Abstract
Purpose
Identification of predictive biomarkers is critically needed to improve selection of patients who derive the most benefit from platinum-based chemotherapy. We hypothesized that decreased expression of SMARCA4/BRG1, a known regulator of transcription and DNA repair, is a novel predictive biomarker of increased sensitivity to adjuvant platinum-based therapies in NSCLC.
Experimental Design
The prognostic value was tested using a gene expression microarray from the Director’s Challenge Lung Study (n=440). The predictive significance of SMARCA4 was determined using a gene expression microarray (n=133) from control and treatment arms of the JBR.10 trial of adjuvant cisplatin/vinorelbine. Kaplan-Meier method and log-rank tests were used to estimate and test the differences of probabilities in overall survival (OS) and disease-specific survival (DSS) between expression groups and treatment arms. Multivariate Cox regression models were used while adjusting for other clinical covariates.
Results
In the Director’s Challenge Study, reduced expression of SMARCA4 was associated with poor OS compared to high and intermediate expression (P<0.001 and P=0.009, respectively). In multivariate analysis, compared to low, high SMARCA4 expression predicted a decrease in risk of death (HR=0.6, 95% CI: 0.4–0.8, P=0.002). In the JBR.10 trial, improved five-year DSS was noted only in patients with low SMARCA4 expression when treated with adjuvant cisplatin/vinorelbine (HR=0.1, 95% CI: 0.0–0.5, P=0.002 [low]; HR 1.0, 95% CI: 0.5–2.3, P=0.92 [high]). An interaction test was highly significant (P=0.01).
Conclusions
Low expression of SMARCA4/BRG1 is significantly associated with worse prognosis; however, it is a novel significant predictive biomarker for increased sensitivity to platinum-based chemotherapy in NSCLC.
Keywords: SMARCA4, BRG1, NSCLC, predictive, cisplatin
INTRODUCTION
Lung cancer is the most deadly cancer in the world and 85% of lung cancers are non-small cell lung cancer (NSCLC) (1). Chemotherapy remains a major treatment modality and the only therapy proven to prolong survival of early stage patients after surgery. Although in recent years there have been major advancements in early detection and targeted therapies, the five-year survival gains have remained relatively small (2). High mortality is due to advanced stage detection of the disease together with the absence of targetable driver mutations in most tumors leaving systemic chemotherapy as the only first line therapeutic option. These therapies are toxic, and while biomarkers can inform the selection of targeted therapies, biomarkers that enable the identification of patients who would benefit from chemotherapy have remained elusive.
The mechanisms responsible for drug resistance include increased efflux and/or inactivation of drugs, defects in apoptosis, and activation of DNA repair pathways (3). Studies on DNA repair pathways to date have been disappointing. ERCC1, a critical component of nucleotide excision repair (NER), has been one of the most well-studied genes in NSCLC in regards to cisplatin sensitivity, albeit with conflicting results attributed to issues regarding detection techniques in clinical tissues (4). The majority of studies have shown that low ERCC1 levels are associated with cisplatin sensitivity (5). However, effect sizes have been small, prospective studies have failed to confirm this association, and these markers are not used in clinical practice.
Overall, the predictive effect of driver mutations with drug sensitivity in tumors with “oncogene addiction”, such as ALK and EGFR is now clear. Numerous studies have attempted to evaluate the impact of tumor suppressors on clinical outcome in lung cancer, but none have produced clinically impactful results. For example, the effect of TP53 mutations on prognosis and chemotherapy sensitivity is unclear and inconsistent. Similarly RB and LKB1 mutations have not had clinical utility. However, little has been done to fully understand how loss of other tumor suppressors, such as SMARCA4/BRG1, affects treatment sensitivities to drugs in standard of care regimens, such as platinum-based therapies as well as emerging therapeutics.
The SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeling complex, which functions as a fundamental regulatory component of transcription, plays a critical role in DNA repair (8). SWI/SNF is frequently abnormal in lung cancer, but has not been previously studied for chemotherapy prediction in resected NSCLC. Notably, BRG1 (SMARCA4), one of two catalytic subunits of SWI/SNF, is a tumor suppressor and mutations have been identified in approximately 10% of NSCLC (9–11). SMARCA4 mutations and/or decreased expression have also been identified in other tumor cell lines and tissues (12, 13). Furthermore, alterations in other components of SWI/SNF, including the other catalytic subunit BRM (SMARCA2) and ARID1A, have been recently identified in cancer (8, 14). Even though somatic missense mutations appear to be the most common mutations, other mechanisms such as insertions, partial and complete deletions, and promoter methylation may have been less well studied but also contribute to the loss of BRG1 in lung cancer (11, 15). Interestingly, although SNF5-deficient rhabdoid tumors and SMARCA4/SMARCA2-deficient small cell carcinoma of the ovary, hypercalcemic type (SCCOHT) tumors do not exhibit genomic instability (16, 17), loss of SMARCA4/BRG1 function in lung cancer may lead to genomic instability as evidenced by a recent publication (18).
The SWI/SNF chromatin remodeling complex has recently been implicated in double-strand break (DSB) repair and NER, two DNA repair pathways inherently involved in resistance toward DNA-damaging agents (19–22). Recently, multiple in vitro studies have shown that reduced expression of BRG1 can enhance sensitivity to cisplatin (23), radiation (24), and the combination of EZH2/TopoII inhibitors (25). Thus, more studies are required not only to validate SMARCA4/BRG1 as a prognostic factor for overall survival (OS), but also as a potential predictive factor in well controlled patient populations with complete clinical treatment data.
Due to BRG1’s apparent role as a tumor suppressor in lung cancer, it has been demonstrated that loss of BRG1 is associated with poor prognosis; however, these studies lack treatment data and have small sample sizes (26, 27). The goal of this study was to characterize the predictive effect of SMARCA4/BRG1 expression on adjuvant cisplatin therapy using patient specimens from a clinical trial (JBR.10; NCT00002583) (28). Specifically, the decreased DNA repair capacity in lung cancer that SMARCA4- and SMARCA2-deficient tumors harbor may in fact be an “Achilles heel” if this type of repair deficiency can be exploited using specific DNA-damaging or targeted agents (8). Therefore, based on in vitro data on the regulation of drug sensitivity by BRG1 and its involvement in DNA repair, we hypothesized that decreased expression of SMARCA4/BRG1 is a predictive biomarker that promotes sensitivity to platinum-based therapies in NSCLC. To address this, we evaluated the association between gene expression and clinical outcomes of both the Director’s Challenge Lung Study (29) (prognostic effect) and the JBR.10 trial (28) (predictive effect). In addition, using the JBR.10 trial we also evaluated the predictive role of SMARCA2. Importantly, herein, we are the first to report that both SMARCA4 and SMARCA2 are predictive biomarkers of cisplatin-based chemotherapy using NSCLC patient specimens.
METHODS
Study Design and Patient Cohorts
The Director’s Challenge Study (n=440) was the first large-scale study to combine high-throughput gene expression data with clinical outcomes in NSCLC from multiple institutions (University of Michigan, Memorial Sloan-Kettering Cancer Center, the H. Lee Moffitt Cancer Center and Research Institute, the Dana-Farber Cancer Institute, and the National Cancer Institute of Canada Clinical Trials Group) (29). Enrollment criteria included diagnosis of lung adenocarcinoma with stage I–III disease and frozen surgical specimen collection. Approximately 60% of the patients had stage I disease and a proportion of the patients were treated with a mixture of adjuvant therapies (chemotherapy and radiation). However, none of the patients received pre-operative chemotherapy or radiation and at least two years of follow-up information was required. The JBR.10 trial (NCT00002583) was a phase III randomized trial of observation (OBS) versus adjuvant cisplatin and vinorelbine (ACT) in completely resected stage IB (T2N) or II (T1-2N1) NSCLC. Patients were stratified by participating institution, nodal status (N0 vs N1), and Ras mutation status of the primary tumor. Four cycles of adjuvant cisplatin were given (cisplatin (50 mg/m2) on days 1 and 8 every 4 weeks and vinorelbine (25 mg/m2) weekly for 16 weeks. In addition, post-operative radiation was not permitted. A subset of patients enrolled on JBR.10 (n=133; 62 OBS, 71 ACT) had frozen surgical specimens collected for gene expression analysis (30). The Director’s Challenge Lung Study (29) and JBR.10 (30) (GSE14814, latest update December 2014) gene expression profiling data were downloaded from the National Cancer Institute Center for Bioinformatics and the National Center for Biotechnology Information GEO database. Consent was obtained for all subjects as part of the clinical studies and the protocols were approved by each institution’s respective Institutional Review Board.
Statistical Analysis
Microarray-based gene expression (Affymetrix U133A, Santa Clara, CA) data from both the Director’s Challenge Lung Study and JBR.10 trial were normalized by the RMA method (31). All probe sets (n=8) for SMARCA4 were tested for both studies. Each probe set was treated individually due to prior recommendations and evidence that unique probe sets for the same gene can have different hybridization signals and sometimes opposite trends likely due to detection of different or multiple splice variants of the gene as shown previously (32–34). For the Director’s Challenge study (n=440), patients were classified into three groups for each probe set based on their tertile expression levels, while for the JBR.10 study (n=133), patients were classified into two groups only for each probe set based on the median expression levels due to the small patient cohort. We used OS and disease-specific survival (DSS) as the time-to-event outcomes. Kaplan-Meier product-limit method and log-rank tests were used to estimate and test the differences of probabilities in OS and DSS between expression groups and treatment arms, and hazard ratios (HR) and 95% confidence intervals (CI) were generated by the univariate Cox regression model. Multivariate Cox regression models were used to validate the prognostic and predictive effects of probes on OS and DSS, respectively while adjusting for other baseline clinical covariates. The interaction test of treatment and SMARCA4 expression group was performed to assess treatment effect differences (HR of ACT and OBS) between the high and low SMARCA4 expression groups in the JBR.10 trial. All analyses were performed using SAS 9·4 (SAS, Inc; Cary, NC) and STATA 13 (StataCorp LP; College Station, Texas).
RESULTS
Prognostic Significance of SMARCA4 Expression
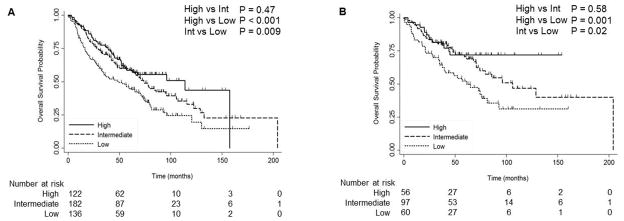
To determine the prognostic significance of SMARCA4, we analyzed the gene expression microarray dataset from the Director’s Challenge Study. This dataset contained 440 adenocarcinoma (NSCLC) samples with associated clinical data. Patients were classified into tertiles: (High (expression > 70%); Intermediate (30% ≤ expression ≤ 70%); and Low (expression < 30%)). Clinical characteristics of this dataset are shown in Table 1 using the most significant probe set (212520_s_at) in relation to survival. Poor OS was noted following low expression of SMARCA4 compared to high and intermediate expression (P<0.001 and P=0.009, respectively) for the most significant probe set (212520_s_at) (Fig. 1A). However, no significant differences in OS was observed between high and intermediate levels of SMARCA4 expression (P=0.47). Decreased OS was observed with low expression of SMARCA4 both with stage I (High vs Low P=0.01) and stages II–III (High vs Low P=0.01) of the disease (Supplementary Fig. S1). Multivariate analysis suggested that patients with high SMARCA4 expression had a decreased risk of death compared to patients with low SMARCA4 expression (high vs low: HR=0.6; 95% CI: 0.4–0·8, P=0.002; intermediate vs low: HR=0.7, 95% CI 0.5–0.9, P=0.01; Table 2) independent of age, stage, gender and differentiation grade. Data utilizing an additional probe set (214360_at) demonstrated a similar trend and statistical significance between high vs low expression (P=0.03; Supplementary Fig. S2). Further, prognostic effects of SMARCA4 were examined in patients who did not receive adjuvant chemotherapy or radiation in the Director’s Challenge study, similar to the entire cohort, low expression of SMARCA4 was significantly correlated with decreased OS (212520_s_at; high vs low P=0.001; intermediate vs low P=0.02; Fig. 1B). However, no significant differences in OS were observed between high and intermediate level of SMARCA4 expression (P=0.58). Univariate analysis results are shown in the Supplement (212520_s_at; Supplementary Table S1). In addition, in the multivariate analysis of patients who did not receive adjuvant treatment, high expression was also a significant independent prognostic marker and correlated with better prognosis (high vs low: HR=0.4; 95% CI: 0.2–0.8, P=0.01; intermediate vs low: HR=0.7, 95% CI 0.4–1.1, P=0.09; Table 2).
Table 1.
Clinical Characteristics of Patients Analyzed for SMARCA4 from the Director’s Challenge Study & JBR.10 Trial
| Director’s Challenge Study SMARCA4 Expression | JBR.10 SMARCA4 Expression | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Clinical Factor | Low | Int | High | P-value | Clinical Factor | Low | High | P-value |
| Age | 0.52 | Age | 0.76 | |||||
| <=65 (n=230) | 66 (29%) | 100 (43%) | 64 (28%) | <=65 (n=87) | 44 (51%) | 43 (49%) | ||
| >65 (n=210) | 70 (33%) | 82 (39%) | 58 (28%) | >65 (n=66) | 22 (48%) | 24 (52%) | ||
|
| ||||||||
| Gender | 0.02 | Gender | 0.42 | |||||
| Female (n=219) | 57 (26%) | 90 (41%) | 72 (33%) | Female (n=42) | 23 (55%) | 19 (45%) | ||
| Male (n=221) | 79 (36%) | 92 (42%) | 50 (22%) | Male (n=91) | 43 (47%) | 48 (53%) | ||
|
| ||||||||
| Treatment | 0.19 | Treatment | 0.78 | |||||
| Adjuvant treatment (n=45) | 17 (38%) | 20 (44%) | 8 (18%) | Adjuvant treatment (n=71) | 36 (51%) | 35 (49%) | ||
| No Adjuvant treatment (n=213) | 60 (28%) | 97 (46%) | 56 (26%) | No adjuvant treatment (n=62) | 30 (48%) | 32 (52%) | ||
|
| ||||||||
| Stage | 0.46 | Stage | 0.53 | |||||
| IA (n=113) | 33 (29%) | 48 (42%) | 32 (28%) | |||||
| IB (n=162) | 49 (30%) | 67 (41%) | 46 (29%) | IB (n=73) | 38 (52%) | 35 (48%) | ||
| IIA+IIB (n=94) | 26 (28%) | 39 (41%) | 29 (31%) | IIA+IIB (n=60) | 28 (47%) | 32 (53%) | ||
| IIIA+IIIB (n=68) | 28 (41%) | 26 (38%) | 14 (21%) | |||||
|
| ||||||||
| Histology | Histology | 0.22 | ||||||
| ADC (n=440) | 136 (31%) | 182 (41%) | 122 (28%) |
ADC (n=71) SQCC (n=52) LCUC (n=10) |
40 (56.3%) 21 (40.4%) 5 (50%) |
31 (43.7%) 31 (59.6%) 5 (50%) |
||
P-values were calculated using Χ2 Test
The probe sets that were the most statistically significant in correlation with clinical outcomes are shown for each study. Probe set 212520_s_at was used for the Director’s Challenge Study and probe set (213719_s_at) was used for the JBR.10 study.
ADC = adenocarcinoma, SQCC = squamous cell carcinoma, LCUC = large cell carcinoma
Figure 1.
Overall survival curves for patients with high, intermediate, and low levels of SMARCA4 (212520_s_at) expression in the Director’s Challenge Study. (A) all patients; (B) patients without adjuvant treatment. Log-rank P-values are shown.
Table 2.
Multivariate analysis of SMARCA4 expression in the Director’s Challenge Study
| Multivariate Analysis | Multivariate Analysis (no adjuvant treatment) | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
| SMARCA4 (212520_s_at) | ||||||
| Low expression | 1.0 | 1.0 | ||||
| Intermediate expression | 0.7 | (0.5, 0.9) | 0.01 | 0.7 | (0.4, 1.1) | 0.09 |
| High expression | 0.6 | (0.4, 0.8) | 0.002 | 0.4 | (0.2, 0.8) | 0.01 |
|
| ||||||
| Age | ||||||
| <65 years | 1.0 | 1.0 | ||||
| ≥65 years | 1.6 | (1.2, 2.1) | <0.001 | 1.8 | (1.1, 2.9) | 0.01 |
|
| ||||||
| Stage | ||||||
| I | 1.0 | 1.0 | ||||
| II–III | 3.1 | (2.4, 4.1) | <0.001 | 3.8 | (2.5, 5.9) | <0.001 |
|
| ||||||
| Gender | ||||||
| Female | 1.0 | 1.0 | ||||
| Male | 1.3 | (1.0, 1.7) | 0.05 | 1.2 | (0.8, 1.9) | 0.40 |
|
| ||||||
| Differentiation | ||||||
| Well | 1.0 | 1.0 | ||||
| Moderate | 0.9 | (0.6, 1.3) | 0.51 | 0.9 | (0.5, 1.6) | 0.60 |
| Poorly | 1.1 | (0.7, 1.6) | 0.84 | 1.0 | (0.5, 1.9) | 0.90 |
Predictive Significance of SMARCA4 in Resectable NSCLC
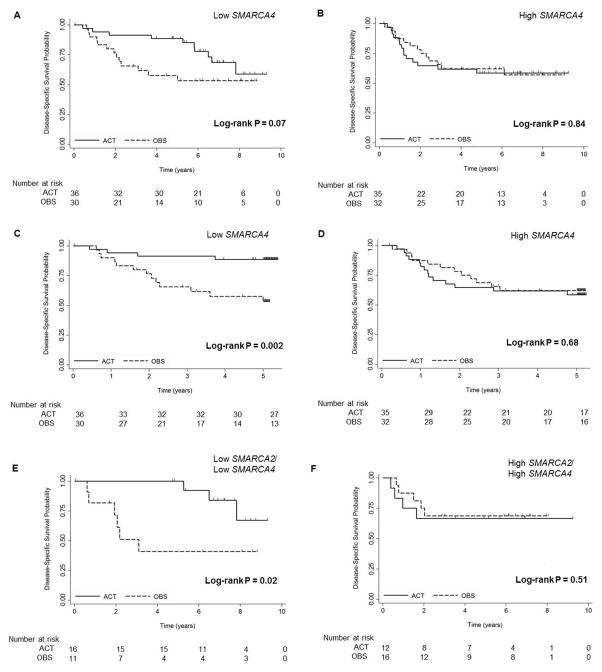
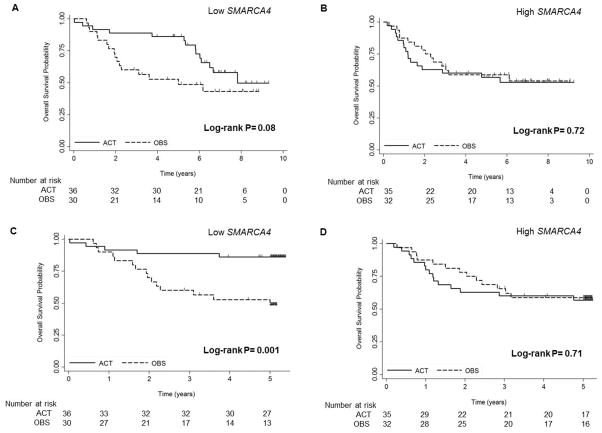
To determine the predictive significance of SMARCA4, gene expression profiling microarray data from the JBR.10 trial were analyzed. Clinical and sample characteristics of these 133 patients have been previously reported (30) and are shown split by SMARCA4 (213719_s_at) expression (Table 1). Two probe sets (208794_s_at and 213719_s_at) showed significantly greater five-year DSS in the SMARCA4 low patient population after the treatment (both P<0.05) (Supplementary Tables S2–3). Kaplan-Meier curves are shown for the most significant probe set (213719_s_at) (Fig. 2–3). Patients with low (Fig. 2A,C) and high (Fig. 2B,D) SMARCA4 expression are plotted comparing two treatment arms (OBS vs. ACT) in Figure 2. Patients with low SMARCA4 expression demonstrated improved DSS with ACT suggesting this subgroup derives a significant benefit from adjuvant cisplatin-based therapy (5-YR DSS P=0.002; Fig. 2C), whereas patients with high SMARCA4 expression did not show DSS advantage after treatment (Fig. 2B,D). In contrast to the low SMARCA4 expression group, HRs were approximately 1 in the high SMARCA4 expression group suggesting this subgroup derives minimal benefit from ACT. Similarly to DSS, patients at 5-YR OS with low SMARCA4 expression levels derived significant benefit to ACT (5-YR OS P=0.001; Fig. 3). This benefit is also demonstrated at ten years and trended towards significance for both DSS (10-YR DSS P=0.07; Fig. 2A) and OS (10-YR OS P=0.08; Fig. 3A) and although the curves get closer together they are still split even after ten years. Five probe sets for SMARCA4 are shown in the Supplementary Fig. S3–5 and although some of the probe sets did not reach significance, all data support the conclusion that patients expressing low levels of SMARCA4 derive a large benefit from cisplatin-based adjuvant therapy, while the patients with high SMARCA4 expression did not show this benefit.
Figure 2.
Overall and five-year disease-specific survival curves by treatment arm (ACT or OBS) for patients with low (A and C) and high (B and D) levels of SMARCA4 (213719_s_at) expression, and low (E) and high (F) levels of both SMARCA4 (213719_s_at) and SMARCA2 (206543_at) expression in the JBR.10 trial. OBS, observation. ACT, adjuvant chemotherapy.
Figure 3.
Comparison of overall survival and five-year overall survival by treatment arm for patients with low (A and C) and high (B and D) level of SMARCA4 (213719_s_at) in the JBR.10 trial. OBS, observation. ACT, adjuvant chemotherapy.
Upon univariate analysis, two probe sets (213719_s_at and 208794_s_at) were statistically significant (P<0.05) (Supplementary Tables 2–3) and four other probe sets trended toward improved benefit for the low SMARCA4 expression group with ACT (Supplementary Fig. S5). Upon multivariate analysis (Table 3), in the low SMARCA4 patient subset, independent of age, stage, and histology, patients have improved five-year DSS after treatment (213719_s_at (ACT vs OBS HR=0.1, 95% CI: 0.0–0.5, P=0.002); 208794_s_at (HR=0.3, 95% CI: 0.1–0·9, P=0.03)). Thus, low expression of SMARCA4 mRNA was statistically associated with improved disease-specific survival with adjuvant cisplatin/vinorelbine in completely resectable stage IB/II NSCLC patients. Importantly, multivariate analysis showed in the low SMARCA4 expression patients that overall survival was also improved after treatment (Table 3). No probe sets approached significance in the high SMARCA4 expression group demonstrating this subgroup did not associate with improved survival with cisplatin/vinorelbine. An interaction test was performed comparing HRs of ACT and OBS for five-year DSS and OS between the high and low SMARCA4 expression groups. The testing results revealed that the ACT treatment effect was affected significantly by SMARCA4 expression in one probe set and trended toward significance in another (5-YR DSS: 213719_s_at; P=0.01; 5-YR OS: 213719_s_at; P=0.007, Table 3).
Table 3.
Multivariate analysis of SMARCA4 expression and treatment arm (Observation vs Cisplatin/Vinerolbine) in the JBR.10 Trial
| Multivariate Analysis (DSS) | Multivariate Analysis (5YR DSS) |
Multivariate Analysis (OS) |
Multivariate Analysis (5YR OS) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe | Variable | Comparison | HR | 95% CI | P-value | HR | 95% CI | P-value | Interaction P-value |
HR | 95% CI | P-value | HR | 95% CI | P-value | Interaction P-value* |
| 213719_s_at | Low SMARCA4 | ACT vs OBS | 0.3 | (0.1, 0.8) | 0.02 | 0.1 | (0.0, 0.5) | 0.002 | 0.01 | 0.4 | (0.2, 0.9) | 0.02 | 0.1 | (0.0, 0.5) | <0.001 | 0.007 |
| Stage: II vs I | 5.3 | (2.0, 14.1) | <0.001 | 5.9 | (1.8, 18.7) | 0.003 | 3.1 | (1.4, 6.9) | 0.005 | 5.0 | (1.8, 14.4) | 0.003 | ||||
| Age: ≥65 vs <65 | 2.6 | (1.0, 7.0) | 0.05 | 4.2 | (1.3, 13.8) | 0.02 | 2.3 | (1.0, 5.2) | 0.05 | 4.2 | (1.5, 12.1) | 0.008 | ||||
| Histology: SQCC vs ADC | 0.6 | (0.2, 1.6) | 0.29 | 0.7 | (0.2,2.3) | 0.60 | 0.7 | (0.3,1.6) | 0.43 | 0.9 | (0.3,2.5) | 0.83 | ||||
| Histology: LCUC vs ADC | 4.3 | (1.1,17.5) | 0.04 | 8.1 | (1.8,37.0) | 0.007 | 2.6 | (0.7,9.5) | 0.16 | 6.8 | (1.6,29.3) | 0.01 | ||||
| High SMARCA4 | ACT vs OBS | 1.0 | (0.4, 2.1) | 0.91 | 1.0 | (0.5, 2.3) | 0.92 | 1.0 | (0.5, 2.1) | 0.99 | 1 | (0.5, 2.2) | 0.96 | |||
| Stage: II vs I | 1.5 | (0.7, 3.2) | 0.33 | 1.4 | (0.6, 3.0) | 0.42 | 1.4 | (0.7, 2.8) | 0.40 | 1.4 | (0.6, 2.9) | 0.41 | ||||
| Age: ≥65 vs <65 | 2.5 | (1.1, 5.4) | 0.03 | 2.5 | (1.1,5.7) | 0.02 | 2.7 | (1.3, 5.7) | 0.01 | 2.6 | (1.2,5.5) | 0.02 | ||||
| Histology: SQCC vs ADC | 0.3 | (0.1,0.6) | 0.003 | 0.3 | (0.1,0.7) | 0.005 | 0.3 | (0.1,0.7) | 0.004 | 0.3 | (0.1,0.7) | 0.005 | ||||
| Histology: LCUC vs ADC | 1.1 | (0.3,4.0) | 0.83 | 1.2 | (0.3,4.2) | 0.78 | 1.1 | (0.3,3.7) | 0.93 | 1.1 | (0.3,4) | 0.84 | ||||
| 208794_s_at | Low SMARCA4 | ACT vs OBS | 0.4 | (0.2, 0.9) | 0.03 | 0.3 | (0.1, 0.9) | 0.03 | 0.22 | 0.5 | (0.2, 1.2) | 0.13 | 0.4 | (0.2, 1.0) | 0.04 | 0.37 |
| Stage: II vs I | 3.6 | (1.4, 8.9) | 0.007 | 2.8 | (1.0, 7.6) | 0.05 | 2.7 | (1.2, 6.0) | 0.02 | 3.0 | (1.1, 8.1) | 0.03 | ||||
| Age: ≥65 vs <65 | 2.3 | (0.9, 5.5) | 0.07 | 2.4 | (0.9, 6.4) | 0.09 | 1.8 | (0.8, 1.0) | 0.17 | 2.1 | (0.8, 5.4) | 0.14 | ||||
| Histology: SQCC vs ADC | 0.4 | (0.1,1.0) | 0.05 | 0.5 | (0.2,1.4) | 0.18 | 0.3 | (0.1,0.7) | 0.01 | 0.4 | (0.2,1.3) | 0.13 | ||||
| Histology: LCUC vs ADC | 4.9 | (0.9,26.0) | 0.06 | 6.6 | (1.2,37.0) | 0.03 | 2.8 | (0.6,13.5) | 0.21 | 5.9 | (1.1,32.4) | 0.04 | ||||
| High SMARCA4 | ACT vs OBS | 0.9 | (0.4, 1.9) | 0.70 | 0.7 | (0.3, 1.7) | 0.46 | 0.7 | (0.4, 1.5) | 0.43 | 0.7 | (0.3, 1.4) | 0.29 | |||
| Stage: II vs I | 2.0 | (0.9, 4.4) | 0.07 | 2.1 | (0.9, 4.7) | 0.08 | 1.6 | (0.8, 3.2) | 0.17 | 1.9 | (0.9, 4.0) | 0.09 | ||||
| Age: ≥65 vs <65 | 2.0 | (0.9, 4.6) | 0.09 | 2.3 | (1.0, 5.4) | 0.05 | 2.8 | (1.3, 5.8) | 0.007 | 2.8 | (1.3, 6.1) | 0.01 | ||||
| Histology: SQCC vs ADC | 0.5 | (0.2, 1.3) | 0.15 | 0.6 | (0.2, 1.4) | 0.23 | 0.7 | (0.3, 1.6) | 0.45 | 0.7 | (0.3,1.6) | 0.39 | ||||
| Histology: LCUC vs ADC | 1.7 | (0.6,5.3) | 0.34 | 2.0 | (0.7,6.3) | 0.22 | 1.7 | (0.6,5.1) | 0.36 | 1.9 | (0.6,6.0) | 0.25 | ||||
interaction P-values displayed are for 5-YR disease-specific survival (DSS) or overall survival (OS) ADC = adenocarcinoma, SQCC = squamous cell carcinoma, LCUC = large cell carcinoma
Since SMARCA2 is another catalytic subunit of SWI/SNF, it is of interest to determine if SMARCA2 loss is also associated with improved survival with cisplatin-based chemotherapy and increases the predictive power of SMARCA4 in the JBR.10 trial. The benefit of adjuvant chemotherapy was determined in patients with low expression values of SMARCA2 (206543_at) individually and combined with SMARCA4 (Supplementary Fig. S6 and Fig. 2E–F). As shown, patients with low levels of SMARCA2 showed a trend toward improved survival with adjuvant chemotherapy, but did not demonstrate the same predictive significance of SMARCA4 (Supplementary Fig. S6A). High SMARCA2 did not show improvement (Supplementary Fig. S6B). Strikingly, patients with low levels of both SMARCA2 and SMARCA4 (Fig. 2E) seemed to achieve a dramatic benefit (HR 0.3, 95% CI: 0.1–0.9, log-rank P=0.02) in DSS upon treatment with adjuvant cisplatin/vinorelbine compared to observation after surgery. The patients with high expression of both probes did not show a difference (Fig. 2F).
DISCUSSION
Adjuvant cisplatin-based chemotherapy in NSCLC patients reduces the risk of recurrence after complete resection in unselected stage IB, II, and IIIA patients; however, while all patients experience toxicity, not all receive benefit. Thus, predictive biomarkers of adjuvant chemotherapy in NSCLC are desperately needed to determine which patients derive the most benefit. Conversely, identification of those unlikely to benefit opens the opportunity for novel approaches to adjuvant therapy in these patients. Individualizing chemotherapy based on multiple candidate biomarkers in lung cancer has recently failed to demonstrate significant clinical benefit in several clinical trials (35, 36), underscoring the need for better markers.
Common alterations in SWI/SNF in NSCLC have only been recently elucidated. No predictive studies of SMARCA4 using clinical tissues have been published to date and very few studies have been published analyzing the prognostic effect. Importantly, this study is the first to demonstrate the predictive effects of SMARCA4/BRG1 in NSCLC using patient samples from the JBR. 10 trial. In this study, we validated in a large cohort that decreased SMARCA4 is associated with worse prognosis in patients harboring lung adenocarcinomas using the Director’s Challenge Lung Study. Notably, this study also demonstrated for the first time that low SMARCA4/BRG1 expression is associated with increased benefit from cisplatin-based chemotherapy in resectable NSCLC using specimens from the JBR.10 trial.
The connections between DNA repair and chromatin remodeling have only recently begun to be explored. In particular, SWI/SNF remodeling complexes have also been implicated in NER, a critical pathway involved in cisplatin resistance. Recently, BRG1 has been shown to affect the stability of XPC protein as well as the recruitment of XPG and PCNA, which are all essential proteins within NER (22). In a recent paper, knockdown of BRG1 or BRM in H460 lung cancer cells increased cisplatin sensitivity and showed reduced repair of both intrastrand and interstrand adducts suggesting that the mechanism of sensitivity is primarily due to defects in DNA repair (23). In addition, this previous study suggested that BRG1 is important for ERCC1 recruitment, a well known important mediator of NER (23). Of importance, the phenotype of cisplatin resistance was not as pronounced for the BRG1 or BRM knockdowns as previously shown for XPF and ERRC1 demonstrating the different roles of chromatin remodeling and repair proteins in cisplatin sensitivity (23). Given the complexity of the data which has arisen from ERCC1 as a potential biomarker and the known connection of ERCC1 and SMARCA4/BRG1, a panel of molecular biomarkers comprised of both epigenetic regulators and DNA repair/response genes to assess activity may be necessary to accurately select patients for platinum-based regimens in NSCLC and other cancers. In addition, an alternative mechanism of sensitivity to cisplatin in tumors that have loss of SMARCA4 and/or SMARCA2 is loss of Rb activity leading to inhibition of a DNA damage-induced cell cycle checkpoint. This could be of particular importance in patients that have concomitant loss of both SMARCA4 and SMARCA2 which is demonstrated by a previous in vitro study where cancer cells that have loss of both BRG1/SMARCA4 and BRM/SMARCA2 showed loss of the Rb-dependent cisplatin-induced cell cycle checkpoint (37). Due to the growing evidence of the role SMARCA4 on DNA damage response, DNA repair, and drug sensitivity in vitro, it is imperative that the effects of SMARCA4 as a predictive biomarker using clinical specimens is further investigated. Moreover, the best detection method for its predictive value still needs to be determined specifically in regards to mutation vs expression vs protein analysis. Even for expression analysis in this study it is clear that unique probe sets result in slightly different results likely due to hybridization to different areas of the gene (Supplementary Fig. S7) and expression of multiple transcripts. A limitation of this study was that only mRNA expression datasets were analyzed as these were publicly available from both the Director’s Challenge Study and JBR.10. Although mutations are common in clinical specimens, there is evidence that some patients lack expression but have no mutations as we and others have previously found (11, 15). Therefore, a multi-platform approach for detection of SMARCA4/BRG1 along with other epigenetic regulators (including SMARCA2) and DNA repair/response proteins may be in order.
Importantly, our study is the first to show that SMARCA4/BRG1 can be used as a predictive biomarker in clinical specimens. Specifically, our results utilizing expression data from the JBR.10 trial demonstrated SMARCA4 expression levels depict efficacy of cisplatin and vinorelbine in the setting of stage IB–II resectable NSCLC independent of age, stage, and histology. Thus, patients (even those older than 65) with low levels of SMARCA4/BRG1 expression appear to be excellent candidates for platinum-based chemotherapy regimens based on an overall survival advantage in the JBR.10 trial. Further research on the predictive effect of SMARCA4/BRG1 on cisplatin therapy and other DNA repair targeted therapies as well as other SWI/SNF components and methods of detection is warranted to assess their potential to serve as a companion diagnostic in NSCLC.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Predictive biomarkers of chemotherapy response in NSCLC are needed to better characterize patients who derive the greatest benefit. We hypothesized that SMARCA4/BRG1 could be such a marker due to its established role in cisplatin sensitivity and DNA repair in vitro. No studies on the predictive effect of SMARCA4/BRG1 using clinical tissues have been published to date and very few prognostic studies with limited sample sizes have been published. We analyzed data available from both the Director’s Challenge Lung Study and the JBR.10 phase III randomized trial in a hypothesis-driven manner to determine the prognostic and predictive role of SMARCA4/BRG1 from two prospective studies. Importantly, this study is the first to demonstrate the predictive effects with a highly significant interaction test of SMARCA4/BRG1 in NSCLC using patient samples from a randomized trial with an untreated control. We also validated in a large cohort that decreased SMARCA4 is associated with worse prognosis.
Acknowledgments
The authors thank S. Jaharul Haque for assistance in preparation of the manuscript.
GRANT SUPPORT:
This work was supported by the Lung Cancer Research Foundation (EHB), the Paul P. Carbone Memorial Foundation (EHB), Lungevity (DPC), the NIH/NCI (R01CA108633 (AC) and RC2CA148190 (AC) and the OSU Comprehensive Cancer Center (AC).
Footnotes
DISCLOSURE OF POTENTIAL CONFLICT OF INTEREST:
Dr. Carbone reports personal fees from Bayer HealthCare, personal fees from Biothera, personal fees from Boehringer Ingelheim, grants and personal fees from Bristol Myers-Squibb, personal fees from Clovis Oncology, personal fees from Genentech/Roche, personal fees from Merck, personal fees from Novartis, personal fees from Peregrine Pharmaceuticals, personal fees from Pfizer, and personal fees from Synta Pharmaceuticals Corp., during the conduct of the study. No other disclosures are reported.
AUTHORS’ CONTRIBUTIONS:
Conception and design: E.H. Bell, A.R. Chakraborty, X. Mo, D.P. Carbone, A. Chakravarti
Development of methodology: E.H. Bell, X. Mo
Acquisition of data: E.H. Bell, A.R. Chakraborty, X. Mo
Analysis and interpretation of data: E.H. Bell, A.R. Chakraborty, X. Mo, Z. Liu, K. Shilo, S. Kirste, P. Stegmaier, M. McNulty, N. Karachaliou, R. Rosell, G. Bepler, D.P Carbone, A. Chakravarti
Writing, review, and/or revision of the manuscript: E.H. Bell, A.R. Chakraborty, X. Mo, Z. Liu, K. Shilo, S. Kirste, P. Stegmaier, M. McNulty, N. Karachaliou, R. Rosell, G. Bepler, D.P Carbone, A. Chakravarti
Administrative, technical, or material support: E.H. Bell, A.R. Chakraborty, X. Mo, Z. Liu, K. Shilo, S. Kirste, P. Stegmaier, M. McNulty, N. Karachaliou, R. Rosell, G. Bepler, D.P Carbone, A. Chakravarti
Study supervision: E.H. Bell
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: a cancer journal for clinicians. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DH, Schiller JH, Bunn PA. Recent clinical advances in lung cancer management. J Clin Oncol. 2014;32:973–82. doi: 10.1200/JCO.2013.53.1228. [DOI] [PubMed] [Google Scholar]
- 3.Shanker M, Willcutts D, Roth JA, Ramesh R. Drug resistance in lung cancer. Lung Cancer: Targets and Therapy. 2010;1:23–6. [PMC free article] [PubMed] [Google Scholar]
- 4.Besse B, Olaussen KA, Soria JC. ERCC1 and RRM1: ready for prime time? J Clin Oncol. 2013;31:1050–60. doi: 10.1200/JCO.2012.43.0900. [DOI] [PubMed] [Google Scholar]
- 5.Rose MC, Kostyanovskaya E, Huang RS. Pharmacogenomics of cisplatin sensitivity in non-small cell lung cancer. Genomics Proteomics Bioinformatics. 2014;12:198–209. doi: 10.1016/j.gpb.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrski T, Dent R, Blecharz P, Foszczynska-Kloda M, Gronwald J, Huzarski T, et al. Results of a phase II open-label, non-randomized trial of cisplatin chemotherapy in patients with BRCA1-positive metastatic breast cancer. Breast Cancer Res. 2012;14:R110. doi: 10.1186/bcr3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mutze K, Langer R, Schumacher F, Becker K, Ott K, Novotny A, et al. DNA methyltransferase 1 as a predictive biomarker and potential therapeutic target for chemotherapy in gastric cancer. Eur J Cancer. 2011;47:1817–25. doi: 10.1016/j.ejca.2011.02.024. [DOI] [PubMed] [Google Scholar]
- 8.Helming KC, Wang X, Roberts CW. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell. 2014;26:309–17. doi: 10.1016/j.ccr.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, et al. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008;29:617–22. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 11.Araujo LH, Timmers C, Bell EH, Shilo K, Lammers PE, Zhao W, et al. Genomic Characterization of Non-Small-Cell Lung Cancer in African Americans by Targeted Massively Parallel Sequencing. J Clin Oncol. 2015 doi: 10.1200/JCO.2014.59.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witkowski L, Carrot-Zhang J, Albrecht S, Fahiminiya S, Hamel N, Tomiak E, et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet. 2014;46:438–43. doi: 10.1038/ng.2931. [DOI] [PubMed] [Google Scholar]
- 13.Wong AK, Shanahan F, Chen Y, Lian L, Ha P, Hendricks K, et al. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60:6171–7. [PubMed] [Google Scholar]
- 14.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Oike T, Ogiwara H, Tominaga Y, Ito K, Ando O, Tsuta K, et al. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 2013;73:5508–18. doi: 10.1158/0008-5472.CAN-12-4593. [DOI] [PubMed] [Google Scholar]
- 16.McKenna ES, Sansam CG, Cho YJ, Greulich H, Evans JA, Thom CS, et al. Loss of the epigenetic tumor suppressor SNF5 leads to cancer without genomic instability. Mol Cell Biol. 2008;28:6223–33. doi: 10.1128/MCB.00658-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamwell LF, Gambaro K, Merziotis M, Crane C, Arcand SL, Bourada V, et al. Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics. Orphanet J Rare Dis. 2013;8:33. doi: 10.1186/1750-1172-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang HT, Chen SM, Pan LB, Yao J, Ma HT. Loss of function of SWI/SNF chromatin remodeling genes leads to genome instability of human lung cancer. Oncol Rep. 2015;33:283–91. doi: 10.3892/or.2014.3584. [DOI] [PubMed] [Google Scholar]
- 19.Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29:1434–45. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006;25:3986–97. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Chen H, Gong M, Gong F. The chromatin remodeling protein BRG1 modulates BRCA1 response to UV irradiation by regulating ATR/ATM activation. Front Oncol. 2013;3:7. doi: 10.3389/fonc.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Q, Wang QE, Ray A, Wani G, Han C, Milum K, et al. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J Biol Chem. 2009;284:30424–32. doi: 10.1074/jbc.M109.044982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kothandapani A, Gopalakrishnan K, Kahali B, Reisman D, Patrick SM. Downregulation of SWI/SNF chromatin remodeling factor subunits modulates cisplatin cytotoxicity. Experimental cell research. 2012;318:1973–86. doi: 10.1016/j.yexcr.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JH, Park EJ, Hur SK, Kim S, Kwon J. Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair (Amst) 2009;8:29–39. doi: 10.1016/j.dnarep.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520:239–42. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–6. [PubMed] [Google Scholar]
- 27.Fukuoka J, Fujii T, Shih JH, Dracheva T, Meerzaman D, Player A, et al. Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin Cancer Res. 2004;10:4314–24. doi: 10.1158/1078-0432.CCR-03-0489. [DOI] [PubMed] [Google Scholar]
- 28.Winton T, Livingston R, Johnson D, Rigas J, Johnston M, Butts C, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–97. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 29.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417–24. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 32.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stalteri MA, Harrison AP. Interpretation of multiple probe sets mapping to the same gene in Affymetrix GeneChips. BMC Bioinformatics. 2007;8:13. doi: 10.1186/1471-2105-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaksik R, Polanska J, Herok R, Rzeszowska-Wolny J. Calculation of reliable transcript levels of annotated genes on the basis of multiple probe-sets in Affymetrix microarrays. Acta Biochim Pol. 2009;56:271–7. [PubMed] [Google Scholar]
- 35.Tiseo M, Bordi P, Bortesi B, Boni L, Boni C, Baldini E, et al. ERCC1/BRCA1 expression and gene polymorphisms as prognostic and predictive factors in advanced NSCLC treated with or without cisplatin. Br J Cancer. 2013;108:1695–703. doi: 10.1038/bjc.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bepler G, Williams C, Schell MJ, Chen W, Zheng Z, Simon G, et al. Randomized international phase III trial of ERCC1 and RRM1 expression-based chemotherapy versus gemcitabine/carboplatin in advanced non-small-cell lung cancer. J Clin Oncol. 2013;31:2404–12. doi: 10.1200/JCO.2012.46.9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strobeck MW, Reisman DN, Gunawardena RW, Betz BL, Angus SP, Knudsen KE, et al. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J Biol Chem. 2002;277:4782–9. doi: 10.1074/jbc.M109532200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.