Abstract
Purpose
In prostate cancer (PCa) cells, there is CD24-dependent inactivation of mutant p53, but the mechanism and its significance remain largely unknown. Here, we validated this observation and explored the therapeutic potential of targeting CD24 in TP53 mutant PCa cells.
Experimental Design
Overall, 543 PCas (522 formalin-fixed paraffin-embedded and 21 frozen tissues) were assessed for protein or mRNA expression of CD24 and TP53. The effects of CD24 on p53-dependent transcriptional regulation, cancer cell growth, the cell cycle, apoptosis, and mutant p53 restoration were also determined.
Results
As determined with three sample cohorts, CD24 and p53 were not expressed in prostate epithelial cells but in PCa cells in 48% of cases for CD24 and 16% of cases for p53 (mutant form). Expressions of CD24 and mutant p53 were more frequently observed in late-stage and metastatic prostate tumors. Mutant p53 accompanied with CD24 was expressed in most cases (91.6%, 76/83). Silencing of CD24 increased the transcriptional activity of p53 target genes, such as CDKNA1, VDR, and TP53INP1, leading to suppression of p53-dependent cell growth, cell cycle arrest, and apoptosis in most TP53-mutant PCa cells. Silencing of CD24 enhanced restoration of PRIMA-1-induced mutant p53 in endogenous TP53P223L/V274F DU145 cells and in PC3 cells transfected with TP53R273H.
Conclusions
In human PCas, there is CD24-dependent inactivation of mutant p53. The co-expression of CD24 and p53 may help identify aggressive cancers. Targeting CD24 provides a strategy to enhance mutant p53-restoring therapies, especially in patients with TP53R273H PCa.
Keywords: CD24, TP53, prostate cancer, tumor progression
Introduction
The signal transducer CD24 is a cell surface molecule anchored by glycosylphosphatidylinositol (GPI). CD24 is not expressed in most cancer stem cells but is expressed in 68% of clinical samples across all major cancer types, especially late-stage cancers (1). Notably, CD24 serves as a marker for poor prognosis of human cancers (1), including prostate cancers (PCas) (2, 3). Analyses involving ectopic and/or inducible expression (4–7), targeted mutations (6, 8, 9), gene silencing (4, 6, 7, 10), and antibody blocking (11) demonstrate the oncogenic function of CD24. Our recent study showed that intracellular CD24 is sufficient to promote cell proliferation in PCa cells (6). Furthermore, the effects of CD24 on tumor progression and metastasis have been demonstrated with xenogenic (4–7, 9) and transgenic tumor models (6, 8). To validate the role of CD24 in tumor progression and metastasis in PCa, we crossed the CD24-null allele (12) into transgenic adenocarcinoma mouse prostate (TRAMP) mice, a strain that develops spontaneous PCas (13). The results showed that targeted mutation of CD24 retards the growth, progression, and metastasis of PCa cells (6), supporting a role of CD24 in tumor progression and metastasis. In addition, CD24 is highly expressed in hematopoietic cells and is involved in both adaptive and innate immunity (14), which may contribute to tumor progression and metastasis. However, our transplantation of bone marrow-derived CD24+/+ or CD24−/− cells had no impact on tumor progression and metastasis in a TRAMP model (6), suggesting that CD24 in non-hematopoietic cells contributes to tumor progression and metastasis.
CD24 is expressed in half of PCa cases but is not expressed in prostate epithelial cells (2, 3). High CD24 expression levels are associated with lymph node metastases, advanced clinical stages, and shortened overall survival of patients with PCa (2, 3). Although CD24 overexpression is implicated in tumor progression and metastasis (7, 9, 10, 15–18), a causative relationship has not been established. Recently, we identified, in PCa cells, a new function of CD24, inhibition of the ARF-NPM interaction. This inhibition causes ARF degradation, resulting in increased MDM2 levels and subsequently reduced p53 and levels of its target p21/CDKN1A (6). Further, we observed that most of the missense TP53 mutations in PCa cells inactivate p53 functionally only if the cells also express CD24, but silencing of CD24 prevents mutational inactivation of TP53 in its p53-dependent transcriptional activity and inhibition of tumor growth (6). In support of the functional interaction between CD24 and p53, our in silico analysis revealed that TP53 mutates at a higher rate among PCas with higher levels of CD24 mRNA (6). Thus, CD24 is necessary for inactivating TP53 mutations, suggesting that, in PCas, CD24-dependent inactivation of mutant p53 may contribute to tumor progression and metastasis. However, this observation needs to be validated in human PCas.
TP53, a frequently mutated gene, is mutated in about 30% of PCas (19). TP53 mutation or loss of function promotes the invasion and metastasis of PCa cells (20, 21), and accumulation of mutant p53 is related to an increased risk of tumor progression and disease-specific death and, in patients with PCa, to development of distant metastasis at year 5 (21–24). Under conditions of homeostasis, wild-type (WT) p53 is unstable, with a half-life of less than 20 minutes, mainly due to degradation by its E3 ubiquitin ligase, MDM2 (25). Thus, within cells, WT p53 is maintained at low concentrations (26). However, mutant p53 proteins are generally modified by post-translational modifications at specific sites, such as phosphorylation, acetylation, and SUMOylation, which stabilize p53, leading to a nuclear accumulation of mutant p53 as an oncogene. These post-translational modifications may alter the conformation of p53 to affect interaction with cofactors or binding to promoters (27). Mutant p53 proteins may also inhibit remaining WT p53 and thereby transform mutant p53 into a dominant oncogene (28). However, the mechanisms underlying the oncogenic function of mutant p53 remain elusive. Since our recent data showed that silencing CD24 restores at least part of the tumor suppressor activity of mutant p53 in PCa cells (6), CD24 may be a modulator of p53-driven tumor progression. In the present study, we assessed the expressions of CD24 and p53 and their associations with tumor progression and metastasis and explored the therapeutic potential of targeting CD24 in TP53-mutant PCa cells.
Materials and Methods
Human tissue specimens
We studied 697 formalin-fixed paraffin-embedded (FFPE) tissue specimens, including 522 PCa tissues, 34 benign prostatic hyperplasia tissues, and 141 normal prostate tissues. Of the FFPE tissues, 309 were collected at the University of Alabama at Birmingham (UAB) Hospital between 2000 and 2014; 388 FFPE tissue microarray (TMA) samples were purchased from US Biomax, Inc. (Rockville, MD) and ISU ABXIS, Co. (Seoul, South Korea). Brief clinical and pathological characteristics of the subjects are presented in Table 1. All patients were diagnosed by histological examination of specimens obtained from surgical resections. The pathological stage of PCa at the time of diagnosis was determined based on the Tumor-Node-Metastasis (TNM) system for UAB and US Biomax specimens and the American Joint Committee on Cancer (AJCC) Cancer stage grouping (29) for ISU ABXIS specimens. Pathological grading was based on specimens corresponding to Gleason scores of 2–6, 7, and 8–10, respectively. In the UAB cohort, there were three races (140 Caucasians, 35 African-Americans, and 6 Asians) but only Asians in the US Biomax and ISU ABXIS cohorts.
Table 1.
Human Subject Characteristics
| Groups | UAB a | US Biomax b | ISU ABXIS b |
|---|---|---|---|
| Age years, median (range) | 62 (45–89) | 69 (20–89) | 64 (43–80) |
| PSA (ng/ml), median (range) | 21.8 (1.1–169) | 17.5 (0–161) | 11.0 (0–135) |
| Race | |||
| Caucasians | 231 | - | - |
| African-Americans | 63 | - | - |
| Asians | 13 | 237 | 151 |
| Others | 2 | - | - |
| Tissue Type | |||
| Normal Prostate | 106 | 19 | 16 |
| Hyperplasia | 22 | 12 | - |
| Malignant | 181 | 206 | 135 |
| Tumor stage (TNM) | |||
| T1-2 | 68 | 72 | - |
| T3 | 45 | 56 | - |
| T4 | 10 | 18 | - |
| N+ or M+ | 58 | 60 | - |
| Tumor stage c | |||
| II | - | - | 76 |
| III | - | - | 59 |
| Gleason score | |||
| G2-6 | 45 | 40 | 11 |
| G7 | 70 | 67 | 60 |
| G8-10 | 66 | 94 | 64 |
Formalin-fixed samples and some frozen samples
Formalin-fixed TMA samples
II: There is no evidence that the cancer has spread outside the prostate. The tumor involves more tissue within the prostate, can be felt during rectal exam, or is found with a biopsy that is performed because of a high PSA level.
III: The cancer has spread outside the prostate to nearby tissues.
Frozen PCa specimens were selected from the Tissue Procurement Shared Facility at the Comprehensive Cancer Center, UAB. The specimens were collected from 31 patients with PCa who underwent primary surgery between January 2012 and June 2015. All had histologically-confirmed PCa with information on tumor stage (TNM) and grade (Gleason). For all specimens, informed consent was obtained from all subjects in accordance with the requirements of the Institutional Review Board of UAB.
Cell lines, antibodies, DNA constructs, and reagents
Human PCa cell lines DU145, PC3, and LNCaP were obtained from the American Type Culture Collection (ATCC). Cell lines were authenticated by examination of morphology and growth characteristics and confirmed to be mycoplasma-free. Cells were maintained in Dulbecco’s modified Eagle’s Medium supplemented with 10% fetal bovine serum (Life Technologies). Antibodies used for this study were human CD24 (ML5, BD Biosciences), p53 (DO-1, Santa Cruz Biotechnology), and p21 (DCS60, Cell Signaling). CD24 and TP53 expression or shRNA constructs were created as described in our previous study (6). PRIMA-1 (Sigma) was used for restoring mutant p53. All constructs were verified by nucleotide sequencing.
Immunohistochemistry (IHC) analysis
The ABC detection system (Vectastain Elite ABC) was used for immunostaining according to the manufacturer’s protocol as described previously (30). Specific primary antibodies were used to detect CD24 (ML5, 1:100) and p53 (DO-1, 1:200). Protein expressions of CD24 in the plasma membrane and cytoplasm and p53 in nuclei were classified as negative or positive. The results were determined to be negative if <10% of cells within tumor areas were stained or positive if 10%-100% were stained. All slides were examined by two pathologists (S.W. and R.J.G.) in a blinded fashion.
Laser capture microdissection
Frozen tissue sections were used for laser capture microdissection to obtain prostate epithelial and cancer cells, as described previously (30, 31). For analysis of gene expression, 5000 cells were microdissected from target tissues.
Quantitative real-time PCR (qPCR)
mRNA expression was analyzed using qPCR on the Real-Time PCR System (Roche) with SYBR Green dye (Promega) in accordance with the manufacturer’s protocol. Relative expression levels were determined using the comparative method (2− Δ ΔCt) against endogenous GAPDH controls. The primer sequences are listed in Table S1.
Western blots and quantitative chromatin immunoprecipitation (ChIP)
Western blotting was performed as described previously (6, 31). WT or mutant p53-transfected PC3 cells were used for ChIP assays, as described previously (6, 31). Briefly, cells were sonicated and fixed with 1% paraformaldehyde. The anti-p53 (DO-1) and anti-IgG antibodies were used to pull down chromatin associated with WT or mutant p53. The amounts of the specific DNA fragments were quantitated by real-time PCR and normalized against the genomic DNA preparation from the same cells. The ChIP qPCR primers are listed in Table S1.
Cell proliferation, cell cycle progression, and apoptosis
1x104 CD24+ (CD24-transduced) or CD24− (CD24 shRNA-transduced) PC3 cells were transfected with TP53R175H or TP53R273H using Lipofectamine 2000 (Life Technologies) and treated with PBS or PRIMA-1 (50 μM) for 5 days. After starvation for 48 hours, cell morphology, viability, and number were monitored microscopically. At 0 and 24 hours, cell cycle progression was determined by propidium iodide (PI) staining and flow cytometry, as described previously (6). At 5 days, apoptosis was detected by flow cytometry based on cell binding to Annexin V (561012, BD Biosciences) and 7-aminoactinomycin (7-AAD; 555816, BD Biosciences) (32).
Experimental animals
IL2Rcg−/−Nod.Scid (NSG) mice were purchased from The Jackson Laboratory (JAX). All mice were bred at the animal facilities of the Animal Resources Program at the UAB. All experiments were conducted in accordance with accepted standards of animal care and approved by the Institutional Animal Care and Use Committee of UAB.
Xenogeneic transplantation
100 μL (5x106) of CD24+ or CD24- TP53R273H stably-transfected PC3 cells were subcutaneously injected into the left flanks of 8-week-old male NSG mice. After 2 weeks, mice were treated every other day with intraperitoneal injections of 100 μL of PRIMA-1 (100 mg/kg) or PBS for 2 weeks. Tumor size and weight were measured as described previously (30, 31).
Statistical analyses
Comparisons between groups were performed using Chi-square or Fisher exact tests for categorical data. Logistic regression analyses were performed, and odds ratios (OR) and confidence intervals (CI) were used for quantifying differences between groups in IHC protein expression. For quantitative data, the distribution of data for each group was evaluated using a one-sample Kolmogorov-Smirnov test. In samples with normal distributions, the means of the variables were compared using a two-tailed t test between two groups. In samples with non-normal distributions, the medians of the variable between two groups were compared using a Mann-Whitney test. One-way analysis of variance (ANOVA) or two-way ANOVA were used to test for overall differences followed by Fisher’s protected least significant difference (PLSD) test. The Benjamini-Hochberg method was used to adjust for false discovery rates. All data were entered into an Access database using Excel 2010 and analyzed with SPSS (version 20; IBM) and Stat View (version 5.0.1; SAS).
Results
CD24 is a positive marker of tumor progression and metastasis in PCa
Analysis of data cohorts of Grasso et al. (33) and Wallance et al. (34) deposited in Oncomine.com revealed that CD24 mRNA is overexpressed in human PCa tissues compared with matched normal tissues (6). To substantiate this observation, we analyzed the independent data cohorts of Yu et al. (35) and Singh et al. (36) from Oncomine.com and validated the overexpression of CD24 mRNA in PCa tissues (Fig. S1A), suggesting that CD24 transcripts are likely to be associated with prostate tumorigenesis. Furthermore, to investigate changes of CD24 expression in prostate tumor progression, we analyzed the Gene Expression Omnibus (GEO) database and found that expression of CD24 is increased stepwise in tumor-adjacent normal tissues (1.14-fold change), localized tumor tissues (2.00-fold), and metastatic tumor tissues (2.33-fold), compared with normal prostate tissues (Fig. S1B). To validate our observation, we analyzed an independent dataset stored in The Cancer Genome Atlas (TCGA) and found that, in human PCas, expression of CD24 is also increased stepwise with increasing TNM tumor stage (T2 vs. T1: 1.43-fold ; T3 vs. T1: 2.03-fold; T3 vs. T2: 1.43-fold) (Fig. S1C). These data show overexpression of CD24 mRNA in human prostate tumors, particularly in high-stage and metastatic tumors.
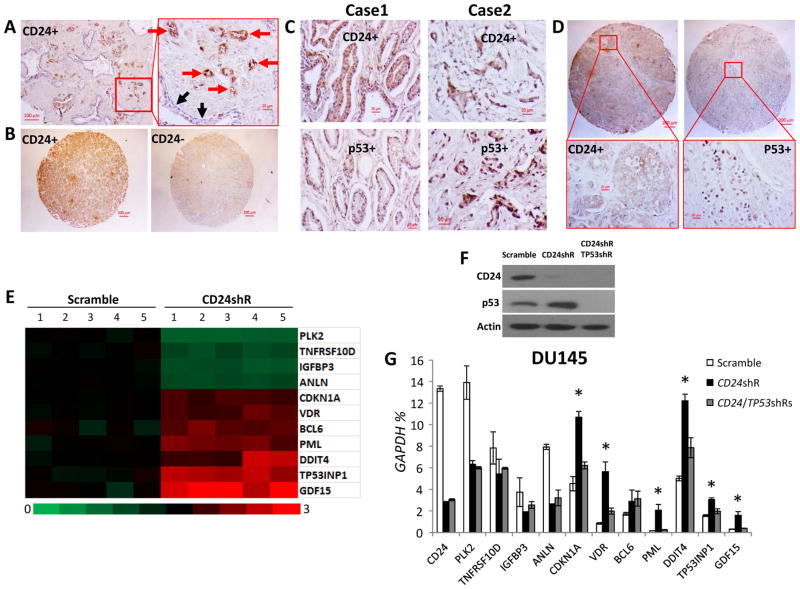
CD24 expression was determined by IHC in FFPE tissues, including 522 PCa samples, 34 benign prostatic hyperplasia samples, and 141 normal prostate samples (Table 2). Overall, CD24 was expressed in 48% of the PCa tissues but not in benign prostatic hyperplasia or normal prostate tissues (Table 2, Fig. 1A and 1B). In particular, PCa cells stained with CD24 antibody, but tumor-adjacent normal prostate epithelial cells did not stain (Fig. 1A). Notably, more CD24 was expressed in the cytoplasm than in the plasma membrane (Fig. 1A and 1B). Since intracellular CD24 is likely to be responsible for its oncogenic function (6), intracellular CD24 in human PCas was included in our expression analyses. However, we also observed nuclear CD24 staining in PC cells in 2.3% of the PCa cases, especially in metastatic cases (Table S2 and Figure S2).
Table 2.
Association of CD24 and p53 expressions with tumor stage and grade
| CD24
|
p53
|
|||||
|---|---|---|---|---|---|---|
| Tumor stage or grade | Negative n (%) | Positive n (%) | p-valuea | Negative n (%) | Positive n (%) | p-valuea |
| Tumor stage (UAB) | ||||||
| T1-2 | 41 (60.3) | 27 (39.7) | 0.007 b | 61 (89.7) | 7 (10.3) | 0.139 b |
| T3 | 19 (42.2) | 26 (57.8) | 38 (84.4) | 7 (15.6) | ||
| T4 | 3 (30.0) | 7 (70.0) | 8 (80.0) | 2 (20.0) | ||
| N+ or M+ | 17 (29.3) | 41 (70.7) | 43 (74.1) | 15 (25.9) | ||
| Tumor stage (US Biomax) | ||||||
| T1-2 | 50 (69.4) | 22 (30.6) | 0.004 | 65 (90.3) | 7 (9.7) | 0.104 b |
| T3 | 29 (51.8) | 27 (48.2) | 48 (85.7) | 8 (14.3) | ||
| T4 | 8 (44.4) | 10 (55.6) | 14 (77.8) | 4 (22.2) | ||
| N+ or M+ | 23 (38.3) | 37 (61.7) | 45 (75.0) | 15 (25.0) | ||
| Tumor stage (ISU ABXIS) | ||||||
| II | 52 (68.4) | 24 (31.6) | 0.038 | 70 (92.1) | 6 (7.9) | 0.035 |
| III | 30 (50.8) | 29 (49.2) | 47 (79.7) | 12 (20.3) | ||
| Gleason grade (UAB) | ||||||
| G2-6 | 27 (60.0) | 18 (40.0) | 0.021 | 43 (95.6) | 2 (4.4) | 0.014 b |
| G7 | 31 (44.3) | 39 (55.7) | 58 (82.9) | 12 (17.1) | ||
| G8-10 | 22 (33.3) | 44 (66.7) | 49 (74.2) | 17 (25.8) | ||
| Gleason grade (US Biomax) | ||||||
| G2-6 | 28 (70.0) | 12 (30.0) | 0.082 | 37 (92.5) | 3 (7.5) | 0.073 b |
| G7 | 34 (49.3) | 33 (49.3) | 59 (88.1) | 8 (11.9) | ||
| G8-10 | 47 (50.0) | 47 (50.0) | 72 (76.6) | 22 (23.4) | ||
| Gleason grade (ISU ABXIS) | ||||||
| G2-6 | 7 (63.6) | 4 (36.4) | 0.596 b | 11 (100.0) | 0 (0.0) | 0.371 b |
| G7 | 39 (65.0) | 21 (35.0) | 52 (86.7) | 8 (13.3) | ||
| G8-10 | 36 (56.2) | 28 (43.8) | 54 (84.4) | 10 (15.6) | ||
Pearson Chi-square test
Fisher’s Exact test
Figure 1.
CD24 and mutant p53 and their regulated gene expressions in human PCa cells. (A) A representative histology of PCas shows CD24 staining in PCa cells but no staining in normal prostate epithelial cells from the same patient. The red and black arrows indicate PCa cells and normal prostate epithelial cells, respectively. (B) A representative CD24 staining of TMA PCa samples. (C and D) Representative IHC data showing co-expression of CD24 and mutant p53 in primary cancer samples from two representative cases and TMA samples from the same case, using monoclonal anti-CD24 and p53 antibodies, respectively. (E) A heat-map depiction of alterations in p53 target gene expression in DU145 cells following CD24 gene silencing was generated from Affymetrix Human U133 Plus 2.0 microarrays. The microarray data were submitted to NCBI GEO (accession No. GSE46708). (F) Silencing of CD24 and TP53 by shRNAs was validated by Western blots. (G) The mRNA expression of p53 target genes was validated by qPCR. Data are presented as the means and SD of triplicates. The relative amounts are expressed as percentages of GAPDH. * p < 0.05 when CD24 shRNA was compared with scramble control and CD24/TP53 shRNAs (two-tailed t test). shR, shRNA. All experiments were repeated three times.
To assess the contribution of CD24 to progression of human PCas, the protein expression of CD24 was compared with pathological parameters, including tumor stage and grade. CD24 expression positively correlated with tumor stages in the three sample cohorts: UAB (p=0.007), US Biomax (p=0.004), and ISU ABXIS (p=0.038) (Table 2). CD24 expression was > 2 times more likely to be present in metastatic tumors than in localized tumors: UAB subjects (OR=2.63, 95% CI: 1.34–5.16, p=0.005) and US Biomax subjects (OR=2.37, 95% CI=1.27–4.40, p=0.007] (Table 3). Although there was a significant association in CD24 expression with tumor grades (Gleason scores) in the UAB cohort (p=0.021), this observation was not confirmed in the other two cohorts (Table 2).
Table 3.
Age-adjusted OR according to CD24 and p53 expressions for tumor metastasis
| Negative n (%) | Positive n (%) | OR a | 95% CI b | p-value | |
|---|---|---|---|---|---|
| UAB | |||||
| CD24 | |||||
| Metastatic (N+ or M1+) | 17 (29.3) | 41 (70.7) | 2.63 | 1.34–5.16 | 0.005 |
| Localized (pT1-4, N0, M0) | 63 (51.2) | 60 (48.8) | |||
| p53 | |||||
| Metastatic (N+ or M1+) | 43 (74.1) | 15 (25.9) | 2.17 | 0.98–4.82 | 0.056 |
| Localized (pT1-4, N0, M0) | 107 (87.0) | 16 (13.0) | |||
| US Biomax | |||||
| CD24 | |||||
| Metastatic (N+ or M1+) | 23 (38.3) | 37 (61.7) | 2.37 | 1.27–4.40 | 0.007 |
| Localized (pT1-4, N0, M0) | 87 (59.6) | 59 (40.4) | |||
| p53 | |||||
| Metastatic (N+ or M1+) | 45 (75.0) | 15 (25.0) | 2.27 | 1.06–4.87 | 0.036 |
| Localized (pT1-4, N0, M0) | 127 (87.0) | 19 (13.0) | |||
Odds ratio (OR) was adjusted by age
CI: confidence interval
CD24 is a partner of mutant p53 protein in tumor progression of PCa
WT p53 proteins are unstable (25), but missense mutant p53 proteins often accumulate at high levels in tumor tissues (37). Thus, accumulation in tumor tissues of missense mutants, but not WT p53 protein, can be detected by IHC. Most TP53 mutations are missense and thus allow production of full-length mutant p53 proteins (38). Thus, as determined in IHC analysis, detectable p53 protein expression in tumor cells is essentially from missense mutant p53. In the present study, p53 protein accumulation was detected in the nuclei of PCa cells. p53 protein was expressed in 16% of the PCa tissues (Table 2), but not in benign prostatic hyperplasia or normal prostate tissues. Although p53 expression significantly correlated with an increased tumor stage in the ISU ABXIS cohort (p=0.035), no significant differences were found in other two cohorts: UAB (p=0.139) and US Biomax (p=0.104) (Table 2). Furthermore, in the US Biomax cohort, p53 expression was significantly associated with tumor metastasis after adjustment for age (OR=2.27, 95% CI=1.06–4.87, p=0.036). Although p53 expression is more than two times likely to be present in tumor metastases, no significant association was found in the UAB cohort (OR=2.17, 95% CI=0.98–4.82, p=0.056) (Table 3). In addition, p53 expression significantly correlated with an increased tumor grade in the UAB cohort (p=0.014), but no significant differences were found in other two cohorts, US Biomax (p=0.073) and ISU ABXIS (p=0.371) (Table 2).
To validate the CD24-dependent inactivation of mutant p53 in human PCas, the relationship between CD24 and p53 protein expressions were investigated in the three cohorts. Although the frequency of p53 expression was less than CD24 expression in all samples, p53 accumulation was accompanied with CD24 expression in most cases (27/31 in the UAB cohort; 32/34 in the US Biomax cohort; and 17/18 in ISU ABXIS cohort) (Fig. 1C and 1D, Table S3). Only a few cases showed expression of p53 without CD24, and all of these were associated with low-stage tumors (T1-3 or II) (Table S3).
CD24-dependent inactivation of mutant p53 and transcriptional regulation of p53 target genes in PCa cells
CD24 mRNA is expressed variably in human PCa cell lines DU145, PC3, and LNCaP (Table S4). DU145 expressed the highest levels of CD24 mRNA, whereas LNCaP did not express detectable levels. A moderate level of CD24 was observed in PC3 cells. Conversely, p53 is a bi-allelic mutant in DU145 cells, null in PC3 cells, and shows WT expression in LNCaP cells. Functional analyses identified a CD24-dependent inactivation of mutant p53 in PCa cells (6), but this observation is has not been validated for human PCas. Likewise, the molecular mechanism of CD24-dependent p53 transcriptional regulation remains undefined. In the present investigation, we analyzed microarray data (6) to identify CD24- and p53-regulated or target genes in DU145 cells. DU145 cells harbor bi-allelic missense mutations of TP53 (p53P223L and p53V274F) (42), and both mutants exhibit tumor suppressor activity based on cellular context and culture conditions. We first identified the eleven p53 target genes (43) that were also downregulated or upregulated after CD24 silencing (Fig. 1E). TP53 shRNA was then used to silence mutant TP53 (Fig. 1F) and determine if p53 is required for these CD24-regulated genes. By qPCR analysis, we determined six candidate genes that were regulated by CD24 through mutant p53P223L/V274F: CDKN1A, VDR, PML, DDIT4, TP53INP1, and GDF15 (Fig. 1G). Although expressions of other p53 target genes, such as PLK2, TNFRSF10D, IGFBP3, ANLN, and BCL6, were regulated by CD24, they were more likely to be independent of mutant p53P223L/V274F (Fig. 1G).
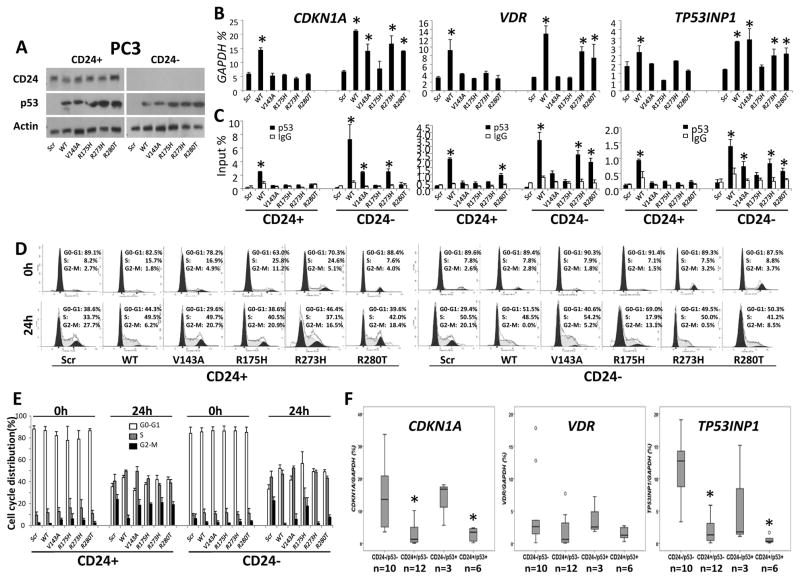
PC3 cells express a moderate level of CD24 but are p53-null (6). To validate the role of CD24 in mutant p53-dependent reactivation, a CD24-overexpressing (CD24+) model was established in PC3 cells by transduction with CD24, and a CD24-silenced (CD24−) cell model was produced by transduction with shRNA (Fig. 2A). WT TP53 and mutant TP53 (TP53R175H, TP53R273H, TP53V143A, or TP53R280T) were transfected into CD24+ PC3 cells and CD24− PC3 cells (Fig. 2A). Using the two models, we addressed the CD24-dependent transcriptional regulation of p53 target genes, such as CDKN1A, VDR, and TP53INP1 (43). Although there was no significant difference in expression of CDKN1A, VDR, and TP53INP1 between CD24+ PC3 cells and CD24− PC3 cells, the transfections of WT TP53, mutants TP53R273H, TP53V143A, and TP53R280T, but not mutant TP53R175H, induced the expression of these p53 target genes in CD24− PC3 cells but not in CD24+ PC3 cells (Fig. 2B). To determine if these genes were upregulated by enhanced p53-binding, quantitative ChIP analyses were performed for CD24+ or CD24− TP53-transfected PC3 cells (Fig. S3). The binding of WT p53, mutant p53R273H, and part of mutants p53V143A and p53R280T, but not mutant p53R175H, in the promoter region of those p53 target genes in CD24− PC3 cells, but not in CD24+ PC3 cells, was noted (Fig. 2C). Since CDKN1A is essential for cell cycle regulation (44), the effect of CD24-dependent p53 transcriptional regulation on cell cycle progression was assessed in PC3 cells. After CD24 silencing, this arrest was observed in cells transfected with WT TP53 and mutants TP53R273H and TP53R280T (Fig. 2D and 2E). Although partial cell cycle arrest was also seen in TP53V143A-transfected cells, there was no change in mutant TP53R175H-transfected cells, regardless of CD24 expression (Fig. 2D and 2E).
Figure 2.
CD24-dependent transcriptional regulation of p53 target genes in TP53 mutant human PCa cells. (A) Transfection of WT or mutant TP53 was validated by Western blots in CD24+ (CD24-transduced) or CD24− (CD24shRNA-transduced) PC3 cells. p53 mutants: p53V143A, p53R175H, p53R273H, and p53R280T. (B) The mRNA expression of p53 target genes was determined in CD24+ or CD24− PC3 cells by qPCR. The relative amounts are expressed as percentages of GAPDH. (C) ChIP analyses show DNA binding of p53 to p53 target genes in CD24+ and CD24− PC3 cells. The relative amounts of DNA binding are expressed as percentages of total input DNAs. Data are presented as the means and SD of triplicates. * p < 0.05, two-tailed t test. (D) After starvation for 48 hours, cell cycle progression in CD24+ or CD24− PC3 cells was monitored by propidium iodide staining and flow cytometry at 0 and 24 hours. (E) Quantitative cell cycle data are presented as the means and SD of triplicates. (F) mRNA expression of p53 target genes measured by qPCR in microdissected samples with or without CD24 and mutant p53. The relative amounts are expressed as percentages of GAPDH. Data are presented as the medians and interquartile ranges. Small hollow circles indicate that the data do not follow a normal distribution. * p < 0.05, a Mann-Whitney test. Scr, scramble; shR, shRNA; WT, wild-type; n, sample size. All experiments were repeated three times.
LNCaP cells do not express CD24 but are WT p53 (6). To validate the role of CD24 in mutant p53-dependent inactivation, a CD24+ model was generated from CD24− LNCaP cells by transduction with CD24 (Fig. S4). WT TP53 and mutant TP53 (TP53R175H and TP53R273H) were transfected into CD24+ and CD24− endogenous TP53-silenced LNCaP cells (Fig. S4A). With the two models, we confirmed the CD24-dependent transcriptional expression and regulation of the p53 target gene CDKN1A in TP53WT and TP53R273H-transfected cells but not in TP53R175H-transfected cells (Fig. S4B and S4C). Likewise, accelerated cell cycle progression was observed in CD24+ cells compared with CD24− cells when the cells were transfected with WT TP53 and mutant TP53R273H but not with TP53R175H (Fig. S4D and S4E).
To address the CD24-dependent transcriptional regulation of mutant p53 in primary PCas, IHC analysis was used with laser capture microdissection to obtain PCa cells from CD24−/p53−, CD24+/p53−, CD24−/p53+, and CD24+/p53+ frozen tissues of 21 primary PCa samples. The expressions of CDKN1A and TP53INP1 were significantly higher in CD24−/p53− and CD24−/p53+ cells compared with those in CD24+/p53− or CD24+/p53+ cells, regardless of p53+ expression (Fig. 2F). Although a difference was found in the expression of VDR (Fig. 2F), there was a marginal statistical difference between CD24− cells compared with CD24+ cells (p=0.0498).
Silencing of CD24 enhances PRIMA-1-induced restoration of mutant p53 and p53-dependent cell growth suppression, cell cycle arrest, and apoptosis in PCa cells
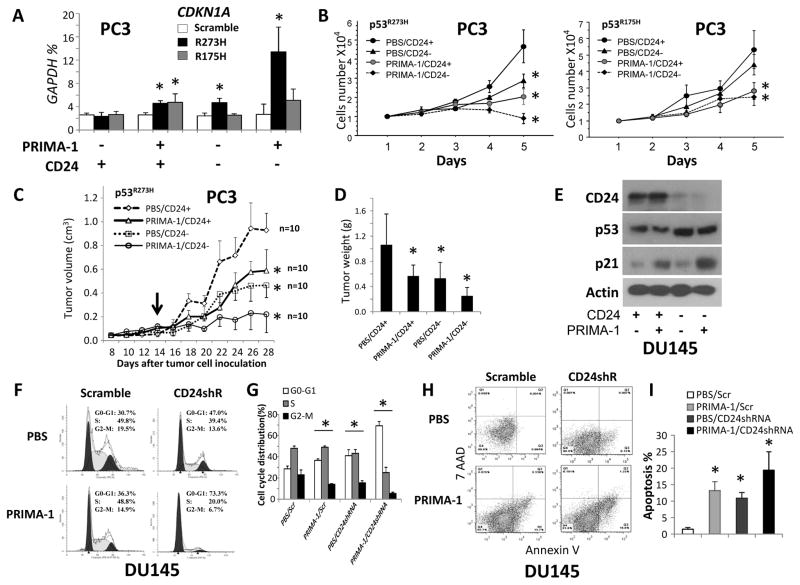
PRIMA-1 is a small molecule that induces p53-dependent apoptosis and activates p53 transcriptional activity in human tumor cells carrying mutant p53, including p53R175H and p53R273H (45, 46). To establish the function of CD24 during treatment of cells with PRIMA-1 for restoring mutant p53, p53-null PC3 cells with different status of TP53 and CD24 were treated with PRIMA-1. An approximately 2-fold induction of CDKN1A expression was observed in TP53R175H and TP53R273H-co-transfected CD24+ PC3 cells; in TP53R273H-transfected CD24− PC3 cells, this induction was increased >5-fold (Fig. 3A). However, no significant difference in CDKN1A expression was found between CD24+ and CD24− TP53R175H-transfected PC3 cells (Fig. 3A). Further, we investigated the interaction of genetic CD24 silencing with PRIMA-1 treatment during cell proliferation in the mutant TP53-transfected PC3 cells. After PRIMA-1 treatment, there was suppression of growth of TP53R273H or TP53R175H-transfected CD24+ PC3 cells in vitro (Fig. 3B) and in vivo (Fig. 3C and 3D). However, after PRIMA-1 treatment, additional growth inhibition was observed for TP53R273H-transfected PC3 CD24− cells, but not for TP53R175H-transfected CD24− PC3 cells (Fig. 3B–D).
Figure 3.
Effect of CD24 in PRIMA-1-induced restoration of mutant p53 and p53-denpendent cell growth suppression, cell cycle arrest, and apoptosis. (A) Quantification of CDKN1A expression after transfection of scramble, TP53R175H, or TP53R273H with treatment of PRIMA-1 in CD24+ and CD24− PC3 cells. The relative amounts are expressed as percentages of GAPDH. Data are presented as the means and SD of triplicates. (B) Cell proliferation after transfection of scramble, TP53R175H, or TP53R273H with treatment of PRIMA-1 in CD24+ or CD24− PC3 cells. Left panel: growth curves of TP53R273H-transfected PC3 cells. Right panel: growth curves of TP53R175H-transfected PC3 cells. *p <0.05, Fisher’s PLSD test. (C) Tumor growth in NSG mice subcutaneously injected with TP53R273H-transfected CD24+ or CD24− PC3 cells with or without PRIMA-1. The black arrow indicates the starting time of treatment with PBS or PRIMA-1 (every other day for two weeks). Data are presented as the means and SD of the tumor volumes; the experiments were repeated 2 times. * p < 0.05, Fisher’s PLSD test. n, sample size. (D) Weights of tumors at day 28 after injection. Data are presented as the means and SD of the tumor weights. * p < 0.05, two-tailed t test. (E) p21 and p53 expressions in CD24+ or CD24− (CD24shRNA-transduced) DU145 cells treated with or without PRIMA-1 for 48 hours were measured by Western blots. The DU145 cells carry endogenous mutant p53P223L/V274F. (F) After starvation of cells for 48 hours, cell cycle progression in CD24+ or CD24− DU145 cells was determined by propidium iodide staining and flow cytometry at 24 hours. (G) Quantitative cell cycle data are presented as the means and SD of triplicates. * p < 0.05 against PBS/Scr group, Fisher’s PLSD test. (H) Apoptosis in CD24+ or CD24− DU145 cells was detected by Annexin V/7-AAD staining at 5 days after treatment with PRIMA-1. (I) Quantitative apoptosis data are presented as the means and SD of triplicates. * p < 0.05 against the PBS/Scr group, two-tailed t test. All above in vitro experiments were repeated three times.
DU145 cells overexpress CD24 and also harbor bi-allelic endogenous p53P223L and p53V274F (6, 42). Thus, the functional context of CD24 and PRIMA-1 in DU145 cells was investigated. Expression of p21 protein, but not CD24 or p53, was induced by PRIMA-1, but silencing of CD24 induced p53 expression in DU145 cells (Figure 3E). Notably, in DU145 cells, silencing of CD24 enhanced PRIMA-1-induced p21 expression (Fig. 3E). In addition, the effects of these administrations on p53-denpendent cell cycle arrest and apoptosis were addressed in DU145 cells. Either silencing of CD24 or treatment with PRIMA-1 induced cell cycle arrest and apoptosis; these effects were enhanced by their combination (Fig. 3F–I).
Discussion
Here, our observation of CD24-dependent mutant p53 inactivation (6) was confirmed for human PCa cells. The co-expression of CD24 and mutant p53, frequently found in aggressive PCas, was established. Functional analyses demonstrated that silencing of CD24 induced expression of p53 target genes, such as CDKN1A, VDR, and TP53INP1, through an increased binding of mutant p53 to specific p53-response elements on these genes in TP53 mutant PCa cells. Notably, CD24-denpendent restoration of mutant p53 was validated in both endogenous mutant p53 in DU145 cells and transfected mutant p53 in PC3 cells. CD24-denpendent mutant p53 restoration was observed in most p53 mutants, especially in a commonly occurring, cancer-associated p53R273H, but not in p53R175H. The molecular mechanism for the restoration of mutant p53 by silencing of CD24 remains to be elucidated. Furthermore, silencing of CD24 increased p53 transcriptional activity, leading to cell cycle arrest in most TP53 mutant PCa cells. Of note, silencing of CD24 enhanced the PRIMA-1-induced restoration of mutant p53 and p53-denpendent inhibition of cell growth, cell cycle arrest, and apoptosis in DU145 cells with TP53P223L/V274F and in TP53R273H-transfected PC3 cells.
Co-expression of CD24 and mutant p53 is a characteristic marker for identifying aggressive PCas. In the present study, we established, by examination of three public datasets, that CD24 mRNA is overexpressed in high-stage and metastatic PCas. We validated the expression of CD24 protein in half of human PCas but not in normal prostate or benign prostatic hyperplasia (2, 3). We discovered that, in high-stage and metastatic PCas, mutant p53 is almost always accompanied with overexpressed CD24. While the mechanism underlying a functional interaction of CD24 with mutant p53 in tumor progression remains to be understood, their combination could refine the differentiation of patients with aggressive PCas, especially metastatic cancers, from patients with indolent PCas. Such information could affect the diagnosis and treatment of patients with aggressive PCas. Further, although CD24 is mainly expressed on the cytoplasmic membrane and cytoplasm, nuclear staining was observed in few cases, especially in metastatic cases. Likewise, CD24 has been detected in nuclear fractions of DU145 cells (6). Thus, CD24 may interact with p53 directly in nuclei, but additional studies are required for validation of this possibility.
TP53 is a tumor suppressor gene that is frequently mutated in PCas. In PCa cells, however, CD24 is a genetic modifier for restoration of mutant p53. More than 80% of the mutations are missense mutations that lead to less p53 transcriptional activity (19–21). Strategies targeting p53 have been developed, including gene therapies that restore the function of p53, but restoration of mutant p53 has been challenging (48). In the present study, CD24 was silenced to release mutant p53 tumor suppression and enhance PRIMA-1-induced restoration of mutants of p53. This approach should improve the current therapeutic strategies for enhancing restoration of mutant p53 for cancer therapy. However, silencing of CD24 does not release or enhance restoration of all mutants of p53, such as p53R175H. These observations are expected to be relevant in patient selection for restoration of mutant p53. For example, PRIMA-1 has been used for a clinical trial in cancer patients (49). CD24 and p53 status may help identify appropriate patients for targeting mutant p53 therapy.
Silencing of CD24 prevents functional inactivation of mutant p53 (6) and, in PCa cells, enhances PRIMA-1-induced restoration of mutant p53, especially p53R273H. In particular, expression of the p53 target genes CDKN1A, VDR, and TP53INP1 were increased in CD24− tumor cells relative to CD24+ tumor cells, except for cells with p53R175H, suggesting a functional context of CD24 and mutant p53 in PCa cells. The mechanism underlying the interaction of CD24 with mutant p53 remains largely unknown. We recently demonstrated that intracellular CD24 competitively inhibits ARF binding to NPM, resulting in decreased ARF, increased MDM2, and decreased levels of WT p53 and the p53 target p21/CDKN1A (6). The association between CD24 and NPM was most prominent during the metaphase and anaphase of mitosis. However, a CD24-mutant p53 interaction could occur transiently during mitosis. In the present study, we observed that silencing of CD24 enhanced the PRIMA-1-induced restoration of mutant p53 and its tumor suppressing function, whereas treatment with PRIMA-1 did not change the CD24 expression in DU145 cells. Thus, silencing of CD24-induced p53 restoration appears to be independent of PRIMA-1-induced restoration of mutant p53. However, in PCa cells, there was a synergistic restoration of mutant p53 by a combination of silencing of CD24 and treatment of PRIMA-1, suggesting a functional interaction between the two. In addition, p53R273H is a DNA contact mutant, whereas p53R175H is a conformational mutant (50). Our data showed that silencing of CD24 results in functional restoration of p53R273H but not p53R175H, suggesting that, during CD24-dependent restoration of mutant p53, silencing of CD24 influences DNA contact but not the conformation of mutant p53.
In conclusion, this is the first characterization of the functional interaction of CD24 and p53 in human PCas, which enhances understanding of PCa pathogenesis. Our data also suggest that a combination of CD24 and p53 may help identify aggressive PCas. In addition, targeting of CD24 is expected to improve pharmacological p53-restoring therapies, such as PRIMA-1, in patients with TP53 R273H PCas.
Supplementary Material
Statement of translational relevance (required).
First, our data indicate that CD24 with mutant p53 is an integrated biomarker to refine the stratification of patients with aggressive prostate cancer, especially metastatic prostate cancer, from patients with indolent prostate cancer. This will affect the diagnosis and treatment of patients with aggressive prostate cancer. Second, TP53 is a tumor suppressor gene that is frequently mutated in prostate cancers. More than 80% of the mutations are missense mutations that lead to lack of p53 transcriptional activity. Strategies targeting p53 have been developed, including gene therapies that restore the function of p53, but restoration of mutant p53 has been challenging. In the present study, we silenced CD24 to release mutant p53 tumor suppression and enhance PRIMA-1-induced restoration of mutant p53 in prostate cancer cells. Our findings provide a promising approach to restore mutant p53 function for aggressive prostate cancer therapy.
Acknowledgments
We thank Dr. Donald Hill for editorial assistance in preparing this manuscript.
Grant Support
Research reported in this publication was supported by the National Institutes of Health/National Cancer Institute (CA179282 and CA199586 for L.W.; CA118948 for L.W. and S.B.; CA013148 for R.L. and S.B.), the Department of Defense (PC140308 for R.L. and PC130594 for L.W. and W.H.Y.), the UAB Pittman Scholar Award (for L.W.), and a Mercer University Seed Grant (for W.H.Y.).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: R. Liu, L. Wang, X. Zhang
Development of methodology: R. Liu, J.W. Lillard, W.H. Yang, D. Chen
Acquisition of data (provided animals, provided facilities, etc.): W. Zhang, B. Yi, C. Wang
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): D. Chen, S. Bae, L. Wang, R. Liu, L.L.H. Nguyen, S. Wei, R.J. Guo, W. Zhang, B. Yi, C. Wang
Writing, review, and/or revision of the manuscript: R. Liu, L. Wang, X. Zhang
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): W. Zhang, B. Yi, C. Wang, X. Zhang, J.W. Lillard, C. Lu
Study supervision: R. Liu, L. Wang
References
- 1.Lee JH, Kim SH, Lee ES, Kim YS. CD24 overexpression in cancer development and progression: a meta-analysis. Oncol Rep. 2009;22:1149–56. doi: 10.3892/or_00000548. [DOI] [PubMed] [Google Scholar]
- 2.Kristiansen G, Pilarsky C, Pervan J, Sturzebecher B, Stephan C, Jung K, et al. CD24 expression is a significant predictor of PSA relapse and poor prognosis in low grade or organ confined prostate cancer. Prostate. 2004;58:183–92. doi: 10.1002/pros.10324. [DOI] [PubMed] [Google Scholar]
- 3.Kristiansen G, Pilarsky C, Wissmann C, Kaiser S, Bruemmendorf T, Roepcke S, et al. Expression profiling of microdissected matched prostate cancer samples reveals CD166/MEMD and CD24 as new prognostic markers for patient survival. J Pathol. 2005;205:359–76. doi: 10.1002/path.1676. [DOI] [PubMed] [Google Scholar]
- 4.Smith SC, Oxford G, Wu Z, Nitz MD, Conaway M, Frierson HF, et al. The Metastasis-Associated Gene CD24 Is Regulated by Ral GTPase and Is a Mediator of Cell Proliferation and Survival in Human Cancer. Cancer research. 2006;66:1917–22. doi: 10.1158/0008-5472.CAN-05-3855. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez SJ, Rojas JI, Redal MA, Patrucco L, Correale J, Argibay PF, et al. CD24 as a genetic modifier of disease progression in multiple sclerosis in Argentinean patients. J Neurol Sci. 2011;307:18–21. doi: 10.1016/j.jns.2011.05.032. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Liu R, Ye P, Wong C, Chen GY, Zhou P, et al. Intracellular CD24 disrupts the ARF-NPM interaction and enables mutational and viral oncogene-mediated p53 inactivation. Nat Commun. 2015;6:5909. doi: 10.1038/ncomms6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas S, Harding MA, Smith SC, Overdevest JB, Nitz MD, Frierson HF, et al. CD24 is an effector of HIF-1-driven primary tumor growth and metastasis. Cancer research. 2012;72:5600–12. doi: 10.1158/0008-5472.CAN-11-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D, Zheng L, Jin L, Zhou Y, Li H, Fu J, et al. CD24 polymorphisms affect risk and progression of chronic hepatitis B virus infection. Hepatology. 2009;50:735–42. doi: 10.1002/hep.23047. [DOI] [PubMed] [Google Scholar]
- 9.Overdevest JB, Knubel KH, Duex JE, Thomas S, Nitz MD, Harding MA, et al. CD24 expression is important in male urothelial tumorigenesis and metastasis in mice and is androgen regulated. Proc Natl Acad Sci U S A. 2012;109:E3588–96. doi: 10.1073/pnas.1113960109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Overdevest JB, Thomas S, Kristiansen G, Hansel DE, Smith SC, Theodorescu D. CD24 offers a therapeutic target for control of bladder cancer metastasis based on a requirement for lung colonization. Cancer research. 2011;71:3802–11. doi: 10.1158/0008-5472.CAN-11-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sagiv E, Kazanov D, Arber N. CD24 plays an important role in the carcinogenesis process of the pancreas. Biomedicine & Pharmacotherapy. 2006;60:280–4. doi: 10.1016/j.biopha.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen PJ, Lorenz B, Muller AM, Wenger RH, Brombacher F, Simon M, et al. Altered erythrocytes and a leaky block in B-cell development in CD24/HSA-deficient mice. Blood. 1997;89:1058–67. [PubMed] [Google Scholar]
- 13.Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92:3439–43. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang X, Zheng P, Tang J, Liu Y. CD24: from A to Z. Cell Mol Immunol. 2010;7:100–3. doi: 10.1038/cmi.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumann P, Cremers N, Kroese F, Orend G, Chiquet-Ehrismann R, Uede T, et al. CD24 expression causes the acquisition of multiple cellular properties associated with tumor growth and metastasis. Cancer research. 2005;65:10783–93. doi: 10.1158/0008-5472.CAN-05-0619. [DOI] [PubMed] [Google Scholar]
- 16.Baumann P, Thiele W, Cremers N, Muppala S, Krachulec J, Diefenbacher M, et al. CD24 interacts with and promotes the activity of c-src within lipid rafts in breast cancer cells, thereby increasing integrin-dependent adhesion. Cell Mol Life Sci. 2012;69:435–48. doi: 10.1007/s00018-011-0756-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W, Monahan KB, Pfefferle AD, Shimamura T, Sorrentino J, Chan KT, et al. LKB1/STK11 Inactivation Leads to Expansion of a Prometastatic Tumor Subpopulation in Melanoma. Cancer cell. 2012;21:751–64. doi: 10.1016/j.ccr.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mierke CT, Bretz N, Altevogt P. Contractile forces contribute to increased glycosylphosphatidylinositol-anchored receptor CD24-facilitated cancer cell invasion. J Biol Chem. 2011;286:34858–71. doi: 10.1074/jbc.M111.245183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ecke TH, Schlechte HH, Hubsch A, Lenk SV, Schiemenz K, Rudolph BD, et al. TP53 mutation in prostate needle biopsies--comparison with patients follow-up. Anticancer Res. 2007;27:4143–8. [PubMed] [Google Scholar]
- 20.Wang Y, Zhang YX, Kong CZ, Zhang Z, Zhu YY. Loss of P53 facilitates invasion and metastasis of prostate cancer cells. Mol Cell Biochem. 2013;384:121–7. doi: 10.1007/s11010-013-1789-1. [DOI] [PubMed] [Google Scholar]
- 21.Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–41. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 22.Aprikian AG, Sarkis AS, Fair WR, Zhang ZF, Fuks Z, Cordon-Cardo C. Immunohistochemical determination of p53 protein nuclear accumulation in prostatic adenocarcinoma. J Urol. 1994;151:1276–80. doi: 10.1016/s0022-5347(17)35231-x. [DOI] [PubMed] [Google Scholar]
- 23.Navone NM, Troncoso P, Pisters LL, Goodrow TL, Palmer JL, Nichols WW, et al. p53 protein accumulation and gene mutation in the progression of human prostate carcinoma. J Natl Cancer Inst. 1993;85:1657–69. doi: 10.1093/jnci/85.20.1657. [DOI] [PubMed] [Google Scholar]
- 24.Visakorpi T, Kallioniemi OP, Heikkinen A, Koivula T, Isola J. Small subgroup of aggressive, highly proliferative prostatic carcinomas defined by p53 accumulation. J Natl Cancer Inst. 1992;84:883–7. doi: 10.1093/jnci/84.11.883. [DOI] [PubMed] [Google Scholar]
- 25.Gu J, Chen D, Rosenblum J, Rubin RM, Yuan ZM. Identification of a sequence element from p53 that signals for Mdm2-targeted degradation. Mol Cell Biol. 2000;20:1243–53. doi: 10.1128/mcb.20.4.1243-1253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–31. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 27.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nature reviews. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 28.Soussi T, Lozano G. p53 mutation heterogeneity in cancer. Biochem Biophys Res Commun. 2005;331:834–42. doi: 10.1016/j.bbrc.2005.03.190. [DOI] [PubMed] [Google Scholar]
- 29.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–4. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Liu R, Li W, Chen C, Katoh H, Chen GY, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer cell. 2009;16:336–46. doi: 10.1016/j.ccr.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu R, Yi B, Wei S, Yang WH, Hart KM, Chauhan P, et al. FOXP3-miR-146-NF-kappaB Axis and Therapy for Precancerous Lesions in Prostate. Cancer research. 2015;75:1714–24. doi: 10.1158/0008-5472.CAN-14-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu R, Liu C, Chen D, Yang WH, Liu X, Liu CG, et al. FOXP3 Controls an miR-146/NF-kappaB Negative Feedback Loop That Inhibits Apoptosis in Breast Cancer Cells. Cancer research. 2015;75:1703–13. doi: 10.1158/0008-5472.CAN-14-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer research. 2008;68:927–36. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 35.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22:2790–9. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 36.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer cell. 2002;1:203–9. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 37.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nature reviews. 2006;6:909–23. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Q, Rammohan K, Lin S, Robinson N, Li O, Liu X, et al. CD24 is a genetic modifier for risk and progression of multiple sclerosis. Proc Natl Acad Sci U S A. 2003;100:15041–6. doi: 10.1073/pnas.2533866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Lin S, Rammohan KW, Liu Z, Liu JQ, Liu RH, et al. A dinucleotide deletion in CD24 confers protection against autoimmune diseases. PLoS Genet. 2007;3:e49. doi: 10.1371/journal.pgen.0030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Liu R, Li D, Lin S, Fang X, Backer G, et al. A hypermorphic SP1-binding CD24 variant associates with risk and progression of multiple sclerosis. Am J Transl Res. 2012;4:347–56. [PMC free article] [PubMed] [Google Scholar]
- 42.Isaacs WB, Carter BS, Ewing CM. Wild-type p53 suppresses growth of human prostate cancer cells containing mutant p53 alleles. Cancer research. 1991;51:4716–20. [PubMed] [Google Scholar]
- 43.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–12. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 44.Warfel NA, El-Deiry WS. p21WAF1 and tumourigenesis: 20 years after. Curr Opin Oncol. 2013;25:52–8. doi: 10.1097/CCO.0b013e32835b639e. [DOI] [PubMed] [Google Scholar]
- 45.Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–8. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 46.Bykov VJ, Zache N, Stridh H, Westman J, Bergman J, Selivanova G, et al. PRIMA-1(MET) synergizes with cisplatin to induce tumor cell apoptosis. Oncogene. 2005;24:3484–91. doi: 10.1038/sj.onc.1208419. [DOI] [PubMed] [Google Scholar]
- 47.Martin DN, Starks AM, Ambs S. Biological determinants of health disparities in prostate cancer. Curr Opin Oncol. 2013;25:235–41. doi: 10.1097/CCO.0b013e32835eb5d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lehmann BD, Pietenpol JA. Targeting mutant p53 in human tumors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:3648–50. doi: 10.1200/JCO.2012.44.0412. [DOI] [PubMed] [Google Scholar]
- 49.Lehmann S, Bykov VJ, Ali D, Andren O, Cherif H, Tidefelt U, et al. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:3633–9. doi: 10.1200/JCO.2011.40.7783. [DOI] [PubMed] [Google Scholar]
- 50.Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer research. 2000;60:6788–93. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.