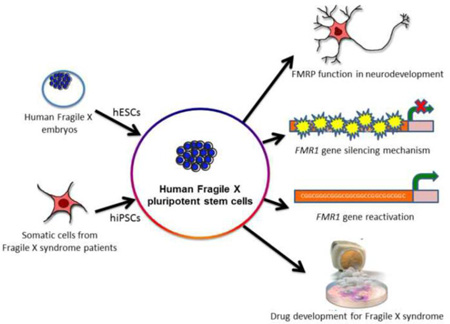
Graphical abstract
I. Introduction
Fragile X syndrome (FXS) is the most common inherited cause of intellectual disability with a prevalence of 1 in 5000 (Coffee et al., 2009). Characteristics include learning deficits and IQ between 20 and 60 as well as hyperactivity, attention deficit disorder, and autistic-like behavior (Hagerman and Hagerman, 2002). At least 25% of individuals with FXS meet the diagnostic criteria for autism (Cohen et al., 1991; Fisch et al., 1986; Hagerman et al., 2005; Hatton et al., 2006; Kaufmann et al., 2004; Lathe, 2009; Reiss et al., 1986). Individuals with FXS also have an increased incidence of seizures and reduced motor coordination. FXS features are not limited to the nervous system and include connective tissue dysplasia, facial dysmorphia, hyperextensible joints, mitral valve prolapse and macro-orchidism.
FXS is caused by a mutation in a single gene, the Fragile X Mental Retardation Gene 1 (FMR1), resulting in lack of the Fragile X Mental Retardation Protein (FMRP) (Pieretti et al., 1991; Verkerk et al., 1991). As in many neuropsychiatric disorders, animal models have been useful in studying characteristics and potential mechanisms underlying FXS. FXS models have shown that FMRP is an RNA binding protein that binds to specific mRNAs to control their location and protein translation (Eberhart et al., 1996). This function implies that FMRP plays a crucial role in neuronal development, function and synaptic plasticity (Liu-Yesucevitz et al., 2011; Sidorov et al., 2013). The absence of FMRP results in increased protein synthesis, leading to enhanced signaling in a number of intracellular pathways, including the mTOR, mGLuR5, ERK, Gsk3β, PI3K, and insulin pathways. Studies in animal models demonstrate that FMRP also plays a crucial role in neuronal development. FMRP deficiency leads to fate switch of neural precursor cells from neuron to glia lineages and increased death of immature neurons, leading to reduced neuronal production (Guo et al., 2011; Guo et al., 2012b; Luo et al., 2010). FMRP-deficient neurons also exhibit impaired morphological development of neuronal dendrites and spines. Data from mouse models has informed our understanding of FXS and several drug trials have been instituted in FXS patients as a direct result of these studies.
Yet, there are several critical reasons why it is necessary to use human cells to define underlying mechanisms that lead to FXS characteristics, particularly those affecting the nervous system. First, the epigenetic silencing of the FMR1 gene that causes FXS occurs only in human. The causal mutation in FXS is a trinucleotide CGG repeat expansion. When the mutational expansion of the CGG repeats exceeds 200 in humans, it leads to methylation of the repeats and the FMR1 promoter, chromatin condensation, and a loss of FMRP expression. Mice engineered to mimic the human mutation in the FMR1 gene do not show methylation and silencing characteristic of the gene in humans (Brouwer et al., 2007). These results indicate that epigenetic mechanisms in human and mice are different and preclude the ability to study epigenetic mechanisms of FMR1 silencing in mouse models of FXS.
Differences between the formation and structure of the brain in mice and humans also present challenges to understanding the mechanisms of abnormal brain development and function in FXS. The formation of the brain is prolonged in humans, taking months compared to weeks in mice. More importantly, the human brain is more reliant on the role of interneurons and astrocytes and so FXS mouse models may not adequately reveal differences in these particular systems. For example, human interneuron development occurs over a protracted period of time and integrates unique mechanisms to generate more numerous and more elaborate interneurons (Hansen et al., 2013; Hansen et al., 2010; LaMonica et al., 2012; Lui et al., 2011; Marin, 2013; Tyson and Anderson, 2013). Thus, it is important to study the cause and consequences of FMR1 silencing in neural development and function in the human context.
II. FXS is caused by a human specific mutation
II. A. Epigenetic regulation of gene expression
Epigenetic mechanisms, mediated by DNA methylation, histone modification, and noncoding RNAs are known to play significant roles in regulating stem cells and development as well as adult neuroplasticity (Jobe et al., 2012).
II. A. 1. DNA methylation
DNA methylation is catalyzed by methyl transferases including de novo Dnmt3a and Dnmt3b that add methyl groups onto unmethylated DNA and Dnmt1 that recognizes hemi-methylated DNA and maintains DNA methylation. A majority of genomic DNA methylation, particularly in the brain, is at cytosine residues in the context of CpG dinucleotide (mCG), with additional methylation at non-CpG sites (mCA, mCT, mCC or collectively called mCH). Genome-wide DNA methylation studies have demonstrated a drastic increase in DNA methylation levels in neurons during postnatal development that coincides with neuronal maturation, suggesting a critical role for DNA methylation during neuronal development. Active DNA demethylation involves multi-step chemical reactions by several groups of proteins and through the production of 5-hydroxymethylation of cytosine (5hmC) (Piccolo and Fisher, 2014).
II. A. 2. Histone modification
Chromatin comprised of nucleosome repeats of 147 base pairs (bp) of DNA sequence wrapped around two copies each of histone proteins, H2A, H2B, H3 and H4 can exist in either highly condensed heterochromatin associated with gene silencing or loosely packed euchromatin associated with gene expression. The amino (N)-terminal tails of core histones are subject to a variety of covalent modifications including acetylation, methylation, ubiquitination, phosphorylation, ribosylation, SUMOylation, etc. The combination of these histone modifications are called "the histone code" and binding of modified histones to specific genomic regions control the activation or repression of the associated genes (Bernstein et al., 2007). Some histone modifications like acetylation of lysine 9 and 14 (Ace-H3) and di- or tri-methylation of lysine 4 (H3K4) are signatures of actively expressed chromatin and referred to as “active histone marks”. Other marks, such as di- or tri-methylation of lysine 9 (H3K9) or lysine 27 (H3K27) on histone H3 are associated with silent chromatin domains and are referred to as “repressive histone marks”. These histone modifications are catalyzed by enzymes such as histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyl transferases (HMTs), and histone demethylases (HdMTs).
II. A. 3. Non-coding RNAs
A large portion of the genome is transcribed into non-protein-coding RNA called noncoding RNA (ncRNA). Among them, the best studied are small ncRNAs including microRNAs. Recently, the involvement of long ncRNAs (lncRNAs) is increasingly recognized as an important aspect of regulation (Jobe et al., 2012).
All three epigenetic mechanisms are, to some extent, involved in FMR1 gene expression and shutdown. DNA methylation of the expanded CGGs in the FMR1 gene is the major, if not only, cause of FMR1 gene inactivation in FXS. Associated with this striking DNA methylation change, histone marks in the FMR1 gene locus also shift from an active to a repressive state. Several lncRNAs within the FMR1 gene locus also undergo inactivation, but it is unclear how much they contribute to FXS and whether they are involved in the gene inactivation process. As detailed below, extensive efforts have been devoted to understanding their roles in FMR1 gene silencing and activation so as to better understand the underlying cause of FXS.
II. B. FMR1 Gene Silencing
The causal mutation in FXS is a trinucleotide CGG repeat expansion in the FMR1 gene. Expansion of the CGG repeats over 200 leads to FMR1 silencing and is thought to occur about 11 weeks of gestation in vivo (Willemsen et al., 2002).
II. B. 1. CGG repeats and FMR1 silencing
CGG trinucleotide repeats are normally present in the FMR1 gene of all humans. Population genetics studies have shown that the modal number of CGG repeats is 30, with significant numbers of individuals with repeats both below and above this number (Mailick et al., 2014). In most people with the number of near modal CGG repeat numbers, the CGG repeat usually remains stable through generations. However, due to reasons that are not understood, the CGG repeats sometimes expand through the germ line leading to CGG repeat lengths between 55 to 200, termed pre-mutation. Once considered unaffected, individuals with a pre-mutation FMR1 gene have more recently been shown to exhibit some mild to moderate pathological presentations termed Fragile X-associated disorders, even though FMR1 gene transcription is not reduced (Lozano et al., 2014). These disorders include premature ovarian insufficiency (POI) and Fragile X-associated tremor/ataxia syndrome (FXTAS). Mutational expansion of the CGG repeats beyond 200 triggers methylation of the repeats and the FMR1 promoter, chromatin condensation, and a loss of transcription, resulting in FXS. The mechanism that prompts FMR1 gene inactivation remains unclear and is an active field of study.
The importance of FMR1 methylation in gene silencing is illustrated by the existence of individuals who are mosaic for FMR1 gene methylation. These FXS “methylation mosaics” have an absence of methylation in a subpopulation of cells, reduced methylation in different cells, or absence of methylation in one allele in females (Stoger et al., 2011). As described in Section V, rare males with FMR1 full-length CGG expansion mutations show no or only mild symptoms because their CGG and FMR1 gene are unmethylated (Hagerman et al., 1994; Loesch et al., 1993; Loesch et al., 2004; Loesch et al., 2012).
II. B. 2. Genetic and epigenetic signatures of FMR1
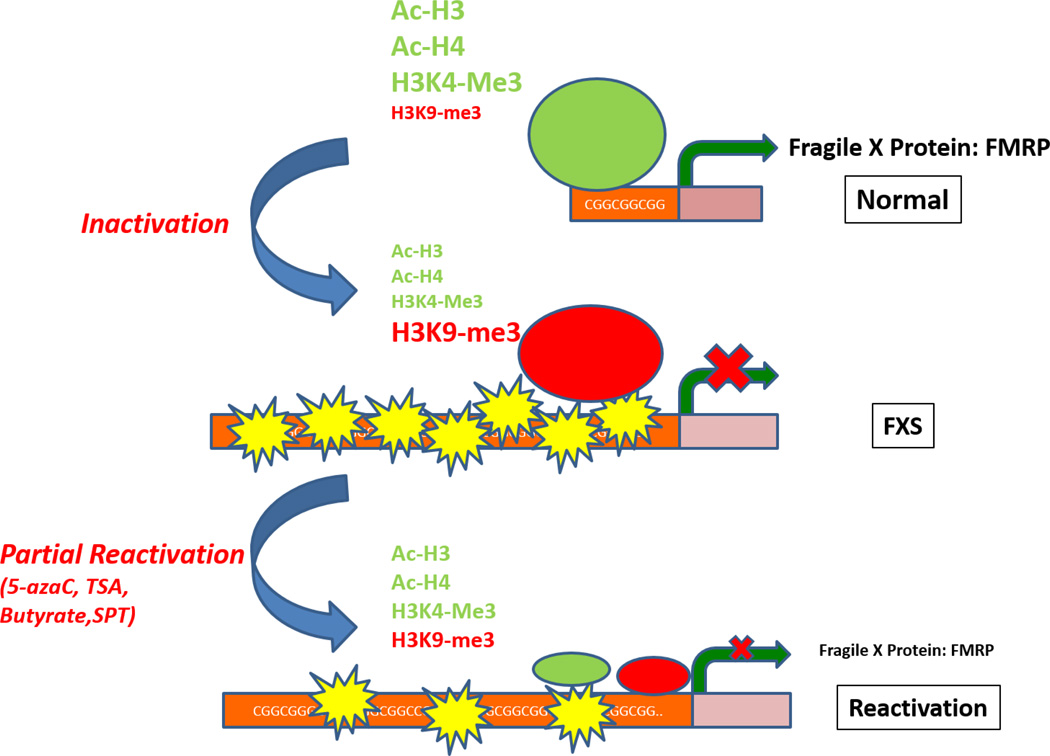
Extensive studies have compared the genetic and epigenetic signatures of active and repressed FMR1 genes (Figure 1). DNA footprinting studies have identified four footprints in the FMR1 gene promoter that correspond to the consensus binding site of four transcription factors, α-PAL/NRF1, Sp1, H4TF1/Sp1-like, and c-myc. These footprints are found in FMR1 in several different cell types derived from normal individuals but absent in FMR1 of cells derived from FXS individuals (Drouin et al., 1997; Schwemmle, 1999; Schwemmle et al., 1997). Therefore FMR1 gene repression is correlated with the absence of transcription factor binding. Drouin et al., also made an interesting observation that the same footprint sites are present in both FMR1 gene and the gene of huRNP-A2, a ribonucleoprotein. The authors predicted that FMRP might have similar a RNA transport function as huRNA-A2. This function for FMRP was later confirmed by FMRP regulation of activity-dependent RNA transport during neuronal dendritic spine development (Dictenberg et al., 2008).
Figure 1. Epigenetic profiles of the human FMR1 gene.
The FMR1 gene promoter in normal cells is unmethylated and is enriched with active chromatin markers such as acetylated Histone H3 (Ac-H3), acetylated Histone H4 (Ac-H4), trimethylated H3 at lysine 4 (H3K4-me3) but low in inhibitory chromatin markers such as trimethylated histone H3 at lysine 9 (H3K9-me3). The FMR1 gene promoter in cells from FXS patients is methylated and is enriched with inactive chromatin marker such as H3K9-me3 but low in active chromatin markers. Treatment with DNA methyltransferase inhibitors such as 5azaC or HDAC inhibitors such as trichostatin A (TSA), butyrate or splitomicin (SPT) can partially change the histone signature and reactivate the FMR1 gene.
Using “3C” chromosome conformation analysis of a 170 kb locus encompassing the human FMR1 gene, Gheldof et al., discovered a significant difference in chromosome conformation between FMR1-expressing versus non-expressing cells. The FMR1 gene promoter is at the center of a 50 kb chromosome domain exhibiting less interactions among each other in FMR1-expressing cells compared to in FXS cells (Gheldof et al., 2006). These results suggest that silencing of the FMR1 gene is associated with broader changes at the chromosome level than previously anticipated.
Several studies have compared epigenetic signatures of active versus repressed FMR1 promoters (Figure 1). Chromatin immunoprecipitation studies have shown that in normal cells with FMR1 gene transcription, the 5’ region of FMR1 gene is associated with acetylated H3 and H4 that are associated with actively transcribed genes (Coffee et al., 1999). However in FXS cells, there is a significantly reduced level of acetylated H3 or H4 associated of the FMR1 gene. The levels of acetylated H3 on the FMR1 promoter are inversely correlated with repeat sizes (Coffee et al., 2002). In addition, H3K4 methylation is decreased in FXS cells whereas H3K9 methylation is increased, consistent with the inactive status of FMR1 gene in FXS cells. These changes in histone modifications are restricted to FMR1 promoter regions (Gheldof et al., 2006). Furthermore, the silencing of FMR1 gene in human embryonic stem cells is associated with loss of active chromatin markers including H3K4me2 and gain of H3K9me3 (Avitzour et al., 2014). However, studies of full mutation males with an unmethylated FMR1 promoter show increase in deacetylated H3 and H4 and methylated H3K9 (Pietrobono et al., 2005; Tabolacci et al., 2008b) suggesting that histone modification might be independent of DNA methylation in FMR1 gene shutdown. The seemingly contradictory messages from these studies suggest increasingly complex mechanisms regulate FMR1 gene expression. To that point, FMRP has been shown to interact with microRNAs (Liu et al., 2014) and the human FMR1 gene locus encodes several lncRNAs (Pastori et al., 2014; Peschansky et al., 2015). Further other genes such as FAM 11A (Shaw et al., 2002) and several long noncoding RNAs such as antisense FMR1 (ASFMR1) (Ladd et al., 2007) and FMR6 (Pastori et al., 2014) in the locus are also methylated and silenced in addition to FMR1.
II. B. 3. Potential mechanisms of FMR1 silencing
Several potential mechanisms underlying FMR1 gene inactivation have been proposed and investigated.
A DNA methylation boundary was discovered upstream of the FMR1 gene at about 685 to 800 nucleotides from the CGG repeats (Naumann et al., 2009) and several nuclear proteins, including insulator protein CTCF, bind to this region. Binding of CTCF to this region is lost in FXS cells with FMR1 silenced (Ladd et al., 2007). A later report (Lanni et al., 2013) confirmed that CTCF binding is needed to prevent FMR1 gene silencing, but not to prevent general DNA methylation. The authors speculate that CTCF may function by modulating chromosome conformation. Another study using human embryonic stem cells suggests that changes in histone markers precede DNA methylation changes (Eiges et al., 2007). Further, CGG containing FMR1 mRNA was found to inhibit its own expression. Demethylation of the FMR1 promoter leads to increased repressive chromatin marker H3K27 methylation binding to the promoter that is dependent on the presence of mutant mRNA (Kumari and Usdin, 2014). Recently, FMR1 silencing was shown to be mediated by FMR1 mRNA containing long CGG repeats (Colak et al., 2014). Therefore, several mechanisms may be at play in silencing FMR1 expression.
Recapitulation of the FMR1 silencing mutation has been unsuccessful in animal models, particularly mouse, because epigenetic silencing does not occur in the same way. Therefore, it is necessary to use human cells to define the mechanisms of FMR1 silencing.
III. Human pluripotent stem cells as a model to study FXS
Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and particularly induced PSCs (hiPSCs), offer a model system to reveal cellular and molecular events underlying normal and abnormal neural development. hESCs are isolated from preimplantation embryos and retain the two characteristics unique to stem cells: self-renewal and pluripotency (Thomson et al., 1998). hiPSCs are reprogrammed from somatic cells by forced expression of stem cell genes and have the characteristics of hESCs (Takahashi et al., 2007; Yu et al., 2007). Patient-derived hiPSCs provide a paradigm to understand neurological disease pathogenesis, including FXS, in the human genetic background.
While providing an unparalleled tool for the study of early human brain development, there are multiple factors in hPSC studies that introduce variability and affect the ability to compare data from multiple studies. Variability can be introduced through patient differences, iPSC reprogramming methods, and neuronal differentiation paradigms. Many of these problems can be overcome by using cells from enough different individuals to enable statistically meaningful results. Alternatively, either engineered or spontaneously-generated isogenic cell lines can provide a more practical alternative to limit genetic diversity.
III. A. Embryonic Stem Cells
FXS hESCs were first isolated and reported from preimplantation embryos carrying the FXS mutation by Verlinsky in 2005 (Verlinsky et al., 2005). The heritability of FXS enables the identification of affected embryos through the use of preimplantation genetic diagnosis during the in vitro fertilization process. hESCs can be isolated from the inner cell mass (ICM) of these embryos for research purposes (Ben-Yosef et al., 2008; Kuliev et al., 2005; Pickering et al., 2003; Stephenson et al., 2009). Since this initial report, other FXS hESCs have been published (Eiges et al., 2007; Gerhardt et al., 2014). Yet only a very few FXS hESC lines are approved and listed on the NIH Human Embryonic Stem Cell Registry, thereby hindering the use of these cells by NIH-funded researchers.
Eiges et al., provided the first detailed characterization of a single human FXS hESC line (Eiges et al., 2007). The data confirmed that the cells derived from a FXS preimplantation embryo met the criteria for hESCs and that the cells retained the full length mutation of the FMR1 gene. Surprisingly, however, the FMR1 gene was unmethylated and expressed in these cells. Epigenetic silencing did not occur until these cells were differentiated and thus provided a new paradigm in which to study the mechanisms of FMR1 silencing (Figure 2). More recent evidence from multiple FXS hESC lines, however, suggests that the epigenetic silencing in FXS hESCs may occur more easily than initially observed (Avitzour et al., 2014). These results also suggest that the initial embryonic ICM cells may have different methylation of the FMR1 gene at the time of stem cell derivation. Therefore, epigenetic silencing can occur in the undifferentiated state. As more human FXS hESCs are reported and characterized, the prevalence of this phenomenon will be revealed.
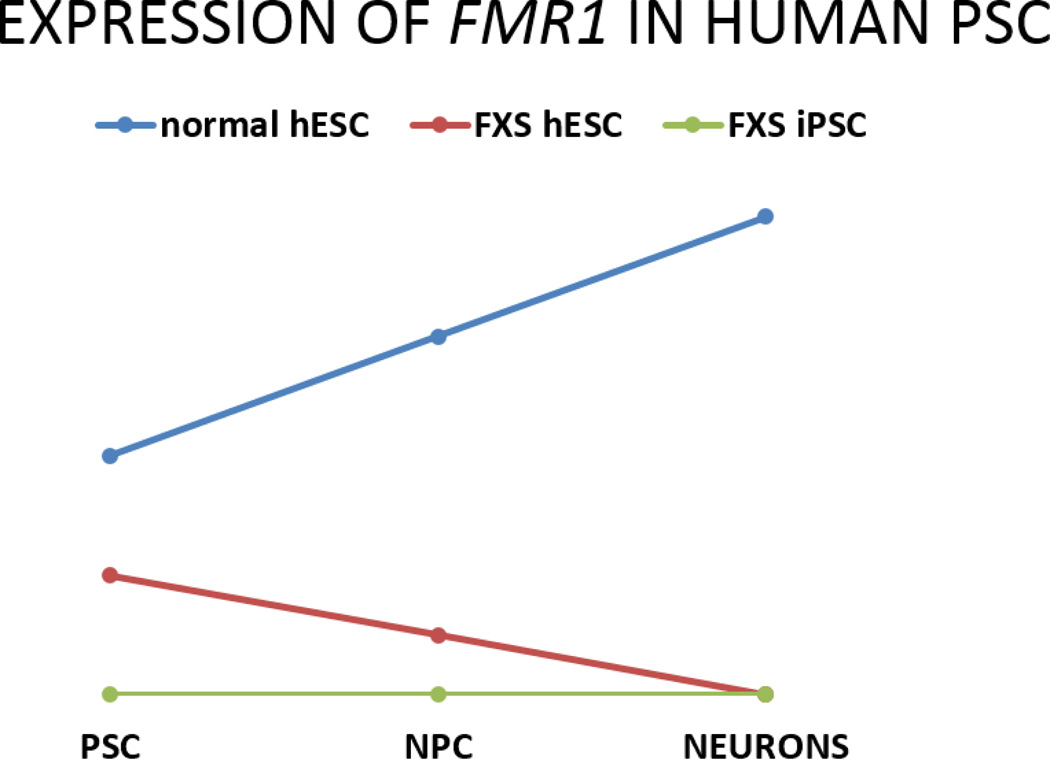
Figure 2. Neuronal differentiation-dependent FMR1 gene inactivation.
Graph depicts the FMR1 gene expression in hPSCs during neuronal differentiation. The FMR1 gene is expressed in normal hESCs and its expression levels increase during neuronal differentiation (blue line). FMR1 is expressed in FXS hESCs but at lower levels than normal due to partial inactivation and is gradually silenced during differentiation (red line). The FMR1 gene is silenced in FXS hiPSCs and throughout neural differentiation. (PSC=pluripotent stem cells, NPC=neural progenitor cells).
III. B. Induced pluripotent stem cells
The ability to generate hiPSCs from somatic cells of FXS individuals has enabled the generation of FXS hiPSC lines from patient fibroblasts (Bar-Nur et al., 2012; Brick et al., 2014; de Esch et al., 2014; Doers et al., 2014; Halevy et al., 2015; Kaufmann et al., 2015). Without exception, the methylated, silenced FMR1 mutation in the patient fibroblasts is retained through the reprogramming process (Urbach et al., 2010). Reprogramming also causes a rare unmethylated full mutation in patient somatic cells to be silenced (de Esch et al., 2014). Therefore, it is not yet possible to isolate full mutation FXS hiPSCs that have the unsilenced gene (Figure 2). These cells are useful to test mechanisms of epigenetic reactivation. These cells are also valuable for studying the effect of FMR1 loss as cells differentiate from the undifferentiated state.
IV. Neural differentiation of FXS hPSCs
Most of what we know about the role of FMRP in neural development comes from FXS mouse models. FMRP is an RNA binding protein that binds to specific mRNAs to control the location and protein translation of these mRNAs (Eberhart et al., 1996). FMRP plays a crucial role in neuronal development, function and synaptic plasticity (Liu-Yesucevitz et al., 2011; Sidorov et al., 2013). How the loss of FMRP manifests in the human nervous system is unknown. Although higher density but immature long and thin neuronal dendritic spines are consistently found in FXS postmortem brains (Hinton et al., 1991; Irwin et al., 2001; Wisniewski et al., 1991), the underlying mechanisms of this phenotype is not well defined. Mouse models have revealed that neural cells that lack FMRP exhibit neurogenesis and neuronal maturation deficits (Li and Zhao, 2014). Further, altered synaptic plasticity has been established in the FXS mouse model and is thought to be due to the lack of FMRP’s role as a negative regulator of translation (Liu-Yesucevitz et al., 2011). It is important to define whether human FXS neural cells have similar deficits so that therapeutics to affect neural development and function in FXS can be more intelligently designed.
IV. A. Neural differentiation from hPSCs
To take advantage of the power of hPSCs to model human brain development and to define the steps that go awry in FXS, it is critical to differentiate hPSCs into the specific neural subtypes that are affected in FXS (Kim et al., 2014a). This likely includes all neural subtypes, but hPSC neural research in general is focused on cortical excitatory neurons and astrocytes, with emerging work on interneurons and other glial subtypes. The differentiation of neural cells from hPSCs was pioneered by Su-Chun Zhang beginning with the initial report of neuron differentiation from hESCs (Zhang et al., 2001). The Zhang lab and others developed core methods for the generation of dorsal forebrain derived (cortical-like) neurons (Chambers et al., 2009; Eiraku et al., 2008; Pankratz et al., 2007; Watanabe et al., 2005). These methods are effective for hiPSCs as well although there is variable efficiency between different hiPSC lines that can affect the interpretation of results (Hu et al., 2010). Neurons generated through these and similar methods neurons have characteristics that correspond primarily to excitatory projection neurons in the cortex (Hansen et al., 2011; Mariani et al., 2012; Shi et al., 2012). Therefore they are of value in the study of FXS, where dendritic spines of excitatory neurons are altered. Other neuronal subtypes as well as astrocytes (Emdad et al., 2012; Krencik et al., 2011; Ruiz et al., 2010; Shaltouki et al., 2013) and oligodendrocytes (Czepiel et al., 2011; Hu et al., 2009; Liu et al., 2011; Pouya et al., 2011; Sharp et al., 2011) can be differentiated from hPSCs. More recently, the directed differentiation of interneurons from hPSCs has been accomplished (Kim et al., 2014b; Liu et al., 2013; Maroof et al., 2013; Nicholas et al., 2013), thus providing the ability to generate a range of human neural subtypes for disease modeling.
FMRP is expressed in all neural cells (Bakker et al., 2000; Devys et al., 1993; Willemsen et al., 2004), so the characterization of many cells would be valuable to better understand FXS. While the focus of FXS neurobiology has been on excitatory neurons, emerging evidence suggests that inhibitory neurons are also dysfunctional in FXS (Cea-Del Rio and Huntsman, 2014). Astrocytes express FMRP and evidence from mouse suggests that the lack of FMRP in astrocytes may be detrimental (Jacobs et al., 2012; Jacobs et al., 2010; Pacey and Doering, 2007). Recent human neuroimaging studies suggest that individuals with FXS have white matter defects that may link development and function of oligodendrocytes to FXS neuropathology (Green et al., 2015; Villalon-Reina et al., 2013)
IV. B. Neural differentiation from FXS hPSCs
Despite the establishment of FXS hESCs and hiPSCs, there is relatively limited data on the development and phenotypic characterization of human FXS neurons derived from PSCs and no reports of glial cells specifically differentiated from FXS hPSCs.
The most comprehensive description of the neural development from FXS PSCs comes from hESCs by Dalit Ben-Yosef’s lab, the first to characterize FXS hESC-derived neural cells (Telias et al., 2013). The authors differentiated three FXS hESC lines into neurons and assessed neuronal development and function. The differentiation paradigm was fairly standard, although the subtype of neurons generated was neither defined nor optimized and may have been a ventrally derived neuronal subtype given the addition of sonic hedgehog (SHH), a ventralizing morphogen. The study showed that the FXS hESCs expressed less FMR1 as they differentiated, consistent with the previous report (Eiges et al., 2007). FXS hESCs, when differentiated, had reduced expression of neural induction genes and a delay in neurogenesis, although neurons could be generated. The neurons had deficits in neuronal maturation as evidenced by decreased neuronal gene expression, increased progenitor gene expression and immature electrophysiological properties. The authors conclude from their study that FXS hESCs can successfully differentiate into neurons in vitro and FMR1 silencing mimics that during embryogenesis.
Only limited studies to date have addressed the neuronal development of human neurons derived from FXS iPSCs (Doers et al., 2014; Halevy et al., 2015; Sheridan et al., 2011). Similar to results from FXS hESCs, dorsal forebrain neurons can be generated from FXS hiPSCs, although progenitor characteristics were not well-defined in any study. The gene expression patterns of hiPSC-derived FXS neurons suggest defects in neuronal differentiation (Sheridan et al., 2011) and maturation (Halevy et al., 2015) similar to what have been shown in mouse models (Guo et al., 2011; Guo et al., 2012a; Guo et al., 2015). We have found that hiPSC-derived FXS neurons exhibit defective neurite outgrowth (Doers et al., 2014). A recent study using a novel micro-raft culture method, showed that FXS hiPSC-derived neurons exhibit reduced pre-synaptic vesicle recycling (Niedringhaus et al., 2015), recapitulating what has been shown in mouse models (Deng et al., 2011).
Therefore, the limited studies using human FXS hESCs and hiPSCs suggest that FMRP is important for neural precursor differentiation and neuronal maturation. Further experiments are acutely needed on more human hPSCs lines to delineate: 1) specific phenotypes in neural progenitors (e.g. cell cycle abnormalities, cell death); 2) phenotypes in differentiation (e.g. developmental delay, fate switch); 3) phenotypes affecting synaptic development and synaptic plasticity; and 4) electrophysiological properties of mature human FXS neurons. The achievement of these goals will enable critical studies to dissect the mechanisms of FMRP’s actions in human cortical neurons.
V. Reactivation of the FMR1 gene in hPSCs
V. A. Rationale for FMR1 gene restoration as a potential therapy
Since the coding sequence of the silenced FMR1 gene is normal, a possible therapeutic strategy is to restore the transcription of FMR1 in FXS (Figure 1). In fact, there are now several reports of males with FMR1 full-length CGG expansion mutations who show no or mild symptoms because their FMR1 genes are unmethylated (Hagerman et al., 1994; Loesch et al., 1993; Loesch et al., 2004; Loesch et al., 2012; Pietrobono et al., 2005; Smeets et al., 1995; Tabolacci et al., 2008b). Therefore an unmethylated FMR1 gene carries out normal functions resulting in near normal intelligence instead of intellectual disability associated with FXS.
The mouse genetics studies shed more hope of this notion. We have discovered that restoring FMR1 in adult-born new neurons using inducible genetics restore several adult neurogenesis-dependent learning and memory in mice (Guo et al., 2011). These data suggest that the neuronal developmental deficits seen in FMRP-deficient neurons might be reversible.
V. B. FMR1 gene restoration strategies
The identification of effective methods to reactivate the FMR1 gene and restore FMRP expression has been extremely challenging. A number of studies have shown that treatment of human FXS lymphoblastoid cell lines with a DNA methyltransferase (DNMT) inhibitor 5-azacytidine (5azaC) or 5-azadeoxycytidine (5azadC) results in partial reactivation of the FMR1 gene and FMRP expression. Upon treatment, the FMR1 promoter becomes passively unmethylated through cell division (Chiurazzi et al., 1998; Pietrobono et al., 2002). Importantly, the increase in FMR1 mRNA production is associated with increased active chromatin marker binding and decreased repressive chromatin marker binding to the FMR1 promoter (Brendel et al., 2013; Kumari and Usdin, 2014; Pietrobono et al., 2002; Tabolacci et al., 2008a) (Figure 1). In contrast, methotrexate, a folate antagonist that acts by inhibiting dihydrofolate reductase (DHFR) and has some DNA methylation inhibition activity does not reduce DNA methylation in the FMR1 promoter. DHFR does lead to some mRNA but not protein expression (Brendel et al., 2013). These results suggest that reversing DNA methylation of CGG repeats might be a promising method for gene restoration therapy.
Chemicals affecting histone modification have also been explored for FMR1 reactivation. Most studies have so far focused on Class I, II, and IV HDAC inhibitors include butyrate and trichostatin A (TSA). Using human FXS lymphoblastoid cell lines, Chiurazzi et al., have shown that treatment with HDAC inhibitors, phenylbutyrate, sodium butyrate and TSA, leads to moderately increased FMR1 gene transcription, yet less than compared to the effect of 5azaC or 5-azadC (Chiurazzi et al., 1999). However other studies show no FMR1 transcription after TSA treatment (Coffee et al., 2002; Coffee et al., 1999) or VPA treatment (Tabolacci et al., 2008a). Interestingly, combined treatment with 5azaC and HDAC inhibitor leads to 2–5 fold higher reactivation compared to 5azaC treatment alone (Chiurazzi et al., 1999). Recently, more effective reactivation, comparable to that by 5azaC, has been achieved by using splitomicin (SPT), an inhibitor of SIRT1 a class III HDAC (Biacsi et al., 2008; Kumari and Usdin, 2014). Knockdown of SIRT1 in either lymphoblastoid cell lines or FXS patient-derived fibroblasts leads to increased deacetylation of H4K16 and increased FMR1 gene transcription without significantly affecting DNA methylation. This study suggests that inhibition of certain key HDACs may be able to reactivate the FMR1 gene without altering DNA methylation.
The limited success of using known epigenetic reagents to reactivate FMR1 genes has prompted studies to explore novel chemical reagents and molecules to reactivate the FMR1 gene. A major challenge is to establish a screening method that can effectively and efficiently report FMR1 gene expression. Several reports of screening technologies have been recently published. In one study, FXS hiPSCs were differentiated into neural progenitor cells (NPCs), immunostained with an FMRP antibody, and analyzed by high content imaging for FMRP levels (Kaufmann et al., 2015). Using this system 50,000 compounds covering epigenetic targets and known FMRP regulated pathways were screened and several compounds (identity not revealed) that induced weak reactivation were identified (Kaufmann et al., 2015). In another screen, dual FMRP antibodies were used to establish a time-resolved fluorescence resonance energy transfer (TR-FRET) dual antibody assay to increase specificity. Human iPSC-derived NPCs were used to screen ~5000 compounds including a FDA-approved drug library. Six hits were identified that enhanced FMR1 gene transcription modestly, although no significant FMRP was detected (Kumari et al., 2015). Interestingly, one of the identified compounds is SB216763 that we have previously found to rescue learning deficits in FMR1-null mice through enhancing Wnt signaling (Guo et al., 2012a). Yet, none of the compounds identified so far can reactivate FMR1 expression to near normal levels, necessitating new and better strategies.
The discovery and rapid development of gene editing technology have opened new avenue for gene correction-based therapies. The Zinc Finger Protease (ZFN) and Transcription activator-like effector nucleases (TALENs) methods, though promising, were difficult to use and had relatively low specificity (Hsu et al., 2014). Nevertheless, proof of concept experiments have shown that TALEN can be used to correct AT-rich repeats in FATS a common fragile site in mice (Ma et al., 2014). The newest CRISPR/Cas9-based gene editing method is significantly easier to use and exhibits much higher specificity (Zhang et al., 2014). Recently, Park et al used CRISPR/Cas9 to delete the CGG repeats in the silenced FMR1 gene in FXS hiPSCs and demonstrated activation of FMR1 gene (Park et al., 2015). Although the efficiency of this deletion is extremely low and the deletion is not restricted to the CGG repeat, the results of this work suggest that deletion of silenced CGG repeat might be a promising gene reactivation strategy for FXS. In addition to deletion of CGG, Cas9 can also be used to deliver a transcriptional activator (e.g.VP64) to specific silenced genes to reactivate them (Scott et al., 2014). With the fast advancement in gene editing and gene therapy, reactivation of FMR1 using genetic methods may become feasible in the near future.
VI. Future Directions
Although much work has been done using FXS mouse models, how FMRP regulates human neurogenesis and neuronal development remains unclear. In addition, the FMR1 gene inactivation and reactivation studies have not been successfully translated in vivo. These hurdles are due to several reasons. First and foremost, the most-widely used mouse model of FXS has a knockout of the FMR1 gene rather than a repeat expansion in the gene, rendering it useless for studies of reactivation. Additionally, mice engineered with expanded repeats in the FMR1 gene do not show methylation and silencing characteristic of the gene in humans (Brouwer et al., 2007; Ludwig et al., 2014). Further, some of the demethylating agents can be toxic and need to be targeted to the brain where FMRP expression is most relevant. Lastly, DNMT inhibitors inhibit DNA methylation of newly synthesized DNA during cell division; therefore their effect on post-mitotic neurons is unclear. Therefore an in vivo model that recapitulates the pathology of human FXS neurodevelopment will be necessary for testing new therapeutic reactivation strategies. In addition, gene-specific reactivation of FMR1 using novel gene editing methods may provide good alternatives for chemical-based reactivation. However delivery of these genetic reagents to human brains presents major hurdles clinically. Lastly, the questions remain whether certain pathways and functions can be restored after the critical period. Future research in these areas will shed light on both basic mechanisms and therapeutic potentials for FXS.
Highlights.
Fragile X syndrome (FXS) is characterized by intellectual disability and autism.
Human specific epigenetic silencing of the FMR1 gene causes FXS.
Human pluripotent stem cells are a model to study human neural development in FXS.
Human FXS pluripotent stem cells provide insight into FMR1 epigenetic silencing.
Acknowledgements
This work was supported by grants from FRAXA and the John Merck Fund (A.B. and X.Z), the NIH (R01MH080434, R01MH078972, and R21NS095632 to X.Z., 1R21HD085288 to A.B.), and a core grant to the Waisman Center from the NICHD (P30 HD03352).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avitzour M, Mor-Shaked H, Yanovsky-Dagan S, Aharoni S, Altarescu G, Renbaum P, Eldar-Geva T, Schonberger O, Levy-Lahad E, Epsztejn-Litman S, et al. FMR1 epigenetic silencing commonly occurs in undifferentiated fragile X-affected embryonic stem cells. Stem Cell Reports. 2014;3:699–706. doi: 10.1016/j.stemcr.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker CE, de Diego Otero Y, Bontekoe C, Raghoe P, Luteijn T, Hoogeveen AT, Oostra BA, Willemsen R. Immunocytochemical and biochemical characterization of FMRP, FXR1P, and FXR2P in the mouse. Exp Cell Res. 2000;258:162–170. doi: 10.1006/excr.2000.4932. [DOI] [PubMed] [Google Scholar]
- Bar-Nur O, Caspi I, Benvenisty N. Molecular analysis of FMR1 reactivation in fragile-X induced pluripotent stem cells and their neuronal derivatives. J Mol Cell Biol. 2012;4:180–183. doi: 10.1093/jmcb/mjs007. [DOI] [PubMed] [Google Scholar]
- Ben-Yosef D, Malcov M, Eiges R. PGD-derived human embryonic stem cell lines as a powerful tool for the study of human genetic disorders. Mol Cell Endocrinol. 2008;282:153–158. doi: 10.1016/j.mce.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Biacsi R, Kumari D, Usdin K. SIRT1 inhibition alleviates gene silencing in Fragile X mental retardation syndrome. PLoS Genet. 2008;4:e1000017. doi: 10.1371/journal.pgen.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel C, Mielke B, Hillebrand M, Gartner J, Huppke P. Methotrexate treatment of FraX fibroblasts results in FMR1 transcription but not in detectable FMR1 protein levels. Journal of neurodevelopmental disorders. 2013;5:23. doi: 10.1186/1866-1955-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick DJ, Nethercott HE, Montesano S, Banuelos MG, Stover AE, Schutte SS, O'Dowd DK, Hagerman RJ, Ono M, Hessl DR, et al. The Autism Spectrum Disorders Stem Cell Resource at Children's Hospital of Orange County: Implications for Disease Modeling and Drug Discovery. Stem Cells Transl Med. 2014;3:1275–1286. doi: 10.5966/sctm.2014-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer JR, Mientjes EJ, Bakker CE, Nieuwenhuizen IM, Severijnen LA, Van der Linde HC, Nelson DL, Oostra BA, Willemsen R. Elevated Fmr1 mRNA levels and reduced protein expression in a mouse model with an unmethylated Fragile X full mutation. Exp Cell Res. 2007;313:244–253. doi: 10.1016/j.yexcr.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cea-Del Rio CA, Huntsman MM. The contribution of inhibitory interneurons to circuit dysfunction in Fragile X Syndrome. Front Cell Neurosci. 2014;8:245. doi: 10.3389/fncel.2014.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiurazzi P, Pomponi MG, Pietrobono R, Bakker CE, Neri G, Oostra BA. Synergistic effect of histone hyperacetylation and DNA demethylation in the reactivation of the FMR1 gene. Hum Mol Genet. 1999;8:2317–2323. doi: 10.1093/hmg/8.12.2317. [DOI] [PubMed] [Google Scholar]
- Chiurazzi P, Pomponi MG, Willemsen R, Oostra BA, Neri G. In vitro reactivation of the FMR1 gene involved in fragile X syndrome. Hum Mol Genet. 1998;7:109–113. doi: 10.1093/hmg/7.1.109. [DOI] [PubMed] [Google Scholar]
- Coffee B, Keith K, Albizua I, Malone T, Mowrey J, Sherman SL, Warren ST. Incidence of Fragile X Syndrome by Newborn Screening for Methylated FMR1 DNA. American journal of human genetics. 2009;85:503–514. doi: 10.1016/j.ajhg.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Zhang F, Ceman S, Warren ST, Reines D. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. Am J Hum Genet. 2002;71:923–932. doi: 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Zhang F, Warren ST, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nature genetics. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- Cohen IL, Sudhalter V, Pfadt A, Jenkins EC, Brown WT, Vietze PM. Why are autism and the fragile-X syndrome associated? Conceptual and methodological issues. Am J Hum Genet. 1991;48:195–202. [PMC free article] [PubMed] [Google Scholar]
- Colak D, Zaninovic N, Cohen MS, Rosenwaks Z, Yang WY, Gerhardt J, Disney MD, Jaffrey SR. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science. 2014;343:1002–1005. doi: 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czepiel M, Balasubramaniyan V, Schaafsma W, Stancic M, Mikkers H, Huisman C, Boddeke E, Copray S. Differentiation of induced pluripotent stem cells into functional oligodendrocytes. Glia. 2011;59:882–892. doi: 10.1002/glia.21159. [DOI] [PubMed] [Google Scholar]
- de Esch CE, Ghazvini M, Loos F, Schelling-Kazaryan N, Widagdo W, Munshi ST, van der Wal E, Douben H, Gunhanlar N, Kushner SA, et al. Epigenetic characterization of the FMR1 promoter in induced pluripotent stem cells from human fibroblasts carrying an unmethylated full mutation. Stem Cell Reports. 2014;3:548–555. doi: 10.1016/j.stemcr.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng PY, Sojka D, Klyachko VA. Abnormal presynaptic short-term plasticity and information processing in a mouse model of fragile X syndrome. J Neurosci. 2011;31:10971–10982. doi: 10.1523/JNEUROSCI.2021-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Developmental cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doers ME, Musser MT, Nichol R, Berndt ER, Baker M, Gomez TM, Zhang SC, Abbeduto L, Bhattacharyya A. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev. 2014;23:1777–1787. doi: 10.1089/scd.2014.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin R, Angers M, Dallaire N, Rose TM, Khandjian EW, Rousseau F. Structural and functional characterization of the human FMR1 promoter reveals similarities with the hnRNP-A2 promoter region. Hum Mol Genet. 1997;6:2051–2060. doi: 10.1093/hmg/6.12.2051. [DOI] [PubMed] [Google Scholar]
- Eberhart DE, Malter HE, Feng Y, Warren ST. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum Mol Genet. 1996;5:1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, Yaron Y, Eden A, Yanuka O, Benvenisty N, et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Emdad L, D'Souza SL, Kothari HP, Qadeer ZA, Germano IM. Efficient differentiation of human embryonic and induced pluripotent stem cells into functional astrocytes. Stem Cells Dev. 2012;21:404–410. doi: 10.1089/scd.2010.0560. [DOI] [PubMed] [Google Scholar]
- Fisch GS, Cohen IL, Wolf EG, Brown WT, Jenkins EC, Gross A. Autism and the fragile X syndrome. Am J Psychiatry. 1986;143:71–73. doi: 10.1176/ajp.143.1.71. [DOI] [PubMed] [Google Scholar]
- Gerhardt J, Tomishima MJ, Zaninovic N, Colak D, Yan Z, Zhan Q, Rosenwaks Z, Jaffrey SR, Schildkraut CL. The DNA replication program is altered at the FMR1 locus in fragile X embryonic stem cells. Mol Cell. 2014;53:19–31. doi: 10.1016/j.molcel.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheldof N, Tabuchi TM, Dekker J. The active FMR1 promoter is associated with a large domain of altered chromatin conformation with embedded local histone modifications. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12463–12468. doi: 10.1073/pnas.0605343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T, Barnea-Goraly N, Raman M, Hall SS, Lightbody AA, Bruno JL, Quintin EM, Reiss AL. Specific effect of the fragile-X mental retardation-1 gene (FMR1) on white matter microstructure. Br J Psychiatry. 2015 doi: 10.1192/bjp.bp.114.151654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Allan AM, Zong R, Zhang L, Johnson EB, Schaller EG, Murthy AC, Goggin SL, Eisch AJ, Oostra BA, et al. Ablation of Fmrp in adult neural stem cells disrupts hippocampus-dependent learning. Nat Med. 2011;17:559–565. doi: 10.1038/nm.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Murthy AC, Zhang L, Johnson EB, Schaller EG, Allan AM, Zhao X. Inhibition of GSK3beta improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum Mol Genet. 2012a;21:681–691. doi: 10.1093/hmg/ddr501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Murthy AC, Zhang L, Johnson EB, Schaller EG, Allan AM, Zhao X. Inhibition of GSK3beta improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum Mol Genet. 2012b;21:681–691. doi: 10.1093/hmg/ddr501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Polich ED, Su J, Gao Y, Christopher DM, Allan AM, Wang M, Wang F, Wang G, Zhao X. Fragile X Proteins FMRP and FXR2P Control Synaptic GluA1 Expression and Neuronal Maturation via Distinct Mechanisms. Cell Rep. 2015;11:1651–1666. doi: 10.1016/j.celrep.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Hagerman PJ. Fragile X syndrome. 2nd. Baltimore MD: Johns Hopkins University Press; 2002. [Google Scholar]
- Hagerman RJ, Hull CE, Safanda JF, Carpenter I, Staley LW, O'Connor RA, Seydel C, Mazzocco MM, Snow K, Thibodeau SN, et al. High functioning fragile X males: demonstration of an unmethylated fully expanded FMR-1 mutation associated with protein expression. American journal of medical genetics. 1994;51:298–308. doi: 10.1002/ajmg.1320510404. [DOI] [PubMed] [Google Scholar]
- Hagerman RJ, Ono MY, Hagerman PJ. Recent advances in fragile X: a model for autism and neurodegeneration. Curr Opin Psychiatry. 2005;18:490–496. doi: 10.1097/01.yco.0000179485.39520.b0. [DOI] [PubMed] [Google Scholar]
- Halevy T, Czech C, Benvenisty N. Molecular mechanisms regulating the defects in fragile X syndrome neurons derived from human pluripotent stem cells. Stem Cell Reports. 2015;4:37–46. doi: 10.1016/j.stemcr.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Flandin P, Yoshikawa K, Rubenstein JL, Alvarez-Buylla A, Kriegstein AR. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16:1576–1587. doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Rubenstein JL, Kriegstein AR. Deriving excitatory neurons of the neocortex from pluripotent stem cells. Neuron. 2011;70:645–660. doi: 10.1016/j.neuron.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton DD, Sideris J, Skinner M, Mankowski J, Bailey DB, Jr, Roberts J, Mirrett P. Autistic behavior in children with fragile X syndrome: prevalence, stability, and the impact of FMRP. Am J Med Genet A. 2006;140a:1804–1813. doi: 10.1002/ajmg.a.31286. [DOI] [PubMed] [Google Scholar]
- Hinton VJ, Brown WT, Wisniewski K, Rudelli RD. Analysis of neocortex in three males with the fragile X syndrome. Am J Med Genet. 1991;41:289–294. doi: 10.1002/ajmg.1320410306. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BY, Du ZW, Zhang SC. Differentiation of human oligodendrocytes from pluripotent stem cells. Nat Protoc. 2009;4:1614–1622. doi: 10.1038/nprot.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, Zhang SC. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Jacobs S, Cheng C, Doering LC. Probing astrocyte function in fragile X syndrome. Results Probl Cell Differ. 2012;54:15–31. doi: 10.1007/978-3-642-21649-7_2. [DOI] [PubMed] [Google Scholar]
- Jacobs S, Nathwani M, Doering LC. Fragile X astrocytes induce developmental delays in dendrite maturation and synaptic protein expression. BMC Neurosci. 2010;11:132. doi: 10.1186/1471-2202-11-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe EM, McQuate AL, Zhao X. Crosstalk among Epigenetic Pathways Regulates Neurogenesis. Frontiers in neuroscience. 2012;6:59. doi: 10.3389/fnins.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann M, Schuffenhauer A, Fruh I, Klein J, Thiemeyer A, Rigo P, Gomez-Mancilla B, Heidinger-Millot V, Bouwmeester T, Schopfer U, et al. High-Throughput Screening Using iPSC-Derived Neuronal Progenitors to Identify Compounds Counteracting Epigenetic Gene Silencing in Fragile X Syndrome. J Biomol Screen. 2015 doi: 10.1177/1087057115588287. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Cortell R, Kau AS, Bukelis I, Tierney E, Gray RM, Cox C, Capone GT, Stanard P. Autism spectrum disorder in fragile X syndrome: communication, social interaction, and specific behaviors. Am J Med Genet A. 2004;129:225–234. doi: 10.1002/ajmg.a.30229. [DOI] [PubMed] [Google Scholar]
- Kim DS, Ross PJ, Zaslavsky K, Ellis J. Optimizing neuronal differentiation from induced pluripotent stem cells to model ASD. Front Cell Neurosci. 2014a;8:109. doi: 10.3389/fncel.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TG, Yao R, Monnell T, Cho JH, Vasudevan A, Koh A, Peeyush KT, Moon M, Datta D, Bolshakov VY, et al. Efficient specification of interneurons from human pluripotent stem cells by dorsoventral and rostrocaudal modulation. Stem Cells. 2014b;32:1789–1804. doi: 10.1002/stem.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krencik R, Weick JP, Liu Y, Zhang ZJ, Zhang SC. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat Biotechnol. 2011;29:528–534. doi: 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuliev A, Rechitsky S, Tur-Kaspa I, Verlinsky Y. Preimplantation genetics: Improving access to stem cell therapy. Ann N Y Acad Sci. 2005;1054:223–227. doi: 10.1196/annals.1345.028. [DOI] [PubMed] [Google Scholar]
- Kumari D, Swaroop M, Southall N, Huang W, Zheng W, Usdin K. High-Throughput Screening to Identify Compounds That Increase Fragile X Mental Retardation Protein Expression in Neural Stem Cells Differentiated From Fragile X Syndrome Patient-Derived Induced Pluripotent Stem Cells. Stem cells translational medicine. 2015;4:800–808. doi: 10.5966/sctm.2014-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari D, Usdin K. Polycomb group complexes are recruited to reactivated FMR1 alleles in Fragile X syndrome in response to FMR1 transcription. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, Hagerman RJ, Tassone F, Tapscott SJ, Filippova GN. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- LaMonica BE, Lui JH, Wang X, Kriegstein AR. OSVZ progenitors in the human cortex: an updated perspective on neurodevelopmental disease. Curr Opin Neurobiol. 2012;22:747–753. doi: 10.1016/j.conb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni S, Goracci M, Borrelli L, Mancano G, Chiurazzi P, Moscato U, Ferre F, Helmer-Citterich M, Tabolacci E, Neri G. Role of CTCF protein in regulating FMR1 locus transcription. PLoS Genet. 2013;9:e1003601. doi: 10.1371/journal.pgen.1003601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathe R. Fragile X and autism. Autism. 2009;13:194–197. doi: 10.1177/13623613090130020402. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhao X. Concise review: Fragile X proteins in stem cell maintenance and differentiation. Stem cells. 2014;32:1724–1733. doi: 10.1002/stem.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chen J, Ehrlich S, Walton E, White T, Perrone-Bizzozero N, Bustillo J, Turner JA, Calhoun VD. Methylation patterns in whole blood correlate with symptoms in schizophrenia patients. Schizophrenia bulletin. 2014;40:769–776. doi: 10.1093/schbul/sbt080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Jiang P, Deng W. OLIG gene targeting in human pluripotent stem cells for motor neuron and oligodendrocyte differentiation. Nat Protoc. 2011;6:640–655. doi: 10.1038/nprot.2011.310. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu H, Sauvey C, Yao L, Zarnowska ED, Zhang SC. Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat Protoc. 2013;8:1670–1679. doi: 10.1038/nprot.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesch DZ, Huggins R, Hay DA, Gedeon AK, Mulley JC, Sutherland GR. Genotype-phenotype relationships in fragile X syndrome: a family study. Am J Hum Genet. 1993;53:1064–1073. [PMC free article] [PubMed] [Google Scholar]
- Loesch DZ, Huggins RM, Hagerman RJ. Phenotypic variation and FMRP levels in fragile X. Mental retardation and developmental disabilities research reviews. 2004;10:31–41. doi: 10.1002/mrdd.20006. [DOI] [PubMed] [Google Scholar]
- Loesch DZ, Sherwell S, Kinsella G, Tassone F, Taylor A, Amor D, Sung S, Evans A. Fragile X-associated tremor/ataxia phenotype in a male carrier of unmethylated full mutation in the FMR1 gene. Clinical genetics. 2012;82:88–92. doi: 10.1111/j.1399-0004.2011.01675.x. [DOI] [PubMed] [Google Scholar]
- Lozano R, Rosero CA, Hagerman RJ. Fragile X spectrum disorders. Intractable Rare Dis Res. 2014;3:134–146. doi: 10.5582/irdr.2014.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig AL, Espinal GM, Pretto DI, Jamal AL, Arque G, Tassone F, Berman RF, Hagerman PJ. CNS expression of murine fragile X protein (FMRP) as a function of CGG-repeat size. Hum Mol Genet. 2014;23:3228–3238. doi: 10.1093/hmg/ddu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Shan G, Guo W, Smrt RD, Johnson EB, Li X, Pfeiffer RL, Szulwach KE, Duan R, Barkho BZ, et al. Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 2010;6:e1000898. doi: 10.1371/journal.pgen.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Wang J, Shen B, Qiu L, Huang X, Li Z. Efficient targeting of FATS at a common fragile site in mice through TALEN-mediated double-hit genome modification. Biotechnol Lett. 2014;36:471–479. doi: 10.1007/s10529-013-1387-z. [DOI] [PubMed] [Google Scholar]
- Mailick MR, Hong J, Rathouz P, Baker MW, Greenberg JS, Smith L, Maenner M. Low-normal FMR1 CGG repeat length: phenotypic associations. Front Genet. 2014;5:309. doi: 10.3389/fgene.2014.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, Horvath TL, Vaccarino FM. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:12770–12775. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O. Human cortical interneurons take their time. Cell Stem Cell. 2013;12:497–499. doi: 10.1016/j.stem.2013.04.017. [DOI] [PubMed] [Google Scholar]
- Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, Liu B, Goulburn A, Stanley EG, Elefanty AG, et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann A, Hochstein N, Weber S, Fanning E, Doerfler W. A distinct DNA-methylation boundary in the 5'- upstream sequence of the FMR1 promoter binds nuclear proteins and is lost in fragile X syndrome. Am J Hum Genet. 2009;85:606–616. doi: 10.1016/j.ajhg.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, Arnold CM, Chen YJ, Stanley EG, Elefanty AG, et al. Functional Maturation of hPSC-Derived Forebrain Interneurons Requires an Extended Timeline and Mimics Human Neural Development. Cell Stem Cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedringhaus M, Dumitru R, Mabb AM, Wang Y, Philpot BD, Allbritton NL, Taylor AM. Transferable neuronal mini-cultures to accelerate screening in primary and induced pluripotent stem cell-derived neurons. Sci Rep. 2015;5:8353. doi: 10.1038/srep08353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacey LK, Doering LC. Developmental expression of FMRP in the astrocyte lineage: implications for fragile X syndrome. Glia. 2007;55:1601–1609. doi: 10.1002/glia.20573. [DOI] [PubMed] [Google Scholar]
- Pankratz MT, Li XJ, Lavaute TM, Lyons EA, Chen X, Zhang SC. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25:1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Halevy T, Lee DR, Sung JJ, Lee JS, Yanuka O, Benvenisty N, Kim DW. Reversion of FMR1 Methylation and Silencing by Editing the Triplet Repeats in Fragile X iPSC-Derived Neurons. Cell Rep. 2015;13:234–241. doi: 10.1016/j.celrep.2015.08.084. [DOI] [PubMed] [Google Scholar]
- Pastori C, Peschansky VJ, Barbouth D, Mehta A, Silva JP, Wahlestedt C. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Hum Genet. 2014;133:59–67. doi: 10.1007/s00439-013-1356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschansky VJ, Pastori C, Zeier Z, Motti D, Wentzel K, Velmeshev D, Magistri M, Bixby JL, Lemmon VP, Silva JP, et al. Changes in expression of the long non-coding RNA FMR4 associate with altered gene expression during differentiation of human neural precursor cells. Front Genet. 2015;6:263. doi: 10.3389/fgene.2015.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo FM, Fisher AG. Getting rid of DNA methylation. Trends in cell biology. 2014;24:136–143. doi: 10.1016/j.tcb.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Pickering SJ, Braude PR, Patel M, Burns CJ, Trussler J, Bolton V, Minger S. Preimplantation genetic diagnosis as a novel source of embryos for stem cell research. Reprod Biomed Online. 2003;7:353–364. doi: 10.1016/s1472-6483(10)61877-9. [DOI] [PubMed] [Google Scholar]
- Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Pietrobono R, Pomponi MG, Tabolacci E, Oostra B, Chiurazzi P, Neri G. Quantitative analysis of DNA demethylation and transcriptional reactivation of the FMR1 gene in fragile X cells treated with 5-azadeoxycytidine. Nucleic acids research. 2002;30:3278–3285. doi: 10.1093/nar/gkf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrobono R, Tabolacci E, Zalfa F, Zito I, Terracciano A, Moscato U, Bagni C, Oostra B, Chiurazzi P, Neri G. Molecular dissection of the events leading to inactivation of the FMR1 gene. Hum Mol Genet. 2005;14:267–277. doi: 10.1093/hmg/ddi024. [DOI] [PubMed] [Google Scholar]
- Pouya A, Satarian L, Kiani S, Javan M, Baharvand H. Human induced pluripotent stem cells differentiation into oligodendrocyte progenitors and transplantation in a rat model of optic chiasm demyelination. PLoS One. 2011;6:e27925. doi: 10.1371/journal.pone.0027925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss AL, Feinstein C, Rosenbaum KN. Autism and genetic disorders. Schizophr Bull. 1986;12:724–738. doi: 10.1093/schbul/12.4.724. [DOI] [PubMed] [Google Scholar]
- Ruiz S, Brennand K, Panopoulos AD, Herrerias A, Gage FH, Izpisua-Belmonte JC. High-efficient generation of induced pluripotent stem cells from human astrocytes. PLoS One. 2010;5:e15526. doi: 10.1371/journal.pone.0015526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemmle S. In vivo footprinting analysis of the FMR1 gene: proposals concerning gene regulation in high-functioning males. American journal of medical genetics. 1999;84:266–267. [PubMed] [Google Scholar]
- Schwemmle S, de Graaff E, Deissler H, Glaser D, Wohrle D, Kennerknecht I, Just W, Oostra BA, Doerfler W, Vogel W, et al. Characterization of FMR1 promoter elements by in vivo-footprinting analysis. Am J Hum Genet. 1997;60:1354–1362. doi: 10.1086/515456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JN, Kupinski AP, Boyes J. Targeted genome regulation and modification using transcription activator-like effectors. Febs J. 2014;281:4583–4597. doi: 10.1111/febs.12973. [DOI] [PubMed] [Google Scholar]
- Shaltouki A, Peng J, Liu Q, Rao MS, Zeng X. Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells. 2013;31:941–952. doi: 10.1002/stem.1334. [DOI] [PubMed] [Google Scholar]
- Sharp J, Hatch M, Nistor G, Keirstead H. Derivation of oligodendrocyte progenitor cells from human embryonic stem cells. Methods Mol Biol. 2011;767:399–409. doi: 10.1007/978-1-61779-201-4_29. [DOI] [PubMed] [Google Scholar]
- Shaw MA, Chiurazzi P, Romain DR, Neri G, Gecz J. A novel gene, FAM11A, associated with the FRAXF CpG island is transcriptionally silent in FRAXF full mutation. European journal of human genetics : EJHG. 2002;10:767–772. doi: 10.1038/sj.ejhg.5200881. [DOI] [PubMed] [Google Scholar]
- Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF, Haggarty SJ. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- Sidorov MS, Auerbach BD, Bear MF. Fragile X mental retardation protein and synaptic plasticity. Molecular brain. 2013;6:15. doi: 10.1186/1756-6606-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets HJ, Smits AP, Verheij CE, Theelen JP, Willemsen R, van de Burgt I, Hoogeveen AT, Oosterwijk JC, Oostra BA. Normal phenotype in two brothers with a full FMR1 mutation. Hum Mol Genet. 1995;4:2103–2108. doi: 10.1093/hmg/4.11.2103. [DOI] [PubMed] [Google Scholar]
- Stephenson EL, Mason C, Braude PR. Preimplantation genetic diagnosis as a source of human embryonic stem cells for disease research and drug discovery. Bjog. 2009;116:158–165. doi: 10.1111/j.1471-0528.2008.02009.x. [DOI] [PubMed] [Google Scholar]
- Stoger R, Genereux DP, Hagerman RJ, Hagerman PJ, Tassone F, Laird CD. Testing the FMR1 promoter for mosaicism in DNA methylation among CpG sites, strands, and cells in FMR1-expressing males with fragile X syndrome. PLoS One. 2011;6:e23648. doi: 10.1371/journal.pone.0023648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabolacci E, De Pascalis I, Accadia M, Terracciano A, Moscato U, Chiurazzi P, Neri G. Modest reactivation of the mutant FMR1 gene by valproic acid is accompanied by histone modifications but not DNA demethylation. Pharmacogenetics and genomics. 2008a;18:738–741. doi: 10.1097/FPC.0b013e32830500a1. [DOI] [PubMed] [Google Scholar]
- Tabolacci E, Moscato U, Zalfa F, Bagni C, Chiurazzi P, Neri G. Epigenetic analysis reveals a euchromatic configuration in the FMR1 unmethylated full mutations. European journal of human genetics : EJHG. 2008b;16:1487–1498. doi: 10.1038/ejhg.2008.130. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Telias M, Segal M, Ben-Yosef D. Neural differentiation of Fragile X human Embryonic Stem Cells reveals abnormal patterns of development despite successful neurogenesis. Dev Biol. 2013;374:32–45. doi: 10.1016/j.ydbio.2012.11.031. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tyson JA, Anderson SA. The protracted maturation of human ESC-derived interneurons. Cell Cycle. 2013;12:3129–3130. doi: 10.4161/cc.26351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Verlinsky Y, Strelchenko N, Kukharenko V, Rechitsky S, Verlinsky O, Galat V, Kuliev A. Human embryonic stem cell lines with genetic disorders. Reprod Biomed Online. 2005;10:105–110. doi: 10.1016/s1472-6483(10)60810-3. [DOI] [PubMed] [Google Scholar]
- Villalon-Reina J, Jahanshad N, Beaton E, Toga AW, Thompson PM, Simon TJ. White matter microstructural abnormalities in girls with chromosome 22q11.2 deletion syndrome, Fragile X or Turner syndrome as evidenced by diffusion tensor imaging. Neuroimage. 2013;81:441–454. doi: 10.1016/j.neuroimage.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci. 2005;8:288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- Willemsen R, Bontekoe CJ, Severijnen LA, Oostra BA. Timing of the absence of FMR1 expression in full mutation chorionic villi. Hum Genet. 2002;110:601–605. doi: 10.1007/s00439-002-0723-5. [DOI] [PubMed] [Google Scholar]
- Willemsen R, Oostra BA, Bassell GJ, Dictenberg J. The fragile X syndrome: from molecular genetics to neurobiology. Ment Retard Dev Disabil Res Rev. 2004;10:60–67. doi: 10.1002/mrdd.20010. [DOI] [PubMed] [Google Scholar]
- Wisniewski KE, Segan SM, Miezejeski CM, Sersen EA, Rudelli RD. The Fra(X) syndrome: neurological, electrophysiological, and neuropathological abnormalities. Am J Med Genet. 1991;38:476–480. doi: 10.1002/ajmg.1320380267. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet. 2014;23:R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]