Abstract
Meiosis presents many important mysteries that await elucidation. Here we discuss two such aspects. First, we consider how the current meiotic program might have evolved. We emphasize the central feature of this program: how homologous chromosomes find one another ("pair") so as to create the connections required for their regular segregation at Meiosis I. Points of emphasis include the facts that: (i) the classical "bouquet stage" is not required for initial homolog contacts in the current evolved meiotic program; and (ii) diverse observations point to commonality between molecules that mediate meiotic inter-homolog interactions and molecules that are integral to centromeres and/or to microtubule organizing centers (a.k.a. spindle pole bodies or centrosomes). Second, we provide an overview of the classical phenomenon of crossover (CO) interference in an effort to bridge the gap between description on the one hand versus logic and mechanism on the other.
Keywords: meiosis, pairing, bouquet, crossover interference
1. Introduction
Meiosis is complicated and always admits to many points of view, even for features that are generally agreed-upon. This is particularly true for several of the subjects covered here, for which there is a paucity of solid information. However, it can be useful to have a way of thinking about unexplained findings in advance of confirmation or rejection. The comments below thus represent a mixture of: (a) "facts" that are currently generally acknowledged by the community to be true and (b) "preferences" or "ideas" that we find interesting, attractive, provocative of synthetic interpretations and/or generally unappreciated by many. We have tried to make clear which points fall into these two categories.
2. Evolution of Meiosis and the Role of the Bouquet
2.1 Background
No one knows how meiosis evolved (e.g. 1, 2). Moreover, the current evolved process differs substantially in different organisms, likely with more variants as yet to be discovered. Nonetheless, in nearly all organisms, DNA replication is followed by two rounds of chromosome segregation, with homologous chromosomes (homologs) segregating at the first division and sister chromatids segregating at the second division (MI and MII). [One question, then, is why the haploid state is achieved in this manner rather than by skipping DNA replication and carrying out one round of homolog segregation (e.g. 3) or by inverted meiosis as observed in some organisms (4,5)
Regular segregation of homologs at the MI division requires that homologs be physically connected. Given a connection, proper bipolar alignment on the spindle results in tension on centromere/kinetochore complexes, which, in turn, licenses onset of anaphase.
If homologs are physically connected, they must have previously come together in space, i.e. "pair", in order to acquire those connections. Moreover, pairing must have occurred without entanglements among unrelated chromosomes. Thus, evolution must solve these "pairing problems" along with mechanisms for creating connections and for modification of the segregation apparatus to give two appropriate meiotic divisions.
Interestingly, while presence of a link between homologs is almost always crucial, the overall program by which homologs pair varies from one organism to the other. Further, we need to be aware that we know only a handful of organisms. Thus, the range of programs for pairing may be much more variable than is currently appreciated.
2.2 The "canonical" program
In the so-called "canonical" program for pairing that occurs in plants, animals and some fungi, recombination plays a central mechanistic role. The critical connections that ensure regular MI homolog segregation are provided by one or a few DNA crossovers (COs), plus associated local structural changes, in combination with connections (cohesion) between sister chromatids along their lengths. Moreover, at earlier stages, recombination mediates homolog pairing (review in 6). In this process, homologs are specifically recognized and brought together in space, ultimately via juxtaposition of their structural axes, as a specialized byproduct of the early stages of recombination, which creates periodic linkages all along the lengths of the chromosomes. This process yields distance-coalignment of homologs. COs then arise at a small subset of these linkages, accompanied by closer juxtaposition of axes via the synaptonemal complex (SC).
A variety of features ensure that recombination-mediated pairing/coalignment occurs in a regular and efficient way. (i) Early clustering of centromeres at/near the nuclear microtubule organizing center, (MTOC; centrosome) or its fungal equivalent spindle pole body (SPB) as a remnant of the Rabl orientation of the previous mitosis. This configuration tends to place corresponding regions of homologs at similar positions ("latitudes") relative to the centromere cluster thereby reducing the topological complexity of the homology search process. (ii) Recombination-independent pairing, which often occurs locally between homolog telomeres in early prophase and sometimes occurs globally throughout the chromosomes as a pre-meiotic (as in budding yeast, S. pombe and, in modified form, Drosophila) or early meiotic condition (e.g. in mouse, 7). (iii) Specific features of DSB-mediated pairing, which promote propagation of DSB-mediated pairing (e.g. via Mer3; 8). (iv) Processes that eliminate inappropriate connections or entanglements when they do occur, apparently including but not limited to release of constraining recombinational linkages and, potentially, dynamic chromosome movements involving clustering of nuclear envelope-associated telomeres during the "bouquet stage" (further discussion below).
2.3 Evolution of connectedness at prophase and MI
It is generally assumed that meiotic recombination evolved from mitotic recombinational repair of spontaneous or exogenously-induced double-strand breaks (DSBs), given prominent functional participation of general repair proteins in the meiotic recombination process (review in 9). We have previously suggested a more extreme possibility, that meiosis evolved in a diploid organism in response to an environment involving large amounts of radiation-induced damage (10). In such a case, recombinational repair would tend, at some frequency, to give rise to crossover (CO) recombination products between homologs, rather than between sisters as is usually the case in mitosis. CO recombination, in turn, will lead to segregation of homologs during mitosis as well as creating interlockings among unrelated chromosomes, both of which would be inappropriate to regular mitotic sister segregation (8, 10, 11). In contrast, evolution of appropriate centromere/kinetochore modifications (i.e. a reductional configuration) would permit regular homolog segregation. That is, certain features of meiosis, at least, could have evolved initially as a pathway for rescuing mitotic diploid cells from "disaster".
However, connectedness of homologs at prophase can be mediated by DSB-independent pairing in diverse situations (review in 12). Moreover, while the basis for such pairing is not established, two DNA molecules can pair in a homology-dependent fashion in vitro, in the absence of aggregating agents, leading to the suggestion that DNA/DNA pairing is actually the "default option" for the in vivo situation and is specifically precluded in general by programmed features. DSB-independent pairing could well be a primordial mechanism for prophase, with other features (topological catenations? redirected cohesin interactions (13)? centromere linkages? (14; below) serving to maintain those connections into MI as a primordial mechanism for homolog segregation.
2.4 "Non-canonical" programs
Several organisms have a somewhat different pairing programs. Among these examples: some organisms lack SC; some exhibit both SCs and COs in localized regions; some lack recombination altogether and use other types of connections (including the SC) to replace COs/chiasmata; and in others, global recombination-independent pairing precedes a recombination program that ultimately yields COs/chiasmata.
-
(1)
The fission yeast Schizosaccharomyces pombe forms COs but lacks SC. It exhibits a classical bouquet but strikingly the elongated nucleus moves back and forth between the cell poles during the entire meiotic prophase I “horsetail stage”) dragged by microtubules and associated motors. This movement facilitates pairing (absent in mutants deficient for the telomere attachment to the SPB or motor proteins; 15–18). Movement tends to enforce parallel alignment of homolog arms, but interspersed with periods of disrupted alignment upon change of direction (17,19–21).
-
(2)
The freshwater ciliate Tetrahymena thermophila has a polyploid macronucleus (soma) and a micronucleus (germ-line). During meiotic pairing, the micronucleus elongates into a tube (with intranuclear microtubules) in which chromosomes are arrayed in a parallel, elongated state with their telomeres grouped at one pole and centromeres at the other, implying a role for homolog pairing. Interestingly, DNA events appear to occur after this stage. The nucleus returns to a round shape after the events of pairing and recombination, at diplotene (22,23, see also Loidl and Lorenz, this issue: A general overview about meiosis in Tetrahymena).
-
(3, 4)
In Drosophila melanogaster female and in the nematode Caenorhabditis elegans, pairing and SC formation can occur independently of recombination (e.g. 24,25). Unexpectedly, however, in Drosophila female, meiotic DSB-independent pairing is not a continuation of somatic pairing (26,27). In Drosophila, SC is seen initially at centromeres, which occur at the ends of their respective chromosomes, but also nucleates interstitially along the chromosomes (28,29). In C. elegans, pairing/SC formation occurs preferentially at specific «pairing centers» (PCs), also localized at the end of chromosomes and chromosomes lacking these regions fail to synapse (30,31). PCs are characterized by repeated DNA, with different chromosome ends having different families of repeated sequences. Also, the six PCs are recognized by different zinc-finger proteins with the interesting exception that chromosomes I and IV plus chromosomes II and III share the same protein, implying some further discriminatory feature(s) (32–34 review in 25). C. elegans PCs become associated with the nuclear envelope and interact with a pair of SUN/KASH domain proteins that span both NE membranes and connect the ends of the chromosomes to the microtubule-based dynein motors in the cytoplasm. Active motor-driven movement involving cytoskeletal elements appear to have two effects: first, to promote juxtaposition of homologous ends; and second, as a stringency factor to eliminate inappropriate connections, e.g. between PCs that share a common zinc finger protein. Interestingly, however, the SC is required to maintain pairing at later stages of prophase (review in 25; 35–40).
In both female Drosophila and in C. elegans, SC formation precedes initiation of recombination by DSBs, with this and ensuing steps of recombination occurring in the context of the SC. Interestingly, the number of DSBs appears to be much lower in both of these organisms than in organisms that undergo the canonical program. Perhaps this is because, in these two organisms, DSBs are required only to give rise to COs (for genetic diversity and for use in ensuring homolog segregation at MI) whereas, in the canonical program, many DSBs are required for recombination-mediated homolog coalignment.
-
(5)
In organisms that make an SC, the presence of this structure along the lengths of the homologs is required for correct MI segregation, even when CO/chiasma positions are highly localized to specific positions along the chromosomes. Perhaps the SC has a global role in maintaining chromosome order within the nucleus or in interlock resolution (review in 6). However, some species exhibit regular segregation at MI despite only partial SC formation and extremely restricted COs/chiasmata to the ends of the chromosomes. For example: (a) In the orthoptan Paratettix meridiolasis, axis formation, DSBs (defined cytologically by Rad51 and γH2AX foci), SC formation and diplotene chiasmata are restricted to non-centromeric distal regions of the telocentric chromosomes (41). (b) In the planarian flat worm Mesostoma ehrenbergii ehrenbergii (2×=10) only three pairs of chromosomes form bivalents, each with a single chiasma (and thus a single CO) at MI and restricted SC short stretches with one late (CO-correlated) recombination nodule in a lobe of the nucleus at prophase; the two other pairs remain unsynapsed (42).
-
(6)
Drosophila melanogaster male. This organism is among the handful known that exhibits neither recombination nor an SC, although well-aligned bivalents at MI. The X and Y chromosomes become connected via a specific "pairing site" that comprises repeats of specialized rDNA bound by stromalin and Modifier of Mdg4 protein, which are required for pairing (31, 43–45). No PCs are known on Drosophila autosomes except for the very small chromosome 4, which remains paired in 90% of the meiocytes, suggesting a PC-like structure in this chromosome (review in 31).
-
(7)
Regular MI homolog segregation can also be obtained in the absence of recombination by using the SC in a novel way: in the silk worm (Bombyx mori), female meiosis lacks COs/chiasmata but nonetheless makes an SC, which remains until AI to provide the connection necessary for proper homolog segregation (46; review in 47 for other examples). Meiosis in male Bombyx mori occurs by the canonical program.
2.5 A common factor of both the “canonical” and "non-canonical" programs is the presence of the well-conserved meiotic configuration called “the bouquet”
At onset of meiotic prophase, the initial Rabl clustering of centromeres in the vicinity of the MTOC is released. It is followed by a reorganization of the chromosomes such that now the telomeres are clustered in the vicinity of the MTOC/SPB/ centrosome and adopt a conformation called the bouquet (e.g. review in 48). However, a defined MTOC is not required for telomere clustering: bouquet form in plants where the entire nuclear envelope organizes the cytoplasmic microtubules during both mitotic and meiotic cycles (discussion in 49). Bouquet formation is accompanied by dynamic telomere-led movements mediated by their linkage through the nuclear envelope to the cytoskeleton, whose dynamics drive telomere movement along the nuclear periphery (above).
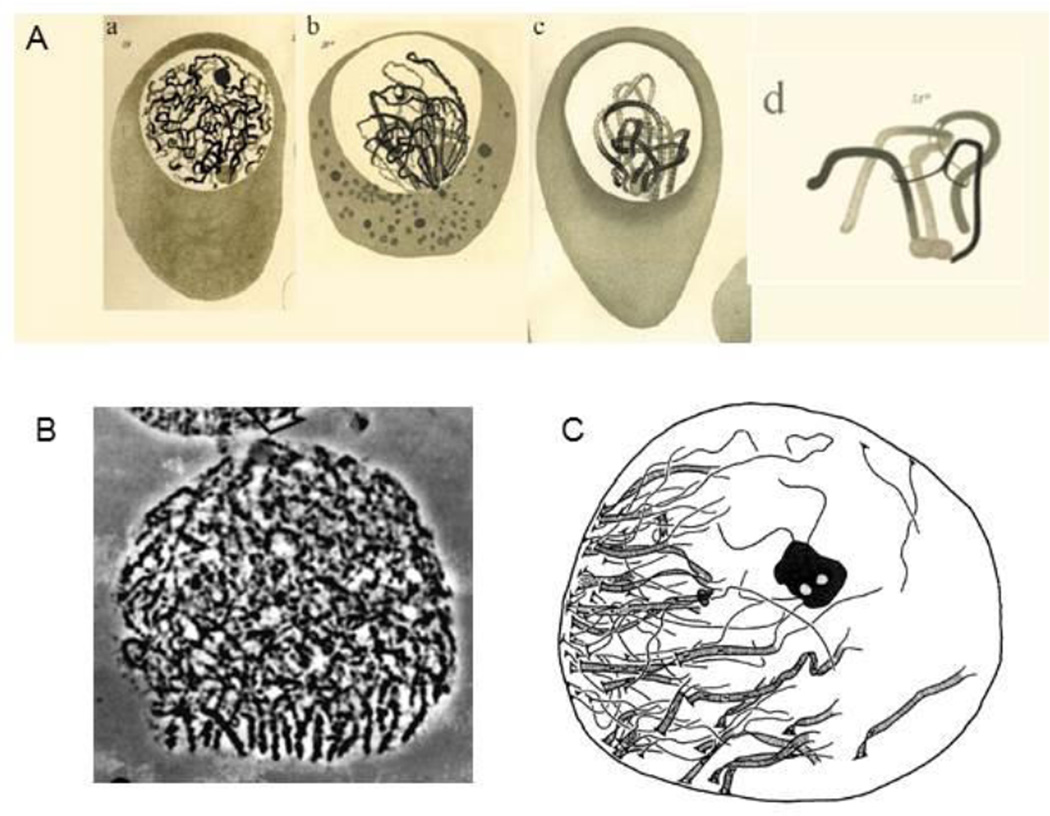
The bouquet was originally described by meticulous analysis of flat-worm meiosis (50). The original bouquet images of Gelei plus images in other organisms are shown in Figure 1. Gelei showed that the bouquet is seen from early zygotene to pachytene and that homologs are synapsing from the clustering area on. This and ensuing observations lead to the assumption that the role of the bouquet was to reduce the complexity of the homology search process required for pairing/synapsis (e.g. review in 48). However, subsequent studies revealed that homologs are already coaligned prior to synapsis in several organisms, thus removing the imperative for such a role (review in 54; 55, 25). We propose that occurrence of this bouquet configuration is a central nucleating feature of early meiotic evolution and that its role has progressively changed in concert with progressive evolution of the meiotic process. This view can accommodate diverse otherwise disparate observations, including the conservation of this feature in organisms with well-evolved non-canonical programs (above).
Figure 1. Bouquet Configurations.
A. Gelei’s original discovery of the bouquet: a= leptotene, b=zygotene (thick threads= synapsis), c=pachytene all ends are still clustered; d = bivalent interlocking (50). B, C. Bouquets in salamander (from 51) and human spermatocyte (52). [B and C reprinted from (53).]
2.6 Is the presence of the bouquet a relic of the first steps of meiotic evolution?
The ideas below are one way to think about a possible path for evolution of meiosis. These ideas are motivated by trying to understand why all organisms (known so far) have kept the bouquet in their meiotic program, even despite the diverse details of those programs as discussed above.
One key consideration in our thinking concerns the dynamics of telomere reorientation from Rabl into the bouquet during the meiotic program. While orientation towards, and possible link with, the SPB/centrosome is not always required (e.g. plants, above), clustering is conserved. Since clustering could easily have been lost by evolution, it would seem that it has, per se, an important role(s).
A second consideration is that, while pairing is mediated by DSBs in the canonical program, bouquet formation and thus chromosome movement during leptotene are both independent of DSBs’ formation and the downstream process of recombination in Sordaria and plants (8,56,57). Thus, as DSB-deficient mutants are severely defective in spatial juxtaposition and synapsis of homologs, bouquet formation does not require interactions between homologs. And conversely, pairing failure cannot be attributed to a failure to make a bouquet. However, while not necessary for its formation, the recombination proteins are required for timely bouquet resolution in Sordaria (8,56), budding yeast, worm and mouse (e.g. 58,59), indicating a possible role of the bouquet as a checkpoint sensor to evaluate the status of the chromosome and/or the CO recombination process before allowing completion of synapsis.
A third consideration, which emerges in part from our own work, is that some proteins that are integral components of the SC are also SPB/centrosome components. For example: Sordaria Sme4 (with Zip1-like functions) and Hei10, budding yeast, fission yeast Mps3 and Sad1, and the mouse Death Inducer-Obliterator DIDO3 are also SPB/centrosome components (15,60–63). One common factor in these molecules is that they are all coiled-coil proteins, in accord with their participation in important structures. For example, Sordaria Sme4 has very similar functions in the two structures, i.e. to keep the two homologs together via the SC and to mediate the juxtaposition of two parts of the SPB, giving a change in the shape that allows formation of a larger microtubule array as required for specialized roles during ascospore formation (60). Interestingly also, meiotic HORMA-domain proteins and cohesin Rec8 of C. elegans, which are regular components of chromosomal axes, are also implicated in correct centriole separation during division II (64).
A fourth triggering consideration is the fact that in S. pombe and C. elegans, movements directed by clustered telomeres might have a mixture of roles, both to promote regular homolog juxtaposition and pairing and also as a stringency factor to eliminate inappropriate linkages or entanglements (e.g. 25,35).
Integrating these considerations, we propose that homolog pairing during meiosis began with evolution of the bouquet configuration. In this first primordial state, chromosome ends would be anchored through the nuclear envelope to/near the nuclear MTOC with regular spatial coalignment of homologs ensured either by nuclear movement as in S. pombe or by a specialized nuclear shape as in Tetrahymena. The next step in evolution could be a situation analogous to that in C. elegans, with evolution of "pairing centers" that mediate a more specialized DSB -independent pairing process with juxtaposition (and stringency) still mediated by end-led cytoskeleton-directed movements. Then, some proteins that initially evolved as SPB/centrosome components would evolve into SC components (as seen by roles in both features by Sme4, Mps3 and DIDO3 (above)). Regular spatial coalignment of homologs would still be mediated by telomere/NE/SPB/cytoskeleton localization and movement while the newly recruited proteins would now be used to link homologues via synapsis.
This progression would permit recombination-independent pairing and recombination-mediated CO formation (likely from mitotic-repair like processes) to initially evolve independently. Finally, in most organisms this primordial process would then have diminished in importance, with recombination (via multiple interstitial DSB-mediated interactions) evolved from mitotic repair, now becoming predominant and thus also used for pairing and synapsis.
By this idea, since telomere regions would be, since the beginning of evolution, already the first to undergo DSB-independent pairing and synapsis, recombination might also initiate firstly or preferentially in those regions as seen in orthoptans (41), in barley (65) and in mouse (66).
Motion and clustering of telomeres, no longer used for pairing per se, would now be kept because recruited instead to solve the newly created problem of resolution of entanglements or other DNA links like ectopic interactions between non-homologous chromosomes (e.g. 67) and would therefore explain the conservation of the bouquet formation.
3. Crossover (CO) Interference
One of the most interesting aspects of meiosis from a mechanistic point of view is the highly regulated process that determines the number and positions of COs along chromosomes through the effects of CO interference. We focus on COs that arise in this spatially-patterned way. A minority of COs that occur outside of this pathway (so-called Type II COs; e.g. 68) will not be extensively discussed. For an excellent recent general review, see Hunter (9); for earlier reviews, see 69 and 70; and for a review of the beam-film model and logic, see 71).
3.1 Introduction to CO Interference
Discovery
CO interference was discovered by Sturtevant (72), and further elucidated by Muller (73), by genetic analysis of Drosophila recombination, in the context of experiments designed to show that genes occur in a linear order, in accord with their localization to chromosomes. The fundamental observation was that when a chromosome is divided into intervals by genetic markers, the frequency of cases in which a single gamete exhibits a CO in both of two given intervals (the number of "double COs " for those intervals) is often less than the frequency expected from their occurrence in each of the intervals independently (as defined by the product of the frequencies of COs in the two intervals considered independently). That is, one CO appears to "interfere" with formation of another CO. The effect of interference is particularly strong for two intervals that are very close together.
Assessment
Further genetic analysis in Drosophila led to a classical way of defining CO interference: Coefficient of Coincidence (CoC) analysis (74) (Figure 2AB). Following Sturtevant and Muller, a chromosome is divided into intervals and all possible pairs of intervals are examined to determine, for each pair, the "observed" frequency of gametes with double COs versus the "expected" frequency. The ratio of observed/expected frequencies is the CoC. CoC values for each pair of intervals is then plotted as a function of the distance between the two intervals. Universally, in organisms that exhibit CO interference, the CoC is small or zero at very short inter-interval distance (reflecting essentially "absolute" interference) and rises progressively to ~CoC=1 (reflecting absence of interference). At even larger distances, the CoC often fluctuates up and down at values above CoC=1. This feature reflects a tendency for COs to be evenly-spaced along chromosomes. CoC analysis provides a powerful, logically correct way of describing CO interference and can be extremely accurate for any type of CO pattern, as long as the dataset is sufficiently large (discussion in 75; 76).
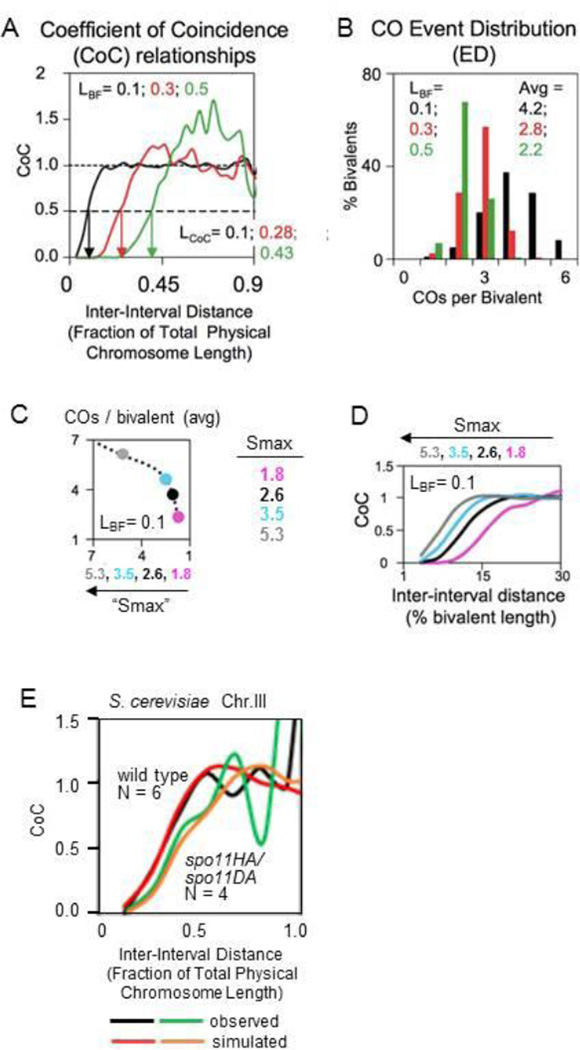
Figure 2. Coefficient of Coincidence (CoC) Analyses.
A, B. CoC relationships (A) and the density distribution of different numbers of COs per bivalent (B) from data sets created by simulations using the beam-film model under different values of relevant parameters. Among these, (L) or (LBF) denotes the distance over which the interference signal spreads. (LCoC) denotes the inter-interval distance at which CoC = 0.5. [reprinted from 75] C, D Beam-film simulations illustrate the fact that when the strength of CO-designation increases, the number of COs per bivalent increases (C) and the CoC curves shift to the left, even though there is no change in the distance over which the interference signal spreads [adapted from 77.]. In such simulations, the strength of CO-designation is given by parameter Smax and interference distance is given by parameter L. Simulation parameter values are defined relativistically. Smax can range from zero (i.e. no CO-designation) to non-zero values that give appropriate outcomes. In simulations of several experimental datasets, the values of Smax range from ~1 to ~3.5 (75). Parameter L is defined as a fraction of bivalent length. In the simulation shown, Smax is varied from 1.8 to 5.3 while L remains constant at L=0.1 (10% of bivalent length). E. Reduction in the average number of DSBs per chromosome resulting from a spo11 mutant condition shifts the CoC curve to the right as observed for S. cerevisiae chromosome III (compare black and green). Beam-film simulations under best-fit conditions for this organism demonstrate that this shift is attributable solely to a decrease in the number of DSB-mediated precursor interactions (N) from N=6 in the wild-type to N=4 in the mutant with no change in the spreading interference signal or any other feature of CO formation/patterning (data and rationale in 75; and 78). [Figure is by L. Zhang and N.K. unpublished].
Other methods for describing CO interference are used. Many studies utilize so-called "gamma distribution analysis" (e.g. 79). The shape parameter of the gamma distribution is a measure of evenness and thus an indirect indicator of interference. However, the value of the gamma distribution shape parameter can be affected by changes in CO patterns that arise from factors unrelated to CO interference (e.g. the efficiency with which a CO-designated interaction actually matures into a CO), thus limiting the usefulness of this tool for analysis of variations in interference per se (75). Genetic studies can also use specialized approaches that permit analysis of small chromosomal regions (e.g. "modified CoC analysis" among others (80)). CO patterns can also be defined by analysis of chiasmata, the cytological correlates of COs as seen at the diplotene stage (e.g. 81).
3.2 Known Features of CO Interference
Certain features of CO interference appear to be becoming clear.
Coordinate DNA and chromosomal changes
Meiotic recombination occurs after DNA replication, with all DNA events occurring in recombination complexes that are intimately associated with the chromosome axes (6,12,82,83). Recombination is initiated at many positions along the chromosomes via programmed axis-associated double-strand breaks (DSBs). In most organisms (e.g. mammals, plants and some fungi), the "canonical" program (alluded to above) is as follows. Most DSBs identify an homologous sequence on a DNA molecule of a non-sister chromatid of a homolog chromosome, forming a "nascent D-loop", which also becomes axis-associated. Concomitant events result in spatial coalignment of the two axes, a process that comprises "homolog pairing", and often yields inter-axis bridges which comprise DNA and recombination proteins (discussion in 84, this issue). A few CO sites, usually one to several per chromosome, appear to be designated from among these many DSB-mediated interactions, which are thus describable as "precursors" to CO-designations (and thus eventual COs).
In some organisms, a cytological marker for CO sites is available as early as the leptotene/zygotene transition. Analysis of such organisms strongly suggests that CO-designation with concomitant interference occurs in a single step, at the early stage of leptotene/zygotene, in accord with bridges as the relevant "precursor" (discussion in 61,77,12,6). CO-designation at this early stage triggers an ensuing cascade of biochemical and structural changes that culminates in formation of mature COs at a later stage (pachytene).
It has been suggested that, in some cases, CO-designation might occur in two steps (85,86) but this remains to be validated. For example, in many organisms, at mid-pachytene, cytologically-detectable Msh4 foci occur in a number that is intermediate between that of total recombinational interactions (many) and COs (few), consistent with an intermediate step (85). However, this same phenomenon occurs in Sordaria where the availability of an early CO marker, plus other observations, strongly implies that CO-designation occurs in a single step and, furthermore, raises the distinct possibility that the intermediate number of Msh4 foci reflects sites of SC nucleations, among which only a subset are also sites of COs (61,77; below).
The majority of precursor (DSB) interactions are not fated to become inter-homolog COs; instead, they mature to other fates, usually to "inter-homolog non-crossover products" but also sometimes to other fates including inter-sister interactions and, probably, "Type II" COs (9,68,75).
Axes vs SC
In organisms that use the above-described pathway, CO-designation is followed by installation of the synaptonemal complex (SC), which is not required for CO interference (recent discussions in 9,78). To a first approximation, SC is always nucleated at the sites of all CO-designations. In addition, in most organisms, SC is nucleated at a number of other "precursor" sites. Nucleation at COs sites allows CO recombinational interactions to be associated with the SC in special ways as required for ensuing CO maturation and/or eventual formation of chiasmata at diplotene (e.g. 87). SC nucleation presumably occurs at other sites because the number of COs per bivalent is relatively small, such that CO site nucleation is thus insufficient to ensure that the entire chromosome acquires SC in a reasonable time frame. It can be noted, however, that in some organisms, SC does not form along the entire lengths of the chromosomes (see above). Also, in some organisms, notably Drosophila female and C. elegans, the entire recombination program appears to occur nearly analogously to the "canonical" program, but with fewer DSBs and all DNA events occurring in the context of already-formed SC (see Section I above). In these organisms, the roles of chromosome structure that are carried out by bridge-linked chromosome axes in the "canonical" program are likely carried out instead by SC-linked axes (for analogies, see e.g. 88,89).
Communication
The central feature of CO interference is communication along the chromosomes.
The metric of interference is chromosome length. Interference occurs when chromosomes are well-organized (above) and requires physically intact chromosomes (90). Moreover, the "metric" of CO interference is physical distance along the chromosomes, not genetic distance (cM) or genomic distance (Mb) (discussion in 9,78). A most remarkable feature of CO interference is that the signal emanating from a single CO site can extend for very different distances in different organisms, in accord with the fact that all organisms exhibit one or a few COs per bivalent regardless of their genomic size or meiotic prophase physical (axis) length, e.g. from ~300nm in budding yeast to tens of microns in a higher plant (78 and references therein).
Axes as the conduit? It seems that information travels along the chromosome axes specifically (9). The strongest argument may be the following. Meiotic chromosomes are organized as chromatin loops along the structural axes. In several species, notably human (91) male and female chromosomes differ because one sex has longer axes but shorter loops while the other has shorter axes and longer loops. Importantly, CO interference is the same in both sexes when the metric is µm axis length (e.g. for Arabidopsis and human: 92,93). This result implies directly that chromatin loop size and the total density of DNA per µm chromosome length are not relevant variables. Functional studies of axis component mutants are consistent with this view but are not definitive, since alterations in axis components can affect CO patterns by affecting aspects of the process other than communication per se (e.g. CO-designation; below).
Reaction/diffusion and/or mechanical stress and stress relief. The basis for communication remains to be established. In general, patterns can be achieved in either of two ways (or by a combination of both) (for examples in a bacterial system, see 94–96). At one extreme, the chromosomes per se are an essentially passive substrate upon which biochemical events occur in a "reaction-diffusion" process. By such a mechanism, all events involved in patterning might specifically affect only the recombination complexes, with changes in the chromosomes' axis/SC status (below) as downstream consequences. At the other extreme, the chromosomes are active mechanical participants in the communication process. In any mechanical system, a change in stress at one position will inevitably result in redistribution of that effect for some distance away from that point (for example, if you poke your finger into a balloon, the indentation spreads out away from the pressure point). Mechanical stress, in turn, implies deformation of the chromosomes into a higher energy state. It has been proposed specifically that meiotic chromosome axes are under stress along their lengths and that CO-designation is promoted by such stress (97). Each such event will, by its nature, locally alleviate stress, and that effect could then redistribute along the chromosomes, outward in both directions, thus disfavoring additional stress-promoted CO-designations nearby. An additional feature of mechanical stress redistribution is that, in systems that are not simple elastic objects, stress redistribution dissipates with distance away from its nucleation site, predicting that CO interference would similarly decay, yielding automatically a "zone of interference". In fact, however, mechanical effects and reaction-diffusion effects can interact in complex ways, as first proposed by Turing (96) and recently implied by studies of the MinDE system in E.coli (95 and references therein). For example, mechanical changes might be nucleated locally at a CO site and then propagated along the lengths of the chromosomes by molecular events in a ratchet-like process with a probabilistic decay distance.
We have proposed specifically that stress arises because the entropically-favored state of chromatin loops (in an expanded configuration) is constrained by connections along the chromosomes, including the chromosome axes, with local alleviation of this stress at CO sites then spreading along the chromosomes to confer interference (97,98). Several types of observations are consistent with this view. First, one component implicated in CO interference is Topoisomerase II (78). Topoisomerase II occurs preferentially along/above chromosome axes and its role is to mediate mechanical stress along chromosomes resulting from aberrant DNA/chromosome topology. Second, axis disruption and increased chromatin diffuseness occur at sites of COs in Sordaria (99), and local chromosome extension occurs at sites of COs in C. elegans (89), in accord with release of constrained chromatin expansion stress at these sites. Third, the period when CO-designation is triggered is a period of global chromosome expansion whereas the ensuing period is a period of global chromosome compaction.
3.3 Mechanistic Components and Implications for Functional Dissection
Implicit in the above description of events is the fact that CO patterning can be separated into several component parts, more or less independent of any particular model or mechanism.
Precursor array
One component is the array of "precursor" interactions upon which patterning is imposed (e.g. DSB-mediated inter-homolog "bridges", as discussed above). Experimental and simulation studies of CO patterns reveal that these precursors are sometimes themselves evenly-spaced (discussions in 84 this issue; 75), suggesting that patterning of CO sites is preceded by some earlier type of patterning along the chromosomes. Other analyses further suggest that the number of precursors along a given genetic chromosome is relatively similar from one nucleus to another (75; N.K. L.Zhang and S.Wang unpublished).
Patterning features
The second component is the CO patterning process itself. Three additional features are implicit in the above description. (a) Precursor recombinational interactions will have some average sensitivity to the CO designation process. (b) Different precursors will necessarily not be identical, so there will be some particular distribution of relative sensitivities around the average value, which may be broader or narrower according to the situation. Since there will always be some threshold to the sensitivity required for CO-designation, differences in this distribution will affect the fraction of precursors that are sensitive enough to actually undergo CO-designation. This seemingly esoteric feature can, in fact, have important effects (L.Zhang, S. Wang and NK unpublished). (c) The effect of CO interference will be to make affected precursor interactions less sensitive to the CO-designation process and thus less likely to undergo CO-designation.
CO maturation
The third component is CO maturation, which determines whether a particular CO-designated interaction will go through all of the ensuing steps necessary to create a mature CO (or its corresponding experimentally-detected correlate). Defects in CO maturation do not alter CO interference as defined by CoC analysis or genetic analysis, in accord with the fact that they arise after patterns are already established.
Separation of CO patterning into these different aspects can, in principle, allow to assign mutant phenotypes to certain specific effects. For example, absence of Mlh1 can be shown to affect CO maturation, rather than CO interference (discussion in 75). Other types of distinctions can be identified with appropriate mathematical simulations (e.g. 75,71,77,61). However, in practice, these distinctions may sometimes be too subtle to detect and/or mutations may coordinately alter more than one aspect, thus precluding simple interpretation.
3.4 Transiting to a Mechanistic Definition of "Interference": Examples
CO interference can be defined by diverse criteria. Phenomenologically, any change detected by one of these methods is said to reflect a change in "CO interference". More subtly, it is implicitly assumed that changes in these relationships represent changes in the "communication" aspect of CO interference. For example, when CoC relationships are shifted to lower values, interference is said to be "weaker" as if the process of spreading of the interference signal is somehow less robust; oppositely, if CoC relationships are shifted to higher values, interference is said to be "stronger". However, when specific mechanistic aspects are considered, it becomes clear that these effects can result from changes in features other than the "spreading interference signal" per se.
A general case that often occurs is as follows. Given CO-designation and interference as separate components, if more and more CO-designations can occur, COs will tend to occur closer together, even if each CO sets up exactly the same type of spreading interference signal (Figure 2CD). That is, by any applied conventional criterion, CO interference will be said to be "weaker" whereas, in fact, the spreading of the "interference signal" involved in communication is, per se, unchanged and all that has happened is a change in the frequency of CO-designation. This conundrum suggests that, where possible, more precise language, and more precise thinking, will be useful in describing observed effects.
The importance of accommodating this particular conundrum is illustrated by several specific examples.
Nested designation of CO sites and SC nucleation sites
It is well known, particularly in plants, that SC nucleates at the sites of inter-axis bridges and that the number of SC nucleations far exceeds the number of COs along any given bivalent (e.g. 100). Correlations between SC nucleations and sites of recombination-correlated "nodules" have also been reported in other organisms, notably Sordaria (101). A recent analysis of data for Sordaria shows that SC nucleations and CO patterns both arise as part of a single interference process (77). Each designation of an SC nucleation site sets up interference; however, the earliest nucleations concomitantly result in CO-designation at the same site whereas later nucleations result only in SC nucleation and not in CO-designation. "CoC" curves for total nucleations, and for nucleations that give rise to COs, are offset, at lower and higher inter-interval distances respectively, even though the length of the spreading interference signal is the same for both outcomes. Interestingly, in budding yeast, there seems to be essentially a 1:1 relationship between the number of CO-designation sites and the number of SC nucleations and on the very strong dependence of SC formation on CO-designation in some situations. This is likely related to (and is the basis for?) the fact that budding yeast has many more COs per bivalent than other organisms despite its diminutive genome content. More generally, this mechanism can give nested sets comprising patterns for multiple entities, not just two (77).
CO homeostasis as manifested in CO-correlated Zip3 foci
In yeast, when the number of total recombinational interactions is reduced by reducing the total number of DSBs, the number of COs (marked by Zip3 foci) does not decrease commensurately, a phenomenon known as CO homeostasis (102). At reduced DSB levels, the average distance between COs increases and CoC curves shift to higher values (75,78 Figure 2E). Viewed "phenomenologically", this would seem to suggest that "CO interference is increased". However, by quantitative analysis, the observed results are explained simply by the decrease in the number of precursor interactions, with no change in the strength of CO-designation or CO interference. In fact, the phenomenon of “CO homeostasis” can be fully explained by the interplay of the reduced number of precursors and the normal CO-designation/interference process (9, 75,78). This is understandable as follows: the lower the number of precursors, the less likely it is that a given precursor will be affected by interference coming in from a nearby precursor and thus the more likely that precursor is to give a CO. Thus, the reduction in COs expected from a reduced number of DSBs/precursors is partially compensated by an increase in the probability that each DSB/precursor will give a CO. These two opposing effects are sufficient to quantitatively explain CO homeostasis of programmed COs.
A CO driving force for structurally discontinuous bivalents
Several "mutant" situations have been described in which only a subset of genomic chromosome length is available for CO-designation and, concomitantly, the available regions acquire more than the usual number of COs. A classical example is the "inter-chromosomal effect" in Drosophila in which the presence of a gross chromosomal rearrangement (e.g. inversion) triggers both prolongation of prophase and a higher number of COs on the regular regions of the genome, but still with the presence of detectable interference (by standard criteria) (e.g. 103 and references therein). More recent examples are provided by mutants in Arabidopsis (104) and Sordaria (asy2; D.Z. unpublished) in which, on a per-nucleus basis, pre-SC homolog pairing is defective but with concomitant occurrence of a higher than normal number of COs in the regions that do manage to pair. Other examples of such situations likely exist. Such situations will be manifested in decreased CO interference by standard criteria. It remains to be determined whether these effects imply a specific tendency for COs to occur in a particular number per nucleus per se (104) and/or an effective tendency in this direction because structurally aberrant regions trigger checkpoint surveillance responses which increase either the time available for CO-designation and/or the nature of that process itself. The point of relevance here is that there is no particular reason to invoke a change in the nature of the spreading interference signal per se although, of course, this cannot be excluded.
3.5 The Logic of CO Patterning
The phenomena of the two previous sections are qualitatively compatible not only with different specific mechanisms of CO patterning but with different basic logics for the underlying events. Three specific models have been presented: the first such model as proposed by King and Mortimer (105); the counting model of Stahl and colleagues (106); and our own "beam-film" model (71,75,97). The three models are based on different logic (as well as having different mechanisms). Notably, also, the beam-film model was originally formulated in the context of a stress-and-stress relief mechanism (above), but is in fact mathematically applicable to any mechanism that works by the same logic. In addition, mathematical models have been presented that describe outcomes without regard to underlying mechanism. When we actually know how CO interference works, some yet unenvisioned and/or more complex logic may emerge. Nonetheless, for the moment, several points seem clear.
First, all three specific models assume "precursors" and "CO-designation". Precursors are presumptively DSB-mediated inter-homolog interactions (e.g. bridges, discussed above). "CO-designation" is the process which determines that a particular precursor becomes fated to eventually become a CO. CO interference infuences the pattern of CO-designation sites. Precursors that do not undergo CO-designation end up maturing to some other fate. Also, in all three models, "CO maturation" can be a factor.
Second, the counting model envisioned that some process progresses along the chromosomes from one precursor interaction to the next and determines a CO-designation and then skips some roughly integer number of precursors and then carries out another CO-designation. Despite the fact that this model does a spectacular job of explaining experimental data (106), it seems not to be correct (102).
Third, the mathematics of the beam-film model make it possible to specify experimentally-verifiable features. This applies most notably to precursors, i.e. their number and whether the precursors are evenly or randomly (Poisson) distributed along chromosomes and whether they occur in similar numbers in different nuclei or are randomly distributed. Most other models assume a Poisson distribution of precursors among different chromosomes within and among nuclei and/or along an individual chromosome, which is likely not usually the in vivo situation.
Fourth, the King and Mortimer model specifies that, following CO-designation, the interference signal will continue indefinitely until/unless it runs in an interference signal coming from the other direction from an adjacent CO-designation. The implication of this situation is that the number of COs will depend on the kinetics of the situation, i.e., the relative rates of CO-designation and spreading of the interference signal. This would not be true either of a counting model or of the logic of the beam-film model.
Fifth, the logic of the beam-film model is different in logic from the other models and has several other unique features.
CO interference spreads outward in both directions from each CO-designation site, automatically dissipating with distance (see also above). This implies that each CO-designation automatically sets up a "zone of interference" which disfavors further CO-designations in the affected region, to decreasing extent with increasing distance from the designation site.
CO-designations occur sequentially. As a result, a second CO designation will tend to occur away from previous the first interference zone and ensuing COs, if any, will "fill in the holes" between prior CO-designation/interference zones, thus giving even spacing of COs. In this logic, kinetics plays no role.
The CO-designation process is very strong (or, equivalently, precursor interactions are very sensitive to CO-designation). This has two major consequences. First, it can ensure that every bivalent acquires at least one CO-designation as required for regular MI homolog segregation (further discussion below). Second, it implies that CO-designations can continue to occur until no more precursors are sensitive enough to respond; and since the effect of CO interference is to reduce the sensitivity of precursors, CO-designations will keep occurring until they are limited by the effects of interference. Thus, the "strength of CO interference", and more specifically the length over which the interference signal spreads (the "interference distance") is very important for determining not only the tendency for even spacing of COs but also the number of COs.
Finally: the logic of the beam-film can be captured mathematically; and simulations based on these mathematical expressions can explain CO patterns very accurately in many different situations, including predictions as to the nature of the precursor array and maturation efficiency, as well as features of CO patterning per se, and provide a very useful tool in revealing and/or appreciating complex features of CO patterns (e.g. 75,77,61; above).
3.6 The Obligatory CO (and Crossover Assurance)
Regular segregation of homologs at the MI division requires that they be connected (above). In the canonical program, this connection requires at least one CO plus sister connections distal to the most centromere-proximal CO. As a result, as an observed fact, virtually every bivalent acquires at least one CO, even when the average number of COs is very low, including one and only one per bivalent (e.g. 107). This observed fact is referred to operationally as "the obligatory CO".
Three types of explanations for ensuring the obligatory CO have been considered. First, there is some special mechanism to give the first CO and then another mechanism takes care of CO interference and later COs. Second, interference is per se required to ensure the first CO and thus keep the frequency of zero-CO bivalents to an acceptably low level. This possibility has arisen, historically, in the context of the King and Mortimer model (above). In that consideration, precursors were assumed to be Poisson-distributed among chromosomes (e.g. for a particular chromosome in different nuclei or among different chromosomes within a given nucleus). In such a case, a particular bivalent would sometimes fail to acquire even one precursor and the consequence would be an unacceptably high frequency of zero-CO bivalents. For this reason, the King and Mortimer postulated that passage of interference through a region resulted in release of precursors in that region which, in turn would rebind in regions that had not yet "experienced" interference. As a result, bivalents that lacked even a single precursor (and thus could not have experienced interference) would eventually acquire at least one precursor and thus could acquire a CO. This prediction is not supported by current data (78). Third, the obligatory CO is achieved as an intrinsic feature of the overall CO patterning process. The logic of the beam-film model places it in this category. Qualitatively, the obligatory CO can be ensured by making the strength of the CO-designation process (or the sensitivity of precursors to that process) sufficiently high that, as a practical matter, every bivalent virtually always acquires at least one CO. (In the original "stress hypothesis", it would be necessary that the level of stress, or the sensitivity of precursors to that stress, would be high enough to ensure that one precursor would undergo stress-promoted CO designation.) However, additional features must also be in place for this scenario to work. (a) Each CO-designated interaction must successfully mature to a CO/chiasma. In the absence of efficient maturation, a bivalent could acquire a single obligatory "CO-designation" but the effect would then be lost because that CO-designated event would not actually become a CO. (b) Each bivalent must always acquire not only at least one precursor but a sufficient number of precursors that at least one is sufficiently sensitive to undergo CO-designation. This requirement is influenced by several variables whose effects can contribute in various combinations to ensure an appropriate final outcome (75; N.K. and L.Zhang, unpublished). These factors include: the average number of precursors for a given bivalent; the extent to which a given chromosome always acquires the same/similar number of precursors in all nuclei (i.e. deviation from a random/Poisson distribution of precursors on that chromosome for different nuclei); the absolute sensitivities of the precursors to CO-designation or, equivalently, the strength of the CO designation process; and finally, the distribution of precursor sensitivities around the average, with a narrower distribution implying a smaller fraction of precursors that will not be adequately sensitive to the CO designation process. Importantly, the one variable that is NOT required for the obligatory CO is CO interference (75).
It can also be noted that the average number of precursors per bivalent is proportional to chromosome length. It is this effect which underlies the well-known fact that (all other things being equal), shorter chromosomes have higher frequencies of zero-CO bivalents (e.g. for yeast Chromosome III, see 75).
Recently, the term "CO assurance" has been introduced to convey, in a mechanistically general way, the fact that various features of the recombination process are required to ensure the observed final outcome of at least one CO per bivalent, thereby distinguishing the "observational phenomenon" from the "mechanistic requirements" (108,9). Specifically: "Crossover assurance comprises temporally and functionally distinct designation/commitment and implementation/execution steps" (108). This term will thus encompass any/all of the specific features described above.
3.7 Functional Coupling between Recombinosome and Axis Changes During and After CO Patterning
A prominent feature of meiotic recombination is the intimate relationship between the biochemical components required for DNA events and the chromosome structural axes. Both features are specified by a combination of meiosis-specific components and components that participate (also) in the corresponding non-meiotic processes, i.e. general DNA double-strand break repair and prophase chromosome structure in mitotic cells (to which meiotic prophase structure is highly analogous; 109). Classical examples are mitotic/meiotic RecA homologs Rad51/Dmc1 and cohesin kleisin subunits Scc1-Mcd1/Rec8 and Rad21L.
What underlies CO-designation and CO Interference?
At present, there is almost no concrete information about which molecules are involved in these processes. A recent study identifies a SUMO-dependent pathway involved in these processes, for which known targets are TopoII and meiotic axis component Red1, with others likely remaining to be identified. However, this pathway enhances, but is not essential for, the patterning process. Also, Qiao et al (110), based on studies in mouse, propose that recombinosome differentiation (into CO and NCO types) involves a balance between the opposing effects of SUMOylation and ubiquitination as mediated, in that organism, by Zip3 and Hei10. The primary target of CO-designation should be the recombination complex itself, although direct targeting of the recombinosome/structure interface is also possible. Thus, analysis of components likely present at the critical stage should be fruitful. But what about structure components that might be essential for this process? Analysis of these molecules is complicated because they are almost all involved in setting up prophase structure and/or even earlier events, and thus affect assembly of the array of total "precursor" recombinational interactions before CO patterning stage, as well as having key roles in downstream steps that are required for generating a detectable signal. And most critically, what is the molecular basis for spreading of information along the chromosomes? And if the interference signal spreads along the chromosome axes, how might a signal be transmitted from the recombination complex to the structures to initiate this effect? (For previous discussions of the difficulties in dissecting CO interference, see 9,69). New clues and/or new approaches are needed to answer these questions.
Post-interference events
In the canonical program, CO-designation and accompanying interference are followed by prominent changes at both the DNA and structural levels. CO recombination proceeds via two specific long-lived intermediates that progress to mature COs via discrete sequential stages (9). Concomitant with initiation of these events, SC is installed all along the chromosomes, with nucleation at sites of COs and, in most organisms, sites of other precursors (above). SC installation can also be accompanied by other prominent changes, e.g. removal or remodeling of HORMAD proteins (e.g. 111).
The existence of these coordinate downstream changes are indicated by many observations. Among these, "ZMM" proteins are specifically required in budding yeast for both progression of CO recombination and SC formation; however, at least two of these proteins, the SC central region component Zip1 and MutS-homolog Msh4/5, are not required for CO interference (112,78). Instead, these factors likely mediate functional and physical linkages between CO-specific recombination complexes and underlying SC at the nucleation and/or later stages. In some cases, e.g. Arabidopsis (113), ZMM proteins are required for formation of "regulated" COs but not for SC formation. This may reflect the fact that in plants, with their long chromosomes, many SC nucleations normally occur at sites other than CO-designated sites, so loss of normal CO-designation has little effect.
Two aspects of SC formation seem interesting in this context.
Is spreading of the SC licensed by spreading of the interference signal? There are hints that SC spreading might be directly licensed by the CO interference signal per se. This would be true not only at the point of nucleation of interference (where CO-designation would nucleate onset of interference and thus local nucleation of SC, as in the nested interference model described above) but also during propagation of the SC away from those positions along the chromosomes, which would occur in the wake of the spreading interference signal. One hint to this effect is that in Sordaria, when DSBs are induced artificially by gamma-irradiation, only a limited amount of SC is formed at each of a few positions, implying that SC cannot spread without limit along the chromosome lengths (114). Similarly, in yeast, reduction in the number of CO-designations concomitantly reduces the number of SC nucleations and does not lead to full-length SC (115). A second hint is that SC installation is often accompanied by chromosome shortening (a characteristic of the zygotene stage) (47). It is easy to envision that the interference signal might result in longitudinal shortening and thus, in some way, license SC transverse filament installation. (This is, in fact, a predicted effect of relief of chromatin expansion stress in the stress model described above; 97,98).
Two-phase synapsis. In the normal meiotic program, SC formation along chromosome arms is precluded until it is licensed by CO-designation and interference, where such licensing involves nucleation at particular positions followed by spreading (above). This regulated installation limits SC formation to homologous chromosomes. In contrast, nearly universally, SC can be installed promiscuously at a later time in the relevant stage, where it is installed irrespective of underlying chromosomal homology and without programmed nucleation, e.g. as seen in "fold-back synapsis" and other configurations (so-called "two-phase synapsis"; 116,47). In accord with the idea that an expanded chromosome stage might limit SC installation, with chromosome compaction then permitting SC installation: chromatin expands globally at leptotene (prior to CO-designation/interference), compacts at zygotene (e.g. as a global effect that would presumably complement interference-based compaction), expands at early pachytene and then compacts again at mid/late pachytene, which is the time of promiscuous SC installation (97).
Also, SC nucleates at centromeres at/before the leptotene stage in both yeast and Drosophila and, in yeast, does not require normal "nucleation factors" that are essential for interstitial SC nucleation at CO sites (e.g. Zip3) (117,28,29). Perhaps centromeres have a more compact chromatin structure, and permit SC formation, analogously to the second, non-specific phase of synapsis (118).
3.8 The evolutionary driving force for CO interference?
In general, CO interference could be evolutionarily advantageous for either (or both) of two reasons: because of its genetic consequences or for some mechanistic reason (previous discussions in 69,9,10, 87).
Genetic advantages. With respect to genetic consequences, CO interference clearly directly determines the spacing of COs along bivalents. However, the considerations above imply that it also strongly influences the number of COs, with CO-designations occurring sequentially until limited by the effects of CO interference. If this is true, a shorter or longer "interference distance" implies more or fewer COs. Both of these effects (lower number of COs and the tendency for COs to be spaced out along the chromosomes) will tend to promote continued linkage of genes (and thus their resultant traits) located near one another on a chromosome, working against the tendency of COs to disrupt such linkages. This is one widely-suggested evolutionary rationale for CO interference. Naturally, other mechanisms for achieving this effect can be envisioned, thus perhaps explaining why some organisms appear to lack CO interference altogether (most notably S. pombe [119] and Aspergillus nidulans [120]).
A second possible evolutionary role might be considered. In many organisms, notably but not exclusively higher plants, whole genome duplications or other events can yield genomes containing more than two identical (or closely similar) copies of each chromosome. In such situations pairwise early recombinational interactions can lead to complex patterns of COs among different combinations of homologs. These complexities will, in principle, be reduced if CO interference extends along much or all of the lengths of the chromosomes. To the extent that polyploidy is both an advantage and a risk for sexual reproduction, CO interference might again be ameliorating the disadvantageous risks (121).
It is also interesting to consider that CO interference might be used as part of a process that limits CO formation to certain regions. For example, in many organisms (notably plants), COs/chiasmata tend to occur preferentially near the ends of chromosomes. A recent exploration of this situation in barley (65) has shown that the entire program of recombination/SC nucleation occurs earlier at ends than in the middle of the chromosome. It was proposed that this timing difference, coupled with CO interference could explain the paucity of COs in the middles of chromosomes: if CO-designation occurs first near the ends of a bivalent, spreading of the interference signal inward will preclude/disfavor formation of later COs in the central region. Implicit in this scenario is a greater sensitivity of precursors to CO-designation in regions near ends. Given that CO-designation is nested within the SC nucleation program, this difference will permit also SC formation in centrally-located regions without accompanying CO-designation while concomitantly ensuring a timing difference such that SC formation in central regions occurs later than in near-end regions. A similar situation likely occurs in other organisms, e.g. human male meiosis (e.g. 66).
Mechanistic advantages. With respect to mechanistic advantages, one possibility that having fewer and/or more evenly-spaced COs, and thus fewer and/or more evenly-spaced diplotene chiasmata, may be important for creating a chromosome structure that permits regular segregation of homologs at Meiosis I (10, 87). A second possible advantage of interference arises from the fact that homolog pairing/coalignment at prophase necessarily (and, in fact) involves some risk of unwanted entanglements. By diplotene, the only connections among chromosomes that remain to enforce such entanglements are COs/chiasmata. Thus, fewer COs and/or a tendency for COs to be far from centromeres will minimize the risk that entanglements generated earlier in prophase will persist and compromise the process of homolog segregation at MI. A third hypothesis is that CO interference is required to ensure that at least one CO occurs along each bivalent. This hypothesis seems not to be correct (see third scenario for the basis of the obligatory CO above).
3.9 Summary
Despite significant progress in recent years, the phenomenon of CO interference remains substantially enigmatic, particularly as regards mechanism and biological significance, thus providing fertile ground for future research.
Acknowledgments
Research in the laboratories of the authors was supported by grants to D.Z. from the Centre National de la Recherche Scientifique (Unité Mixte de Recherche 8621, now I2BC) and to N.K (including a sub-contract collaboration with D.Z.) from the NIH RO1-GM044794. We are grateful to Jim Henle and Beth Weiner for help in manuscript preparation and to members of our laboratories for their many contributions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Niklas KJ, Cobb ED, Kutschera U. Did meiosis evolve before sex and the evolution of eukaryotic life cycles? Bioessays. 2014 Nov;36(11):1091–1101. doi: 10.1002/bies.201400045. [DOI] [PubMed] [Google Scholar]
- 2.Wilkins AS, Holliday R. The evolution of meiosis from mitosis. Genetics. 2009 Jan;181(1):3–12. doi: 10.1534/genetics.108.099762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurst LD, Nurse P. A note on the evolution of meiosis. J Theor Biol. 1991 Jun 21;150(4):561–563. doi: 10.1016/s0022-5193(05)80447-3. [DOI] [PubMed] [Google Scholar]
- 4.Cabral G, Marques A, Schubert V, Pedrosa-Harand A, Schlögelhofer P. Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat Commun. 2014 Oct 8;5:5070. doi: 10.1038/ncomms6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heckmann S, Jankowska M, Schubert V, Kumke K, Ma W, Houben A. Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nat Commun. 2014 Oct 8;5:4979. doi: 10.1038/ncomms5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zickler D, Kleckner N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb Perspect Biol. 2015 May 18;7(6):a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boateng KA, Bellani MA, Gregoretti IV, Pratto F, Camerini-Otero RD. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev Cell. 2013 Jan 28;24(2):196–205. doi: 10.1016/j.devcel.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storlazzi A, Gargano S, Ruprich-Robert G, Falque M, David M, Kleckner N, Zickler D. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell. 2010 Apr 2;141(1):94–106. doi: 10.1016/j.cell.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter N. Meiotic Recombination: The Essence of Heredity. Cold Spring Harb Perspect Biol. 2015 Oct 28;7(12):a016618. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleckner N. Meiosis: how could it work? Proc Natl Acad Sci U S A. 1996 Aug 6;93(16):8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beumer KJ, Pimpinelli S, Golic KG. Induced chromosomal exchange directs the segregation of recombinant chromatids in mitosis of Drosophila. Genetics. 1998 Sep;150(1):173–188. doi: 10.1093/genetics/150.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleckner N, Zhang L, Weiner B, Zickler D. Meiotic chromosome dynamics. In: Rippe K, editor. Genome Organization and Function in the Mammalian Cell Nucleus. New York: John Wiley and Sons; 2012. pp. 487–533. [Google Scholar]
- 13.Tsai JH, Yan R, McKee BD. Homolog pairing and sister chromatid cohesion in heterochromatin in Drosophila male meiosis I. Chromosoma. 2011 Aug;120(4):335–351. doi: 10.1007/s00412-011-0314-0. [DOI] [PubMed] [Google Scholar]
- 14.Kurdzo EL, Dawson DS. Centromere pairing--tethering partner chromosomes in meiosis I. FEBS J. 2015 Jul;282(13):2458–2470. doi: 10.1111/febs.13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006 Apr 7;125(1):59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 16.Chikashige Y, Yamane M, Okamasa K, Tsutsumi C, Kojidani T, Sato M, Haraguchi T, Hiraoka Y. Membrane proteins Bqt3 and −4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J Cell Biol. 2009 Nov 2;187(3):413–427. doi: 10.1083/jcb.200902122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding DQ, Yamamoto A, Haraguchi T, Hiraoka Y. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev Cell. 2004 Mar;6(3):329–341. doi: 10.1016/s1534-5807(04)00059-0. [DOI] [PubMed] [Google Scholar]
- 18.Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell. 2009 Nov;17(5):598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994 Apr 8;264(5156):270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- 20.Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998 Mar;111(Pt 6):701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- 21.Ding DQ, Haraguchi T, Hiraoka Y. From meiosis to postmeiotic events: alignment and recognition of homologous chromosomes in meiosis. FEBS J. 2010 Feb;277(3):565–570. doi: 10.1111/j.1742-4658.2009.07501.x. [DOI] [PubMed] [Google Scholar]
- 22.Howard-Till RA, Lukaszewicz A, Loidl J. The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. PLoS Genet. 2011 Mar;7(3):e1001359. doi: 10.1371/journal.pgen.1001359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loidl J, Lukaszewicz A, Howard-Till RA, Koestler T. The Tetrahymena meiotic chromosome bouquet is organized by centromeres and promotes interhomolog recombination. J Cell Sci. 2012 Dec 1;125(Pt 23):5873–5880. doi: 10.1242/jcs.112664. [DOI] [PubMed] [Google Scholar]
- 24.Lake CM, Hawley RS. The molecular control of meiotic chromosomal behavior: events in early meiotic prophase in Drosophila oocytes. Annu Rev Physiol. 2012;74:425–451. doi: 10.1146/annurev-physiol-020911-153342. [DOI] [PubMed] [Google Scholar]
- 25.Rog O, Dernburg AF. Chromosome pairing and synapsis during Caenorhabditis elegans meiosis. Curr Opin Cell Biol. 2013 Jun;25(3):349–356. doi: 10.1016/j.ceb.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cahoon CK, Hawley RS. Flies get a head start on meiosis. PLoS Genet. 2013;9(12):e1004051. doi: 10.1371/journal.pgen.1004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christophorou N, Rubin T, Huynh JR. Synaptonemal complex components promote centromere pairing in pre-meiotic germ cells. PLoS Genet. 2013;9(12):e1004012. doi: 10.1371/journal.pgen.1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeo S, Lake CM, Morais-de-Sá E, Sunkel CE, Hawley RS. Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr Biol. 2011 Nov 8;21(21):1845–1851. doi: 10.1016/j.cub.2011.09.044. [DOI] [PubMed] [Google Scholar]
- 29.Tanneti NS, Landy K, Joyce EF, McKim KS. A pathway for synapsis initiation during zygotene in Drosophila oocytes. Curr Biol. 2011 Nov 8;21(21):1852–1857. doi: 10.1016/j.cub.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 30.MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005 Dec 16;123(6):1037–1050. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai JH, McKee BD. Homologous pairing and the role of pairing centers in meiosis. J Cell Sci. 2011;Jun124(Pt 12):1955–1963. doi: 10.1242/jcs.006387. [DOI] [PubMed] [Google Scholar]
- 32.Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009 Nov 25;139(5):907–919. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips CM, Dernburg AF. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev Cell. 2006 Dec;11(6):817–829. doi: 10.1016/j.devcel.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 34.Phillips CM, Meng X, Zhang L, Chretien JH, Urnov FD, Dernburg AF. Identification of chromosome sequence motifs that mediate meiotic pairing, synapsis in C. elegans. Nat Cell Biol. 2009 Aug;11(8):934–942. doi: 10.1038/ncb1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rog O, Dernburg AF. Direct Visualization Reveals Kinetics of Meiotic Chromosome Synapsis. Cell Rep. 2015 Mar 10; doi: 10.1016/j.celrep.2015.02.032. pii: S2211-1247(15)00178-3. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, Gruenbaum Y, Schweizer D, Loidl J, Jantsch V. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell. 2007 Jun;12(6):873–885. doi: 10.1016/j.devcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 37.Wynne DJ, Rog O, Carlton PM, Dernburg AF. Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol. 2012 Jan 9;196(1):47–64. doi: 10.1083/jcb.201106022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lui DY, Colaiácovo MP. Meiotic development in Caenorhabditis elegans. Adv Exp Med Biol. 2013;757:133–170. doi: 10.1007/978-1-4614-4015-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woglar A, Jantsch V. Chromosome movement in meiosis I prophase of Caenorhabditis elegans. Chromosoma. 2014 Mar;123(1-2):15–24. doi: 10.1007/s00412-013-0436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labella S, Woglar A, Jantsch V, Zetka M. Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev Cell. 2011 Nov 15;21(5):948–958. doi: 10.1016/j.devcel.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 41.Viera A, Santos JL, Rufas JS. Relationship between incomplete synapsis and chiasma localization. Chromosoma. 2009 Jun;118(3):377–389. doi: 10.1007/s00412-009-0204-x. [DOI] [PubMed] [Google Scholar]
- 42.Croft JA, Jones GH. Meiosis in Mesostoma ehrenbergii ehrenbergii IV. Recombination nodules in spermatocytes and a test of the correspondence of late recombination nodules and chiasmata. Genetics. 1989 Feb;121(2):255–262. doi: 10.1093/genetics/121.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKee BD. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim Biophys Acta. 2004 Mar 15;1677(1-3):165–180. doi: 10.1016/j.bbaexp.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 44.McKee BD, Yan R, Tsai JH. Meiosis in male Drosophila. Spermatogenesis. 2012 Jul 1;2(3):167–184. doi: 10.4161/spmg.21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas SE, Soltani-Bejnood M, Roth P, Dorn R, Logsdon JM, Jr, McKee BD. Identification of two proteins required for conjunction and regular segregation of achiasmate homologs in Drosophila male meiosis. Cell. 2005 Nov 18;123(4):555–568. doi: 10.1016/j.cell.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 46.Rasmussen SW. Meiosis in Bombyx mori females. Philos Trans R Soc Lond B Biol Sci. 1977 Mar 21;277(955):343–350. doi: 10.1098/rstb.1977.0022. [DOI] [PubMed] [Google Scholar]
- 47.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 48.Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol. 2001 Aug;2(8):621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- 49.Cowan CR, Carlton PM, Cande WZ. Reorganization and polarization of the meiotic bouquet-stage cell can be uncoupled from telomere clustering. J Cell Sci. 2002 Oct 1;115(Pt 19):3757–3766. doi: 10.1242/jcs.00054. [DOI] [PubMed] [Google Scholar]
- 50.Gelei J. Weitere Studien über die Oogenese des Dendrocoelum lacteum. Die Konjugation der Chromosomen in der Literatur unde meine Befunde. Archiv f Zellforsch. 1921;16:300–365. [Google Scholar]
- 51.Kezer J, Sessions SK, Leon P. The meiotic structure and behavior of the strongly heteromorphic X/Y sex chromosomes of neotropical plethodontid salamanders of the genus Oedipina. Chromosoma. 1989;98:433–442. [Google Scholar]
- 52.Rasmussen SW, Holm PB. Human meiosis II. Chromosome pairing and recombination nodules in human spermatocytes. Carlsberg Res. Comm. 1978;43:275–327. [Google Scholar]
- 53.Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 54.Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006 Jun;115(3):158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
- 55.Lee CY, Conrad MN, Dresser ME. Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet. 2012;8(5):e1002730. doi: 10.1371/journal.pgen.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Storlazzi A, Tessé S, Gargano S, James F, Kleckner N, Zickler D. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 2003 Nov 1;17(21):2675–2687. doi: 10.1101/gad.275203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harper L, Golubovskaya I, Cande WZ. A bouquet of chromosomes. J Cell Sci. 2004 Aug 15;117(Pt 18):4025–4032. doi: 10.1242/jcs.01363. [DOI] [PubMed] [Google Scholar]
- 58.Scherthan H. Factors directing telomere dynamics in synaptic meiosis. Biochem Soc Trans. 2006 Aug;34(Pt 4):550–553. doi: 10.1042/BST0340550. [DOI] [PubMed] [Google Scholar]
- 59.MacQueen AJ, Colaiácovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002 Sep 15;16(18):2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Espagne E, Vasnier C, Storlazzi A, Kleckner NE, Silar P, Zickler D, Malagnac F. Sme4 coiled-coil protein mediates synaptonemal complex assembly, recombinosome relocalization, and spindle pole body morphogenesis. Proc Natl Acad Sci U S A. 2011 Jun 28;108(26):10614–10619. doi: 10.1073/pnas.1107272108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Muyt A, Zhang L, Piolot T, Kleckner N, Espagne E, Zickler D. E3 ligase Hei10: a multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev. 2014 May 15;28(10):1111–1123. doi: 10.1101/gad.240408.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conrad MN, Lee CY, Wilkerson JL, Dresser ME. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007 May 22;104(21):8863–8868. doi: 10.1073/pnas.0606165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prieto I, Kouznetsova A, Fütterer A, Trachana V, Leonardo E, Alonso Guerrero A, Cano Gamero M, Pacios-Bras C, Leh H, Buckle M, Garcia-Gallo M, Kremer L, Serrano A, Roncal F, Albar JP, Barbero JL, Martínez-A C, van Wely KH. Synaptonemal complex assembly and H3K4Me3 demethylation determine DIDO3 localization in meiosis. Chromosoma. 2009 Oct;118(5):617–632. doi: 10.1007/s00412-009-0223-7. [DOI] [PubMed] [Google Scholar]
- 64.Schvarzstein M, Pattabiraman D, Bembenek JN, Villeneuve AM. Meiotic HORMA domain proteins prevent untimely centriole disengagement during Caenorhabditis elegans spermatocyte meiosis. Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):E898–E907. doi: 10.1073/pnas.1213888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Higgins JD, Osman K, Jones GH, Franklin FC. Factors underlying restricted crossover localization in barley meiosis. Annu Rev Genet. 2014;48:29–47. doi: 10.1146/annurev-genet-120213-092509. [DOI] [PubMed] [Google Scholar]
- 66.Pratto F, Brick K, Khil P, Smagulova F, Petukhova GV, Camerini-Otero RD. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014 Nov 14;346(6211):1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klutstein M, Cooper JP. The Chromosomal Courtship Dance-homolog pairing in early meiosis. Curr Opin Cell Biol. 2014 Feb;26:123–131. doi: 10.1016/j.ceb.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson LK, Lohmiller LD, Tang X, Hammond DB, Javernick L, Shearer L, Basu-Roy S, Martin OC, Falque M. Combined fluorescent and electron microscopic imaging unveils the specific properties of two classes of meiotic crossovers. Proc Natl Acad Sci U S A. 2014 Sep 16;111(37):13415–13420. doi: 10.1073/pnas.1406846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hillers KJ. Crossover interference. Curr Biol. 2004 Dec 29;14(24):R1036–R1037. doi: 10.1016/j.cub.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 70.Berchowitz LE, Copenhaver GP. Genetic interference: don't stand so close to me. Curr Genomics. 2010 Apr;11(2):91–102. doi: 10.2174/138920210790886835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang S, Zickler D, Kleckner N, Zhang L. Meiotic crossover patterns: obligatory crossover, interference and homeostasis in a single process. Cell Cycle. 2015;14(3):305–314. doi: 10.4161/15384101.2014.991185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sturtevant AH. The behavior of the chromosomes as studied through linkage. Z induct Abstamm-u VererbLehre. 1915;13:234–287. [Google Scholar]
- 73.Muller HJ. The mechanism of crossing over, parts I-IV. Am Nat. 1916;50:193–434. [Google Scholar]
- 74.Charles DR. The spatial distribution of cross-overs in X-chromosome tetrads of Drosophila melanogaster. J Genet. 1938;36:103–126. [Google Scholar]
- 75.Zhang L, Liang Z, Hutchinson J, Kleckner N. Crossover patterning by the beam-film model: analysis and implications. PLoS Genet. 2014 Jan 30;10(1):e1004042. doi: 10.1371/journal.pgen.1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.White M, Zhang L, Wang S, Kleckner N. Modeling of CO interference as inspired by the beam-film model. Methods in Microbiology. 2016 in press. [Google Scholar]
- 77.Zhang L, Espagne E, de Muyt A, Zickler D, Kleckner NE. Interference-mediated synaptonemal complex formation with embedded crossover designation. Proc Natl Acad Sci U S A. 2014 Nov 25;111(47):E5059–E5068. doi: 10.1073/pnas.1416411111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang L, Wang S, Yin S, Hong S, Kim KP, Kleckner N. Topoisomerase II mediates meiotic crossover interference. Nature. 2014 Jul 31;511(7511):551–556. doi: 10.1038/nature13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Boer E, Dietrich AJ, Höög C, Stam P, Heyting C. Meiotic interference among MLH1 foci requires neither an intact axial element structure nor full synapsis. J Cell Sci. 2007 Mar 1;120(Pt 5):731–736. doi: 10.1242/jcs.003186. [DOI] [PubMed] [Google Scholar]
- 80.Malkova A, Swanson J, German M, McCusker JH, Housworth EA, Stahl FW, Haber JE. Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics. 2004 Sep;168(1):49–63. doi: 10.1534/genetics.104.027961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jones GH. The control of chiasma distribution. Symp Soc Exp Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- 82.Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002 Dec 13;111(6):791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 83.Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011 Aug 5;146(3):372–383. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 84.Zickler D, Espagne E. Sordaria, a model system to uncover links between meiotic pairing and recombination. THIS ISSUE. 2016 doi: 10.1016/j.semcdb.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Boer E, Stam P, Dietrich AJ, Pastink A, Heyting C. Two levels of interference in mouse meiotic recombination. Proc Natl Acad Sci U S A. 2006 Jun 20;103(25):9607–9612. doi: 10.1073/pnas.0600418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Globus ST, Keeney S. The joy of six: how to control your crossovers. Cell. 2012 Mar 30;149(1):11–12. doi: 10.1016/j.cell.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 87.Qiao H, Chen JK, Reynolds A, Höög C, Paddy M, Hunter N. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet. 2012 Jun;8(6):e1002790. doi: 10.1371/journal.pgen.1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Joyce EF, McKim KS. Drosophila PCH2 is required for a pachytene checkpoint that monitors double-strand-break-independent events leading to meiotic crossover formation. Genetics. 2009 Jan;181(1):39–51. doi: 10.1534/genetics.108.093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Libuda DE, Uzawa S, Meyer BJ, Villeneuve AM. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature. 2013 Oct 31;502(7473):703–706. doi: 10.1038/nature12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hillers KJ, Villeneuve AM. Chromosome-wide control of meiotic crossing over in C. elegans. Curr Biol. 2003 Sep 16;13(18):1641–1647. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 91.Gruhn JR, Rubio C, Broman KW, Hunt PA, Hassold T. Cytological studies of human meiosis: sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PLoS One. 2013 Dec 20;8(12):e85075. doi: 10.1371/journal.pone.0085075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Drouaud J, Mercier R, Chelysheva L, Bérard A, Falque M, Martin O, Zanni V, Brunel D, Mézard C. Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet. 2007 Jun;3(6):e106. doi: 10.1371/journal.pgen.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, Li J, Xu L, Tang F, Xie XS, Qiao J. Genome analyses of single human oocytes. Cell. 2013 Dec 19;155(7):1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 94.Hu L, Vecchiarelli AG, Mizuuchi K, Neuman KC, Liu J. Directed and persistent movement arises from mechanochemistry of the ParA/ParB system. Proc Natl Acad Sci U S A. 2015 Dec 22;112(51):E7055–E7064. doi: 10.1073/pnas.1505147112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vecchiarelli AG, Li M, Mizuuchi M, Mizuuchi K. Differential affinities of MinD and MinE to anionic phospholipid influence Min patterning dynamics in vitro. Mol Microbiol. 2014 Aug;93(3):453–463. doi: 10.1111/mmi.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Turing AM. The chemical basis of morphogenesis. Phil Trans R Soc Lond Ser B. 1952;237(641):37–72. [Google Scholar]
- 97.Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, Hutchinson J. A mechanical basis for chromosome function. Proc Natl Acad Sci U S A. 2004 Aug 24;101(34):12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004 Apr 2;117(1):29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 99.Storlazzi A, Tesse S, Ruprich-Robert G, Gargano S, Pöggeler S, Kleckner N, Zickler D. Coupling meiotic chromosome axis integrity to recombination. Genes Dev. 2008 Mar 15;22(6):796–809. doi: 10.1101/gad.459308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Albini SM, Jones GH. Synaptonemal complex-associated centromeres and recombination nodules in plant meiocytes prepared by an improved surface-spreading technique. Exp Cell Res. 1984 Dec;155(2):588–592. doi: 10.1016/0014-4827(84)90219-2. [DOI] [PubMed] [Google Scholar]
- 101.Zickler D, Moreau PJ, Huynh AD, Slezec AM. Correlation between pairing initiation sites, recombination nodules and meiotic recombination in Sordaria macrospora. Genetics. 1992 Sep;132(1):135–148. doi: 10.1093/genetics/132.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006 Jul 28;126(2):285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Joyce EF, McKim KS. Chromosome axis defects induce a checkpoint-mediated delay and interchromosomal effect on crossing over during Drosophila meiosis. PLoS Genet. 2010 Aug 12;6(8):e1001059. doi: 10.1371/journal.pgen.1001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jahns MT, Vezon D, Chambon A, Pereira L, Falque M, Martin OC, Chelysheva L, Grelon M. Crossover localisation is regulated by the neddylation posttranslational regulatory pathway. PLoS Biol. 2014 Aug 12;12(8):e1001930. doi: 10.1371/journal.pbio.1001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.King JS, Mortimer RK. A polymerization model of chiasma interference and corresponding computer simulation. Genetics. 1990 Dec;126(4):1127–1138. doi: 10.1093/genetics/126.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lande R, Stahl FW. Chiasma interference and the distribution of exchanges in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1993;58:543–552. doi: 10.1101/sqb.1993.058.01.061. [DOI] [PubMed] [Google Scholar]
- 107.Jones GH, Franklin FC. Meiotic crossing-over: obligation and interference. Cell. 2006 Jul 28;126(2):246–248. doi: 10.1016/j.cell.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 108.Shinohara M, Oh SD, Hunter N, Shinohara A. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet. 2008 Mar;40(3):299–309. doi: 10.1038/ng.83. [DOI] [PubMed] [Google Scholar]
- 109.Liang Z, Zickler D, Prentiss M, Chang FS, Witz G, Maeshima K, Kleckner N. Chromosomes Progress to Metaphase in Multiple Discrete Steps via Global Compaction/Expansion Cycles. Cell. 2015 May 21;161(5):1124–1137. doi: 10.1016/j.cell.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qiao H, Prasada Rao HB, Yang Y, Fong JH, Cloutier JM, Deacon DC, Nagel KE, Swartz RK, Strong E, Holloway JK, Cohen PE, Schimenti J, Ward J, Hunter N. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat Genet. 2014 Feb;46(2):194–199. doi: 10.1038/ng.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, McKay MJ, Toth A. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009 Oct;5(10):e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fung JC, Rockmill B, Odell M, Roeder GS. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell. 2004 Mar 19;116(6):795–802. doi: 10.1016/s0092-8674(04)00249-1. [DOI] [PubMed] [Google Scholar]
- 113.Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M. The molecular biology of meiosis in plants. Annu Rev Plant Biol. 2015;66:297–327. doi: 10.1146/annurev-arplant-050213-035923. [DOI] [PubMed] [Google Scholar]
- 114.Tessé S, Storlazzi A, Kleckner N, Gargano S, Zickler D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12865–12870. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Henderson KA, Keeney S. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc Natl Acad Sci U S A. 2004 Mar 30;101(13):4519–4524. doi: 10.1073/pnas.0400843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McClintock Kleckner N. (1933): implications for meiotic chromosome pairing. In: Fedoroff N, Botstein D, editors. The Dynamic Genome: Barbara McClintock's Ideas in the Century of Genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 233–241. [Google Scholar]
- 117.Tsubouchi T, Macqueen AJ, Roeder GS. Initiation of meiotic chromosome synapsis at centromeres in budding yeast. Genes Dev. 2008 Nov 15;22(22):3217–3226. doi: 10.1101/gad.1709408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chuong H, Dawson DS. Meiotic cohesin promotes pairing of nonhomologous centromeres in early meiotic prophase. Mol Biol Cell. 2010 Jun 1;21(11):1799–1809. doi: 10.1091/mbc.E09-05-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994 Jul;137(3):701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Käfer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- 121.Bomblies K, Jones G, Franklin C, Zickler D, Kleckner N. The challenge of evolving stable polyploidy: could an increase in "crossover interference distance" play a central role? Chromosoma. 2016 Jan 12; doi: 10.1007/s00412-015-0571-4. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]