Abstract
During the first division of meiosis, segregation of homologous chromosomes reduces the chromosome number by half. In most species, sister chromatid cohesion and reciprocal recombination (crossing-over) between homologous chromosomes are essential to provide tension to signal proper chromosome segregation during the first meiotic division. Crossovers are not distributed uniformly throughout the genome and are repressed at and near the centromeres. Rare crossovers that occur too near or in the centromere interfere with proper segregation and can give rise to aneuploid progeny, which can be severely defective or inviable. We review here how crossing-over occurs and how it is prevented in and around the centromeres. Molecular mechanisms of centromeric repression are only now being elucidated. However, rapid advances in understanding crossing-over, chromosome structure, and centromere functions promise to explain how potentially deleterious crossovers are avoided in certain chromosomal regions while allowing beneficial crossovers in others.
Keywords: meiosis, homologous recombination, crossing-over, centromeres, chromosome segregation, aneuploidy
Meiosis is a specialized form of cell division that consists of one round of chromosomal replication followed by two rounds of nuclear division to form gametes – eggs and sperm in animals; ovules and pollen in plants; and spores in fungi, such as yeast. It achieves the essential task of reducing the number of chromosomes to half by segregating the centromeres (and the connected chromosome arms) of homologous chromosomes (homologs) at the first meiotic division. The centromeres of sister chromatids, which are generated by chromosomal replication before the first meiotic division, separate subsequently in the second meiotic division. In most species separation of homologs requires the presence of a physical connection between the two chromosomes, which is genetically identified as a crossover. Crossovers are the sites of genetic exchange (recombination), by which shuffling of maternal and paternal alleles generates diversity in the progeny. They aid in breaking haplotypes and therefore can generate diversity within populations. Thus, crossovers have a dual role in meiosis – aiding immediate chromosome segregation and generating novel variants for long-term evolution.
Centromeres are specialized regions on the chromosomes that facilitate binding of a large protein complex (the kinetochore), which in turn attaches the chromosomes to microtubules arranged in an elongated polar structure (the spindle) [1]. Proper attachment of microtubules to centromeres is essential for faithful chromosome segregation. Centromeres can be defined as the regions that bind the centromere-specific histone H3 variant, CENP-A, which promotes kinetochore assembly. The regions in the immediate vicinity of the centromeres, the pericentric regions, in almost all species exist as heterochromatin containing H3 K9 methylated histones (H3 K9me). The pericentric region does not directly bind the kinetochore, but it nevertheless plays a crucial role in proper chromosomal segregation. Due to their indispensable function in segregation, the centromere and pericentric region are repressed for recombination because a crossover in either of these regions can interfere with proper segregation. Missegregation produces a chromosomal imbalance (aneuploidy) which can give rise to genetic disorders such as Down syndrome in humans [2]. In this review we describe how crossovers are formed and how their distribution is properly regulated to aid successful meiosis. In particular, we discuss how crossovers are avoided in and near the centromeres (the pericentric region).
1. Crossover formation and chromosome segregation in meiosis
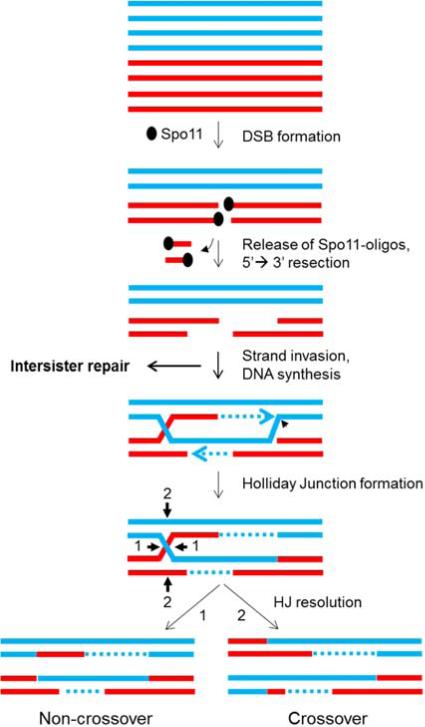
Meiotic recombination is initiated by the induction of programmed DNA double-strand breaks (DSBs) by the meiosis-specific, highly conserved protein Spo11 and several partner proteins that, like Spo11, are essential for DSB formation and recombination [3] (Fig. 1). DSBs as initiators of meiotic recombination have been directly observed by DNA analysis, such as Southern blot hybridization, in both budding and fission yeasts. DSBs have been inferred in other organisms due to the requirement of Spo11 orthologs for recombination or for the formation of foci of the Rad51 and Dmc1 DNA strand-exchange proteins or the γH2AX modified histone; these foci represent sites of DSBs. After introduction of DSBs in a topoisomerase-like manner, Spo11 remains covalently attached to the DSB ends and is later cleaved off by an endonuclease, generating a short oligonucleotide attached to Spo11 [4]. The DSB ends are processed to generate an extended 3’ single-stranded DNA overhang bound by Rad51 strand-exchange protein and (in many species) its meiosis-specific paralog Dmc1. This single-stranded DNA-protein complex can invade an intact duplex (either the homolog or the sister chromatid) to produce a DNA joint molecule. The hybrid DNA intermediate formed with the homolog can be further processed and resolved into either a crossover, where the flanking DNA around the DSB site reciprocally recombines, or a non-crossover, where there is no exchange of the flanking DNA (Fig. 1) [5,6]. In addition, repair can occur such that one but not the other DNA is locally changed to the genotype of the other, resulting in a non-reciprocal exchange called gene conversion; a gene conversion can be part of either a crossover or a non-crossover.
Figure 1. Formation of crossovers and non-crossovers at Spo11-induced DSBs.
Red and blue lines are single DNA strands, and each pair depicts one sister chromatid of each homologous chromosome (the non-interacting sisters are shown only in the top panel); dotted lines are DNA synthesized during DSB repair. After meiotic replication, Spo11 or its ortholog induces DSBs and covalently attaches to the 5’ ends. Spol11 is then cleaved from the ends by an endonuclease to release a short oligonucleotide attached to Spo11. The 5’ ends are further resected to generate long 3’ overhangs, which invade intact duplex DNA, either the homolog or the sister chromatid; this side reaction with the sister produces no genetic recombinants. Holliday junctions, either single (shown) or double (not shown), are formed and then resolved into either crossovers or non-crossovers (respectively with the non-parental or parental DNA configuration flanking the region of DNA exchange). These reactions have been demonstrated by direct DNA analysis in budding and fission yeasts and are inferred, from genetic and cytological data and limited DNA analysis, to occur in other species. In an alternative to Holliday junction formation, strand invasion at the third step can prime limited DNA synthesis; the unwound product can anneal with the other initially broken DNA end and produce a non-crossover. This proposed reaction is called synthesis-dependent strand-annealing.
DSBs are not induced uniformly across the genome in the species tested but tend to be concentrated at specific regions known as hotspots. Similarly, recombination events may also cluster to give rise to recombination hotspots, while some parts of the genome are “cold.” In the species tested, the number of DSBs in meiotic cells exceeds the number of crossovers generated, implying that many DSBs get resolved into non-crossovers or are repaired with the sister chromatid, which cannot generate a genetic recombinant.
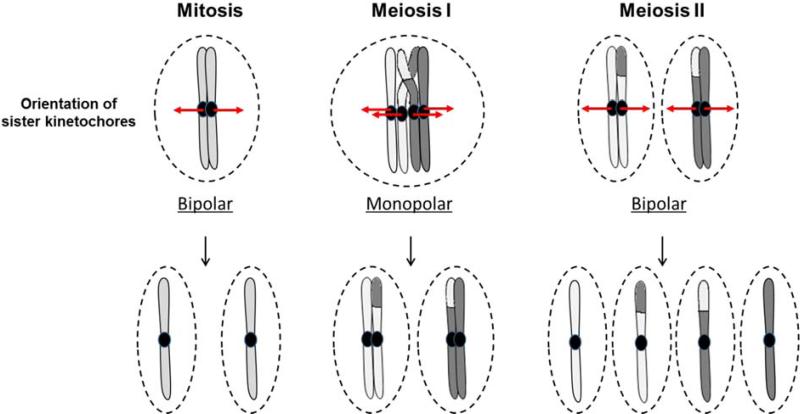
Proper chromosomal segregation involves equal distribution of chromosomes into daughter cells during nuclear division. In mitosis, the kinetochores of sister chromatids, generated by replication, attach to microtubules originating from opposite spindle poles (bipolar orientation) (Fig. 2) [7]. The sister chromatids are joined together by cohesion imparted by a complex of proteins (cohesins). The chromatids are then pulled toward the opposites poles; the proper direction of segregation is facilitated by the tension due to cohesion. Cohesin is cleaved as the sisters are separated into two diploid nuclei. In meiosis, by contrast, during the first meiotic division (MI), the kinetochores of sister chromatids attach to microtubules originating from the same pole (monopolar orientation) (Fig. 2), and homologous centromeres attach to microtubules from opposite poles [7]. Crossovers generated during recombination stabilize the oppositely-oriented pair of homologs (bivalents) and along with sister chromatid cohesion generate tension to allow proper pulling of the homologous centromeres and their attached chromosomal arms to opposite poles. At the onset of segregation in MI, cohesin in the arms is removed while that at the centromere is maintained. The centromeric cohesin facilitates bipolar orientation of sister centromeres during the second meiotic division (MII) (Fig.2) [7]. The end of meiosis generates four haploid nuclei. Hence, crossovers have a beneficial role in both generation of diversity and chromosome segregation, and therefore most species need to ensure at least one crossover per bivalent during meiosis.
Figure 2. Orientation of kinetochores during mitosis and meiosis.
In mitosis, kinetochores attach to the centromeres of the sister chromatids, orient towards opposite poles (bipolar orientation), and segregate into separate daughter nuclei. During the first meiotic division, however, both sister kinetochores orient towards the same pole (monopolar orientation) and segregate into the same daughter nucleus. Proper direction of segregation is ensured by sister chromatid cohesion and the tension generated due to crossovers (light grey-dark grey junction) between homologs. This reductional division results in separation of the homologous centromeres and the attached chromosomal arms and reduces the number of chromosomes by half. The second meiotic division is similar to mitosis, and bipolar orientation of the sister kinetochores results in separation of the chromatids into four haploid nuclei.
2. Harmful roles of crossovers in meiotic segregation
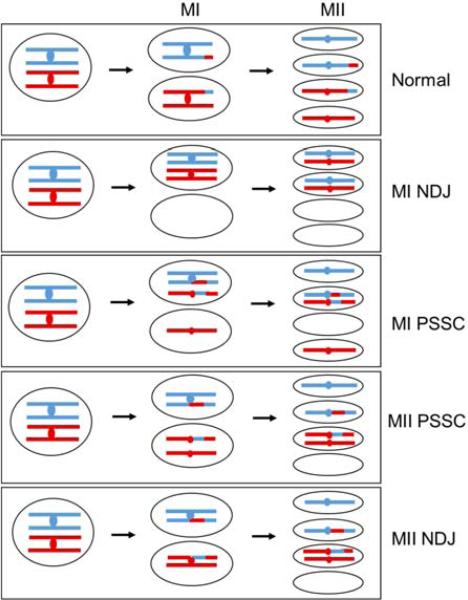
Despite the important function of crossovers in meiosis, crossovers in certain genomic regions, such as near the centromere, can be as harmful as the lack of crossovers and result in missegregation. There are examples of proper segregation of chromosomes lacking crossovers (achiasmate segregation). However, in the absence of such backup systems, failure to form crossovers results in random segregation such that the homologous centromeres can migrate towards either the same or opposite poles. In MI, migration of homologs to the same pole (non-disjunction) followed by equational segregation at MII results in two MI disomes (nuclei with two copies of a chromosome, in this case having different sister centromeres) and two nullisomes (nuclei lacking a chromosome) (Fig. 3). These are rare in wild type but are more frequent in mutants with few or no crossovers. For example, trisomy for chromosome 21 in humans, associated with Down syndrome, often results from reduced levels of recombination during meiosis as analyzed in several conceptuses [8,9]. This is also true for human trisomies of chromosomes 15, 16 and 18, where the genetic maps (determined by recombination frequencies between genes) are significantly shorter, reflecting fewer crossovers, than controls [10].
Figure 3. Meiotic chromosomal missegregation resulting from pericentric crossovers.
Red and blue lines depict homologs (duplex DNA); red and blue dots depict centromeres. Crossovers, depicted by red-blue junctions, in chromosomal arms are essential for proper segregation of chromosomes during meiosis, but crossovers too near the centromere are harmful. Normally during MI, the centromeres of homologs with their attached chromosomal arms segregate to opposite poles, whereas in MII the sister centromeres segregate to opposite poles, giving rise to four haploid nuclei. During non-disjunction (NDJ) of homologs in MI, which primarily arises due to lack of crossovers in the arms, both homologs migrate to the same pole and then segregate properly at MII, giving rise to two nullisomes (nuclei lacking a chromosome) and two disomes (nuclei with two chromosome copies). Precocious separation of sister chromatids (PSSC) occurs when cohesion is lost between the sister chromatids and can occur in either MI or MII. Crossovers too close to the centromeres (pericentric crossovers) are associated with such PSSC events. In MI PSSC, sister centromeres of one homolog segregate to opposite poles at MI followed by proper MII, giving rise to a nullisome and a disome containing homologous centromeres. In MII PSSC, sisters stay in the same nucleus at MI but missegregate at MII, giving rise to a nullisome and a disome containing sister centromeres. MII NDJ has proper MI but aberrant segregation at MII, resulting in a fate similar to MII PSSC. These aberrant events are also linked to the presence of pericentric crossovers. Some of these missegregation events give rise to similar types of aneuploids, but careful tetrad analysis with multiple markers can distinguish them.
Aberrant crossover positioning can also lead to missegregation. Crossovers too close to the centromere affect proper segregation and result in aneuploid inviable gametes. Centromere-proximal crossovers disrupt cohesion in the pericentric region or interfere with proper kinetochore orientation to the spindle poles during segregation. A significant number of maternally derived sex chromosome trisomies in humans possess meiotic exchanges at or close to the centromere [11,12]. Recombination proximal to a centromere is associated with as much as 60% of MI segregation errors in the budding yeast Saccharomyces cerevisiae [13]. In addition, crossovers near a centromere in S. cerevisiae are strongly correlated with precocious separation of sister chromatids (PSSC), whose effects can be seen in both MI and MII (Fig. 3) [14]. It has been proposed that PSSC is involved in the majority of human age-related trisomies and is a major source of missegregation events in humans [8,15,16]. Non-disjunction at MII, which can lead to formation of MII disomes (in this case having identical sister centromeres), are also associated with crossovers near the centromere (Fig. 3). This type represents ~22% of all missegregation events in humans and ~6% in Drosophila [8,15,17]. Because of the deleterious effects of pericentric crossovers, organisms have evolved mechanisms to stringently avoid them, as discussed below.
3. Diverse properties of centromeres
3.1 Repetitive DNA structure
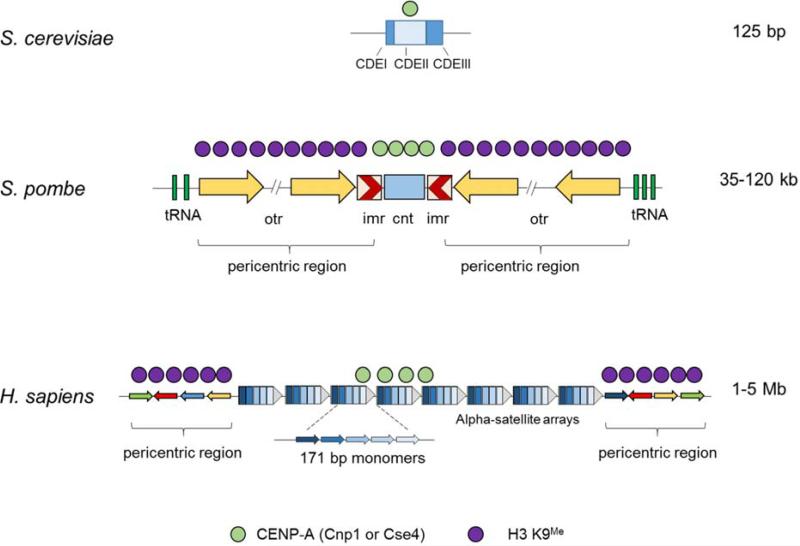
As noted above, centromeres are the sites of kinetochore attachment and play a key role in establishing orientation during chromosomal segregation. Although the functions of centromeres in kinetochore binding, microtubule attachment, and segregation are conserved across species, centromeres and the flanking pericentric regions are structurally diverse. S. cerevisiae has a simple (“point”) centromere, which is approximately 125 bp in length and is subdivided into three conserved DNA elements – CDEI, II and III (Fig. 4) [18,19]. It binds a single nucleosome containing CENP-A (Cse4 in budding yeast) [20]. A single base-pair mutation in CDEIII can abolish centromere function, underscoring the simplicity of budding yeast centromere structure. S. cerevisiae and other budding yeasts do not contain heterochromatin around their centromeres, unlike other organisms.
Figure 4. Structure of centromeres in yeasts and humans.
The budding yeast S. cerevisiae has 125 bp (“point”) centromeres with three conserved regions – CDEI, II and III. A single nucleosome containing the centromere-specific histone H3 variant Cse4 (CENP-A in most other organisms) occupies the whole centromere. The fission yeast S. pombe contains large 35 – 120 kb (“regional”) centromeres consisting of a central core (cnt) surrounded by innermost repeats (imr) and outermost repeats (otr). The central core contains nucleosomes with CENP-A (Cnp1 in S. pombe), while the inverted repeats bear H3 K9me histones that form the pericentric heterochromatin. Human centromeres have large 1 – 5 Mb arrays of repetitive DNA consisting of 171 bp alpha-satellite repeats as the basic repeating unit. An array consists of multiple copies of higher order repeats which themselves are made up of multiple alpha-satellites that are diverged from each other. These arrays contain CENP-A interspersed with H3 K4me histones. The pericentric region contains stretches of monomeric alpha-satellite repeats that are in random orientation and are bound by H3 K9me histones that form the heterochromatin. The complete organization of human centromeres is still being worked out, and the schematic shown here is a model based on current understanding.
In contrast, centromeres and the pericentric region in the distantly related fission yeast Schizosaccharomyces pombe are much larger and more complex (Fig. 4). They span 35 – 120 kb and consist of a unique 4 – 5 kb central core region (cnt) flanked by repetitive DNA elements, the ~5 kb innermost repeat (imr), and multiple 4 – 7 kb outermost repeats (otr) [21]. The cnt binds CENP-A (Cnp1 in S. pombe) and facilitates kinetochore assembly. A portion of the imr and the multiple otr repeats together constitute the pericentric region (Fig. 4). These repeat elements are organized into heterochromatin containing H3 K9me, which is essential for proper centromere function as discussed below.
Centromeres in plants and animals are even more complex and extend up to megabases in length. In humans they consist almost entirely of repetitive sequences, the most common of which is the alpha-satellite DNA [22]. It has a 171 bp consensus sequence that is arranged in complex higher order repeat patterns to form large arrays (Fig. 4). Some of the alpha-satellites are bound by CENP-A along with canonical H3 histones, whereas the pericentric monomeric repeats harbor heterochromatin with H3 K9me and the heterochromatin protein, HP1 [23]. Collectively, these proteins establish the centromere-specific chromatin structure on which other centromere-specific proteins and the microtubule-binding complex assemble to form the functional kinetochore. (Some plants and animals have holocentric chromosomes, to which the spindle microtubules can attach at many points along the entire chromosome rather than to a discrete region, but these holocentrics are not discussed here.)
3.2. Pericentric heterochromatin
The pericentric region contains heterochromatin (in all species examined except budding yeasts, such as S. cerevisiae) and is essential for proper centromeric functions and sister chromatid cohesion. Heterochromatin formation is well-studied in S. pombe and largely depends on the RNA interference (RNAi) machinery [24], although an RNAi-independent pathway involving the histone deacetylase Sir2 has also been described [25]. Native transcripts from the otr repeats are processed by the RNA-dependent RNA polymerase and the Dicer ribonuclease complex into small interfering RNA (siRNA) duplexes that are guided back to the pericentric region via the Argonaute-containing RNA-induced silencing complex. This complex helps in recruiting Clr4, the only histone H3 K9 methyl transferase in S. pombe [26]. H3 K9me is bound by proteins such as Chp1 and the heterochromatin protein (HP1) homologs Swi6 and Chp2 via a protein domain (the chromodomain) that has a high affinity for H3 K9me [27]. Once bound, Swi6 helps to bring in more Clr4, which also contains a chromodomain, and by a positive feed-back loop aids in further establishment and spread of heterochromatin to neighboring regions. Apart from contributing to heterochromatin maintenance, Swi6 also helps recruit to the pericentric region cohesin subunits, such as Psc3 and Rec8, and other meiosis-specific proteins, such as Shugoshin, that preserve pericentric sister chromatid cohesin at meiosis I [28]. Swi6 also recruits Epe1, a protein that antagonizes heterochromatin formation and facilitates transcription of repeats within the pericentric region [29]. Chp2 brings in chromatin modulators, such as the histone deacetylase Clr3, that help maintain the transcriptionally repressed state in the pericentric region. A delicate balance between Epe1 and Clr3 activities ensures proper heterochromatinization at the pericentric regions [30].
In multicellular eukaryotes, heterochromatin is organized in a similar manner, although many steps in the pathway are poorly understood. SUV39H is the major H3 K9 methyl transferase at centromeres that helps in recruiting HP1, whose function is similar to that of its counterpart Swi6 in S. pombe. Histone deactelyation and DNA methylation also play important functions in heterochromatin maintenance in mammals and plants [31-33]. Many of these features bear on the repression of pericentric crossing-over, as discussed below.
4. Pericentric repression of meiotic crossing-over
Around 80 years ago, the centromeres or spindle attachment regions on the chromosomes in Drosophila melanogaster were observed to negatively affect the rates of crossing-over immediately around them [34-36]. This was termed the centromere effect. Apart from reduction of crossovers in the pericentric heterochromatin, the repression extended into the neighboring euchromatin as well. In Drosophila, placement of a segment from chromosome III near the centromere of chromosome IV led to a reduced crossover frequency in the translocated segment. The repressing effect steadily weakened as the distance increased between the region assayed and the centromere [36].
Since these initial discoveries in Drosophila, the phenomenon of pericentric repression of meiotic recombination has been shown in several other species. Early genetic analysis in S. cerevisiae showed a reduced density (frequency per kb of DNA), relative to that in chromosomal arms, of both crossovers (up to 5-fold lower) and non-crossovers (4 – 7-fold lower) near CEN3 [37,38]. More recent genome-wide microarray analyses with high resolution confirm this repression. The nearest crossover event on many chromosomes is at least 2 – 3 kb away from the centromere; on others it ranges from ~5 to 18 kb [39]. The nearest non-crossover event occurs ~2.5 kb from the centromeres. Another analysis exhibited an average 6-fold reduction of both crossover and non-crossover densities within 10 kb on each side of the centromeres [40].
The first evidence for pericentric repression in S. pombe came from studies that mapped the centromere on chromosome 2. A 50 kb centromeric region on chromosome 2 has a lower density of meiotic recombination than the genome average [41]. A more recent study showed that the crossover density across the largest centromere (on chromosome 3), using closely flanking markers, is ~200 fold lower than the genome average of 0.16 cM/kb (<0.1% across the ~120 kb cen3) [42]. There is evidence for pericentric repression in other fungi as well. In Neurospora crassa, crossover density decreases dramatically approaching the centromere [43]. Similarly, in Aspergillus nidulans, genetic mapping showed ~6-fold reduced recombination density in the centromere-proximal regions of chromosome IV [44].
Pericentric repression is also present in plants and animals. Calculation of recombination densities across large, highly repetitive centromeres, such as those in multicellular eukaryotes, can be problematic due to the difficulty of scoring many meiotic progeny, the absence of markers closely flanking the centromere, and the inability to accurately determine physical distances. Nevertheless, genome-wide and cytological data for the distribution of recombination events in plants demonstrate reduced meiotic exchange near centromeres in a number of species such as Arabidopsis thaliana [45,46], tomato [47], rice (Oryza sativa) [48,49], wheat [50] and maize [51]. This repressive effect is not restricted to the immediate vicinity of the centromere but may extend for megabases in plants. In A. thaliana, recombination in the ~2.5 Mb centromeric core regions is ~50-fold less dense (cM/Mb) than in the chromosomal arms [32,33,45]. Among animals, pericentric repression of recombination has been shown in Drosophila (see above), humans [52] and chickens [53]. Early studies on human chromosomes indicated reduced crossover-density near centromeres [54,55]. Integrated physical and genetic maps generated for human centromere 10 showed 9 – 11-fold reduced recombination [56] and ~8-fold reduced meiotic exchange near the human X centromere compared to the rest of the chromosome [52].
More recent studies have focused on understanding the factors causing pericentric repression of meiotic recombination. Possible explanations are centromeric sequence repetitiveness, function in kinetochore assembly, or surrounding highly condensed and inaccessible heterochromatin. CEN8 of rice spans a 750 kb CENP-A rich region. Unlike other rice centromeres, it does not contain DNA repeats but appears otherwise similar to the other DNA repeat-rich centromeres [57]. Despite this difference, it contains a ~2.3 Mb region that lacks detectable crossovers [48], indicating that, at least in this case, repetitive DNA is not necessary for pericentric repression. S. cerevisiae does not contain pericentric heterochromatin, yet centromere-proximal recombination densities are low, as noted above. Thus, at least in this case, heterochromatin is not necessary for pericentric repression. Furthermore, a single base-pair mutation in S. cerevisiae CDEIII that abolishes centromeric function stimulates proximal crossing-over and gene conversion by approximately 2.5-fold relative to wild type [38]. This suggests that centromeric function such as kinetochore assembly may be a factor in repression of recombination, at least in S. cerevisiae. Indeed, kinetochore components, such as the Ctf19 complex, reduce DSB formation within a ~6 kb region surrounding the centromeres [58]. The Ctf19 complex, by promoting enrichment of cohesin at the pericentric regions, may also influence partner choice (sister vs. homolog) for DSB repair. Current understanding of the mechanism of pericentric repression will be discussed later.
5. Gene conversion at centromeres
Although centromeres are often inert for crossovers, their evolution and higher order structure suggest that gene conversion events do occur in centromeres. In order to maintain the human centromeric structure with nearly identical tandem repeat arrays, it has been proposed that centromeres undergo recombination via random unequal exchange and accompanying gene conversion. This repeated recombination can remove variation arising from mutation in individual monomers, resulting in homogenization of the repeats, as well as promote their expansion or contraction. Evidence of such repeated exchanges leading to rearrangements and expansions within core centromeric structures of human alpha-satellites, rice and mouse centromeres has been observed in somatic tissues [59-63]. However, there is no direct evidence that such exchanges occur in the germ line and are stably passed on to the next generation.
Conversion events in the centromere might be too rare to be observed in the usual laboratory experiments but nevertheless be frequent enough to affect populations. Indeed, a population-based genetic analysis of 93 recombinant inbred lines of maize, using a retrotransposon polymorphism at centromeres, revealed two independent cases of gain of markers from one centromeric haplotype to the other [64]. Linkage disequilibrium and analysis of flanking genotypic markers indicated gene conversion rather than crossing-over. The conversion rates were estimated to be ~1 × 10−5 per marker per generation, similar to other regions on the maize chromosome; such low-frequency conversions would be undetectable in standard meiotic crosses. Nevertheless, these results suggest that pericentric recombination occurs at a low evolutionary rate to homogenize repeats but not frequently enough to cause significant missegregation during meiosis.
6. A role for heterochromatin in meiotic recombination repression near centromeres
Heterochromatic regions are transcriptionally inactive for artificially inserted protein coding genes, and therefore heterochromatin may repress recombination as well. As discussed above, the RNAi pathway plays a major role in establishing heterochromatin formation in S. pombe. Recombination across cen3, the largest centromere in S. pombe, is <0.1% in wild type [42]. RNAi and heterochromatin mutants, such as dcr1, ago1, clr4, and chp1, manifest increased crossover density across cen3 ~30 – 90-fold, to nearly that of the genome mean. As expected, the mutants have increased pericentric DSBs, as analyzed by Southern blot hybridization [42]. Surprisingly, recombination is not significantly increased in swi6, chp2 and clr3 mutants, despite the strong effects of these mutations on alleviating repression of gene expression. These results suggest that heterochromatin represses gene expression and recombination by distinct but overlapping mechanisms.
Like S. pombe clr4, the Drosophila Su(var)3-9 gene encodes a histone H3 K9 methyl transferase. Mutations in Su(var)3-9 increase the frequency of DSBs, measured as RAD51 foci, up to 2-fold in heterochromatin of germ cells (the oocytes and the surrounding nurse cells) relative to wild type [65]. The number of foci returns to the wild-type level when the Spo11-homolog MEI-W68 is removed, suggesting that at least half of the DSBs in the Su(var)3-9 mutant are generated during meiotic recombination. A less severe effect is observed in a dcr-2 mutant, disabled for the RNAi pathway that regulates heterochromatin formation in Drosophila. Mutations in Su(var)3-9 and other Su(var) genes (9 out of the 16 studied) that are involved in heterochromatin establishment and propagation also enhance up to 6-fold crossing-over between markers flanking the pericentric-centromeric region on chromosome 3 [66]. Conversely, additional genome copies of Su(var)3-7 and Su(var)2-5 significantly reduce crossing-over in this interval. Therefore, heterochromatin, in particular H3 K9me, seems to repress both meiotic DSB formation and recombination in both Drosophila and S. pombe pericentric regions.
In Tetrahymena as well, heterochromatin appears to regulate DSB formation during meiosis. Tetrahymena cells have two nuclei – a large polyploid macronucleus and a small diploid micronucleus. Meiosis takes place inside the micronucleus, which has highly condensed, transcriptionally silent chromatin for most of its vegetative cell cycle [67]. In the early stages of meiosis, the centromeres and highly-repetitive pericentric regions move to one pole of the micronucleus, while the telomeric and gene-containing regions extend to the opposite pole. Tetrahymena lacks H3 K9me and H3 K4me, but its Ezl3 protein forms H3 K23me, which is characteristic of heterochromatin in meiotic prophase in this ciliate [68]. γH2AX foci generally accumulate in the genic regions during early meiosis and distribute throughout the micronucleus by late prophase. In EZL3Δ cells, γH2AX foci initially accumulate in the centromeric pole, persist throughout the meiotic lag, and finally distribute throughout the micronucleus [68]. This apparent mislocalization of DSBs is not seen in EZL2Δ cells, which lack H3 K27me, indicating specificity of H3 K23me formed by Ezl3 in pericentric repression. H3 K23me-enriched heterochromatin may block DSB formation by Spo11 during meiosis. Hence, histone modifications characteristic of heterochromatin other than H3 K9me also appear to impede meiotic DSB formation.
Apart from histone modifications, DNA methylation can also influence recombination rates during meiosis. A. thaliana met1 mutants show strong defects in DNA cytosine methylation and ~3-fold increase, relative to wild type, in crossover density in the ~2.5 Mb core of the centromere (averaged over all centromeres) [32,33]. The effects of these alterations on chromosome segregation remain to be elucidated.
To summarize, in diverse species heterochromatin appears to play a key but not universal role in repressing meiotic DSBs and recombination in pericentric regions.
7. Non-uniform genome-wide DSB and crossover distributions in meiosis
The distributions across the genome of both DSBs and crossovers are tightly regulated during meiosis. The regulatory mechanisms acting in the chromosomal arms may, with variations, also act in the centromeres to prevent harmful crossovers. Understanding how DSBs and crossovers are distributed throughout the genome may suggest potential mechanisms of pericentric repression, which is largely unknown so far.
As noted in section 1, Spo11 or its ortholog along with multiple accessory proteins is needed in most, if not all, organisms to initiate meiotic recombination (i.e., the formation of DSBs) [3]. The distribution of DSBs throughout the genome varies among species. In S. cerevisiae, DSBs are generally found in nucleosome-depleted regions, and histone H3 K4 methylation influences both DSB distribution and frequency [69,70]. In S. pombe, DSBs occur preferentially in large intergenic regions not necessarily enriched for nucleosome-depleted regions, and transcription factor-binding influences a small subset of hotspots [71,72]. In mice and humans, the chromosomal distribution of bound PRDM9 methyltransferase, which can methylate histone H3 K4, is a strong predictor of DSB formation, but PRDM9 is not essential for formation of DSBs, which are abundant but distributed differently in PRDM9 mutants than in wild type [73-76].
There is ample evidence that crossover distribution across the genome is also non-uniform but not absolutely correlated with DSB distribution. In mice and humans, there is a partial correlation between DSB maps and those generated for recombination [77,78]. Moreover, there are more DSBs than actual crossovers per meiotic cell in several species – the DSB:crossover ratio ranges from ~2:1 in budding and fission yeast to ~10:1 in mice and ~15:1 in A. thaliana. [71,79]. As noted in section 1, DSBs can be repaired into non-crossovers and thereby increase the DSB:crossover ratio and potentially influence crossover distribution. The choice of repairing a DSB with the sister chromatid as opposed to the homolog adds another level of regulation. One might expect interhomolog (IH) repair to be preferred over intersister (IS) repair to increase genetic diversity and to aid chromosome segregation. This is true in S. cerevisiae, with an IS:IH ratio of ~1:5 at the few DSB hotspots tested [80-82], but is reversed in S. pombe, with an IS:IH ratio of ~3 or 4:1 at the two hotspots tested [83,84]. In S. pombe, DSBs in non-hotspot (“cold”) chromosomal arm regions are inferred from genetic evidence to be repaired primarily with the homolog, resulting in a nearly uniform crossover distribution in spite of DSB hotspots (crossover invariance) [83,85]. In addition, in S. cerevisiae there is spatiotemporal regulation of DSB formation that can influence the IS:IH ratio and crossover distribution based on chromosomal position, presence of factors such as the synaptonemal complex-modifying SUMO E3 ligase Zip3, or time of DSB formation [86,87]. Crossover interference, which reduces the occurrence of one crossover near another, also adds to the complexity. Hence, although DSBs are essential for recombination, there are multiple levels of regulation that determine crossover position and frequency. All these factors can have an effect on pericentric crossover repression.
8. Molecular mechanism of pericentric repression in meiosis
Although the paucity of crossovers at and near centromeres was discovered over 80 years ago, the molecular basis for pericentric repression is not yet explained in any organism. Heterochromatin, present at pericentric regions in most species, is often considered “inaccessible,” for example to the transcription machinery, since genes inserted into heterochromatin are often not expressed [27]. But heterochromatic regions are acted on by the replication machinery every cell cycle, just like euchromatic regions, and are evidently accessible to proteins that form heterochromatin, such as histone-modifying enzymes and the RNAi machinery. In S. pombe, the Rec8 cohesin subunit is protected by Shugoshin in MI and phosphorylated by casein kinase homologs in MII specifically in pericentric regions, which are heterochromatic [88,89]. Thus, “inaccessibility” of recombination proteins due to heterochromatin is not a satisfactory explanation at the molecular level for the lack of DSBs and recombination. The limiting molecular step, not yet identified, could be either the absence of a positive effector (activator) or the presence of a negative effector (repressor) of recombination. For example, in wild-type cells heterochromatin could exclude an activator from the pericentric region or recruit a repressor to it. Distinguishing these possibilities is not simple, since a repressor could inactivate an activator that has a more immediate role in regulating the process than does the repressor. These possibilities must be kept in mind as one tries to determine the molecular mechanism of crossover repression in and near centromeres.
More generally, one can ask, is crossing-over limited by DSB formation or by the choice of repair mechanism? DSB formation could be limited by a lack of either Spo11 binding or Spo11 activation. If repair plays a role, is crossing-over limited by the choice of template (sister vs. homolog) or by resolution of recombination intermediates into crossovers vs. non-crossovers? Evidence for each of these mechanisms of repression is discussed next.
8.1. Blocking of meiotic DSB formation at centromeres
Formation of DSBs is a major determinant of where recombination events can occur. When centromeres are devoid of crossovers, they may lack DSBs as well. In S. cerevisiae DSBs are 2 – 3-fold less dense (number per kb per meiosis) than the genome-wide average in 5 – 10 kb intervals surrounding the centromeres [90]. A more recent, nucleotide-resolution DSB map estimates a 7-fold reduction in 3 kb intervals surrounding the centromeres [91]. S. pombe centromeres and pericentric regions lack detectable meiotic DSBs, observed both by direct physical analysis (Southern blot hybridization) and genome-wide mapping of Rec12 (Spo11 ortholog)-oligonucleotides [42,71]. γH2AX foci are limited in meiotic heterochromatin regions in Drosophila as well as in the Tetrahymena micronucleus, as noted above [65,68]. These data imply that the mechanism of repression of recombination around centromeres can be controlled at an early stage, the formation of DSBs.
Absence of pericentric DSBs might result from failure of Spo11 (or its ortholog) either to bind or to be activated for DSB formation in the pericentric regions. There is evidence for both possibilities. ChIP analysis for Rec12-binding following protein-DNA crosslinking showed strong Rec12 binding to all three centromeres in S. pombe [92]. Catalytically inactive Rec12 (Y98F) mutant protein also binds to the centromeres [93]. In S. cerevisiae, Spo11 binds first to centromeres during premeiotic replication without immediate recruitment of other essential partner proteins such as Mre11, part of the MRN complex also containing Rad50 and Nbs1 (Xrs2 in S. cerevisiae) [94]. Artificial tethering of Spo11 to centromere-proximal regions (and other cold regions) via fusion to the GAL4 DNA-binding domain in S. cerevisiae does not result in DSBs although in other regions it does [95,96]. These data from two diverse yeasts suggest that activation of Spo11 or Rec12, not their binding, can be limiting for DSB formation. In A. thaliana, SPO11 foci appear earlier than the actual γH2AX foci, indicative of a significant delay between SPO11 loading and break formation, which is seen in S. cerevisiae as well [97,98]. Thus, the mere presence of Spo11, perhaps without its partner proteins, is not sufficient to induce DSBs. Repression of pericentric meiotic recombination may be due to lack of Spo11-activating proteins rather than by lack of the DSB-inducing protein itself.
8.2. Choice of DSB repair mechanism
Crossover formation could also be limited by the mechanism of DSB repair. For example, DSB repair with the sister chromatid does not reassort parental alleles or produce crossovers joining homologs. In addition, resolution of repair intermediates could be limited to non-crossover outcomes. A few studies show pericentric DSBs, suggesting repair as the limiting step for crossing-over. In S. cerevisiae, centromere-proximal DSB hotspots are observed within 5 kb of the centromere [91,99], although their strength and density are lower than the genome average [91]. DSB hotspots near CEN2, CEN4, and CEN15 were confirmed by Southern blot hybridization of DNA from dmc1 mutants, which accumulate broken DNA [99]. Therefore, in pericentric regions with infrequent recombination, these DSBs may undergo intersister repair or be resolved as non-crossovers.
There are conflicting reports on the distribution of non-crossovers specifically around the centromeres. In S. cerevisiae, absence of Zip1, a synaptonemal complex component, alleviates pericentric repression during meiosis. Genome-wide analysis showed a significant increase in crossover density within 10 kb of the centromeres, up to the genome-wide level in zip1 mutants [40]. A comparable increase in non-crossover levels suggests no alteration in the crossover/non-crossover ratio. Tetrad analyses for gene conversion and crossover events, using standard genetic markers both proximal and distal to CEN3, confirmed the genome-wide analysis [40]. Since neither an increase in DSBs nor a change in crossover/non-crossover ratio is observed, Zip1 may increase intersister over interhomolog repair during meiosis. In contrast, genome-wide high resolution maps for crossovers and non-crossovers in wild-type Drosophila show, for centromere-proximal regions, non-crossovers at the genome mean but crossovers at a considerably lower level [100]. This outcome indicates non-crossover products as a legitimate fate for DSBs formed in such crossover-cold regions. Therefore, regulation at the level of either DSB formation or repair, or both, could explain pericentric repression of meiotic recombination in different organisms.
9. Diversity among species
Although the basic mechanism of meiotic recombination (DSB formation by Spo11 or its ortholog followed by DSB repair) appears conserved, its regulation is highly diverse. For example, the MRN complex, which is required for DSB repair, is essential for DSB formation in S. cerevisiae, butnot in S. pombe or A. thaliana [101-103]. In S. cerevisiae the MRN complex is recruited last to the Spo11 binding site, perhaps to ensure its presence for efficient repair [104]. The cohesin subunit Rec8 is needed for DSB formation at most hotspots and for most recombination in S. pombe [93,105]. In S. cerevisiae rec8Δ mutants, however, DSBs occur at the wild-type (or higher) level at the artificial HIS4:LEU2 hotspot as assayed by Southern blot hybridization [106]. A more comprehensive genome-wide analysis in rec8Δ shows a severe reduction of Spo11 binding and DSB formation at several intervals on different chromosomes including those near the centromeres [94,107]. Such species- and region-specific requirements for DSB formation indicate multiple levels of regulation of DSB formation. Therefore, it is quite tempting to speculate that there are several mechanisms to explain pericentric repression of meiotic recombination in different species. This view is supported by the wide range in the levels of crossover repression seen among various species.
10. Conclusions
Crossovers are important for proper segregation of meiotic chromosomes but are harmful when they occur too close to the centromeres. Crossovers at the centromere or in the pericentric region have a high propensity to cause missegregation, which can give rise to aneuploidy and cause genetic disorders such as Down syndrome. The phenomenon of pericentric repression of meiotic recombination was discovered more than 80 years ago and is now well established in several species. Yet the mechanistic basis for this repression is still unknown – the rate-limiting step at the molecular level has not been identified in any species. The role of heterochromatin in causing repression has been a major focus in many studies. However, not all mutants affecting heterochromatin function with respect to transcriptional silencing are similarly derepressed for recombination. Hence, there could be a different threshold or mechanism for heterochromatic repression of transcription and recombination. DSBs are not seen or indirectly inferred in the pericentric regions in the majority of organisms tested. Regulation of DSB formation by controlling activation of Spo11 or its homolog may be the rate-limiting step to prevent DSB formation, given that Spo11 and its homologs can bind to the pericentric regions in some species. But control at the level of DNA repair, by choice of partner or resolution mechanism for DSB repair, has also been inferred in a few cases. In the future, it will be essential to take into account the interplay between proteins recruited specifically to the pericentric heterochromatin and those of the recombination machinery in mediating repression of crossovers that would otherwise be deleterious for the organism.
Acknowledgments
We are grateful to Sue Amundsen, Bernard de Massy, Ines Anna Drinnenberg, Greg Copenhaver, Raphael Mercier, Sarah Zanders, and an anonymous reviewer for helpful comments on the manuscript. Our research is supported by grants R01 GM031693 and R01 GM032194 from the National Institutes of Health of the United States of America.
Abbreviations
- DSB
DNA double-strand break
- MI
first meiotic division
- MII
second meiotic division
- PSSC
precocious separation of sister chromatids
- H3 K9me
histone H3 methylated on lysine 9
- siRNA
small interfering RNA
- IH
interhomolog
- IS
intersister
- MRN
Mre11-Rad50-Nbs1
- NDJ
non-disjunction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Przewloka MR, Glover DM. The kinetochore and the centromere: a working long distance relationship. Annu Rev Genet. 2009;43:439–465. doi: 10.1146/annurev-genet-102108-134310. [DOI] [PubMed] [Google Scholar]
- 2.Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age old problem. Nat Rev Genet. 2012;13:493–504. doi: 10.1038/nrg3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam I, Keeney S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol. 2015;7:a016634. doi: 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. In: Egel R, Lankenau D-H, editors. Recombination and meiosis: crossing-over and disjunction. Springer-Verlag; Berlin: 2007. pp. 81–123. [Google Scholar]
- 5.Smith KN, Nicolas A. Recombination at work for meiosis. Current Opinion in Genetics and Development. 1998;8:200–211. doi: 10.1016/s0959-437x(98)80142-1. [DOI] [PubMed] [Google Scholar]
- 6.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hauf S, Watanabe Y. Kinetochore orientation in mitosis and meiosis. Cell. 2004;119:317–327. doi: 10.1016/j.cell.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Lamb NE, Freeman SB, Savage-Austin A, Pettay D, Taft L, et al. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat Genet. 1996;14:400–405. doi: 10.1038/ng1296-400. [DOI] [PubMed] [Google Scholar]
- 9.Savage AR, Petersen MB, Pettay D, Taft L, Allran K, et al. Elucidating the mechanisms of paternal non-disjunction of chromosome 21 in humans. Hum Mol Genet. 1998;7:1221–1227. doi: 10.1093/hmg/7.8.1221. [DOI] [PubMed] [Google Scholar]
- 10.Lynn A, Ashley T, Hassold T. Variation in human meiotic recombination. Annual Rev Genomics Hum Genet. 2004;5:317–349. doi: 10.1146/annurev.genom.4.070802.110217. [DOI] [PubMed] [Google Scholar]
- 11.May KM, Jacobs PA, Lee M, Ratcliffe S, Robinson A, et al. The parental origin of the extra X chromosome in 47,XXX females. Am J Hum Genet. 1990;46:754–761. [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas NS, Ennis S, Sharp AJ, Durkie M, Hassold TJ, et al. Maternal sex chromosome non-disjunction: evidence for X chromosome-specific risk factors. Hum Mol Genet. 2001;10:243–250. doi: 10.1093/hmg/10.3.243. [DOI] [PubMed] [Google Scholar]
- 13.Sears DD, Hegemann JH, Shero JH, Hieter P. Cis-acting determinants affecting centromere function, sister-chromatid cohesion and reciprocal recombination during meiosis in Saccharomyces cerevisiae. Genetics. 1995;139:1159–1173. doi: 10.1093/genetics/139.3.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rockmill B, Voelkel-Meiman K, Roeder GS. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics. 2006;174:1745–1754. doi: 10.1534/genetics.106.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 16.Ottolini CS, Newnham LJ, Capalbo A, Natesan SA, Joshi HA, et al. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat Genet. 2015;47:727–735. doi: 10.1038/ng.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koehler KE, Boulton CL, Collins HE, French RL, Herman KC, et al. Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nat Genet. 1996;14:406–414. doi: 10.1038/ng1296-406. [DOI] [PubMed] [Google Scholar]
- 18.Hegemann JH, Fleig UN. The centromere of budding yeast. Bioessays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- 19.Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 20.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, et al. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willard HF. Evolution of alpha satellite. Curr Opin Genet Dev. 1991;1:509–514. doi: 10.1016/s0959-437x(05)80200-x. [DOI] [PubMed] [Google Scholar]
- 23.Schueler MG, Sullivan BA. Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet. 2006;7:301–313. doi: 10.1146/annurev.genom.7.080505.115613. [DOI] [PubMed] [Google Scholar]
- 24.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 25.Buscaino A, Lejeune E, Audergon P, Hamilton G, Pidoux A, et al. Distinct roles for Sir2 and RNAi in centromeric heterochromatin nucleation, spreading and maintenance. EMBO J. 2013;32:1250–1264. doi: 10.1038/emboj.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 27.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 28.Sakuno T, Watanabe Y. Studies of meiosis disclose distinct roles of cohesion in the core centromere and pericentromeric regions. Chromosome Res. 2009;17:239–249. doi: 10.1007/s10577-008-9013-y. [DOI] [PubMed] [Google Scholar]
- 29.Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- 32.Yelina NE, Lambing C, Hardcastle TJ, Zhao X, Santos B, et al. DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Genes Dev. 2015;29:2183–2202. doi: 10.1101/gad.270876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yelina NE, Choi K, Chelysheva L, Macaulay M, de Snoo B, et al. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet. 2012;8:e1002844. doi: 10.1371/journal.pgen.1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sax K. Chromosome structure and the mechanism of crossing over. Journal Arnold Arb. 1932;11:193–220. [Google Scholar]
- 35.Beadle GW. A Possible Influence of the Spindle Fibre on Crossing-Over in Drosophila. Proc Natl Acad Sci U S A. 1932;18:160–165. doi: 10.1073/pnas.18.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mather K. Crossing over and Heterochromatin in the X Chromosome of Drosophila Melanogaster. Genetics. 1939;24:413–435. doi: 10.1093/genetics/24.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambie EJ, Roeder GS. Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics. 1986;114:769–789. doi: 10.1093/genetics/114.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambie EJ, Roeder GS. A yeast centromere acts in cis to inhibit meiotic gene conversion of adjacent sequences. Cell. 1988;52:863–873. doi: 10.1016/0092-8674(88)90428-x. [DOI] [PubMed] [Google Scholar]
- 39.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen SY, Tsubouchi T, Rockmill B, Sandler JS, Richards DR, et al. Global analysis of the meiotic crossover landscape. Development Cell. 2008;15:401–415. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO Journal. 1986;5:1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, et al. RNAi and heterochromatin repress centromeric meiotic recombination. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8701–8705. doi: 10.1073/pnas.0914160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis CR, Kempainen RR, Srodes MS, McClung CR. Correlation of the physical and genetic maps of the centromeric region of the right arm of linkage group III of Neurospora crassa. Genetics. 1994;136:1297–1306. doi: 10.1093/genetics/136.4.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aleksenko A, Nielsen ML, Clutterbuck AJ. Genetic and physical mapping of two centromere-proximal regions of chromosome IV in Aspergillus nidulans. Fungal Genet Biol. 2001;32:45–54. doi: 10.1006/fgbi.2001.1251. [DOI] [PubMed] [Google Scholar]
- 45.Haupt W, Fischer TC, Winderl S, Fransz P, Torres-Ruiz RA. The centromere1 (CEN1) region of Arabidopsis thaliana: architecture and functional impact of chromatin. Plant J. 2001;27:285–296. doi: 10.1046/j.1365-313x.2001.01087.x. [DOI] [PubMed] [Google Scholar]
- 46.Copenhaver GP, Browne WE, Preuss D. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc Natl Acad Sci U S A. 1998;95:247–252. doi: 10.1073/pnas.95.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherman JD, Stack SM. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics. 1995;141:683–708. doi: 10.1093/genetics/141.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harushima Y, Yano M, Shomura A, Sato M, Shimano T, et al. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Si W, Yuan Y, Huang J, Zhang X, Zhang Y, et al. Widely distributed hot and cold spots in meiotic recombination as shown by the sequencing of rice F2 plants. New Phytol. 2015;206:1491–1502. doi: 10.1111/nph.13319. [DOI] [PubMed] [Google Scholar]
- 50.Saintenac C, Falque M, Martin OC, Paux E, Feuillet C, et al. Detailed recombination studies along chromosome 3B provide new insights on crossover distribution in wheat (Triticum aestivum L.). Genetics. 2009;181:393–403. doi: 10.1534/genetics.108.097469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson LK, Doyle GG, Brigham B, Carter J, Hooker KD, et al. High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics. 2003;165:849–865. doi: 10.1093/genetics/165.2.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahtani MM, Willard HF. Physical and genetic mapping of the human X chromosome centromere: repression of recombination. Genome Res. 1998;8:100–110. doi: 10.1101/gr.8.2.100. [DOI] [PubMed] [Google Scholar]
- 53.Rahn MI, Solari AJ. Recombination nodules in the oocytes of the chicken, Gallus domesticus. Cytogenet Cell Genet. 1986;43:187–193. doi: 10.1159/000132319. [DOI] [PubMed] [Google Scholar]
- 54.Hulten M. Chiasma distribution at diakinesis in the normal human male. Hereditas. 1974;76:55–78. doi: 10.1111/j.1601-5223.1974.tb01177.x. [DOI] [PubMed] [Google Scholar]
- 55.Hulten MA, Palmer RW, Laurie DA. Chiasma derived genetic maps and recombination fractions: chromosome 1. Ann Hum Genet. 1982;46:167–175. doi: 10.1111/j.1469-1809.1982.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 56.Jackson MS, See CG, Mulligan LM, Lauffart BF. A 9.75-Mb map across the centromere of human chromosome 10. Genomics. 1996;33:258–270. doi: 10.1006/geno.1996.0190. [DOI] [PubMed] [Google Scholar]
- 57.Yan H, Jin W, Nagaki K, Tian S, Ouyang S, et al. Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell. 2005;17:3227–3238. doi: 10.1105/tpc.105.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vincenten N, Kuhl LM, Lam I, Oke A, Kerr AR, et al. The kinetochore prevents centromere-proximal crossover recombination during meiosis. Elife. 2015;4 doi: 10.7554/eLife.10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pertile MD, Graham AN, Choo KH, Kalitsis P. Rapid evolution of mouse Y centromere repeat DNA belies recent sequence stability. Genome Res. 2009;19:2202–2213. doi: 10.1101/gr.092080.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roizes G. Human centromeric alphoid domains are periodically homogenized so that they vary substantially between homologues. Mechanism and implications for centromere functioning. Nucleic Acids Res. 2006;34:1912–1924. doi: 10.1093/nar/gkl137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma J, Bennetzen JL. Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc Natl Acad Sci U S A. 2006;103:383–388. doi: 10.1073/pnas.0509810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schueler MG, Dunn JM, Bird CP, Ross MT, Viggiano L, et al. Progressive proximal expansion of the primate X chromosome centromere. Proc Natl Acad Sci U S A. 2005;102:10563–10568. doi: 10.1073/pnas.0503346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schindelhauer D, Schwarz T. Evidence for a fast, intrachromosomal conversion mechanism from mapping of nucleotide variants within a homogeneous alpha-satellite DNA array. Genome Res. 2002;12:1815–1826. doi: 10.1101/gr.451502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi J, Wolf SE, Burke JM, Presting GG, Ross-Ibarra J, et al. Widespread gene conversion in centromere cores. PLoS Biol. 2010;8:e1000327. doi: 10.1371/journal.pbio.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng JC, Karpen GH. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 2009;5:e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Westphal T, Reuter G. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics. 2002;160:609–621. doi: 10.1093/genetics/160.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Collins K, Gorovsky MA. Tetrahymena thermophila. Curr Biol. 2005;15:R317–318. doi: 10.1016/j.cub.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 68.Papazyan R, Voronina E, Chapman JR, Luperchio TR, Gilbert TM, et al. Methylation of histone H3K23 blocks DNA damage in pericentric heterochromatin during meiosis. Elife. 2014;3:e02996. doi: 10.7554/eLife.02996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tischfield SE, Keeney S. Scale matters: The spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell Cycle. 2012;11:1496–1503. doi: 10.4161/cc.19733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borde V, Robine N, Lin W, Bonfils S, Geli V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO Journal. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Research. 2014;24:1650–1664. doi: 10.1101/gr.172122.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wahls WP, Smith GR. A heteromeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes Dev. 1994;8:1693–1702. doi: 10.1101/gad.8.14.1693. [DOI] [PubMed] [Google Scholar]
- 73.Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pratto F, Brick K, Khil P, Smagulova F, Petukhova GV, et al. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Serrentino ME, Borde V. The spatial regulation of meiotic recombination hotspots: are all DSB hotspots crossover hotspots? Exp Cell Res. 2012;318:1347–1352. doi: 10.1016/j.yexcr.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 80.Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 81.Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 82.Lao JP, Cloud V, Huang CC, Grubb J, Thacker D, et al. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 2013;9:e1003978. doi: 10.1371/journal.pgen.1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hyppa RW, Smith GR. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell. 2010;142:243–255. doi: 10.1016/j.cell.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, et al. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Molecular Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
- 86.Serrentino ME, Chaplais E, Sommermeyer V, Borde V. Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet. 2013;9:e1003416. doi: 10.1371/journal.pgen.1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Joshi N, Brown MS, Bishop DK, Borner GV. Gradual implementation of the meiotic recombination program via checkpoint pathways controlled by global DSB levels. Mol Cell. 2015;57:797–811. doi: 10.1016/j.molcel.2014.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol. 2010;12:500–506. doi: 10.1038/ncb2052. [DOI] [PubMed] [Google Scholar]
- 89.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 90.Buhler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, et al. A Hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ludin K, Mata J, Watt S, Lehmann E, Bahler J, et al. Sites of strong Rec12/Spo11 binding in the fission yeast genome are associated with meiotic recombination and with centromeres. Chromosoma. 2008;117:431–444. doi: 10.1007/s00412-008-0159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fowler KR, Gutiérrez-Velasco S, Martín-Castellanos C, Smith GR. Protein determinants of meiotic DNA break hotspots. Molecular Cell. 2013;49:983–996. doi: 10.1016/j.molcel.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kugou K, Fukuda T, Yamada T, Ito M, Sasanuma H, et al. Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol Biol Cell. 2009;13:3064–3076. doi: 10.1091/mbc.E08-12-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fukuda T, Kugou K, Sasanuma H, Shibata T, Ohta K. Targeted induction of meiotic double-strand breaks reveals chromosomal domain-dependent regulation of Spo11 and interactions among potential sites of meiotic recombination. Nucleic Acids Res. 2008;36:984–997. doi: 10.1093/nar/gkm1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robine N, Uematsu N, Amiot F, Gidrol X, Barillot E, et al. Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:1868–1880. doi: 10.1128/MCB.02063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Prieler S, Penkner A, Borde V, Klein F. The control of Spo11's interaction with meiotic recombination hotspots. Genes Dev. 2005;19:255–269. doi: 10.1101/gad.321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanchez-Moran E, Santos JL, Jones GH, Franklin FC. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007;21:2220–2233. doi: 10.1101/gad.439007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Current Biology. 2007;17:2003–2012. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 100.Comeron JM, Ratnappan R, Bailin S. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 2012;8:e1002905. doi: 10.1371/journal.pgen.1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Young JA, Hyppa RW, Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Puizina J, Siroky J, Mokros P, Schweizer D, Riha K. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell. 2004;16:1968–1978. doi: 10.1105/tpc.104.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 104.Borde V, Lin W, Novikov E, Petrini JH, Lichten M, et al. Association of Mre11p with double-strand break sites during yeast meiosis. Mol Cell. 2004;13:389–401. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- 105.Ellermeier C, Smith GR. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10952–10957. doi: 10.1073/pnas.0504805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Klein F, Mahr P, Galova M, Buonomo SBC, Michaelis C, et al. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 107.Sun X, Huang L, Markowitz TE, Blitzblau HG, Chen D, et al. Transcription dynamically patterns the meiotic chromosome-axis interface. Elife. 2015;4 doi: 10.7554/eLife.07424. [DOI] [PMC free article] [PubMed] [Google Scholar]