872(14)60259-9. [DOI] [PubMed] [Google Scholar]
- 122.Johns N, Tan BH, MacMillan M, Solheim TS, Ross JA, Baracos VE, et al. Genetic basis of interindividual susceptibility to cancer cachexia: selection of potential candidate gene polymorphisms for association studies. J Genet. 2014;93:893–916. doi: 10.1007/s12041-014-0405-9. [DOI] [PubMed] [Google Scholar]
- 123.Ruzzo A, Catalano V, Canestrari E, Giacomini E, Santini D, Tonini G, et al. Genetic modulation of the interleukin 6 (IL-6) system in patients with advanced gastric cancer: a background for an alternative target therapy. BMC Cancer. 2014;14:357. doi: 10.1186/1471-2407-14-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tan BH, Fearon KC. Cytokine gene polymorphisms and susceptibility to cachexia. Curr Opin Support Palliat Care. 2010;4:243–8. doi: 10.1097/SPC.0b013e32833e4a5d. [DOI] [PubMed] [Google Scholar]
- 125.Sandri M. Protein breakdown in cancer cachexia. Semin Cell Dev Biol. 2015 doi: 10.1016/j.semcdb.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 126.Talbert EE, Guttridge DC. Impaired regeneration: A role for the muscle microenvironment in cancer cachexia. Semin Cell Dev Biol. 2015 doi: 10.1016/j.semcdb.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tsoli M, Swarbrick MM, Robertson GR. Lipolytic and thermogenic depletion of adipose tissue in cancer cachexia. Semin Cell Dev Biol. 2015 doi: 10.1016/j.semcdb.2015.10.039. [DOI] [PubMed] [Google Scholar]
- 128.Burfeind K, Michaelis KA, Marks DL. The central role of hypothalamic inflammation in the acute illness response and cachexia. Semin Cell Dev Biol. 2015 doi: 10.1016/j.semcdb.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kalantar-Zadeh K, Rhee C, Sim JJ, Stenvinkel P, Anker SD, Kovesdy CP. Why cachexia kills: examining the causality of poor outcomes in wasting conditions. J Cachexia Sarcopenia Muscle. 2013;4:89–94. doi: 10.1007/s13539-013-0111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Molnar MZ, Streja E, Kovesdy CP, Budoff MJ, Nissenson AR, Krishnan M, et al. High platelet count as a link between renal cachexia and cardiovascular mortality in end-stage renal disease patients. Am J Clin Nutr. 2011;94:945–54. doi: 10.3945/ajcn.111.014639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Di Sebastiano KM, Yang L, Zbuk K, Wong RK, Chow T, Koff D, et al. Accelerated muscle and adipose tissue loss may predict survival in pancreatic cancer patients: the relationship with diabetes and anaemia. Br J Nutr. 2013;109:302–12. doi: 10.1017/S0007114512001067. [DOI] [PubMed] [Google Scholar]
- 132.Black K, Garrett IR, Mundy GR. Chinese hamster ovarian cells transfected with the murine interleukin-6 gene cause hypercalcemia as well as cachexia, leukocytosis and thrombocytosis in tumor-bearing nude mice. Endocrinology. 1991;128:2657–9. doi: 10.1210/endo-128-5-2657. [DOI] [PubMed] [Google Scholar]
- 133.Cramer L, Hildebrandt B, Kung T, Wichmann K, Springer J, Doehner W, et al. Cardiovascular function and predictors of exercise capacity in patients with colorectal cancer. J Am Coll Cardiol. 2014;64:1310–9. doi: 10.1016/j.jacc.2014.07.948. [DOI] [PubMed] [Google Scholar]
- 134.Arthur ST, Noone JM, Van Doren BA, Roy D, Blanchette CM. One-year prevalence, comorbidities and cost of cachexia-related inpatient admissions in the USA. Drugs Context. 2014;3:212265. doi: 10.7573/dic.212265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kazemi-Bajestani SM, Becher H, Fassbender K, Chu Q, Baracos VE. Concurrent evolution of cancer cachexia and heart failure: bilateral effects exist. J Cachexia Sarcopenia Muscle. 2014;5:95–104. doi: 10.1007/s13539-014-0137-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Milner JD, Vogel TP, Forbes L, Ma CA, Stray-Pedersen A, Niemela JE, et al. Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood. 2015;125:591–9. doi: 10.1182/blood-2014-09-602763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shum AM, Fung DC, Corley SM, McGill MC, Bentley N, Tan TC, et al. Cardiac and Skeletal Muscles Show Molecularly Distinct Responses to Cancer Cachexia. Physiol Genomics 2015:physiolgenomics. 2014:00128. doi: 10.1152/physiolgenomics.00128.2014. [DOI] [PubMed] [Google Scholar]
- 138.Tseng YC, Kulp SK, Lai IL, Hsu EC, He WA, Frankhouser DE, et al. Preclinical Investigation of the Novel Histone Deacetylase Inhibitor AR-42 in the Treatment of Cancer-Induced Cachexia. J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/djv274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.White JP, Baynes JW, Welle SL, Kostek MC, Matesic LE, Sato S, et al. The regulation of skeletal muscle protein turnover during the progression of cancer cachexia in the Apc(Min/+) mouse. PLoS ONE. 2011;6:e24650. doi: 10.1371/journal.pone.0024650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Huang TT, Deoghare HV, Smith BK, Beaver TM, Baker HV, Mehinto AC, et al. Gene expression changes in the human diaphragm after cardiothoracic surgery. J Thorac Cardiovasc Surg. 2011;142:1214–22. 1222.e1. doi: 10.1016/j.jtcvs.2011.02.025. [DOI] [PubMed] [Google Scholar]
- 141.Chen W, Hong YQ, Meng ZL. Bioinformatics analysis of molecular mechanisms of chronic obstructive pulmonary disease. Eur Rev Med Pharmacol Sci. 2014;18:3557–63. [PubMed] [Google Scholar]
- 142.Zhang L, Pan J, Dong Y, Tweardy DJ, Dong Y, Garibotto G, et al. Stat3 activation links a C/EBPδ to myostatin pathway to stimulate loss of muscle mass. Cell Metab. 2013;18:368–79. doi: 10.1016/j.cmet.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Leger B, Derave W, De Bock K, Hespel P, Russell AP. 2008 doi: 10.1089/rej.2007.0588. [DOI] [PubMed] [Google Scholar]
- 144.Jeschke MG, Finnerty CC, Herndon DN, Song J, Boehning D, Tompkins RG, et al. Severe injury is associated with insulin resistance, endoplasmic reticulum stress response, and unfolded protein response. Ann Surg. 2012;255:370–8. doi: 10.1097/SLA.0b013e31823e76e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Merritt EK, Cross JM, Bamman MM. Inflammatory and protein metabolism signaling responses in human skeletal muscle after burn injury. J Burn Care Res. 2012;33:291–7. doi: 10.1097/BCR.0b013e3182331e4b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.White AT, LaBarge SA, McCurdy CE, Schenk S. Knockout of STAT3 in skeletal muscle does not prevent high-fat diet-induced insulin resistance. Mol Metab. 2015;4:569–75. doi: 10.1016/j.molmet.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Preston T, Slater C, McMillan DC, Falconer JS, Shenkin A, Fearon KC. Fibrinogen synthesis is elevated in fasting cancer patients with an acute phase response. J Nutr. 1998;128:1355–60. doi: 10.1093/jn/128.8.1355. [DOI] [PubMed] [Google Scholar]
- 148.He WA, Berardi E, Cardillo VM, Acharyya S, Aulino P, Thomas-Ahner J, et al. NF-κB-mediated Pax7 dysregulation in the muscle microenvironment promotes cancer cachexia. J Clin Invest. 2013 doi: 10.1172/JCI68523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Penna F, Costamagna D, Fanzani A, Bonelli G, Baccino FM, Costelli P. Muscle wasting and impaired myogenesis in tumor bearing mice are prevented by ERK inhibition. PLoS ONE. 2010;5:e13604. doi: 10.1371/journal.pone.0013604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sun L, Ma K, Wang H, Xiao F, Gao Y, Zhang W, et al. JAK1-STAT1-STAT3, a key pathway promoting proliferation and preventing premature differentiation of myoblasts. J Cell Biol. 2007;179:129–38. doi: 10.1083/jcb.200703184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang K, Wang C, Xiao F, Wang H, Wu Z. JAK2/STAT2/STAT3 are required for myogenic differentiation. J Biol Chem. 2008;283:34029–36. doi: 10.1074/jbc.M803012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tierney MT, Aydogdu T, Sala D, Malecova B, Gatto S, Puri PL, et al. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat Med. 2014;20:1182–6. doi: 10.1038/nm.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Price FD, von Maltzahn J, Bentzinger CF, Dumont NA, Yin H, Chang NC, et al. Inhibition of JAK-STAT signaling stimulates adult satellite cell function. Nat Med. 2014;20:1174–81. doi: 10.1038/nm.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–25. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Klöppel G, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–69. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 156.Corcoran RB, Contino G, Deshpande V, Tzatsos A, Conrad C, Benes CH, et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 2011;71:5020–9. doi: 10.1158/0008-5472.CAN-11-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Banerjee K, Resat H. Constitutive activation of STAT3 in breast cancer cells: A review. Int J Cancer. 2015 doi: 10.1002/ijc.29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mora LB, Buettner R, Seigne J, Diaz J, Ahmad N, Garcia R, et al. Constitutive activation of Stat3 in human prostate tumors and cell lines: direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 2002;62:6659–66. [PubMed] [Google Scholar]
- 159.Luwor RB, Stylli SS, Kaye AH. The role of Stat3 in glioblastoma multiforme. J Clin Neurosci. 2013;20:907–11. doi: 10.1016/j.jocn.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 160.Pilati C, Zucman-Rossi J. Mutations leading to constitutive active gp130/JAK1/STAT3 pathway. Cytokine Growth Factor Rev. 2015 doi: 10.1016/j.cytogfr.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 161.Devarajan E, Huang S. STAT3 as a central regulator of tumor metastases. Curr Mol Med. 2009;9:626–33. doi: 10.2174/156652409788488720. [DOI] [PubMed] [Google Scholar]
- 162.Lin L, Hutzen B, Zuo M, Ball S, Deangelis S, Foust E, et al. Novel STAT3 phosphorylation inhibitors exhibit potent growth-suppressive activity in pancreatic and breast cancer cells. Cancer Res. 2010;70:2445–54. doi: 10.1158/0008-5472.CAN-09-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cardoso AA, Jiang Y, Luo M, Reed AM, Shahda S, He Y, et al. APE1/Ref-1 regulates STAT3 transcriptional activity and APE1/Ref-1-STAT3 dual-targeting effectively inhibits pancreatic cancer cell survival. PLoS ONE. 2012;7:e47462. doi: 10.1371/journal.pone.0047462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang X, Yue P, Fletcher S, Zhao W, Gunning PT, Turkson J. A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interactions and Stat3-dependent tumor processes. Biochem Pharmacol. 2010;79:1398–409. doi: 10.1016/j.bcp.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Harada D, Takigawa N, Kiura K. The Role of STAT3 in Non-Small Cell Lung Cancer. Cancers (Basel) 2014;6:708–22. doi: 10.3390/cancers6020708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Svinka J, Mikulits W, Eferl R. STAT3 in hepatocellular carcinoma: new perspectives. Hepat Oncol. 2014;1:107–20. doi: 10.2217/hep.13.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dokduang H, Techasen A, Namwat N, Khuntikeo N, Pairojkul C, Murakami Y, et al. STATs profiling reveals predominantly-activated STAT3 in cholangiocarcinoma genesis and progression. J Hepatobiliary Pancreat Sci. 2014;21:767–76. doi: 10.1002/jhbp.131. [DOI] [PubMed] [Google Scholar]
- 168.Guo C, Yang G, Khun K, Kong X, Levy D, Lee P, et al. Activation of Stat3 in renal tumors. Am J Transl Res. 2009;1:283–90. [PMC free article] [PubMed] [Google Scholar]
- 169.Wen W, Liang W, Wu J, Kowolik CM, Buettner R, Scuto A, et al. Targeting JAK1/STAT3 signaling suppresses tumor progression and metastasis in a peritoneal model of human ovarian cancer. Mol Cancer Ther. 2014;13:3037–48. doi: 10.1158/1535-7163.MCT-14-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xie K, Wei D, Huang S. Transcriptional anti-angiogenesis therapy of human pancreatic cancer. Cytokine Growth Factor Rev. 2006;17:147–56. doi: 10.1016/j.cytogfr.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 171.Mace TA, Bloomston M, Lesinski GB. Pancreatic cancer-associated stellate cells: A viable target for reducing immunosuppression in the tumor microenvironment. Oncoimmunology. 2013;2:e24891. doi: 10.4161/onci.24891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cuenca AG, Cuenca AL, Winfield RD, Joiner DN, Gentile L, Delano MJ, et al. Novel Role for Tumor-Induced Expansion of Myeloid-Derived Cells in Cancer Cachexia. J Immunol. 2014 doi: 10.4049/jimmunol.1302895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gore J, Craven KE, Wilson JL, Cote GA, Cheng M, Nguyen HV, et al. TCGA data and patient-derived orthotopic xenografts highlight pancreatic cancer-associated angiogenesis. Oncotarget. 2015 doi: 10.18632/oncotarget.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nagathihalli NS, Castellanos JA, Shi C, Beesetty Y, Reyzer ML, Caprioli R, et al. STAT3 Mediated Remodeling of the Tumor Microenvironment Results in Enhanced Tumor Drug Delivery in a Mouse Model of Pancreatic Cancer. Gastroenterology. 2015 doi: 10.1053/j.gastro.2015.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Molfino A, Gioia G, Rossi Fanelli F, Laviano A. Contribution of Neuroinflammation to the Pathogenesis of Cancer Cachexia. Mediators Inflamm. 2015;2015:801685. doi: 10.1155/2015/801685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ladyman SR, Grattan DR. JAK-STAT and feeding. JAKSTAT. 2013;2:e23675. doi: 10.4161/jkst.23675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mori H, Hanada R, Hanada T, Aki D, Mashima R, Nishinakamura H, et al. Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nat Med. 2004;10:739–43. doi: 10.1038/nm1071. [DOI] [PubMed] [Google Scholar]
- 178.Gong L, Yao F, Hockman K, Heng HH, Morton GJ, Takeda K, et al. Signal transducer and activator of transcription-3 is required in hypothalamic agouti-related protein/neuropeptide Y neurons for normal energy homeostasis. Endocrinology. 2008;149:3346–54. doi: 10.1210/en.2007-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Loh K, Fukushima A, Zhang X, Galic S, Briggs D, Enriori PJ, et al. Elevated hypothalamic TCPTP in obesity contributes to cellular leptin resistance. Cell Metab. 2011;14:684–99. doi: 10.1016/j.cmet.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Derecka M, Gornicka A, Koralov SB, Szczepanek K, Morgan M, Raje V, et al. Tyk2 and Stat3 regulate brown adipose tissue differentiation and obesity. Cell Metab. 2012;16:814–24. doi: 10.1016/j.cmet.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Park H, Ahima R. Leptin signaling. F1000Prime Rep. 2014:6. doi: 10.12703/P6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Petruzzelli M, Schweiger M, Schreiber R, Campos-Olivas R, Tsoli M, Allen J, et al. A Switch from White to Brown Fat Increases Energy Expenditure in Cancer-Associated Cachexia. Cell Metab. 2014 doi: 10.1016/j.cmet.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 183.Lundholm K, Edström S, Ekman L, Karlberg I, Bylund AC, Scherstén T. A comparative study of the influence of malignant tumor on host metabolism in mice and man: evaluation of an experimental model. Cancer. 1978;42:453–61. doi: 10.1002/1097-0142(197808)42:2<453::aid-cncr2820420212>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 184.Lundholm K, Ekman L, Edström S, Karlberg I, Jagenburg R, Scherstén T. Protein synthesis in liver tissue under the influence of a methylcholanthrene-induced sarcoma in mice. Cancer Res. 1979;39:4657–61. [PubMed] [Google Scholar]
- 185.Ternell M, Edström S, Lundholm K. Transcriptional and translational activity in relation to oxygen consumption in isolated liver cells from sarcoma-bearing mice. Cancer Biochem Biophys. 1983;6:213–9. [PubMed] [Google Scholar]
- 186.Wolf RF, Ng B, Weksler B, Burt M, Brennan MF. Effect of growth hormone on tumor and host in an animal model. Ann Surg Oncol. 1994;1:314–20. doi: 10.1007/BF02303570. [DOI] [PubMed] [Google Scholar]
- 187.Tessitore L, Bonelli G, Baccino FM. Early development of protein metabolic perturbations in the liver and skeletal muscle of tumour-bearing rats. A model system for cancer cachexia. Biochem J. 1987;241:153–9. doi: 10.1042/bj2410153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zimmers TA, McKillop IH, Pierce RH, Yoo JY, Koniaris LG. Massive liver growth in mice induced by systemic interleukin 6 administration. Hepatology. 2003;38:326–34. doi: 10.1053/jhep.2003.50318. [DOI] [PubMed] [Google Scholar]
- 189.Figueroa-Clarevega A, Bilder D. Malignant Drosophila tumors interrupt insulin signaling to induce cachexia-like wasting. Dev Cell. 2015;33:47–55. doi: 10.1016/j.devcel.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kwon Y, Song W, Droujinine IA, Hu Y, Asara JM, Perrimon N. Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev Cell. 2015;33:36–46. doi: 10.1016/j.devcel.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Jones A, Friedrich K, Rohm M, Schäfer M, Algire C, Kulozik P, et al. TSC22D4 is a molecular output of hepatic wasting metabolism. EMBO Mol Med. 2013;5:294–308. doi: 10.1002/emmm.201201869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Jin X, Zhang Z, Beer-Stolz D, Zimmers TA, Koniaris LG. Interleukin-6 inhibits oxidative injury and necrosis after extreme liver resection. Hepatology. 2007;46:802–12. doi: 10.1002/hep.21728. [DOI] [PubMed] [Google Scholar]
- 193.Jin X, Zimmers TA, Perez EA, Pierce RH, Zhang Z, Koniaris LG. Paradoxical effects of short- and long-term interleukin-6 exposure on liver injury and repair. Hepatology. 2006;43:474–84. doi: 10.1002/hep.21087. [DOI] [PubMed] [Google Scholar]
- 194.Moh A, Iwamoto Y, Chai GX, Zhang SS, Kano A, Yang DD, et al. Role of STAT3 in liver regeneration: survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab Invest. 2007;87:1018–28. doi: 10.1038/labinvest.3700630. [DOI] [PubMed] [Google Scholar]
- 195.Wang H, Lafdil F, Wang L, Park O, Yin S, Niu J, et al. Hepatoprotective versus oncogenic functions of STAT3 in liver tumorigenesis. Am J Pathol. 2011;179:714–24. doi: 10.1016/j.ajpath.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Aulino P, Berardi E, Cardillo VM, Rizzuto E, Perniconi B, Ramina C, et al. Molecular, cellular and physiological characterization of the cancer cachexia-inducing C26 colon carcinoma in mouse. BMC Cancer. 2010;10:363. doi: 10.1186/1471-2407-10-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Piegari M, Ortiz S, Díaz MDELP, Eynard AR, Valentich MA. Characterization of a murine lung adenocarcinoma (LAC1), a useful experimental model to study progression of lung cancer. J Exp Ther Oncol. 2011;9:231–9. [PubMed] [Google Scholar]
- 198.Garbers C, Aparicio-Siegmund S, Rose-John S. The IL-6/gp130/STAT3 signaling axis: recent advances towards specific inhibition. Curr Opin Immunol. 2015;34:75–82. doi: 10.1016/j.coi.2015.02.008. [DOI] [PubMed] [Google Scholar]