1. Introduction
The healthy human body contains 10 times more microbial cells than human cells! This proclamation has been repeated many times over the past several years in both the scientific literature, as well as the lay press [169]. Although this declaration has been presented as a scientific fact over the past decade, it may not be entirely accurate in view of a recent reexamination of the published data [169]. Most reviews that focus on host-microbe interactions begin with the statement that the healthy human intestine contains approximately 100 trillion (1014) microbes [55,102,102,176,178,211]. Furthermore, many of these publications state, without reference, that the total number of human cells in the body approximates 10 trillion (1013) cells [169,178]. However, this 10-fold excess of microbial to human cells may need to be reevaluated based upon more recent work that has been largely overlooked during the past few years. For example, the statement that the intestinal tract contains 1014 microbial cells is based upon a 44 year-old report that provides little by way of direct quantitative data for this fecal bacterial estimate [108]. Using more sophisticated technology, Suau and coworkers have determined that the numbers of bacteria that reside within the healthy human intestinal tract range from 3 × 1013 to 40 × 1013 (30 trillion to 400 trillion) [194]. The assertion that humans contain 1013 body cells is based upon one sentence from a 46 year-old book that provides no experimental data nor references for this estimate [37]. A recent study by Bianconi et. al. using systematic quantification of cell numbers in different tissues reports that humans contain, on average, 30–40 trillion body cells [12]. These newer data would suggest a more realistic ratio of microbial to human cells that range from 1:1 to 10:1.
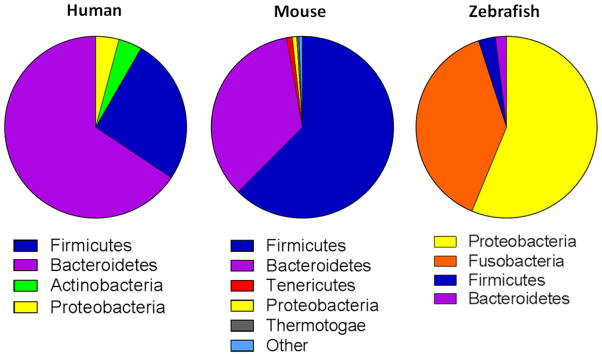
While these more quantitative estimates are, in some cases, quite different from what has been repeatedly stated in scientific and lay publications, they confirm that the gut is home to enormous numbers of bacteria. The large majority (>90%) of intestinal bacteria in the human intestinal tract belongs to the phyla Bacteroidetes and Firmicutes. However, substantial numbers of bacteria belonging to phyla Proteobacteria, Actinobacteria, Fusobacteria, Verrucomicrobia and Cyanobacteria are also observed [38,40,44,102,106,176] (Figure 1). In addition to the enormous population of bacteria, the human gut has also been estimated to contain more than a quadrillion (1015) viruses and bacteriophage, as well as substantial numbers of archaea and fungi [40,55,62,102,106,143,199]. Most of the detailed characterization and bioinformatic analyses of the intestinal microbiota have been performed using human stool and/or mucosal tissue. However, mice have also been extensively used to define the importance of host genetics, microbiota and the immune system in host-microbe interactions in homeostasis and disease [28,141]. The use of mice provide investigators with a small animal model to assess, in a well-controlled environment, the complex host-microbe interactions that occur in vivo. While mice and humans share two major phyla (Bacteroidetes and Firmicutes) and approximately 80 different genera [141,168], major differences exist among bacterial species in these two mammals [141,168].
Figure 1. Relative abundance of the major intestinal phyla in humans, mice and zebrafish.
Deep sequencing of 16S rRNA gene reveals that while the relative abundance of specific bacterial phyla may differ, mammals and teleosts possess similar bacterial communities. Human and zebrafish pie charts were derived from [121,166], respectively (with permission). Mouse pie chart was generated from colonic luminal contents of healthy C57Bl/6 mice housed at the TTUHSC animal facility (unpublished data).
Valuable information has been generated using gnotobiotic and fully colonized mice to assess host-microbiome interactions. However, these studies are limited by the length of time and the high cost associated with the generation of large numbers of genetically-manipulated animals required to yield statistically-powered in vivo studies. In an attempt to shorten the time and cost of new discoveries, investigators have begun to use other vertebrates to model these interactions in healthy and inflamed intestine. For example, zebrafish (Danio rerio) have become increasingly popular for these types of studies given the similarity of their intestinal tract to that of mammals [57,208]. Although major differences exist between teleost and mammal microbiota, zebrafish share many of the major microbial communities that have been identified in rodents and humans [160,161,180,195,198]. Investigators have shown that similar to humans and mice, the teleost gut contains large numbers of Proteobacteria, Firmicutes and Bacteroidetes [30,166,195] (Figure 1). The use of teleosts offer a number of advantages over mice and other rodents due to the relatively low cost to produce and maintain large numbers of larvae and adults, their accelerated development, and their transparent skin that allows for detailed and noninvasive imaging studies [161,216]. Another major advantage of zebrafish is their amenability to produce forward and reverse genetic manipulations [216]. Furthermore, because these vertebrates live in an aqueous environment, the delivery of different chemicals/small molecules, therapeutic agents or microbiota to germ-free or fully colonized zebrafish is relatively a straight forward process [19,47,64,160,161,198].
The continuous exposure of the vertebrate intestine to such large and diverse populations of microorganisms in close proximity to a tissue that contains large numbers of immune cells, makes the gut the largest and most complex component of the immune system. The coexistence of vertebrates with their gut microbiota is a dynamic and mutually beneficial relationship that plays an important role in the well-being of the host [29]. However, the close proximity of potentially harmful/pathogenic microorganisms has forced the intestinal immune system to develop a number of different immune mechanisms to eliminate invading microbes, while suppressing the bystander tissue injury associated with these innate and adaptive immune responses. Failure to properly regulate these protective immune responses may induce chronic inflammatory responses that are thought to be critical immunopathological mechanisms responsible for the development of human inflammatory bowel diseases (IBD; Crohn’s disease, ulcerative colitis). These idiopathic inflammatory diseases affect primarily the small and/or large bowel and are characterized by the infiltration of large numbers of inflammatory leukocytes (e.g. neutrophils, monocytes, and lymphocytes) into the intestinal lamina propria (LP) where they directly or indirectly promote inflammation with tissue injury, loss of goblet cells, fibrosis, erosions and ulcerations. Although the etiology of IBD remains to be defined, it is becoming increasingly appreciated that chronic intestinal inflammation may result from a complex interaction among genetic, immune and microbial factors [73,99,214]. Based upon a large body of experimental and clinical evidence generated over the past 20 years, investigators hypothesize that chronic gut inflammation results from a dysregulated immune response to components of the normal gut flora in genetically-susceptible individuals [31,79,95]. Although mouse models of IBD have been used for more than 20 years and have been instrumental in defining many of the major immunopathological mechanisms responsible for inflammatory tissue injury in these models, progress as been slow for reasons outlined above [95]. Thus, several groups of investigators have turned to the use of zebrafish to model IBD (see below) [19,47,63,64,216]. The objective of this review is to present our current understanding of the role that the intestinal microbiota play in vertebrate intestinal health and inflammation. While our primary focus will be humans and mice, we also present the new and exciting comparative studies being performed in zebrafish to model host-microbe interactions.
2. Development of Bacterial Communities within the Intestinal Tract
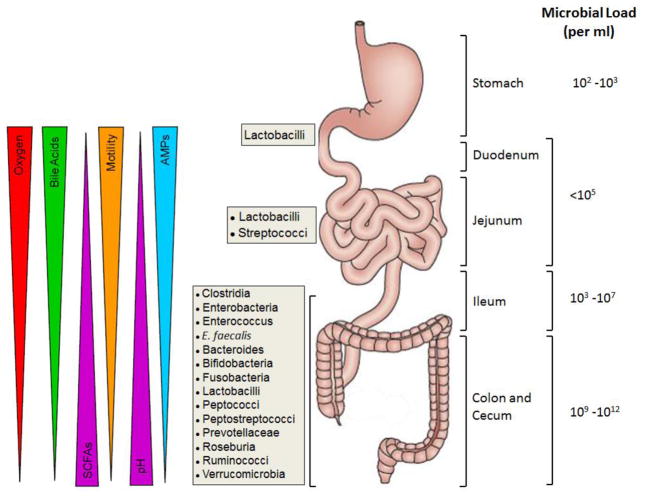
The colonization and development of the intestinal microbiota in all vertebrates is crucial for the generation of a fully functional immune system, production of essential nutrients and vitamins, and metabolism of xenobiotics. While it has been assumed that the development of a stable microbiota in the mammalian gut begins at birth, since in utero the fetus has been thought to be germ-free, more recent reports suggest that this may not be the case as bacteria have been isolated from meconium, umbilical cord and amniotic fluid obtained from healthy pregnancies [81]. Nevertheless, the development of a newborn’s microbiota begins following birth via the colonization of the infant’s intestinal tract with bacteria associated with the mother’s skin, vagina, feces, and breast milk [102,117,190]. During the first three months of life, Bifidobacterium and Lactobacillus colonize the intestinal tract in mammals due to the ingestion of breast milk [97,190]. Early on in the infant’s life, the microbial communities are highly variable and relatively unstable when compared to the adult microbiota which has much greater complexity and phylogenetic diversity [97,149]. It is thought that the stabilization/maturation of the microbiota occurs at approximately 2–3 years of age and that the microbiota can be prepared with genes for the metabolism of food that is not yet being consumed by the infant (i.e. plant polysaccharide metabolism) [6,97,149,217]. Koenig et al., has shown that the assembly of the microbial communities early in life is not random, but instead, occurs by way of specific bacterial successions due to different life events [97]. The fully mature intestinal microbiota is distinctly distributed among the different parts of the gastrointestinal (GI) tract, dictated primarily by the different oxygen concentrations found within the lumen and along the length of the GI tract (Figure 2). It is well-appreciated that the more proximal portions of the GI tract (i.e. stomach, duodenum) contain much higher levels of luminal oxygen and are populated by a higher percentage of aerobic and facultative anaerobic bacteria; although total numbers of bacteria in these regions of the GI tract are much lower than those found in the distal portions of the small and large bowel [42,176]. The distal portion of the small intestine as well as the colon contains little or no oxygen, thereby facilitating the growth of obligate anaerobes [1] (Figure 2).
Figure 2. Spatial organization of microbial communities and physiological gradients along the mammalian gastrointestinal tract.
The numbers and types of bacterial communities, as well as physiological factors vary along the length of the gastrointestinal (GI) tract. It is well-appreciated that the oxygen levels, bile acid concentrations, intestinal motility, antimicrobial peptides (AMPs) and luminal pH in proximal portion of the GI tract (stomach, duodenum, jejunum) play major roles in restricting the numbers and types of microorganisms. In general, aerobic and facultative anaerobic bacteria are found almost exclusively in the proximal portion of the GI tract. The hypoxic nature and more physiological pH of the distal small intestine (ileum) and colon coupled to overall reductions of bile acids, AMPs and gut motility, allows for unfettered growth of large numbers of obligate anaerobic bacteria. These oxygen-sensitive microbes are capable of producing large quantities of short chain fatty acids (SCFAs; acetate, proprionate, butyrate) from complex carbohydrates (fiber) to be used for important colonic and immunologic processes. Figure derived from [38,139,214]. The anatomical structure of the gastrointestinal tract with associated microbial communities and bacterial load were reproduced from reference 139 with permission.
The early development of intestinal microbial communities in mammals contrasts that of teleosts, given their differences with respect to life cycle. For example, zebrafish egg fertilization takes place externally with embryonic development occurring within the sterile chorion. At 2 days post fertilization (d.p.f.) the larvae hatch and are released into the environment where microbial colonization begins [33,219]. As with other vertebrates, microbial colonization of the intestine serves as a potent stimulus to induce the maturation of the gut immune system and digestive functions such that the larvae may begin to ingest food at approximately 5 d.p.f. [8,219]. The development of the intestinal microbiota continues throughout life, continuously responding to changes in the environment as the zebrafish mature from larva to adult. Although little information is currently available describing the intestinal microenvironment, its luminal oxygen tension, the regional microbial composition and total bacterial load in teleosts, it has been reported that the distribution of microbial communities in the hindgut of fish is similar to the distal bowel of mammals as they share similar communities of bacteria such as Proteobacteria, Fusobacteria, Firmicutes and Bacteroidetes [30,166,187,195]. Once established, intestinal microbial communities in both mammals and fish may be continually modified by different factors that include diet, genetics, sex and the environment [36,106,120,195]. Some of these factors may induce stronger and more profound changes in the microbiota, while others such as short term antibiotic use or changes in diet may induce temporary shifts in the different bacterial populations and that they are able to, to a certain point, return to their original state (a term called resilience) [35].
As pointed out earlier, the gut microbiota plays an essential role in the development of a fully functional immune system in both mammals and teleosts [57,124,219]. Studies using germ-free mice reveal that these rodents have under-developed lymphoid tissues, defective T and B cell function, and low numbers of circulating CD4+ T cells and antibody production, all of which can be restored by colonizing mice with microorganisms [28,123]. Similar findings have been described using germ-free zebrafish, which lack specific aspects of gut epithelium differentiation (specification and maturation) and proliferation, decreased protein macromolecule uptake and altered gut motility, all of which can be reversed by the introduction of intestinal microbiota [8,8,161,208]. In addition to helping shape the intestinal and systemic immune system, gut microbiota are essential for providing critical metabolic functions that cannot be accomplished by the host [145]. For example, some intestinal bacteria (obligate anaerobes) in mammals and teleosts are capable of degrading “non-digestible” complex carbohydrates (fiber) to produce short chain fatty acids (SCFA) such as acetate, butyrate and proprionate (Figure 2) [30,55]. These metabolites, rather than glucose, are the preferred energy substrate for colonic epithelial cells. In addition, certain bacteria (i.e. Bacteroides, Bifidobacterium, and Enterococcus) are capable of synthesizing vitamins as well as degrading different xenobiotics as it has been shown in humans [55,97,136].
The development of such dense and complex populations of microorganisms that reside so close to the gut requires that the intestinal immune system maintain tolerance to nonpathogenic/commensal bacteria. This is also true for teleosts whose immune system has to be able to coexist with commensal microorganisms in the gut, gills and skin [46]. Although the mechanisms responsible for tolerance have not been completely defined, there is good evidence to suggest that interactions between commensal bacteria and host mucosa induce highly regulated innate and adaptive immune responses [70,175]. For example, certain commensal bacteria such as segmented filamentous bacteria (SFB) have been shown to induce T helper 17 (Th17) cell differentiation in the small intestine, which protects the host from fungal and bacterial infections [5,76,77]. Interleukin 22 (IL-22), a Th17-derived cytokine, has been shown to enhance tight junctions in epithelial cells and increase mucin and antimicrobial protein production, all of which limit bacterial invasion into the lamina propria. Other bacteria such as the Clostridia clusters IV, XIVa and XVIII and Bacteroidetes have been shown to induce immuno-regulatory T cells called regulatory T cells (Tregs; CD4+Foxp3+ T cells). These T cells are known to suppress immune responses to self and bacterial antigens, promote epithelial repair, and promote tolerance to microbes [4,116,125,176]. In zebrafish, the intestinal microbiota also promotes the regulation of the innate immune responses to infections as reported by Galindo-Villegas et al. [51]. Furthermore, Rawls and coworkers demonstrated that the zebrafish gut microbiota is important for the expression of over 200 genes in the zebrafish intestine, many of which have also been observed in mice and are associated with epithelial proliferation, immune responses and nutrient metabolism [161].
3. Protection Against Invasion of Unwanted Neighbors
In mammals, the intestinal epithelial cell monolayer is composed of absorptive epithelial cells as well as specialized cells (e.g. goblet cells, Paneth cells) that are capable of secreting protective macromolecules and peptides. Immediately underlying the epithelium is the lamina propria which contains large numbers of plasma cells that produce IgA, a signature protective immunoglobulin of mucosal surfaces. Transcellular transport of IgA from the LP to the bowel lumen is mediated by the polymeric immunoglobulin receptor (pIgR), which is localized on the basolateral surface of epithelial cells [82]. Similar to mammals, the teleost intestine consists of three visually distinct anatomical divisions: the rostral intestinal bulb, the mid-intestine and the caudal intestine. In contrast with mammals, zebrafish do not contain a stomach [208]. Gene expression data in zebrafish suggest that the physiological functions of certain segments of its intestine are similar to mammals. Molecular characterization of the zebrafish transcriptome shows that the rostral and mid zebrafish intestine, where most metabolism, transport of nutrients, and energy metabolism take place, are functionally similar to the small intestine in mammals. In turn, the zebrafish caudal intestine is analogous to the mammalian large intestine [209]. It should be noted that crypts and Peyer’s Patches are absent in the zebrafish gut. The intestinal monolayer in zebrafish consists of three principal cell types: enterocytes, enteroendocrine cells, and goblet cells [103,129,216].
Invasion of pathogenic or commensal bacteria into the gut interstitium (i.e. LP) and their dissemination into the systemic circulation has the potential to produce devastating immune responses that could result in systemic inflammation, illness and possibly death. How does the gut limit and/or prevent the invasion of these unwanted neighbors? Essentially, the intestine has evolved four major levels of distinct but overlapping lines of defense that minimize contact of microbes with the intestinal epithelium, as well as eradicate microbes that manage to penetrate the epithelial barrier and gain access to the intestinal tissues [18,69]. The first line of defense in mammals is mediated by goblet cells, which are specialized epithelial cells that produce mucin, a highly glycosylated glycoprotein that polymerizes to form mucus. The macromolecular gel adheres to the apical surface of epithelial cells thereby providing a physical barrier that separates the epithelium from luminal bacteria [150]. The colon contains both an inner and outer layer of mucus. The inner layer of mucus is dense, highly polymerized, tightly adherent to the epithelial cell surface and relatively impermeable to the microbiota due to its resistance to most bacterial proteases [84]. The outer mucus layer is a loose, net-like polymer that occupies approximately 4 times the volume than the inner layer. It is not uncommon to observe some bacteria in the outer layer of mucus that may arise from limited proteolysis of the polymer that is mediated by the host as well as resident bacteria [82,83].
In contrast to the colon, the small intestine contains substantially smaller numbers of bacteria with the epithelium being covered by a single, loosely adherent layer of mucus. However, some pathogenic microorganisms have developed strategies to penetrate the mucus layers, allowing them to gain access to the epithelial cell surface through various strategies that vary among pathogens and commensals alike [38]. For instance, Yghl, an Escherichia coli-derived M60-like protease degrades mucus proteins, thus providing bacteria with access to the mouse small intestinal epithelium [110]. Pic, a secreted protease produced by E. coli and Shigella flexneri, degrades rat ileal mucus and stimulates compensatory mucus hypersecretion by goblet cells [140]. In addition, the food-born pathogen Campylobacter jejuni expresses JlpA surface lipoprotein adhesin, which allows for colonization of the human small intestine and colon [91]. Furthermore, the gastrointestinal pathogen Salmonella enterica penetrates cecal mucus using chemotaxis and flagella [193]. Under the highly acidic conditions of the stomach, Helicobacter pylori may reduce mucus viscosity via alkalization of the environment thereby allowing adhesion of these pathogens to the surface epithelium using SabA adhesin. It is hypothesized that NH3 that causes pH elevation is produced by H. pylori from the hydrolysis of urea [23,115]. The importance of mucus as a protective barrier has been directly demonstrated in genetically-engineered mice that have been rendered devoid of intact colonic mucus via deletion of the Muc2 gene. These Muc2-deficient (Muc2−/−) mice exhibit growth retardation, enhanced bacterial invasion of colonic crypts and development of colonic inflammation by 5 weeks of age [202]. The absence of mucus may also synergize with other genetic defects such that mice devoid of both Muc2 and the IL-10 gene (Muc2−/−/IL-10−/− double deficient animals) develop exacerbated colitis when compared to colitis that develops in Muc2−/− or IL-10−/− animals [201]. These data suggest that chronic colitis in mucus-deficient mice may be due to the adhesion of bacteria to colonic epithelial cells (colonocytes) and invasion of these unwanted neighbors into the lamina propria where they activate innate and adaptive immune responses. In reality, the mechanisms for the induction of inflammation may be more complicated than simple adhesion/invasion of luminal bacteria as it is well-known that certain species of bacteria are capable of adhering to epithelial cells without invading the tissue or launching an intestinal inflammatory response. In fact, commensal bacteria-epithelial cell interactions appear to be required for homeostasis and for mediating tolerance to commensal bacteria [77,159]. The teleost intestinal tract is also endowed with a single protective layer of mucus. Zebrafish possess 5 genes that code expression of gel-forming mucin glycoproteins, which belong to two gene families: Muc2 (Muc2.1 and Muc2.2 genes) and Muc5 (Muc5.1, Muc5.2 and Muc5.3 genes). Sequencing of these genes revealed a great deal of homology to other vertebrates. According to gene sequencing data, the domain architecture of Muc5.1 and Muc5.2 zebrafish mucin proteins is typical for mammalian-secreted mucins [57,80]. The importance of intestinal mucus in zebrafish was demonstrated by Oehlers et al. who found that reduction of mucus secretion via administration of retinoic acid exacerbated enterocolitis induced by the hapten trinitrobenzene sulfonic acid (TNBS) in zebrafish larvae [146].
A second line of defense designed to limit the contact of bacteria with the intestinal epithelium is the secretion of different antimicrobial peptides (AMPs) by specialized epithelial cells (Figure 2). In general, AMPs are restricted to the mucus with only minute amounts diffusing into the gut lumen [131]. Paneth cells, in spite of being a relatively rare cell type located in the intestinal crypts, are essential for limiting access of bacteria to the epithelium [11]. These specialized cells secrete α-defensins (called cryptdins in mice) which bind to bacterial cell membranes and permeabilize them, resulting in death of the microorganism. In contrast to other defensins, human α-defensin 6 (HD6) does not destroy the bacterial membrane. Instead, it forms a nano-net of self-assembling fibrils on the surface of the bacterial cell thereby interfering with its ability to bind to the epithelium [14,149,210]. Of note, mice lack the HD6 analogue [183]. It appears that the secretion of cryptdins as well as cryptdin-related sequences (CRS) antimicrobial peptides is constitutive and does not depend upon the presence of luminal bacteria as it has been found in germ-free mice. In fact, production of cryptdins and CRS1C is increased only modestly in conventional mice challenged with S. enterica or Listeria monocytogenes. In contrast, CRS4C and bactericidal lectin Reg3γ are also produced constitutively by Paneth cells, but their secretion increases significantly following challenge with a pathogen [21,89,156]. Reg3γ is capable of killing Gram positive bacteria by binding to their membrane peptidoglycans and disrupting the bacterial cell wall [21]. In addition to Paneth cells, goblet cells secrete cysteine protease cathepsin K (Ctsk) [144,184]. Cathepsin K contributes to maintaining the normal composition of intestinal microflora as Ctsk−/− mice exhibit dysbiosis [184]. Although it has been demonstrated that other members of the cathepsin proteinase family are capable of lysing bacterial outer membranes [197], the mechanisms responsible for cathepsin K-mediated dysbiosis remain to be defined.
Another AMP produced by intestinal epithelial cells and inflammatory polymorphonuclear leukocytes (PMNs) is Lipocalin 2 (Lcn-2) [93]. This protein binds to the bacterial catecholate-type iron-chelating siderophores thereby preventing the acquisition of iron by bacterial cells, thus limiting bacterial growth [48,56]. Lcn-2 as well as heparin/heparin sulfate interacting protein (HIP/RPL29) expressed on the surface of mouse epithelial cells in the small intestine, are identical to those found in the mouse airway epithelium. These observations suggest that these proteins are part of the innate antibacterial defense of the epithelial surface in mammals [130,215]. Epithelial cell-derived AMPs are quite effective in limiting interactions between luminal bacteria and the epithelium; however, certain strains of bacteria have developed mechanisms whereby they may circumvent some of these antimicrobial properties. For instance, resistance to killing by host AMPs towards Salmonella typhimurium results from modifications of surface lipopolysaccharides (LPS) thereby reducing the efficacy of AMPs to bind to bacteria. In addition, S. typhimurium may secrete the iron-chelating molecule (salmochelin) produced by glycosylation of another siderophore, enterobactin. Salmochelin is no longer bound by Lcn-2 and it is therefore, capable of supplying bacteria with iron [60,158].
Intracellular pattern recognition receptors (PRR) and the nucleotide-binding oligomerization domain receptor (NOD) proteins NOD1 and NOD2 are important regulators of mammalian innate immune responses which are expressed in Paneth cells of the intestinal epithelium as well as in many immune cell types [14,20]. Upon recognition of specific peptidoglycans (PGN) on the bacterial cell surface, NOD1 and NOD2 activate innate immunity in an NF-κB dependent manner [74]. In mice, it was shown that the lack of NOD2 expression causes intestinal dysbiosis [153]. Of interest, the human intracellular pathogen L. monocytogenes avoids recognition by NOD proteins due to the N-deacetylation of its surface peptidoglycan, providing protection from the host lysozyme and allowing it to replicate within host macrophages [16]. Zebrafish orthologs of both mammalian Nod1 and Nod2 genes have been identified and are highly homologous to their mammalian counterparts. Zebrafish Nod2 gene splicing is shown to be similar to human Nod2 [24,101,147]. The ability to control systemic infection is markedly reduced in NOD1−/− as well as NOD2−/− zebrafish larvae infected with S. enterica. Interestingly, the expression of intestinal duox, a gene that encodes for an enzyme that produces H2O2, is decreased in NOD1−/− larvae, but not in NOD2−/− larvae following challenge with S. enterica. This suggests that, unlike in mammals, reactive oxygen species (ROS) production by DUOX in zebrafish is regulated by NOD1 and not by NOD2 [104,147]. Consistent with this hypothesis, increased bacterial load is observed in NOD1−/−, but not in NOD2−/− larvae [147]. In addition to DUOX, other AMPs have been identified in the teleost gut. For instance, Oehlers et al. reported expression of defensin β-like (dfnb) genes which are closely related to mammalian β-defensin genes, as well as expression of peptidoglycan recognition proteins (pglyrp) which exert their bactericidal properties against Gram positive bacteria [147]. Another group of investigators observed increased expression of cathepsin-like protein genes ctsl1a and ctsl1b as well as c-type lectin genes in the intestines of zebrafish infected by Mycobacterium marinum [203]. Brush border-associated alkaline phosphatase (lap) activity localized on zebrafish epithelial cells is also thought to be important for limiting microbe adhesion. Bacterial LPS from mucosa-associated bacteria is thought to induce the expression of Lap that dephosphorylates microbial LPS. Lap also downregulates myeloid differentiation primary response protein (MyD88)-dependent pro-inflammatory signaling in the intestine thereby preventing intestinal inflammation [7]. These data are consistent with the observation that MyD88−/− larvae display reduced survival as well as compromised responses to bacterial flagellin and LPS following challenge with Edwarsiella tarda and S. typhimurium [204].
A third line of defense by which vertebrates limit contact of luminal bacteria with their gut epithelium is by the synthesis and secretion of IgA. In mammals, approximately 70% of the total body production of IgA is induced by the intestinal microbiota [112,114]. IgA class switching by mucosal B cells is maintained by retinoic acid produced constitutively by resident dendritic cells (DC) as well as by transforming growth factor-β (TGF-β) and other compounds provided by the intestinal microenvironment [113]. Numerous CD11c+ DCs are located in the Peyer’s patches and lamina propria of the small bowel. These professional antigen presenting cells continuously sample their immediate surroundings for bacteria and bacterial antigens. If present, DCs will phagocytize whole bacteria or antigens and migrate to the mesenteric lymph nodes (MLNs) where they present microbial peptides to naïve B and T cells. Since antigen- or bacteria-loaded DCs are incapable of migrating beyond the gut draining MLNs and into the circulation, immune responses induced by their interactions with MLN-associated B and/or T cells are restricted to the gut [43,114,142,164]. Following DC-mediated activation, B cells transform into plasma cells, leave the MLNs and enter the systemic circulation. These effector cells then home back to the intestinal lamina propria where they produce bacterial-specific immunoglobulin A (IgA) [68,83,138]. This bacterial specific immunoglobulin is then transported from the intestinal interstitium through epithelial cells via a specific pIgR receptor (see above) and into the gut lumen. Once IgA is deposited onto the luminal surface of the epithelium, it may bind to bacteria in close proximity to the epithelium, thereby preventing invasion of the microorganisms into the gut tissue. In addition, recent studies suggest that IgA coating of luminal bacteria facilitates their uptake by the M cells overlying gut-associated lymphoid tissue (e.g. MLNs, isolated lymphoid follicles), thereby limiting intestinal immune responses to the gut [152,185]. To date, IgA expression has not been detected in fish [22,167] and it is not clear if any particular immunoglobulin class in zebrafish may be considered a functional equivalent of mammalian IgA in mucosal immune responses. The pIgR gene expression in zebrafish gut has been confirmed in a recent study and unlike in mammals, zebrafish plgR is composed of only two domains [98]. It is noteworthy that in another teleost, Takifugu rubripes (fugu), pIgR is capable of binding IgM in the skin mucus [61]. In another study, the relatively primitive immunoglobulin IgT, also known as IgZ, is shown to function as a mucosal antibody and to be protective against intestinal parasites in rainbow trout [221].
The multiple lines of defense that mammals and teleosts have developed to protect against invasion of pathogenic and/or commensal bacteria are quite effective. Occasional breaches in the epithelium may occur, allowing microorganisms to gain access to the intestinal lamina propria. When this occurs, vertebrates mobilize their fourth line of defense: the innate and adaptive immunity. The vertebrate intestine is in actuality, the largest lymphoid tissue in the body which contains substantial numbers of dendritic cells, phagocytic leukocytes and lymphocytes [179]. Microbes which enter the gut lamina propria are rapidly phagocytized and killed by interstitial macrophages [186]. MyD88 signaling in these phagocytes is selectively downregulated insuring that intestinal macrophages produce relatively small amounts of pro-inflammatory cytokines and mediators while possessing potent bactericidal activity. This allows for the removal of unwanted microbes without fully activating an inflammatory response and consequent tissue injury [188,189]. These specific macrophages (called M2 macrophages) are crucial for limiting bystander tissue injury produced by innate immune responses. Intestinal M2 macrophages also contribute greatly to the repair of intestinal damage via their ability to induce the proliferation of epithelial cells to repopulate the injured epithelium [181]. The role of intestinal macrophages in teleosts has not been described in as much detail as their mammalian counterparts. However, their importance was demonstrated in zebrafish following intestinal pathogen S. flexneri infection. In mammals, it was shown that invading bacteria which penetrate the intestinal epithelium are phagocytized by indigenous macrophages and instead of being killed, ingested bacteria leave the phagosome causing macrophage cell pyroptosis [2]. In zebrafish, this leads to a dramatic decrease in macrophage numbers resulting in bacteremia and eventually death of the larvae [137]. This observation is especially valuable because it shows that zebrafish can be a relevant model for studying the pathogenicity of microorganisms such as S. flexneri that are not natural fish pathogens.
Another group of heterogeneous innate immune cells that reside within the gut and provide protection against invading microorganisms, as well as regulate adaptive immunity and wound healing are called innate lymphoid cells (ILCs). ILCs arise from hematolymphoid progenitor cells and may be categorized as cytotoxic ILCs which are represented by natural killer (NK) cells and helper-like ILCs that include ILC1, ILC2 and ILC3 [39]. NK cells and ILC1 produce large amounts of IFN-γ that is used to protect mice against infection by certain bacteria such as L. monocytogenes [165]. ILC2 cells may also protect mice from certain helminths such as Trichinella spiralis via their production of the Th2 cytokine IL-13 [126]. What was considered a subset of NK cells (i.e. NK-22 cells) was shown to protect the host against bacterial invasion as well as maintain epithelial barrier integrity. These IL-22 producing ILCs, which are now considered ILC3, produce little or no IFN-γ and express the retinoic-acid-receptor-related orphan receptor-γt (RORγt) transcription factor [50,107,174,177]. It has been demonstrated that ILC3-derived IL-22 attenuates acute and chronic intestinal inflammation induced in lymphopenic mice (Rag−/− mice) by Citrobacter rodentium and by adoptive transfer of naïve T cells, respectively [177,220]. Another important function of ILCs is anatomical containment of the intestinal microbiota [39,65,191]. Recent studies have demonstrated that immuno-depletion of ILCs resulted in the systemic dissemination of intestinal bacteria that was accompanied by multi-organ inflammation [191]. The loss of containment appeared to be due to the loss of ILC-derived IL-22 as the administration of IL-22 to ILC-depleted mice prevents the dissemination of commensal bacteria [191]. In follow up studies, Hepworth et. al., showed that selective depletion of MHC RORγt+ ILCs expressing major histocompatibility complex II (MHC II) resulted in dysregulated commensal bacteria-dependent CD4+ T cell responses that induced spontaneous and systemic dissemination of commensal bacteria resulting in multi-organ inflammation [65]. Taken together, these data suggest that intestinal homeostasis is mediated by the MHC II-dependent interaction between ILCs and CD4+ T cells. NK cells have currently not been described in zebrafish. It has been suggested that the recently described novel immune-type receptors (NITR) expressing cells in zebrafish and other teleosts may be functionally-equivalent to mammalian NK cells [78,218].
The adaptive immune cells (i.e. lymphocytes) represent another level of defense that synergizes with innate immune cells to maintain homeostasis, as well as protect the host from invasion of pathogenic microbes. One such group of lymphocytes that reside in large numbers within the epithelial monolayer are the intraepithelial lymphocytes (IELs). Natural IELs are either CD8αα+ or CD8αα− and express TCRγδ or TCRαβ [27]. TCRγδ IELs constitute approximately 60% of the total IELs of the small bowel and are generally thought to maintain epithelial integrity and limit immune activation [27]. Along with intestinal macrophages, TCRγδ IELs are capable of producing keratinocyte growth factor (KGF) that mediates intestinal wound healing [26]. It has been shown recently that the ability of TCRγδ-derived KGF to repair acute inflammatory injury in the gut is impaired in mice that lack an important ligand (semaphorin; CD100) for TCRγδ activation [127]. Epithelial cells from CD100-deficient (CD100−/−) or KGF−/− mice exhibit significantly decreased proliferative rates in mouse models of experimental colitis. Due to the expression of the NKG2D NK receptor, TCRγδ IELs are able to recognize the MHC I-like surface markers MICA and MICB expressed widely in the intestines of humans with IBD. NKG2D/MIC interaction is essential for cytotoxic killing of the enterocytes activated by allogeneic ligands [59,72]. TCRγδ IELs stimulated by enterocytes may also respond to bacterial challenge by producing the AMPs Reg3β and Reg3γ [75]. In response to bacterial challenge, IELs migrate to enterocytes in contact with the pathogen, thus providing the host with immediate and essential protection against invading pathogen [41].
If intestinal bacteria evade the multiple layers of defense described above, they will eventually be eliminated by the adaptive immune response called cell mediated immunity. Whole bacteria and/or their antigens will be endocytosed by DCs within the LP. Antigen-loaded DCs will then travel from the LP to the MLNs via the efferent lymphatics. During this process, DCs will process and present the bacterial antigens in the context of their MHC II. Upon arrival to the MLNs, naïve CD4+ T cells will bind to the gut derived DCs via their TCR, whereupon the T cells become primed/activated and polarized and expand to produce larger numbers of antigen-specific Th1 and/or Th17 effector cells. It is during this time that T cells become “imprinted” via the increased expression of surface adhesion molecules such as CCR9, LFA-1, PSGL-1 and VLA-4. Once the effector cells have departed the MLNs and enter the systemic circulation, these imprinted homing proteins will direct migration of the T cells to the intestine where the levels of bacteria and antigens are the greatest [94]. Following extravasation into the gut LP, effector T cells will engage DCs as well as other antigen presenting cells (e.g. macrophages, B cells) that have also processed and presented the same bacterial antigens whereby the effector T cells become rapidly and fully activated to produce large amounts of IFN-γ, IL-2, TNF-α and IL-17. Some of these cytokines will then activate intestinal macrophages to “help” in their phagocytosis and killing of bacteria. These inflammatory cytokines will also enhance the expression of endothelial cell adhesion molecules (e.g. E-selectin, ICAM-1, VCAM-1) thereby promoting the infiltration of additional phagocytic/myeloid leukocytes (e.g. PMNs, monocytes). Lugo-Villarino et al. have described a zebrafish myeloid cell that strongly resembles mammalian DCs morphologically and functionally and have MHC II expression [109]. Because innate and adaptive immune responses produce a number of cytotoxic species such as reactive oxygen and nitrogen metabolites, as well as secrete and activate different hydrolytic enzymes (e.g. metalloproteinases), the host must have ways to limit inflammatory tissue injury during these responses. This is accomplished by the action of thymic-derived and peripheral induced regulatory Tregs [69,79,116,182]. In mice, the majority of Tregs express the Foxp3 transcription factor and CD25. Thymic-derived Tregs (tTregs) acquire expression of Foxp3 during their development in the thymus whereas induced Tregs (iTregs) are produced from conventional CD4+ T cells following their interaction with antigen-loaded DCs in lymphoid and nonlymphoid tissue [13,71]. Both populations of Tregs utilize IL-10 and/or TGF-β to regulate innate and adaptive immune responses [148].
A list of cloned genes for teleost T cell-associated peptides analogous to those expressed in mammals includes α, β, γ, and δ TCR subunits as well as CD3, CD4, CD8, CD28 and recombinase activated gene-1 (Rag-1) [9,17,57,196]. There are very few studies devoted to intestinal T cell function in zebrafish. The genes for zebrafish Foxp3, Stat6, and T-bet transcription factors have been cloned. Foxp3 and T-bet zebrafish genes are identical to their human homologues, while the Stat6 gene has some minor differences [132]. A gene for zebrafish IL-23α, an essential component of IL-23, an interleukin required for Th17 cell differentiation in mammals, has been cloned recently as well [67]. Taken together, these data suggest that zebrafish may possess Th1, Th2, Th17 and Treg cells described in mammals. Mouse T cells, transduced with the zebrafish Foxp3 gene, suppress activation of other T cells in in vitro co-culture assay [157]. Interestingly, two alternatively spliced transcripts have been discovered for the zebrafish Foxp3 gene [132]. According to histologic staining data in rainbow trout, CD8+ cytotoxic T cells can make up more than half of the T cell population in the teleost’s gut [196]. In sea bass, proliferation of T cells by allogeneic stimulation was reduced significantly by addition of cyclosporine A. It suggests that T cell proliferation in teleosts may be controlled by mechanisms similar to those reported for mammals [128].
4. Loss of Tolerance to the Microbiota Induces Intestinal Inflammation
The fact that vertebrates remain healthy despite the continuous presence of enormous numbers of potentially harmful microorganisms, attest to the remarkable ability of the intestinal immune system to maintain tolerance to nonpathogenic/commensal bacteria. It is becoming increasingly appreciated that the loss of tolerance to the commensal microbiota represents an important pathogenetic mechanism responsible for induction of intestinal inflammation [25,73,90,99,176,214]. Based upon their early work performed in rats and mice more than 20 years ago, Powrie and coworkers proposed that the chronic intestinal inflammation observed in patients suffering from IBD results from a dysregulated immune response to intestinal bacteria [3,79,154]. Evidence gathered over the past two decades suggests that IBD most likely arises from a complex interaction between genetic and environmental factors [73,90,99,214].
4.1 Genetic Susceptibility
To date, more than 160 different polymorphisms or susceptibility loci have been identified in patients with diagnosed IBD [25,87,90,206]. As one might expect, a substantial number of these loci common to both CD and UC are associated with innate and adaptive immunity. While the majority of the altered loci are associated with both CD and UC, genome wide associated studies (GWAS) have identified a smaller but significant number of polymorphisms that appear specific for CD. These include NOD2, autophagy related protein 16-like 1 (ATG16L1) and Immunity-Related GTPase Family M (IRGM) genes [87]. Furthermore, genes associated with epithelial barrier function have also been identified [87]. Because these genes express proteins that are important for recognition and processing of intracellular bacteria as well as for limiting the invasion of intestinal bacteria into the gut lamina propria, one could envision a scenario whereby CD may arise from unrelenting bacterial infections [87]. In addition to defects in innate immunity, GWAS have reported several polymorphisms in genes that are crucial for the induction and regulation of adaptive immune responses such IL-17 and IL-23 signaling and IL-10/IL-R interactions [87]. UC-specific susceptibility loci have also been described for genes responsible for HLA class II-mediated antigen presentation (e.g. HLA-DQA1), inflammatory mediator generation (e.g. TNFRSF14, TMFRSF9, IL1R2, IL8Ra, IL7R) and epithelial barrier function (e.g. HNF4A, CHD1, LAMB1 genes) [87,92,206].
Much of our understanding of the immuno-pathogenesis of chronic gut inflammation has come from studies using mouse models of IBD [95,163,200]. Prior to the early 1990s, animal models of IBD required the oral or rectal administration of erosive chemicals to produce colonic mucosal damage that was followed by an acute self-limiting inflammatory response [95]. However, the publication of 4 paradigm-changing studies in 1993 altered dramatically the future of experimental IBD by demonstrating that mice rendered genetically-deficient in certain genes involved in immune regulation developed spontaneous and unrelenting colitis [100,135,154,173]. Since the publication of these ground-breaking studies, numerous other, genetically-engineered mouse models of chronic intestinal inflammation have been generated that continue to provide new and important information related to the immuno-pathogenesis of experimental IBD [85,95,163]. Although none of the mouse models completely recapitulate the clinical and immuno-histopathological features of human disease, data obtained from several studies using chronic mouse models have revealed two important concepts that have served to direct basic and clinical research over the past two decades [95,163]. One basic concept that has come from these studies is that genetics play an important role in the pathogenesis of chronic inflammation. Indeed, numerous studies have shown that chronic inflammation develops in mice that possess one or more defects in genes that are involved in certain pathways related to innate and/or adaptive immune responses such as the T-bet−/− × Rag-2−/− ulcerative colitis (TRUC) [95,163,200].
Another important concept that has come from these mouse studies is that the “environment” (i.e. the intestinal microbiota) is required for induction and perpetuation of chronic gut inflammation in genetically-susceptible mice. Mice that are depleted of their microbiota using antibiotic administration or are delivered by caesarian section and raised under germ-free conditions develop little or no colitis [25,90,96,176]. In addition to mice, zebrafish are also being used as models for investigating the immunopathological mechanisms involved in intestinal inflammation [216]. These teleost models employ the use of chemical haptens [i.e. TNBS, oxazolone or irritants (dextran sodium sulfate (DSS)] to induce acute, self-limiting intestinal inflammation in larvae or adult fish [216]. Although not chronic in nature, these models have revealed several similarities to mouse-based models with respect to inflammation gene expression, leukocyte infiltration, intestinal injury and the requirement for intestinal bacteria [19,63,64,216]. Of note, some investigators have demonstrated that the knock down or deletion of genes that have been shown to be dysfunctional in human IBD or absent in mice that develop IBD, also increase susceptibility of zebrafish to bacterial infection [216].
4.2 Environment: Intestinal Bacteria Drive Chronic Gut Inflammation
There is no question that genetics play an important role in the pathogenesis of IBD. However, genetics alone cannot fully account for the dramatic increase in IBD in modernized societies over the past 50 years. For example, concordance rates for IBD in identical twins is actually quite low with estimates of 30–35% and 10–15% for CD and UC, respectively [10]. In addition, it has been well-documented that countries/societies that have adopted a “Modernized” lifestyle have shown sharp increases in the incidence and prevalence of IBD over the past five decades [10,32,92,134,206]. Taken together, these studies suggest that alterations in intestinal microbiota produced by environmental factors may be responsible for the increased incidence in IBD throughout the world. Multiple studies have demonstrated that increased use (and abuse) of antibiotics, better environmental sanitation, increased personal hygiene, and alterations in diet and lifestyle dramatically alter the composition of intestinal microbiota [10,32,92,134,206]. Moreover, the rapid appearance of IBD in rural-based societies where these chronic diseases were once rare, have increased dramatically over the past 50 years following transition to more modernized communities. Taken together, these genetic and environmental observations support the hypothesis that environmental factors may play an important role in the induction of disease in genetically-susceptible individuals [10,118,134]. This is true not only for human disease but is also observed in mouse and teleost models of intestinal inflammation. As pointed out previously, many of the mouse models of chronic gut inflammation develop little or no disease in the absence of intestinal bacteria. In addition, changes in microbiota may alter dramatically the onset and/or severity of disease in these mice [95,96,163,200].
So, what exactly is the evidence that intestinal bacteria are an important pathogenetic factor that drives gut inflammation in genetically susceptible individuals? In fact, there is really only one study in humans that directly implicates intestinal bacteria in inducing IBD. Studies by Rutgeerts and coworkers showed that when the fecal stream of a segment of the small bowel that has been surgically resected (due to advanced CD) to form a neoterminal ileum was diverted, intestinal inflammation failed to develop [172]; however, when the fecal stream was restored inflammation rapidly appeared, suggesting a role for intestinal microbiota [34,172]. In addition to these direct studies, a number of indirect or association studies have been published demonstrating greater numbers of mucosa-, epithelia- and lamina propria-associated bacteria in histological samples obtained from inflamed vs. healthy intestine [25,25,118,119,122]. As mentioned previously, GWAS have revealed polymorphisms (mutations) in genes that are responsible for recognition and intracellular processing (and killing) of bacteria (i.e. NOD2, ATG16L1 and IRGM) which are strongly associated with patients with CD [87]. Taken together, these data are consistent with the hypothesis that in addition to polymorphisms in genes associated with the adaptive immune system, defects in innate immune responses to intestinal bacteria may represent an important immunopathogenic factor that induces and perpetuates chronic intestinal inflammation [25,92,206]. The beneficial effects of certain antibiotics administered to patients with distal bowel inflammation is consistent with a role for the microbiota in disease pathogenesis [45].
Another important piece of indirect evidence implicating intestinal microbiota in the pathogenesis of human IBD is the observation that gut inflammation appears to always be accompanied by dramatic alterations in the luminal and mucosa-associated composition of intestinal bacteria, a situation called dysbiosis [176,192]. Numerous studies have reported marked dysbiosis in luminal (i.e. fecal) samples collected from humans with active IBD [54,99]. The large majority of these studies have used luminal/fecal samples and have consistently shown a decrease in bacterial diversity and abundance of bacteria within the Firmicutes and Bacteroidetes, whereas the abundance of certain Proteobacteria such as the Enterobacteriaceae (E. coli, AIEC) are increased [25,49,118,119,151,162,176]. However, a more recent study using large numbers of newly diagnosed, treatment-naïve patients with IBD, reports that from the trillions of possible bacteria that reside within the small and large bowel, alterations in abundance of only a select few groups of mucosa-associated bacteria were strongly correlated with presence and quality of disease [54]. This same study also emphasized that alterations observed in luminal/fecal microbial communities did not appear to correlate with disease status. These exciting studies have, for the first time, identified specific communities of mucosa-associated bacteria that may be very important in the induction and progression of chronic intestinal inflammation in genetically-susceptible individuals. Specifically, Gevers and coworkers observed increased abundance of certain pathobionts including Enterobacteriaceae, Pasteurellaceae, Veillonellaceae and Fusobacteriaceae as well as decreased abundance of “protective” Erysipelotrichales, Bacteroidales and Clostridiales [54]. Pathobionts are constituents of the healthy microbiota and are not considered classical pathogens; however these bacteria possess the potential to induce gut inflammation following disruption of the healthy microbiota [171].
Dysbiosis is also commonly observed in several mouse models of IBD [95,96]. Two more recent mouse models have extensively characterized this dysbiosis with the objective of defining the specific populations of pathobionts responsible for induction of disease. One model is the TRUC mouse model. This genetically-engineered mouse develops spontaneous, chronic and communicable colitis that is driven by the interaction between innate immune cells and the microbiota [53]. The fact that colitis is communicable and will develop in wild type mice when housed with TRUC mice suggests that there is an outgrowth of pathogenic bacteria in these mice [53]. Investigators have observed an overabundance of Klebsiella pneumoniae and Proteous mirabilis that correlated well with disease [52]. These two microbes are known to be increased in human IBD [52,53]. Another novel mouse model of IBD that requires the presence of intestinal bacteria and exhibits dysbiosis was created by Kang et. al. [88]. These investigators bred dominant negative TGFβRII (dnTGFβRII) mice with IL-10R2−/− mice to generate dnTGFβRII X IL-10R2−/− offspring which are referred to as dnKO mice. These offspring have defective TGFβ signaling in T cells and defects in IL-10R signaling in all tissues [88]. These dnKO mice develop an accelerated and unrelenting colitis that is abrogated by administration of broad spectrum antibiotics [88]. This same laboratory has also demonstrated that an isolate of Bacteroides, but not Enterobacteriaceae induces robust disease in dnKO but not in controls [15,66]. Taken together, these studies clearly demonstrate the need to assess the colitogenic potential rather than relying solely on 16S rRNA determinations. Despite the potentially important role that dysbiosis may play in the pathogenesis of IBD, it remains unclear whether dysbiosis is a cause or consequence of chronic gut inflammation. There are a few reports showing that dysbiosis may occur in healthy relatives of patients with IBD, suggesting that microbial alterations may precede disease, but little follow up of these family members has been documented [86]. In fact, it has been well-documented that products of the inflammatory reaction are effective at promoting dysbiosis [111]. Defining the role of dysbiosis in IBD may have to await more direct longitudinal studies that can only be accomplished in well-controlled mouse models of IBD. It should be noted that the intestinal microbiota has also been shown to be important for full expression of chemically-induced intestinal inflammation in zebrafish [19,63,64,216].
4.3 Intestinal Microbiota Transplant: A novel therapeutic strategy for treating IBD
Great advances have been made over the past 30 years in understanding the immuno-pathological mechanisms responsible for the induction and perpetuation of IBD. Yet, only a handful of pharmacologic and biologic agents are available to treat these debilitating inflammatory disorders. Furthermore, many of the current immunosuppressive small molecules available for patients may be toxic and the new-generation biologics can be expensive and require infusion in a physician’s office. Thus, it is critical that additional, nontoxic and inexpensive therapies be developed to treat patients with IBD. Given the fact that there is now considerable experimental and clinical evidence demonstrating that the intestinal microbiota may act as a “trigger” to induce and perpetuate chronic gut inflammation in genetically-susceptible individuals, investigators have undertaken different avenues of research in an attempt to restore the dysbiotic gut microbiota that develops in patients with active IBD to a more typical, highly diverse community of microorganisms. A variety of different approaches have been used to manipulate the intestinal microbiota in IBD patients including the use of antibiotics, probiotics, and prebiotics. Manipulating the dysbiotic microbiota to decrease disease-producing pathobionts, while enhancing the numbers and metabolic activity of beneficial bacterial communities, has tremendous potential for therapeutic benefit. The therapeutic efficacy of antibiotic, probiotic and/or prebiotic administration in patients with IBD has not been proven in placebo-controlled, double blind, multicenter clinical studies. However, there are encouraging data suggesting that oral administration of ciprofloxacin and/or metronidazole may be effective in attenuating distal bowel inflammation in patients with CD [105,155,176,207]. Currently, antibiotic therapy is primarily used to treat complications such as fistulas and ulcers in IBD patients.
A great deal of interest has been generated following the publication of a recent clinical study that demonstrated that transplantation (i.e. colonization) of healthy fecal microbiota into patients with recurrent Clostridium difficile colitis essentially cured this recurrent and intractable infection [205]. Van Nood et. al., showed that 94% of the patients that received microbiota transplantation (FMT) had complete resolution of disease whereas only 31% of patients receiving current therapy (e.g. vancomycin) showed significant improvement [205]. These studies produced a great deal of excitement about the possible use of FMT in the treatment of IBD. Although a few open label case/cohort studies have reported tantalizing evidence suggesting a beneficial effect of FMT in the treatment of IBD, by and large, these studies are small, statistically underpowered and/or uncontrolled investigations. Two recent well-controlled clinical studies have been published that assessed the therapeutic efficacy of FMT in patients with UC. In one study, Moayyedi and coworkers reported, in a randomized, double blinded, placebo controlled study that a significantly greater percentage of patients receiving weekly FMT for 6 weeks achieved remission than those patients receiving a placebo (i.e. water; 24% vs. 5%) [133]. In the other study, Rossen et. al., treated one group of patients with active UC with healthy FMT or autologous FMT (infusion of patients own feces to serve as a placebo). These investigators found no significant difference in clinical and endoscopic remission between the two groups [170]. The reasons for the different outcomes in the two studies are not entirely clear at the present time. The two studies did, in fact, contain several major differences including location and frequency of the FMT infusion, concurrent use of anti-TNF therapy in one study, but not the other and possible donor microbial composition for efficacy [58,170]. Nevertheless, both of these studies reconfirm previous observations that FMT is well-tolerated. One potential caveat that has not been adequately addressed is whether the dysbiosis observed in patients with IBD is a cause or consequence of gut inflammation. If in fact dysbiosis is initially created by subclinical inflammation in genetically susceptible individuals leading to the expansion of disease-producing pathobionts such as adherent/invasive Enterobacteriaceae, the use of FMT may not prove as effective as it would be if dysbiosis were the primary trigger of disease [212,213].
5. Conclusions
The vertebrate intestine is home to the vast majority of bacteria present on and within the body. The coexistence of mammals and teleosts with their gut microbiota is a continuously evolving process that provides numerous advantages to both host and bacteria. The intestinal microbiota which is now considered a virtual organ provides the host with the immunologic stimuli, nutrients and vitamins required for developing and maintaining health of the host. In turn, the host provides a physiological environment that is conducive for the steady-state bacterial colonization of the gut. The fact that mammal and teleost homeostasis is maintained in the presence of such large numbers of potentially dangerous microorganisms is testament to the remarkable ability of the intestinal immune system in these land and aquatic animals to maintain tolerance to commensal bacteria while mounting effective immune responses to pathogenic microbes. The inability of the intestinal immune system to maintain tolerance to the intestinal microbiota is thought to be a major immuno-pathogenic mechanism responsible for the induction of IBD in humans. Indeed, there is an accumulating literature suggesting that Crohn’s disease and/or ulcerative colitis results from a dysregulation of immune responses toward intestinal bacteria in genetically-susceptible individuals.
Acknowledgments
Some of the work reported in this manuscript was supported by grants from the DOD (W81XWH-11-1-0666; MBG) and the NIH (R01-DK091269; MBG)
Footnotes
Conflict of interest
The authors declare that there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, Thom SR, Bushman FD, Vinogradov SA, Wu GD. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashida H, Ogawa M, Kim M, Suzuki S, Sanada T, Punginelli C, Mimuro H, Sasakawa C. Shigella deploy multiple countermeasures against host innate immune responses. Curr Opin Microbiol. 2011;14:16–23. doi: 10.1016/j.mib.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Asseman C, Mauze S, Leach MW, Coffman RL, Powrie F. An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J Exp Med. 1999;190:995–1004. doi: 10.1084/jem.190.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aujla SJ, Dubin PJ, Kolls JK. Th17 cells and mucosal host defense. Semin Immunol. 2007;19:377–382. doi: 10.1016/j.smim.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avershina E, Storro O, Oien T, Johnsen R, Pope P, Rudi K. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol Ecol. 2014;87:280–290. doi: 10.1111/1574-6941.12223. [DOI] [PubMed] [Google Scholar]
- 7.Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol. 2006;297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Bernard D, Six A, Rigottier-Gois L, Messiaen S, Chilmonczyk S, Quillet E, Boudinot P, Benmansour A. Phenotypic and functional similarity of gut intraepithelial and systemic T cells in a teleost fish. J Immunol. 2006;176:3942–3949. doi: 10.4049/jimmunol.176.7.3942. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein CN, Shanahan F. Disorders of a modern lifestyle: reconciling the epidemiology of inflammatory bowel diseases. Gut. 2008;57:1185–1191. doi: 10.1136/gut.2007.122143. [DOI] [PubMed] [Google Scholar]
- 11.Bevins CL. Innate immune functions of alpha-defensins in the small intestine. Dig Dis. 2013;31:299–304. doi: 10.1159/000354681. [DOI] [PubMed] [Google Scholar]
- 12.Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, Perez-Amodio S, Strippoli P, Canaider S. An estimation of the number of cells in the human body. Ann Hum Biol. 2013;40:463–471. doi: 10.3109/03014460.2013.807878. [DOI] [PubMed] [Google Scholar]
- 13.Bilate AM, Lafaille JJ. Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu Rev Immunol. 2012;30:733–758. doi: 10.1146/annurev-immunol-020711-075043. [DOI] [PubMed] [Google Scholar]
- 14.Biswas A, Petnicki-Ocwieja T, Kobayashi KS. Nod2: a key regulator linking microbiota to intestinal mucosal immunity. J Mol Med (Berl) 2012;90:15–24. doi: 10.1007/s00109-011-0802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr, Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, Psylinakis E, Bouriotis V, Hugot JP, Giovannini M, Coyle A, Bertin J, Namane A, Rousselle JC, Cayet N, Prevost MC, Balloy V, Chignard M, Philpott DJ, Cossart P, Girardin SE. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci U S A. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boschi I, Randelli E, Buonocore F, Casani D, Bernini C, Fausto AM, Scapigliati G. Transcription of T cell-related genes in teleost fish, and the European sea bass (Dicentrarchus labrax) as a model. Fish Shellfish Immunol. 2011;31:655–662. doi: 10.1016/j.fsi.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol. 2013;14:660–667. doi: 10.1038/ni.2611. [DOI] [PubMed] [Google Scholar]
- 19.Brugman S, Liu KY, Lindenbergh-Kortleve D, Samsom JN, Furuta GT, Renshaw SA, Willemsen R, Nieuwenhuis EE. Oxazolone-induced enterocolitis in zebrafish depends on the composition of the intestinal microbiota. Gastroenterology. 2009;137:1757–1767. doi: 10.1053/j.gastro.2009.07.069. [DOI] [PubMed] [Google Scholar]
- 20.Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castro R, Jouneau L, Pham HP, Bouchez O, Giudicelli V, Lefranc MP, Quillet E, Benmansour A, Cazals F, Six A, Fillatreau S, Sunyer O, Boudinot P. Teleost fish mount complex clonal IgM and IgT responses in spleen upon systemic viral infection. PLoS Pathog. 2013;9:e1003098. doi: 10.1371/journal.ppat.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, Ewoldt RH, McKinley GH, So P, Erramilli S, Bansil R. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci U S A. 2009;106:14321–14326. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang MX, Chen WQ, Nie P. Structure and expression pattern of teleost caspase recruitment domain (CARD) containing proteins that are potentially involved in NF-kappaB signalling. Dev Comp Immunol. 2010;34:1–13. doi: 10.1016/j.dci.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–1728. doi: 10.1053/j.gastro.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R. Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proc Natl Acad Sci U S A. 2002;99:14338–14343. doi: 10.1073/pnas.212290499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol. 2011;11:445–456. doi: 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clements KD, Angert ER, Montgomery WL, Choat JH. Intestinal microbiota in fishes: what’s known and what’s not. Mol Ecol. 2014;23:1891–1898. doi: 10.1111/mec.12699. [DOI] [PubMed] [Google Scholar]
- 31.Coombes JL, Robinson NJ, Maloy KJ, Uhlig HH, Powrie F. Regulatory T cells and intestinal homeostasis. Immunol Rev. 2005;204:184–194. doi: 10.1111/j.0105-2896.2005.00250.x. [DOI] [PubMed] [Google Scholar]
- 32.Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140:1785–1794. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 33.D’costa A, Shepherd IT. Zebrafish development and genetics: introducing undergraduates to developmental biology and genetics in a large introductory laboratory class. Zebrafish. 2009;6:169–177. doi: 10.1089/zeb.2008.0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Haens GR, Geboes K, Peeters M, Baert F, Penninckx F, Rutgeerts P. Early lesions of recurrent Crohn’s disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology. 1998;114:262–267. doi: 10.1016/s0016-5085(98)70476-7. [DOI] [PubMed] [Google Scholar]
- 35.de La Cochetiere MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Dore J. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol. 2005;43:5588–5592. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends Ecol Evol. 2006;21:517–523. doi: 10.1016/j.tree.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 37.Dobzhansky T. Genetics of the evolutionary process. Columbia University Press; 1970. [Google Scholar]
- 38.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2015 doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberl G, Di Santo JP, Vivier E. The brave new world of innate lymphoid cells. Nat Immunol. 2015;16:1–5. doi: 10.1038/ni.3059. [DOI] [PubMed] [Google Scholar]
- 40.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edelblum KL, Singh G, Odenwald MA, Lingaraju A, El BK, McLeod R, Sperling AI, Turner JR. gammadelta Intraepithelial Lymphocyte Migration Limits Transepithelial Pathogen Invasion and Systemic Disease in Mice. Gastroenterology. 2015;148:1417–1426. doi: 10.1053/j.gastro.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Espey MG. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic Biol Med. 2013;55:130–140. doi: 10.1016/j.freeradbiomed.2012.10.554. [DOI] [PubMed] [Google Scholar]
- 43.Farache J, Koren I, Milo I, Gurevich I, Kim KW, Zigmond E, Furtado GC, Lira SA, Shakhar G. Luminal Bacteria Recruit CD103(+) Dendritic Cells into the Intestinal Epithelium to Sample Bacterial Antigens for Presentation. Immunity. 2013;38:581–595. doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fava F, Danese S. Intestinal microbiota in inflammatory bowel disease: friend of foe? World J Gastroenterol. 2011;17:557–566. doi: 10.3748/wjg.v17.i5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feller M, Huwiler K, Schoepfer A, Shang A, Furrer H, Egger M. Long-term antibiotic treatment for Crohn’s disease: systematic review and meta-analysis of placebo-controlled trials. Clin Infect Dis. 2010;50:473–480. doi: 10.1086/649923. [DOI] [PubMed] [Google Scholar]
- 46.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fleming A, Jankowski J, Goldsmith P. In vivo analysis of gut function and disease changes in a zebrafish larvae model of inflammatory bowel disease: a feasibility study. Inflamm Bowel Dis. 2010;16:1162–1172. doi: 10.1002/ibd.21200. [DOI] [PubMed] [Google Scholar]
- 48.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 49.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuchs A, Colonna M. Natural killer (NK) and NK-like cells at mucosal epithelia: Mediators of anti-microbial defense and maintenance of tissue integrity. Eur J Microbiol Immunol (Bp) 2011;1:257–266. doi: 10.1556/EuJMI.1.2011.4.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galindo-Villegas J, Garcia-Moreno D, de OS, Meseguer J, Mulero V. Regulation of immunity and disease resistance by commensal microbes and chromatin modifications during zebrafish development. Proc Natl Acad Sci U S A. 2012;109:E2605–E2614. doi: 10.1073/pnas.1209920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van TW, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, Gonzalez A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 57.Gomez D, Sunyer JO, Salinas I. The mucosal immune system of fish: the evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013;35:1729–1739. doi: 10.1016/j.fsi.2013.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grinspan AM, Kelly CR. Fecal Microbiota Transplantation for Ulcerative Colitis: Not Just Yet. Gastroenterology. 2015;149:15–18. doi: 10.1053/j.gastro.2015.05.030. [DOI] [PubMed] [Google Scholar]
- 59.Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- 60.Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar typhimurium. Infect Immun. 2000;68:6139–6146. doi: 10.1128/iai.68.11.6139-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamuro K, Suetake H, Saha NR, Kikuchi K, Suzuki Y. A teleost polymeric Ig receptor exhibiting two Ig-like domains transports tetrameric IgM into the skin. J Immunol. 2007;178:5682–5689. doi: 10.4049/jimmunol.178.9.5682. [DOI] [PubMed] [Google Scholar]
- 62.Haynes M, Rohwer F. The human virome. In: Nelson KE, editor. Metagenomics of the Human Body. Springer; New York: 2011. pp. 63–78. [Google Scholar]
- 63.He Q, Wang L, Wang F, Li Q. Role of gut microbiota in a zebrafish model with chemically induced enterocolitis involving toll-like receptor signaling pathways. Zebrafish. 2014;11:255–264. doi: 10.1089/zeb.2013.0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He Q, Wang L, Wang F, Wang C, Tang C, Li Q, Li J, Zhao Q. Microbial fingerprinting detects intestinal microbiota dysbiosis in Zebrafish models with chemically-induced enterocolitis. BMC Microbiol. 2013;13:289. doi: 10.1186/1471-2180-13-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, Wherry EJ, Koni PA, Bushman FD, Elson CO, Eberl G, Artis D, Sonnenberg GF. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hickey CA, Kuhn KA, Donermeyer DL, Porter NT, Jin C, Cameron EA, Jung H, Kaiko GE, Wegorzewska M, Malvin NP, Glowacki RW, Hansson GC, Allen PM, Martens EC, Stappenbeck TS. Colitogenic Bacteroides thetaiotaomicron Antigens Access Host Immune Cells in a Sulfatase-Dependent Manner via Outer Membrane Vesicles. Cell Host Microbe. 2015;17:672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holt A, Mitra S, van der Sar AM, Alnabulsi A, Secombes CJ, Bird S. Discovery of zebrafish (Danio rerio) interleukin-23 alpha (IL-23alpha) chain, a subunit important for the formation of IL-23, a cytokine involved in the development of Th17 cells and inflammation. Mol Immunol. 2011;48:981–991. doi: 10.1016/j.molimm.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 68.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 70.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 71.Hori S. Lineage stability and phenotypic plasticity of Foxp3(+) regulatory T cells. Immunol Rev. 2014;259:159–172. doi: 10.1111/imr.12175. [DOI] [PubMed] [Google Scholar]
- 72.Hue S, Mention JJ, Monteiro RC, Zhang S, Cellier C, Schmitz J, Verkarre V, Fodil N, Bahram S, Cerf-Bensussan N, Caillat-Zucman S. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity. 2004;21:367–377. doi: 10.1016/j.immuni.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 73.Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40:843–854. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inohara N, Nunez G. The NOD: a signaling module that regulates apoptosis and host defense against pathogens. Oncogene. 2001;20:6473–6481. doi: 10.1038/sj.onc.1204787. [DOI] [PubMed] [Google Scholar]
- 75.Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, Benjamin JL, Ruhn KA, Hou B, DeFranco AL, Yarovinsky F, Hooper LV. Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci U S A. 2011;108:8743–8748. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ivanov II, Littman DR. Modulation of immune homeostasis by commensal bacteria. Curr Opin Microbiol. 2011;14:106–114. doi: 10.1016/j.mib.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iwanami N. Zebrafish as a model for understanding the evolution of the vertebrate immune system and human primary immunodeficiency. Exp Hematol. 2014;42:697–706. doi: 10.1016/j.exphem.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 79.Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu Rev Immunol. 2009;27:313–338. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- 80.Jevtov I, Samuelsson T, Yao G, Amsterdam A, Ribbeck K. Zebrafish as a model to study live mucus physiology. Sci Rep. 2014;4:6653. doi: 10.1038/srep06653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, Fernandez L, Rodriguez JM. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159:187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 82.Johansen FE, Pekna M, Norderhaug IN, Haneberg B, Hietala MA, Krajci P, Betsholtz C, Brandtzaeg P. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J Exp Med. 1999;190:915–922. doi: 10.1084/jem.190.7.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johansson C, Kelsall BL. Phenotype and function of intestinal dendritic cells. Semin Immunol. 2005;17:284–294. doi: 10.1016/j.smim.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 84.Johansson ME, Sjovall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol. 2013 doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jones-Hall YL, Grisham MB. Immunopathological characterization of selected mouse models of inflammatory bowel disease: Comparison to human disease. Pathophysiology. 2014;21:267–288. doi: 10.1016/j.pathophys.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 86.Joossens M, Huys G, Cnockaert M, De P, Verbeke VK, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 87.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D’Amato M, De JD, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De VM, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kang SS, Bloom SM, Norian LA, Geske MJ, Flavell RA, Stappenbeck TS, Allen PM. An antibiotic-responsive mouse model of fulminant ulcerative colitis. PLoS Med. 2008;5:e41. doi: 10.1371/journal.pmed.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karlsson J, Putsep K, Chu H, Kays RJ, Bevins CL, Andersson M. Regional variations in Paneth cell antimicrobial peptide expression along the mouse intestinal tract. BMC Immunol. 2008;9:37. doi: 10.1186/1471-2172-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kawai F, Paek S, Choi KJ, Prouty M, Kanipes MI, Guerry P, Yeo HJ. Crystal structure of JlpA, a surface-exposed lipoprotein adhesin of Campylobacter jejuni. J Struct Biol. 2012;177:583–588. doi: 10.1016/j.jsb.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kjeldsen L, Cowland JB, Borregaard N. Human neutrophil gelatinase-associated lipocalin and homologous proteins in rat and mouse. Biochim Biophys Acta. 2000;1482:272–283. doi: 10.1016/s0167-4838(00)00152-7. [DOI] [PubMed] [Google Scholar]
- 94.Koboziev I, Karlsson F, Grisham MB. Gut-associated lymphoid tissue, T cell trafficking, and chronic intestinal inflammation. Ann N Y Acad Sci. 2010;1207(Suppl 1):E86–E93. doi: 10.1111/j.1749-6632.2010.05711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Koboziev I, Karlsson F, Zhang S, Grisham MB. Pharmacological intervention studies using mouse models of the inflammatory bowel diseases: translating preclinical data into new drug therapies. Inflamm Bowel Dis. 2011;17:1229–1245. doi: 10.1002/ibd.21557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koboziev I, Reinoso WC, Furr KL, Grisham MB. Role of the enteric microbiota in intestinal homeostasis and inflammation. Free Radic Biol Med. 2014;68:122–133. doi: 10.1016/j.freeradbiomed.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kortum AN, Rodriguez-Nunez I, Yang J, Shim J, Runft D, O’Driscoll ML, Haire RN, Cannon JP, Turner PM, Litman RT, Kim CH, Neely MN, Litman GW, Yoder JA. Differential expression and ligand binding indicate alternative functions for zebrafish polymeric immunoglobulin receptor (pIgR) and a family of pIgR-like (PIGRL) proteins. Immunogenetics. 2014;66:267–279. doi: 10.1007/s00251-014-0759-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 101.Laing KJ, Purcell MK, Winton JR, Hansen JD. A genomic view of the NOD-like receptor family in teleost fish: identification of a novel NLR subfamily in zebrafish. BMC Evol Biol. 2008;8:42. doi: 10.1186/1471-2148-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 103.Lieschke GJ, Trede NS. Fish immunology. Curr Biol. 2009;19:R678–R682. doi: 10.1016/j.cub.2009.06.068. [DOI] [PubMed] [Google Scholar]
- 104.Lipinski S, Till A, Sina C, Arlt A, Grasberger H, Schreiber S, Rosenstiel P. DUOX2-derived reactive oxygen species are effectors of NOD2-mediated antibacterial responses. J Cell Sci. 2009;122:3522–3530. doi: 10.1242/jcs.050690. [DOI] [PubMed] [Google Scholar]
- 105.Looijer-van Langen MA, Dieleman LA. Prebiotics in chronic intestinal inflammation. Inflamm Bowel Dis. 2009;15:454–462. doi: 10.1002/ibd.20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, Chasson L, Hardwigsen J, Anguiano E, Banchereau J, Chaussabel D, Dalod M, Littman DR, Vivier E, Tomasello E. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 108.Luckey TD. Introduction to intestinal microecology. Am J Clin Nutr. 1972;25:1292–1294. doi: 10.1093/ajcn/25.12.1292. [DOI] [PubMed] [Google Scholar]
- 109.Lugo-Villarino G, Balla KM, Stachura DL, Banuelos K, Werneck MB, Traver D. Identification of dendritic antigen-presenting cells in the zebrafish. Proc Natl Acad Sci U S A. 2010;107:15850–15855. doi: 10.1073/pnas.1000494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luo Q, Kumar P, Vickers TJ, Sheikh A, Lewis WG, Rasko DA, Sistrunk J, Fleckenstein JM. Enterotoxigenic Escherichia coli secretes a highly conserved mucin-degrading metalloprotease to effectively engage intestinal epithelial cells. Infect Immun. 2014;82:509–521. doi: 10.1128/IAI.01106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 112.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 113.Macpherson AJ, Slack E. The functional interactions of commensal bacteria with intestinal secretory IgA. Curr Opin Gastroenterol. 2007;23:673–678. doi: 10.1097/MOG.0b013e3282f0d012. [DOI] [PubMed] [Google Scholar]
- 114.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 115.Mahdavi J, Sonden B, Hurtig M, Olfat FO, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson KA, Altraja S, Wadstrom T, Kersulyte D, Berg DE, DuBois A, Petersson C, Magnusson KE, Norberg T, Lindh F, Lundskog BB, Arnqvist A, Hammarstrom L, Boren T. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 117.Mandar R, Mikelsaar M. Transmission of mother’s microflora to the newborn at birth. Biol Neonate. 1996;69:30–35. doi: 10.1159/000244275. [DOI] [PubMed] [Google Scholar]
- 118.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 119.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von BM, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 121.Marsland BJ, Gollwitzer ES. Host-microorganism interactions in lung diseases. Nat Rev Immunol. 2014;14:827–835. doi: 10.1038/nri3769. [DOI] [PubMed] [Google Scholar]
- 122.Martinez-Medina M, Aldeguer X, Lopez-Siles M, Gonzalez-Huix F, Lopez-Oliu C, Dahbi G, Blanco JE, Blanco J, Garcia-Gil LJ, Darfeuille-Michaud A. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflamm Bowel Dis. 2009;15:872–882. doi: 10.1002/ibd.20860. [DOI] [PubMed] [Google Scholar]
- 123.Mathis D, Benoist C. The influence of the microbiota on type-1 diabetes: on the threshold of a leap forward in our understanding. Immunol Rev. 2012;245:239–249. doi: 10.1111/j.1600-065X.2011.01084.x. [DOI] [PubMed] [Google Scholar]
- 124.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 126.McDermott JR, Humphreys NE, Forman SP, Donaldson DD, Grencis RK. Intraepithelial NK cell-derived IL-13 induces intestinal pathology associated with nematode infection. J Immunol. 2005;175:3207–3213. doi: 10.4049/jimmunol.175.5.3207. [DOI] [PubMed] [Google Scholar]
- 127.Meehan TF, Witherden DA, Kim CH, Sendaydiego K, Ye I, Garijo O, Komori HK, Kumanogoh A, Kikutani H, Eckmann L, Havran WL. Protection against colitis by CD100-dependent modulation of intraepithelial gammadelta T lymphocyte function. Mucosal Immunol. 2014;7:134–142. doi: 10.1038/mi.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Meloni S, Zarletti G, Benedetti S, Randelli E, Buonocore F, Scapigliati G. Cellular activities during a mixed leucocyte reaction in the teleost sea bass Dicentrarchus labrax. Fish Shellfish Immunol. 2006;20:739–749. doi: 10.1016/j.fsi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 129.Menke AL, Spitsbergen JM, Wolterbeek AP, Woutersen RA. Normal anatomy and histology of the adult zebrafish. Toxicol Pathol. 2011;39:759–775. doi: 10.1177/0192623311409597. [DOI] [PubMed] [Google Scholar]
- 130.Meyer-Hoffert U, Hornef M, Henriques-Normark B, Normark S, Andersson M, Putsep K. Identification of heparin/heparan sulfate interacting protein as a major broad-spectrum antimicrobial protein in lung and small intestine. FASEB J. 2008;22:2427–2434. doi: 10.1096/fj.07-103440. [DOI] [PubMed] [Google Scholar]
- 131.Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Putsep K, Andersson M. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut. 2008;57:764–771. doi: 10.1136/gut.2007.141481. [DOI] [PubMed] [Google Scholar]
- 132.Mitra S, Alnabulsi A, Secombes CJ, Bird S. Identification and characterization of the transcription factors involved in T-cell development, t-bet, stat6 and foxp3, within the zebrafish, Danio rerio. FEBS J. 2010;277:128–147. doi: 10.1111/j.1742-4658.2009.07460.x. [DOI] [PubMed] [Google Scholar]
- 133.Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Armstrong D, Marshall JK, Kassam Z, Reinisch W, Lee CH. Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology. 2015;149:102–109. doi: 10.1053/j.gastro.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 134.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, Kaplan GG. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 135.Mombaerts P, Mizoguchi E, Grusby MJ, Glimcher LH, Bhan AK, Tonegawa S. Spontaneous development of inflammatory bowel disease in T cell receptor mutant mice. Cell. 1993;75:274–282. doi: 10.1016/0092-8674(93)80069-q. [DOI] [PubMed] [Google Scholar]
- 136.Morowitz MJ, Carlisle EM, Alverdy JC. Contributions of intestinal bacteria to nutrition and metabolism in the critically ill. Surg Clin North Am. 2011;91:771–85. viii. doi: 10.1016/j.suc.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mostowy S, Boucontet L, Mazon Moya MJ, Sirianni A, Boudinot P, Hollinshead M, Cossart P, Herbomel P, Levraud JP, Colucci-Guyon E. The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog. 2013;9:e1003588. doi: 10.1371/journal.ppat.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol. 2003;3:331–341. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]
- 139.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 140.Navarro-Garcia F, Gutierrez-Jimenez J, Garcia-Tovar C, Castro LA, Salazar-Gonzalez H, Cordova V. Pic, an autotransporter protein secreted by different pathogens in the Enterobacteriaceae family, is a potent mucus secretagogue. Infect Immun. 2010;78:4101–4109. doi: 10.1128/IAI.00523-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nguyen TL, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, Littman DR, Reinecker HC. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 143.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, Stappenbeck TS, McGovern DP, Keshavarzian A, Mutlu EA, Sauk J, Gevers D, Xavier RJ, Wang D, Parkes M, Virgin HW. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Novinec M, Lenarcic B. Cathepsin K: a unique collagenolytic cysteine peptidase. Biol Chem. 2013;394:1163–1179. doi: 10.1515/hsz-2013-0134. [DOI] [PubMed] [Google Scholar]
- 145.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Oehlers SH, Flores MV, Hall CJ, Crosier KE, Crosier PS. Retinoic acid suppresses intestinal mucus production and exacerbates experimental enterocolitis. Dis Model Mech. 2012;5:457–467. doi: 10.1242/dmm.009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Oehlers SH, Flores MV, Hall CJ, Swift S, Crosier KE, Crosier PS. The inflammatory bowel disease (IBD) susceptibility genes NOD1 and NOD2 have conserved anti-bacterial roles in zebrafish. Dis Model Mech. 2011;4:832–841. doi: 10.1242/dmm.006122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38:414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 149.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pelaseyed T, Bergstrom JH, Gustafsson JK, Ermund A, Birchenough GM, Schutte A, van der Post S, Svensson F, Rodriguez-Pineiro AM, Nystrom EE, Wising C, Johansson ME, Hansson GC. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260:8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 153.Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, Kobayashi KS. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A. 2009;106:15813–15818. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int Immunol. 1993;5:1461–1471. doi: 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 155.Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology. 2009;136:2015–2031. doi: 10.1053/j.gastro.2009.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Putsep K, Axelsson LG, Boman A, Midtvedt T, Normark S, Boman HG, Andersson M. Germ-free and colonized mice generate the same products from enteric prodefensins. J Biol Chem. 2000;275:40478–40482. doi: 10.1074/jbc.M007816200. [DOI] [PubMed] [Google Scholar]
- 157.Quintana FJ, Iglesias AH, Farez MF, Caccamo M, Burns EJ, Kassam N, Oukka M, Weiner HL. Adaptive autoimmunity and Foxp3-based immunoregulation in zebrafish. PLoS One. 2010;5:e9478. doi: 10.1371/journal.pone.0009478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Raffatellu M, Baumler AJ. Salmonella’s iron armor for battling the host and its microbiota. Gut Microbes. 2010;1:70–72. doi: 10.4161/gmic.1.1.10951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 160.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rehman A, Lepage P, Nolte A, Hellmig S, Schreiber S, Ott SJ. Transcriptional activity of the dominant gut mucosal microbiota in chronic inflammatory bowel disease patients. J Med Microbiol. 2010;59:1114–1122. doi: 10.1099/jmm.0.021170-0. [DOI] [PubMed] [Google Scholar]
- 163.Reinoso, Webb C, Grisham MB. Pharmacological Intervention Studies Using Mouse Models of chronic intestinal inflammation. 2015 In press. [Google Scholar]
- 164.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 165.Reynders A, Yessaad N, Vu Manh TP, Dalod M, Fenis A, Aubry C, Nikitas G, Escaliere B, Renauld JC, Dussurget O, Cossart P, Lecuit M, Vivier E, Tomasello E. Identity, regulation and in vivo function of gut NKp46+RORgammat+ and NKp46+RORgammat- lymphoid cells. EMBO J. 2011;30:2934–2947. doi: 10.1038/emboj.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011;5:1595–1608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rombout JH, Abelli L, Picchietti S, Scapigliati G, Kiron V. Teleost intestinal immunology. Fish Shellfish Immunol. 2011;31:616–626. doi: 10.1016/j.fsi.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 168.Rongvaux A, Takizawa H, Strowig T, Willinger T, Eynon EE, Flavell RA, Manz MG. Human hemato-lymphoid system mice: current use and future potential for medicine. Annu Rev Immunol. 2013;31:635–674. doi: 10.1146/annurev-immunol-032712-095921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rosner JL. Ten times more microbial cells than body cells in humans? Microbe. 2014;9:47. [Google Scholar]
- 170.Rossen NG, Fuentes S, van der Spek MJ, Tijssen JG, Hartman JH, Duflou A, Lowenberg M, van den Brink GR, Mathus-Vliegen EM, de Vos WM, Zoetendal EG, D’Haens GR, Ponsioen CY. Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology. 2015;149:110–118. doi: 10.1053/j.gastro.2015.03.045. [DOI] [PubMed] [Google Scholar]
- 171.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rutgeerts P, Goboes K, Peeters M, Hiele M, Penninckx F, Aerts R, Kerremans R, Vantrappen G. Effect of faecal stream diversion on recurrence of Crohn’s disease in the neoterminal ileum. Lancet. 1991;338:771–774. doi: 10.1016/0140-6736(91)90663-a. [DOI] [PubMed] [Google Scholar]
- 173.Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–261. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 174.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, Diefenbach A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sansonetti PJ. To be or not to be a pathogen: that is the mucosally relevant question. Mucosal Immunol. 2011;4:8–14. doi: 10.1038/mi.2010.77. [DOI] [PubMed] [Google Scholar]
- 176.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 177.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, Di Santo JP. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 178.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 179.Schenk M, Mueller C. Adaptations of intestinal macrophages to an antigen-rich environment. Semin Immunol. 2007;19:84–93. doi: 10.1016/j.smim.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 180.Seedorf H, Griffin NW, Ridaura VK, Reyes A, Cheng J, Rey FE, Smith MI, Simon GM, Scheffrahn RH, Woebken D, Spormann AM, Van TW, Ursell LK, Pirrung M, Robbins-Pianka A, Cantarel BL, Lombard V, Henrissat B, Knight R, Gordon JI. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159:253–266. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Seno H, Miyoshi H, Brown SL, Geske MJ, Colonna M, Stappenbeck TS. Efficient colonic mucosal wound repair requires Trem2 signaling. Proc Natl Acad Sci U S A. 2009;106:256–261. doi: 10.1073/pnas.0803343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shale M, Schiering C, Powrie F. CD4(+) T-cell subsets in intestinal inflammation. Immunol Rev. 2013;252:164–182. doi: 10.1111/imr.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Shanahan MT, Tanabe H, Ouellette AJ. Strain-specific polymorphisms in Paneth cell alpha-defensins of C57BL/6 mice and evidence of vestigial myeloid alpha-defensin pseudogenes. Infect Immun. 2011;79:459–473. doi: 10.1128/IAI.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sina C, Lipinski S, Gavrilova O, Aden K, Rehman A, Till A, Rittger A, Podschun R, Meyer-Hoffert U, Haesler R, Midtling E, Putsep K, McGuckin MA, Schreiber S, Saftig P, Rosenstiel P. Extracellular cathepsin K exerts antimicrobial activity and is protective against chronic intestinal inflammation in mice. Gut. 2013;62:520–530. doi: 10.1136/gutjnl-2011-300076. [DOI] [PubMed] [Google Scholar]
- 185.Slack E, Balmer ML, Macpherson AJ. B cells as a critical node in the microbiota-host immune system network. Immunol Rev. 2014;260:50–66. doi: 10.1111/imr.12179. [DOI] [PubMed] [Google Scholar]
- 186.Smith PD, Smythies LE, Shen R, Greenwell-Wild T, Gliozzi M, Wahl SM. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 2011;4:31–42. doi: 10.1038/mi.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Smriga S, Sandin SA, Azam F. Abundance, diversity, and activity of microbial assemblages associated with coral reef fish guts and feces. FEMS Microbiol Ecol. 2010;73:31–42. doi: 10.1111/j.1574-6941.2010.00879.x. [DOI] [PubMed] [Google Scholar]
- 188.Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, Orenstein JM, Smith PD. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Smythies LE, Shen R, Bimczok D, Novak L, Clements RH, Eckhoff DE, Bouchard P, George MD, Hu WK, Dandekar S, Smith PD. Inflammation anergy in human intestinal macrophages is due to Smad-induced IkappaBalpha expression and NF-kappaB inactivation. J Biol Chem. 2010;285:19593–19604. doi: 10.1074/jbc.M109.069955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Solis G, de los Reyes-Gavilan CG, Fernandez N, Margolles A, Gueimonde M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe. 2010;16:307–310. doi: 10.1016/j.anaerobe.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 191.Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, Tardif MR, Sathaliyawala T, Kubota M, Farber DL, Collman RG, Shaked A, Fouser LA, Weiner DB, Tessier PA, Friedman JR, Kiyono H, Bushman FD, Chang KM, Artis D. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 193.Stecher B, Hapfelmeier S, Muller C, Kremer M, Stallmach T, Hardt WD. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun. 2004;72:4138–4150. doi: 10.1128/IAI.72.7.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Dore J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sullam KE, Essinger SD, Lozupone CA, O’Connor MP, Rosen GL, Knight R, Kilham SS, Russell JA. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol Ecol. 2012;21:3363–3378. doi: 10.1111/j.1365-294X.2012.05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Takizawa F, Dijkstra JM, Kotterba P, Korytar T, Kock H, Kollner B, Jaureguiberry B, Nakanishi T, Fischer U. The expression of CD8alpha discriminates distinct T cell subsets in teleost fish. Dev Comp Immunol. 2011;35:752–763. doi: 10.1016/j.dci.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 197.Thorne KJ, Oliver RC, Barrett AJ. Lysis and killing of bacteria by lysosomal proteinases. Infect Immun. 1976;14:555–563. doi: 10.1128/iai.14.2.555-563.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Toh MC, Goodyear M, Daigneault M, Allen-Vercoe E, Van Raay TJ. Colonizing the embryonic zebrafish gut with anaerobic bacteria derived from the human gastrointestinal tract. Zebrafish. 2013;10:194–198. doi: 10.1089/zeb.2012.0814. [DOI] [PubMed] [Google Scholar]
- 199.Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Valatas V, Vakas M, Kolios G. The value of experimental models of colitis in predicting efficacy of biological therapies for inflammatory bowel diseases. Am J Physiol Gastrointest Liver Physiol. 2013;305:G763–G785. doi: 10.1152/ajpgi.00004.2013. [DOI] [PubMed] [Google Scholar]
- 201.van der Sluis M, Bouma J, Vincent A, Velcich A, Carraway KL, Buller HA, Einerhand AW, van Goudoever JB, Van SI, Renes IB. Combined defects in epithelial and immunoregulatory factors exacerbate the pathogenesis of inflammation: mucin 2-interleukin 10-deficient mice. Lab Invest. 2008;88:634–642. doi: 10.1038/labinvest.2008.28. [DOI] [PubMed] [Google Scholar]
- 202.van der Sluis M, de Koning BA, De Bruijn AC, Velcich A, Meijerink JP, van Goudoever JB, Buller HA, Dekker J, Van S, Renes IIB, Einerhand AW. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 203.van V d, Spaink HP, Meijer AH. Pathogen recognition and activation of the innate immune response in zebrafish. Adv Hematol. 2012;2012:159807. doi: 10.1155/2012/159807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.van V d, van Soest JJ, Spaink HP, Meijer AH. Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system. Dis Model Mech. 2013;6:841–854. doi: 10.1242/dmm.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.van NE, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 206.Ventham NT, Kennedy NA, Nimmo ER, Satsangi J. Beyond Gene Discovery in Inflammatory Bowel Disease: The Emerging Role of Epigenetics. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Walker AW, Lawley TD. Therapeutic modulation of intestinal dysbiosis. Pharmacol Res. 2013;69:75–86. doi: 10.1016/j.phrs.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 208.Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Intestinal growth and differentiation in zebrafish. Mech Dev. 2005;122:157–173. doi: 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 209.Wang Z, Du J, Lam SH, Mathavan S, Matsudaira P, Gong Z. Morphological and molecular evidence for functional organization along the rostrocaudal axis of the adult zebrafish intestine. BMC Genomics. 2010;11:392. doi: 10.1186/1471-2164-11-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Weigmann B, Schughart N, Wiebe C, Sudowe S, Lehr HA, Jonuleit H, Vogel L, Becker C, Neurath MF, Grabbe S, Saloga J, Bellinghausen I. Allergen-induced IgE-dependent gut inflammation in a human PBMC-engrafted murine model of allergy. The Journal of allergy and clinical immunology. 2012;129:1126–1135. doi: 10.1016/j.jaci.2011.11.036. [DOI] [PubMed] [Google Scholar]
- 211.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Winter SE, Baumler AJ. Dysbiosis in the inflamed intestine: chance favors the prepared microbe. Gut Microbes. 2014;5:71–73. doi: 10.4161/gmic.27129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Winter SE, Lopez CA, Baumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wlodarska M, Kostic AD, Xavier RJ. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe. 2015;17:577–591. doi: 10.1016/j.chom.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wu H, Santoni-Rugiu E, Ralfkiaer E, Porse BT, Moser C, Hoiby N, Borregaard N, Cowland JB. Lipocalin 2 is protective against E. coli pneumonia. Respir Res. 2010;11:96. doi: 10.1186/1465-9921-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Yang Y, Tomkovich S, Jobin C. Could a swimming creature inform us on intestinal diseases? Lessons from zebrafish. Inflamm Bowel Dis. 2014;20:956–966. doi: 10.1097/01.MIB.0000442923.85569.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yoder JA. Form, function and phylogenetics of NITRs in bony fish. Dev Comp Immunol. 2009;33:135–144. doi: 10.1016/j.dci.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 219.Zac SW, Burns AR, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJ. The composition of the zebrafish intestinal microbial community varies across development. ISME J. 2015 doi: 10.1038/ismej.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947–957. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, LaPatra SE, Bartholomew J, Sunyer JO. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]