Abstract
Chromosomal inversions are thought to play a major role in climatic adaptation. In D. melanogaster, the cosmopolitan inversion In(3R)Payne exhibits latitudinal clines on multiple continents. Since many fitness traits show similar clines, it is tempting to hypothesize that In(3R)P underlies observed clinal patterns for some of these traits. In support of this idea, previous work in Australian populations has demonstrated that In(3R)P affects body size but not development time or cold resistance. However, similar data from other clines of this inversion are largely lacking; finding parallel effects of In(3R)P across multiple clines would considerably strengthen the case for clinal selection. Here, we have analyzed the phenotypic effects of In(3R)P in populations originating from the endpoints of the latitudinal cline along the North American east coast. We measured development time, egg-to-adult survival, several size-related traits (femur and tibia length, wing area and shape), chill coma recovery, oxidative stress resistance and triglyceride content in homokaryon lines carrying In(3R)P or the standard arrangement. Our central finding is that the effects of In(3R)P along the North American cline match those observed in Australia: standard arrangement lines were larger than inverted lines, but the inversion did not influence development time or cold resistance. Similarly, In(3R)P did not affect egg-to-adult survival, oxidative stress resistance and lipid content. In(3R)P thus seems to specifically affect size traits in populations from both continents. This parallelism strongly suggests an adaptive pattern, whereby the inversion has captured alleles associated with growth regulation and clinal selection acts on size across both continents.
Keywords: Inversions, clines, spatially varying selection, adaptation, body size, life history, D. melanogaster.
Introduction
One of the central goals of evolutionary biology is to understand how organisms adapt to environmental heterogeneity (Hoffmann & Sgrò, 2011; Savolainen et al., 2013). A promising approach towards this end is to investigate systematic, gradual phenotypic and genotypic changes along environmental (e.g., climatic) gradients, so-called “clines”, that are thought to be driven by spatially varying selection (Mayr, 1963; Endler, 1977; de Jong & Bochdanovits, 2003; Charlesworth & Charlesworth, 2010).
A classical model system for studying clinality is Drosophila melanogaster (de Jong & Bochdanovits, 2003; Hoffmann & Weeks, 2007; Adrion et al., 2015), an ancestrally tropical vinegar (fruit) fly that has migrated out of sub-Saharan Africa about 10,000 to 15,000 years ago and subsequently colonized the rest of the world as a human commensal (David & Capy, 1988; Keller, 2007). As a result of its colonization history, this species had to adapt to a wide range of climatic and ecological conditions, including temperate and seasonal habitats. This is evidenced by patterns of clinal differentiation of numerous life history, morphological, and physiological traits across latitude: clinally varying traits include development time (James & Partridge, 1995), body size (Coyne & Beecham, 1987; Imasheva et al., 1994; James et al., 1995, 1997; Zwaan et al., 2000; Gockel et al., 2001; Gibert et al., 2004; Klepsatel et al., 2014; Fabian et al., 2015), wing loading (Stalker, 1980; Azevedo et al., 1998), pigmentation (Telonis-Scott et al., 2011), ovariole number (Capy et al., 1993; Gibert et al., 2004; Klepsatel et al., 2014), diapause propensity (Schmidt et al., 2005; Schmidt & Paaby, 2008), cold and heat resistance (Hoffmann & Shirriffs, 2002), and desiccation resistance (Hoffmann & Parsons, 2009).
Consistent with spatially varying selection, many of these traits exhibit parallel clinal patterns across latitude on multiple continents, even though demography (e.g., admixture) can also contribute to patterns of clinality (Kao et al., 2015; Bergland et al., 2015; Flatt, 2016). For example, qualitatively identical latitudinal clines have been reported across several continents for body size (Coyne & Beecham, 1987; James et al., 1995; van’t Land et al., 1999; Klepsatel et al., 2014; Fabian et al., 2015), pigmentation (David et al., 1985; Munjal et al., 1997; Telonis-Scott et al., 2011), and chill coma recovery time (Gibert et al., 2001; Hoffmann et al., 2002; Ayrinhac et al., 2004).
Despite much work on phenotypic clines in Drosophila, and although several single genetic markers are known to covary latitudinally with trait clines (de Jong & Bochdanovits, 2003; Hoffmann & Weeks, 2007; Adrion et al., 2015; and references therein), little is known about the genetics underlying clinal trait variation (for some exceptions see Schmidt et al., 2008; Paaby et al., 2014) and the mechanisms by which clines are formed and maintained. Recent progress comes from genome-wide studies of the Australian and North American clines that have identified hundreds of clinally varying single nucleotide polymorphisms (SNPs) (Kolaczkowski et al., 2011; Fabian et al., 2012; Bergland et al., 2014; Reinhardt et al., 2014; Bergland et al., 2015; Kapun et al., 2016). While some proportion of these clinal variants is expected to causally contribute to clinal trait variation, other variants might be subject to hitchhiking (genetic draft) or admixture (Fabian et al., 2012; Bergland et al., 2015; Kapun et al., 2016). Thus, identifying the true genic targets of clinal selection remains a considerable challenge (Adrion et al., 2015; Flatt, 2016).
Information on potentially functionally relevant genomic sites or regions might be gleaned from the genome-wide distribution of clinal SNPs. Remarkably, even though clinally varying SNPs occur throughout the genome, the majority of clinal variants is located on the right arm of the third chromosome (3R), especially within the region spanned by a large (~8 Mb), cosmopolitan chromosomal inversion, In(3R)Payne (also called In(3R)P) (Kolaczkowski et al., 2011; Fabian et al., 2012; Kapun et al., 2016).
The In(3R)P inversion is of particular interest for four reasons. First, in several geographic areas (e.g., North American east coast, Australian east coast, India, Japan) this inversion exhibits steep, parallel latitudinal clines: the inverted karyotype reaches intermediate frequencies at low latitudes but is rare or absent at high latitudes (Mettler et al., 1977; Inoue & Watanabe, 1979; Stalker, 1980; Knibb et al., 1981; Knibb, 1982; Das & Singh, 1991; Matzkin et al., 2005; Fabian et al., 2012; Kapun et al., 2014; Rane et al., 2015; Kapun et al., 2016). For example, along the North American cline this arrangement reaches a frequency of ~50% in southern Florida but is absent in Maine (Mettler et al., 1977; Knibb, 1982; Fabian et al., 2012; Kapun et al., 2014, 2016); thus, flies from high-latitude populations are fixed or nearly fixed for the standard arrangement. Second, in Australia and North America, the latitudinal slopes of the In(3R)P clines have remained stable across >40 years of observation, consistent with the clines being maintained by spatially varying selection (Anderson et al., 2005; Umina et al., 2005; Kapun et al., 2014, 2016); in Australia, the intercept of the clinal slope has recently shifted – possibly as a consequence of climate change (Anderson et al., 2005; Umina et al., 2005). Third, recent evidence suggests that the North American cline of In(3R)P is maintained non-neutrally and independent of population structure or admixture (Kapun et al., 2016). Fourth, several inversions in Drosophila have previously been found to be associated with development time, egg-to-adult survival, size-related traits, fecundity and fertility, stress resistance (to cold, heat, starvation), and lifespan (Sperlich & Pfriem, 1986; Hoffmann et al., 2004; Hoffmann & Weeks, 2007; Hoffmann & Rieseberg, 2008; and references therein). Thus, although many alleles within In(3R)P might be in linkage disequilibrium (LD) and thus subject to hitchhiking, the observation that the majority of clinal SNPs resides in the genomic region spanned by this inversion suggests that clinal trait variation might at least partly be driven by In(3R)P (de Jong & Bochdanovits, 2003; Fabian et al., 2012; Kapun et al., 2016).
Indeed, several association mapping studies have linked In(3R)P to clinal size variation among Australian populations (Weeks et al., 2002; Rako et al., 2006; Kennington et al., 2007). Similarly, using quantitative trait locus (QTL) mapping, Calboli et al. (2003) found that the largest QTL peak for body size for the endpoints of the Australian and South American clines overlaps the region of In(3R)P. However, little is known about associations between In(3R)P and clinal phenotypes (including size) for other continents; finding parallel phenotypic effects of In(3R)P across multiple clines would considerably strengthen the case for spatially varying (clinal) selection. Moreover, effects of this inversion polymorphism on clinal fitness-related traits other than size remain largely unknown (cf. Rako et al., 2006).
Here, we investigate – for the first time – the phenotypic effects of In(3R)P in populations that approximate the endpoints of the North American east coastal cline (southern Florida versus Maine). We measured several fitness-related traits thought to be clinal (development time, egg-to-adult survival, proxies of body size [femur length, tibia length, wing area, wing shape], chill coma recovery time, oxidative stress resistance, triglyceride content [a correlate of starvation resistance]) in isochromosomal homokaryon lines carrying In(3R)P or the standard chromosomal arrangement.
Our results for the effects of In(3R)P on several measures of body size mirror those previously observed in populations from the Australian cline (Weeks et al., 2002; Rako et al., 2006; Kennington et al., 2007) – this strongly suggests the existence of parallel adaptive effects of In(3R)P on clinal size variation across both continents that are driven by spatially varying selection.
Materials and Methods
Fly stocks and maintenance
We used isofemale lines collected from populations that approximate the endpoints of the clinal gradient running along the North American east coast: a set of lines from subtropical southern Florida (Homestead and Jacksonville) and one from a temperate population in Maine (Bowdoin) (see Table 1; also see Schmidt et al., 2005; Schmidt & Paaby, 2008; Fabian et al., 2015 for further details on these populations). Since we failed to detect phenotypic differences between the two Florida populations (not shown), we combined lines from both populations for statistical analysis. Isofemale lines were kept for long-term maintenance under constant conditions at 18°C and 60% relative air humidity, at a photoperiod of 12h:12h light:dark.
Table 1.
Summary of samples used in this study and estimates of inversion frequencies. N = number of isofemale lines screened to isolate 3R homokaryons.
| Location | State | N | Latitude | Longitude | Date | Collector | Inversion frequencies | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| In(2L)t | In(2R)NS | In(3L)P | In(3R)K | In(3R)Mo | In(3R)P | |||||||
| Homestead | Florida | 51 | 25.5°N | −71.06 °E | 5/2011 | P. Schmidt | 0.38 | 0.06 | 0.42 | 0.09 | 0.00 | 0.63 |
| Jacksonville | Florida | 32 | 30.3°N | −81.6°E | 8/2011 | R. Cogni | 0.63 | 0.05 | 0.20 | 0.42 | 0.09 | 0.31 |
| Bowdoin | Maine | 35 | 42.3°N | −80.5°E | 10/2012 | P. Schmidt | 0.43 | 0.03 | 0.00 | 0.03 | 0.11 | 0.00 |
All isofemale lines were screened for the presence of six cosmopolitan inversions (In(2L)t, In(2R)NS, In(3L)P, In(3R)K, In(3R)Mo, In(3R)P; see Lemeunier & Aulard, 1992) by extracting DNA from pools of 5–10 individuals from each line with a salt-chloroform extraction protocol and using PCR markers described in Matzkin et al. (2005) and Corbett-Detig et al. (2012). Consistent with previous data (Mettler et al., 1977; Knibb, 1982; Kapun et al., 2016), In(3L)P and In(3R)P segregated at intermediate frequencies in the subtropical samples from Florida but were absent in Maine. In(3R)Mo, in contrast, showed the opposite trend: it segregated at 11% frequency in Maine but was absent in Florida. None of the other inversions showed clinality (Table 1; also see below).
Generation of isochromosomal lines
To isolate wild-type chromosomes either carrying the inverted In(3R)P arrangement or the standard arrangement from isofemale lines (see above), we used a compound (second and third chromosome) balancer (SMB6; TM6B; Bloomington Drosophila Stock Center [BDSC], stock #5687) in an ebony (e1) mutant background (Fig. S1). For a given isofemale line, we crossed a wild-type male from that line to a female carrying the balancer. F1 pupae heterozygous for the balancer were selected visually based on the dominant tubby (Tb1) mutant phenotype. Upon eclosion, F1 adults were backcrossed to the balancer line to amplify the isolated wild-type chromosome. After four days of egg laying, F2 adults were screened for the presence or absence of In(3R)P using PCR markers described in Matzkin et al. (2005). Isochromosomal homokaryon lines were generated by selecting against balancer phenotypes in F3 crosses.
We isolated 41 3R chromosomes carrying In(3R)P (“Florida inverted”, FI) and 30 carrying the standard arrangement (“Florida standard”, FS) from the two Florida populations, and 20 chromosomes carrying the standard arrangement from Maine (“Maine standard”, MS). In total, we were able to generate 14 FI (34.1% of all FI isolates), 13 FS (43.3% of FS isolates) and 6 MS (30% of MS isolates) isochromosomal homokaryon lines for phenotyping (see below). For the remaining isolates we failed to obtain homokaryons, possibly due to recessive deleterious or lethal variants in the wild-type chromosomes; we maintained these lines as heterozygotes over a balancer chromosome but excluded them from the phenotypic assays reported here. We verified 3R karyotype by using PCR on 3–5 single individuals per isolated chromosome, as described above.
During the isolation process we did not control for inversions on chromosomal arms other than 3R: apart from In(2L)t, which segregated in ~30% of isolated lines, other inversions were either absent or present at only very low frequencies. Given that In(2L)t segregated at approximately equal proportions among the three sets of isochromosomal lines, we did not control for its effects in our analyses.
Phenotypic assays
General methods
Isochromosomal lines were used to measure several pre-adult life history traits (development time, egg-to-adult survival), stress-related and physiological traits (chill coma recovery time, oxidative stress resistance, triglyceride content), and proxies of body size (femur length, tibia length, wing area, wing shape) (see below). Isochromosomal lines were assigned randomized identifiers; assays were performed blind with respect to identifiers to eliminate potential bias. Vials or bottles were maintained and experiments performed at 25°C and 60% relative humidity, under a photoperiod of 12h:12h light:dark.
To avoid non-genetic parental and environmental effects assays were performed on flies from the F2 generation. Prior to the assays, we let 100 flies from each line oviposit for 2 days on standard (cornmeal-agar-yeast) medium. Eclosing F1 individuals were distributed into three replicate bottles (~200 flies per bottle) and aged for 5 days; flies were then transferred to new bottles and allowed to lay eggs for 3 hours. For each line, we collected 200 eggs and placed them into bottles containing 25 mL of standard medium. The positions of experimental bottles were randomized once per day to avoid potential effects caused by environmental heterogeneity inside the incubator. Eclosing F2 adults were collected every 6 hours during the day and every 12 hours overnight and aged for 3 days before being used for phenotypic assays (see below).
Pre-adult life history (development time and egg-to-adult survival)
To assess egg-to-adult development time and egg-to-adult survival (proportion viability) we recorded eclosion times for each individual and estimated developmental time in hours relative to the time point of egg laying.
Chill coma recovery
Adults were aged for two days after eclosion prior to the chill coma recovery assay. 24 hours before the start of the assay, we anesthetized flies with CO2 and created new subsets of up to 20 flies per sex and line in new vials with standard medium. To induce chill coma, flies were transferred to empty vials without anesthesia and vials placed on ice at 0°C for 3 hours. Flies were subsequently transferred to petri dishes at room temperature and visually monitored until they woke up. For each individual, the time elapsed between removal from ice and waking was recorded; a fly was deemed “awake” as soon as it was able to stand on all its legs. Flies from this assay were stored for triglyceride measurements at −20°C (see below).
Oxidative stress resistance
Adults were aged for two days after eclosion and split in two replicate subsets of 10 flies per sex and line 24h before the start of the assay. To induce oxidative stress, flies were transferred to media-free vials containing filter paper saturated with 5 mL of 30 mM methyl viologen (paraquat) (Sigma-Aldrich) in 5% sucrose solution (Paaby & Schmidt, 2008). To prevent evaporation, each vial was sealed with parafilm. We monitored mortality every two hours until ~ 90% of all flies had died. We continued monitoring flies in 8 hr intervals until all flies were dead. Corpses were preserved for morphometric measurements in ethanol (see below).
Triglyceride content
Since starvation resistance is often correlated with lipid content (Hoffmann and Harshman, 1999; Schmidt et al., 2005; Goenaga et al., 2013), we measured whole-body triglyceride (triacylglyceride [TAG]) content as a proxy. For each sample, we generated homogenates using 2 pooled flies and estimated serum TAG levels in micrograms per fly from blanks and standards run with each plate, using an enzymatic assay kit (Serum Triglyceride Determination Kit; Sigma-Aldrich) (also see McGowan et al., 1983; Tennessen et al., 2014).
Size-related traits and morphometric analysis
For morphometric measurements we removed the first right leg and right wing of each fly. Both body parts were mounted on slides with CC/Mount™ tissue mounting medium (Sigma-Aldrich) and sealed with cover slips. Images of legs and wings were taken with a digital camera (Leica DFC 290, Leica Microsystems GmbH, Wetzlar, Germany) attached to a stereo dissecting microscope (Leica MZ125). Femur and tibia length were measured as the distance between two sets of landmarks with ImageJ (v.1.47d), following the approach described in Debat et al. (2011).
To minimize measurement error, we repeated all measurements three times and used the average lengths for statistical analysis. For wing measurements, we used ImageJ (v.1.47d) to define two orientation landmarks at the distal side of the humeral break at the posterior end of the costal cell (C) and the notch at the sinus between the alula (Al) and the axillary cell (Ax) of the wing (Fig. S2). These landmarks were used to infer semi-landmarks and to fit B-splines along the outline of the wing and along wing veins with Wings4 and CPR software (van der Linde & Houle, 2009; http://bio.fsu.edu/dhoule/wings.html). Males and females were analyzed separately, and landmark data for every image were processed manually. We applied multivariate outlier detection based on principal components analysis (PCA) of landmark coordinates using CPR and excluded extreme outliers caused by broken wings or images of insufficient quality. As a proxy for wing size we used total wing area, based on spline functions along the wing outline. Wing shape variation was analyzed using LORY software (http://bio.fsu.edu/dhoule/lory.html), following the methods described by Márquez et al. (2012). We obtained point estimates of shape deformation by locally evaluating Jacobian matrices of interpolation functions at pseudo-landmarks using LORY. Log (−log2) - transformed determinants of Jacobian matrices contain information about local space contractions or expansions relative to a reference configuration and can be used as discrete summary variables that describe shape variation.
Deformations of individual configurations were analyzed relative to Procrustes-transformed landmark coordinates, averaged across all individuals for each sex. We fitted elastic body splines (EBS) as interpolation functions at 122 (females) and 124 (males) evenly distributed pseudo-landmarks and calculated log-transformed Jacobian determinants for each individual. To visualize shape differences, we averaged Jacobian determinants across all individuals for each pseudo-landmark, group (FI, FS, MS) and sex. To interpolate shape values between landmarks we performed “kriging” (Gaussian process regression) using the R package kriging and plotted wings by showing interpolated Jacobian determinants for each group and sex using custom software (available upon request from M.K). Finally, to examine variation in allometry between body parts among the three karyotypic groups (FI, FS, MS) we calculated the ratios of (1) femur length to tibia length, (2) femur length versus wing area, and (3) tibia length versus wing area.
Statistical analysis
Statistical analyses were performed using JMP (v.11.1.1) and R (v.3.2.1) software. Given that the In(3R)P is absent in Maine, we could not analyze data with a fully factorial (orthogonal) model, testing the effects of karyotype (standard versus inverted), geography (Florida versus Maine), and the karyotype by geography interaction. We thus created a compound grouping factor g with three levels (“Florida inverted”, FI; “Florida standard”, FS; “Maine standard”, MS) (also see below).
We first performed multivariate analysis of variance (MANOVA) to test the effects of karyotype and geography on multivariate phenotype (i.e., a linear combination of all measured traits, except wing shape [due to its high dimensionality] and size ratios), using the following model: Yi = g +s + g × s, where Yi denotes the matrix of measured individual traits averaged by line and sex for the ith line, g is the nominal fixed grouping factor (with levels FI, FS, MS), s denotes the fixed effect of sex, and g × s denotes the interaction term. We also used MANOVA to analyze multivariate wing shape based on multiple Jacobian determinants, separately for each sex, by using the following model: Yi = g + l(g), where l(g) represents the effect of line nested within the grouping factor g (also see below).
Next, we analyzed each trait (including size ratios; see above) separately using a nested mixed-effects analysis of variance (ANOVA) model of the following form: yi = g +s + g × s + l(g), where yi is the measured phenotype for the ith individual, g denotes the grouping factor, s denotes sex, and l(g) is the random effect of line nested in g, estimated using restricted maximum likelihood (REML). The random line effect was included to account for variation among lines, but we were not primarily interested in the variance component estimates of this effect; we therefore do not report these estimates.
To analyze egg-to-adult survival (proportion viability) we used the following ANOVA model: arcsine squareroot (yi) = g + s + g × s, where yi is the proportion of egg-to adult survival of the ith line and g and s denote the grouping factor and sex, respectively; note that in this analysis “line” was the lowest level of replication.
To tease apart the effects of karyotype and geography we performed post-hoc tests using Tukey’s Honest Significant Difference (HSD) tests implemented in JMP, whenever the effect of the grouping factor g was significant; Tukey’s HSD method corrects for multiple testing (i.e., the family-wise error rate). (For MANOVAs, we used planned contrasts instead since post-hoc tests were not available in JMP.) We were specifically interested in using these tests to determine the effects of In(3R)P karyotype; the effects of geography were only of secondary interest. Significant differences between FI and FS and between FI and MS, with the comparison FS versus MS being non-significant, imply a clear-cut effect of karyotype, and that the standard homokaryons from Florida and Maine have qualitatively identical effects. A pattern where FI versus FS, FI versus MS, and FS versus MS are all significantly different implies that inverted versus standard karyotypes differ in their effect, but that the two standard arrangement genotypes from Florida and Maine differ as well. In this situation, the effects of karyotype and geography can not be completely separated; nonetheless, the significant difference between FI and FS indicates an effect of In(3R)P karyotype. Under either scenario it thus seems safe to conclude that In(3R)P karyotype affects the phenotype of interest.
To compare our results for the differential effects of In(3R)P karyotype on wing area in North America to those from Australia (Queensland; Rako et al. 2006) we calculated Cohen’s standardized effect sizes d (Cohen, 1988) (1) from lines means and standard deviations for the FI and FS lines from Florida (this study) and (2) from approximate values of line means and standard deviations of inverted and standard lines obtained from Fig. 1 in Rako et al. (2006), using the online tool WebPlotDigitizer (Rohatgi, 2015).
In contrast to size data, the assumptions of normality and homoscedasticity underlying ANOVA were not always fulfilled for other traits. Since data for development time, egg-to-adult survival, chill coma recovery and oxidative stress resistance represent failure time or time-to-event data that can violate ANOVA assumptions, we additionally analyzed these traits using mixed-effects Cox (proportional hazards) regression implemented in the R package coxme (Therneau, 2012), following the same model structure as defined above. These analyses yielded outcomes that were qualitatively identical to those based on ANOVA (not shown).
Results
Effects on multivariate phenotype
To account for potential phenotypic correlations among traits we performed MANOVA analysis of the multivariate phenotype, i.e. a linear combination of all measured traits (except wing shape; see below). Examination of contrasts for the grouping factor g (FI versus FS, FI versus MS, FS versus MS) indicated that inverted In(3R)P and standard arrangement differ in their effects on multivariate phenotype (Table S1; also see below and Table S3). The karyotypic effect of In(3R)P was most clearly revealed by the significant difference between the FI and FS groups. Inspection of contrasts also suggested that geographical origin (Florida versus Maine) might affect multivariate phenotype (Table S1). In particular, the significant difference between FS and MS might be consistent with an effect of geography; however, a non-mutually exclusive alternative is that standard arrangements from Florida and Maine differ genotypically in their effects upon phenotype.
Effects on pre-adult life history and stress resistance
Pre-adult life history traits (development time, egg-to-adult survival) were neither affected by In(3R)P karyotype nor by geography (Table 2). Similarly, karyotype and geography had no measurable effect on any of the stress resistance or physiological traits (chill coma recovery time, oxidative stress resistance, triglyceride content) (Table 2).
Table 2.
Mixed-effects ANOVA tables for phenotypic analyses.
| Trait | Factors
|
||
|---|---|---|---|
| group (g) | sex (s) | g × s | |
| Development time (h) | F2,31= 1.07 | F1.3554= 402.52*** | F2,3554= 0.06 |
| Egg-to-adult survival (%) | F2,62= 2.88 | F1,62= 3.12 | F2,62= 0.577 |
| Wing area (mm2) | F2,29= 10.24** | F1,1075= 3551.66*** | F2,1075= 0.89 |
| Femur length (mm) | F1, 29= 6.3** | F1,1053= 525.04*** | F2,1053= 5.1** |
| Tibia length (mm) | F1, 29= 6.39** | F1,1053= 318.66*** | F2,1053= 0.23 |
| Femur-to-tibia ratio | F1, 28= 0.9 | F1,1059= 0.9 | F2,1059= 0.9 |
| Femur-to-wing area ratio | F1, 29= 7.72** | F1,1056= 2268*** | F2,1056= 2.58 |
| Tibia-to-wing area ratio | F1, 29= 5.77** | F1,1055= 2119*** | F2,1055= 3.4* |
| Chill coma recovery (time to recovery, h) | F1, 28= 1.29 | F1,1041= 20.3*** | F2,1040= 9.09** |
| Oxidative stress resistance (age at death, h) | F1, 29= 0.56 | F1,1183=0.65 | F2,1183= 0.03 |
| Triglyceride content (μg) | F1, 29= 0.61 | F1,488=264.76*** | F2,488= 2.68 |
p <0.05;
p < 0.01;
p < 0.001.
Significant among-group effects for the grouping factor g were analyzed using Tukey’s HSD post-hoc tests; results of these tests are shown in Fig. 1. See Materials and Methods and Results sections for further details.
Effects on size, shape and allometry
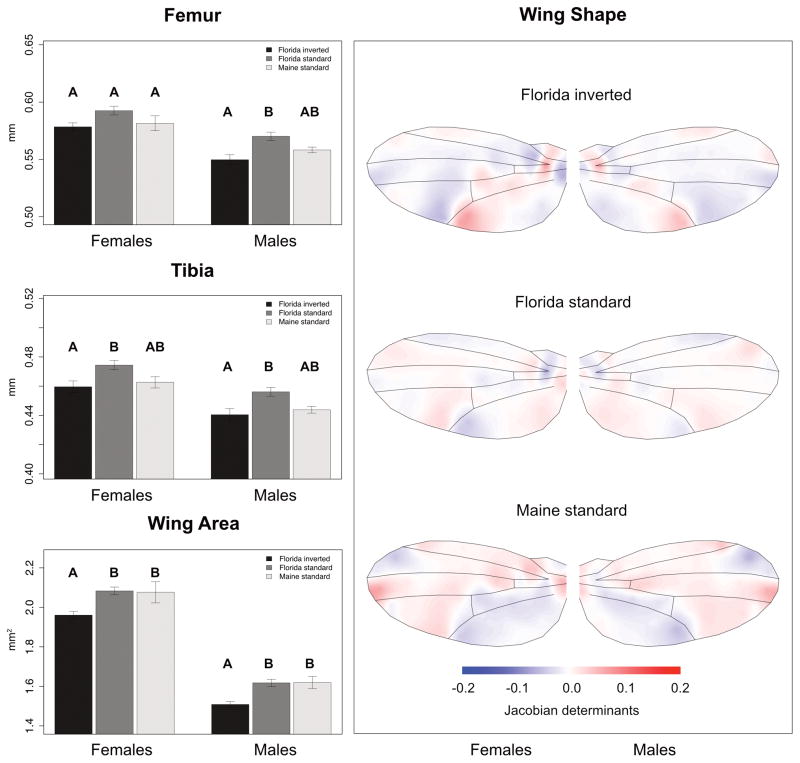
In contrast to life history and stress resistance, inverted and standard chromosomal arrangements differed in their effects on size-related traits. Inverted and standard lines from Florida differed significantly for both femur and tibia length, suggesting an effect of In(3R)P on body size (Table 2). The tibiae of inverted homokaryons were significantly shorter than those of non-inverted lines for both sexes; the same effect was seen for femur length but only in males (Fig. 1, Table 2). Although for both traits standard arrangement lines from Maine did not differ from the two Florida karyotypes (Fig. 1, Table 2), we failed to identify a clear effect of geography when comparing lines from Florida and Maine without accounting for karyotype (not shown). These observations indicate that In(3R)P karyotype affects size, even though geographic differences independent of karyotype might also make a contribution.
Figure 1.
The effects of In(3R)P on size-related traits. The left panel shows trait values averaged across line means for the three different groups differing in In(3R)P karyotype (“Florida inverted”, FI; “Florida standard”, FS; “Maine standard”, MS). Error bars show standard errors. Letters above bars show the outcomes of Tukey’s HSD post-hoc tests, carried out for each sex separately: groups that not containing the same letter are significantly different (p<0.05). The right panel shows average wing outlines and Jacobian determinants for each of the three groups (FI, FS, MS). Jacobian determinants, interpolated with kriging, represent local expansion (positive values; red) or contractions (negative values; blue) relative to the grand mean.
The notion that In(3R)P inverted versus standard arrangements have differential effects on size was clearly confirmed by an analysis of variation in wing size: for both sexes, Florida inverted lines had significantly smaller wings than Florida standard and Maine standard lines, whereas standard arrangement lines from Florida and Maine did not differ from each other (Fig. 1, Table 2). Despite different measurement methods and sample sizes, we found that the effect sizes for wing size differences between inverted and standard karyotypes from low-latitude populations in North America (Florida; our data) and Australia (Queensland; Rako et al., 2006) were large (i. e., Cohen’s d > 1.4) and qualitatively very similar (Florida: d=1.74; Queensland, Australia: d=1.64) across both continents (Table S2).
MANOVA applied to a linear combination of femur length, tibia length and wing area, thus accounting for potential intercorrelations among size-related traits, also revealed significant among-group contrasts consistent with effects of karyotype and geography on size (Table S3).
We next analyzed among-group variation in wing shape. Contrasts from MANOVA performed on Jacobian determinants of pseudo-landmarks showed significant effects of karyotype and geography on wing shape for both sexes (Table S4). Florida inverted and Maine standard lines differed most strongly in their effects on wing shape, with Florida standard lines being intermediate. In both sexes, areas that showed largest variation for wing shape were located at the proximal part of the wing around the humeral break, around the terminal end of the distal (L5) wing vein, and at the distal end of the 1st posterior (1P) wing cell (Fig. 1, Fig. S2).
We also examined whether the three groups differ in allometry by analyzing among-group variation in the size ratios of leg parts (femur length versus tibia length) and different body parts (femur length versus wing area, tibia length versus wing area). While we failed to detect effects for the ratio of femur:tibia length, both group and sex affected the ratios of leg parts to wing area, with the ratios being larger for males than females (Table 2, Fig. S3). This suggests that in males wing size is smaller relative to leg size. For both measures of leg:wing size, Florida inverted lines exhibited larger ratios than Maine standard lines, irrespective of sex. The effect of In(3R)P karyotype was most clear-cut for the femur length:wing area ratio in males: Florida inverted lines had a greater ratio than both Florida and Maine standard lines, while standard lines from Florida and Maine did not differ from each other (Table 2, Fig. S3).
Together, our results indicate that In(3R)P affects multiple aspects of body size, shape and allometry but does not seem to have detectable effects upon pre-adult life history, stress resistance (e.g, chill coma recovery, oxidative stress resistance), and fat content.
Discussion
Chromosomal inversion polymorphisms are commonly found in D. melanogaster populations (Lemeunier & Aulard, 1992) but evidence for selection acting on them is surprisingly scarce (Kapun et al., 2016). In support of a role for selection, In(3R)Payne, a cosmopolitan inversion that is clinally distributed along latitudinal gradients in Australia and North America, has been associated with body size clines in Australian populations (Weeks et al., 2002; Rako et al., 2006; Kennington et al., 2007). However, comparable phenotypic data from other continents are not available, and whether the observations from the Australian cline represent a local phenomenon or a general pattern remains unclear. Moreover, effects of this inversion on traits other than size remain largely unknown (cf. Rako et al., 2006). Here we have investigated the phenotypic effects of In(3R)P in populations originating from the endpoints of the latitudinal cline running along the North American east coast.
In(3R)P has parallel effects on size across the North American and Australian clines
Our study provides the first evidence for an association between In(3R)P and the body size cline (cf. Coyne & Beecham, 1987) in North America. For the endpoints of the Australian cline, Rako et al. (2006) reported that flies carrying In(3R)P had smaller wings than standard arrangement flies. Similarly, for several proxies of body size, we found that inverted flies from the North American cline are smaller than flies carrying the standard chromosomal arrangement. Our findings thus mirror previous observations from the Australian cline (Weeks et al., 2002; Rako et al., 2006; Kennington et al., 2007) and suggest that In(3R)P has parallel – very likely adaptive – effects on body size along both clinal gradients (cf. Kapun et al., 2016).
Another size trait known to exhibit clinal variation on multiple continents – and thus likely to be subject to spatially varying selection – is wing “loading” (the intercept of the relationship between body and wing size) (Azevedo et al., 1998; Gilchrist et al., 2000). Stalker (1980), for example, reasoned that larger wings relative to body size (i.e., low wing loading) might result in increased lift and would thus compensate for lower beat frequencies at lower temperatures experienced at higher latitudes. Perhaps consistent with this prediction, we observed lowest wing loading for standard arrangement lines from Maine, intermediate loading in standard arrangement lines from Florida, and highest loading in inverted lines from Florida. It is noteworthy in this context that QTL mapping has identified a major peak for male flight duration within the region spanned by In(3R)P (Luckinbill et al., 2005; see discussion in Rako et al., 2006).
We also found karyotypic and geographic variation in wing shape. Inverted lines from Florida and standard arrangement lines from Maine differed most strongly in wing shape, while standard lines from Florida showed an intermediate pattern. Consistent with observations by Gilchrist et al. (2000), who investigated wing shape variation along size clines from three continents (albeit without examining In(3R)P), we observed large shape deformations in the anterior distal region between the medial and cubital vein. Moreover, we identified large shape differences at the discal cell and the 3rd posterior cell along the distal vein (L5), indicating shape expansion in Florida inverted lines but shape contraction in Maine standard lines. In contrast, shape differentiation was minimal along the leading edge of the wing. This is in good agreement with kinetic analyses of wing aerodynamics: the anterior-posterior wing region might potentially be functionally constrained since it maintains the rotation axis close to the leading edge (Dickinson et al., 1999; Gilchrist et al., 2000). However, the evolutionary mechanisms that maintain variation in wing shape remain poorly understood; while wing size is subject to directional selection, wing shape seems to be the result of optimizing (stabilizing) selection (potentially due to selection for “canalization” [Flatt, 2005]) rather than directional selection (Gilchrist and Partridge, 2001). Additional data will be required to unravel the potentially adaptive effects of In(3R)P on variation in wing shape.
In(3R)P and the genetic basis of size and shape
Further support for potentially causal links between In(3R)P and size-related traits comes from studies of the genetic basis of size and shape variation in Drosophila (see de Jong & Bochdanovits, 2003; Mirth & Shingleton, 2012; and references therein). Gockel et al. (2002) and Calboli et al. (2003), for example, used QTL analysis to map genetic variation associated with thorax length and wing size and found that the third chromosome accounts for a major proportion of size variation between the endpoints of the Australian and South American clines. Weeks et al. (2002) identified three indel (insertion deletion) and microsatellite polymorphisms within the region spanned by In(3R)P that are strongly associated with body size variation among Australian populations. Similarly, Kennington et al. (2007) found that microsatellite alleles associated with decreased wing size are in strong LD with In(3R)P. Moreover, the gene Dca (Drosophila cold acclimation; also known as smp-30), which is located close to the proximal breakpoint of In(3R)P and likely associated with this inversion through hitchhiking, accounts for approximately 5–10% of natural wing size variation in Australian populations (McKechnie et al., 2010), and a clinal promoter polymorphism in this gene has been shown to decrease wing size (McKechnie et al., 2010; Lee et al., 2011).
In agreement with these findings, the region spanned by In(3R)P harbors several genes known to be important for growth regulation and the determination of body size (de Jong & Bochdanovits, 2003; Fabian et al., 2012; Kapun et al., 2016; see flybase.org for details of gene function and original source references). For example, In(3R)P contains multiple loci involved in insulin/insulin-like growth factor signaling (IIS), a pathway that plays a major role in regulating growth, size and shape, including InR (insulin-like receptor), Tsc1 (tuberous sclerosis complex 1), and Pi3K (Pi3K92E, phosphoinositide 3-kinase at 92E; also known as Dp110) (Brogiolo et al., 2001; de Jong & Bochdanovits, 2003; Oldham & Hafen, 2003; Edgar, 2006; Shingleton et al., 2007; Mirth & Shingleton, 2012; Nässel et al., 2015; also see below). Importantly, InR harbors many alleles that are strongly clinal along the North American east coast (Fabian et al., 2012; Paaby et al., 2014); indeed, a naturally occurring, clinal indel polymorphism in InR (albeit apparently not in LD with In(3R)P) affects body size in North American populations (Paaby et al., 2014).
Whole-genome analyses of clinal variation associated with In(3R)P have also uncovered candidates with known effects on growth, including clinally varying alleles in InR (see above), Tsc1 (see above), Hmgcr (hydroxymethlyglutaryl coenzyme A reductase, known to interact with IIS), Orct2 (organic cation transporter 2 or calderón, involved in IIS as well) and Stat92E (signal-transducer and activator of transcription protein at 92E, a transcription factor involved in JAK/STAT signaling) (Fabian et al., 2012; Kapun et al., 2016). Several of these genes, including InR, Orct2 and Stat92E, also vary clinally along the Australian cline (Kolaczkowski et al., 2011).
Two other interesting candidates are hh (hedgehog) and Dad (Daughters against DPP), both of which harbor clinal alleles associated with In(3R)P in North America (Fabian et al., 2012; Kapun et al., 2016). The hh locus encodes a signaling protein, which forms gradients in the developing wing and controls the placement and spacing of the longitudinal wing veins L3 and L4 (Blair, 2007; Matamoro-Vidal et al., 2015). Perhaps consistent with the involvement of this gene, we identified strong variation in the spacing of these veins among karyotypes (see Fig. 1). Dad encodes a negative regulator of Dpp (Decapentaplegic), a morphogen that modulates the placement of the L2 and L5 wing veins (Tsuneizumi et al., 1997; Matamoro-Vidal et al., 2015); notably, we observed strong shape variation among karyotypes within the 3rd posterior cell along the L5 vein.
Thus, multiple lines of evidence suggest that In(3R)P harbors clinal variants in several major genes known to affect growth, size and shape. Although the causative effects of In(3R)P-linked alleles at these loci on size and shape remain unknown, these variants represent promising candidates for functional testing (cf. Kapun et al., 2016).
In(3R)P has no measurable effects on pre-adult life history or stress resistance
Little is known about whether In(3R)P affects traits other than size. For example, with regard to Australian populations, a study by Anderson et al. (2003) reported an association between cold resistance and In(3R)P, and McColl et al. (1996) found an association between the response to thermal selection and the hsr-omega and hsp68 genes, both located in the region spanned by In(3R)P (Anderson et al., 2003). However, Rako et al. (2006), using a more direct genetic association approach based on In(3R)P homokaryon lines, failed to find an effect of In(3R)P on cold resistance. These findings are in good agreement with ours: we also did not detect any measurable effects of In(3R)P on cold resistance. Although several genes known to be involved in cold resistance are located within the region of In(3R)P (Anderson et al., 2003), it is unknown whether alleles at these loci are in LD with this inversion (cf. Weeks et al. 2002; Rako et al., 2006).
Rako et al. (2006) also found no effects of In(3R)P on development time for the Australian cline, an observation that is again consistent with ours. Given the usually tight physiological and genetic correlations between development time and body size (e.g., in artificial selection or experimental evolution experiments; see de Jong & Bochdanovits, 2003; and references therein), it is perhaps surprising that In(3R)P does not affect development time. However, clinal patterns for this trait often seem to be weak (James & Partridge, 1995) or absent (Fabian et al., 2015); in line with this, development time and body size do not seem to be associated among populations along the Australian cline (James et al., 1995). This raises the interesting but unresolved question of how, in terms of physiological mechanisms, In(3R)P affects size.
We also measured several traits that were not assayed by Rako et al. (2006), including egg-to-adult survival, oxidative stress resistance and triglyceride content; however, again, we could not find any measurable effects of In(3R)P on these traits. For the South American cline, Robinson et al. (2000) also failed to find a cline for fat content (and starvation resistance), albeit without examining In(3R)P. Together with the previous findings from Australia, our results therefore suggest that In(3R)P might have quite specific effects on size-related – but not necessarily other fitness-related – traits; yet, two important caveats remain. First, this inversion might have subtle effects on the non-significant traits we have measured but our statistical power for finding these effects was perhaps insufficient. Secondly, there are other major fitness-related traits known to be clinal (e.g., ovariole number, fecundity, lifespan, reproductive diapause) that we have not measured as a function of In(3R)P karyotype.
The adaptive significance of In(3R)P
The In(3R)P polymorphism exhibits steep, persistent latitudinal frequency clines between subtropical/tropical and temperate, seasonal environments on multiple continents (e.g., North America, Australia, Indian subcontinent, Japan), but – intriguingly – does not seem to be clinal within the tropics proper (e.g. sub-Saharan Africa, Southeast Asia) (Aulard et al., 2002; Glinka et al., 2005). This strongly suggests that the inverted arrangement is selectively favored in warm, low-latitude habitats, whereas the standard arrangement is favored in temperate, seasonal, high-latitude habitats.
Recent findings indeed support the notion that latitudinal clines of In(3R)P are maintained by spatially varying selection: in North America the latitudinal cline of In(3R)P has remained stable for >40 years, deviates from neutral expectation, and is maintained independent of isolation by distance and admixture (Kapun et al., 2016). Moreover, the majority (>90%) of the most strongly clinally varying single nucleotide polymorphisms (SNPs) contained in In(3R)P are shared between the North American and Australian clines, consistent with parallel effects of spatially varying selection across both continents (Kapun et al., 2016).
Interestingly, in areas where In(3R)P is known to be clinal (e.g., North America, Australia, India, Japan), body size also exhibits latitudinal clines (see Introduction). Together with the observation that In(3R)P is associated with body size in both Australia and North America, this suggests that In(3R)P clines might be driven by selection on body size. While the selective forces shaping body size clines still remain largely unknown (Partridge & Coyne, 1997), thermal experimental evolution experiments in Drosophila have shown that adaptation to warm versus cool conditions favors small versus large size (Partridge et al., 1994). Thus, temperature might represent the most parsimonious selective agent underlying latitudinal size clines. As hypothesized by James & Partridge (1995), a possible reason for the existence of a temperature-latitude-size correlation in Drosophila could be that larval food resources might be more ephemeral in the tropical climates due to increased competition, and that this would cause selection to favor rapid development and thus smaller adult size. In temperate habitats, in contrast, resources might be more stable and selection might thus favor longer development time and larger adult size (James & Partridge, 1995). Even though we did not find an effect of In(3R)P on development time, the fact that In(3R)P causes smaller size (through as of yet unknown developmental effects) and that its frequency is much more prevalent in warmer areas might be consistent with such a scenario.
The idea that inversions such as In(3R)P might be shaped by climatic adaptation is underscored by several observations. First, in North America In(3R)P frequency is strongly positively associated with multiple measures of temperature and precipitation, whereas temperature dispersion (range) and seasonality seem to favor higher frequencies of the standard chromosomal arrangement (Kapun et al., 2016; also see Knibb, 1982). Second, along the Australian east coast, the latitudinal cline of In(3R)P has shifted in position (intercept) across a time span of 20 years in response to recent climate change; since no single climatic factor could fully account for this pattern, it is likely that a combination of climatic variables, not temperature alone, has driven this shift (Umina et al., 2005). Third, in support of climatic selection, we have previously found in an experimental evolution experiment that In(3R)Mo and In(3R)C, two inversions that partly overlap with In(3R)P, were selectively favored in replicate populations exposed to cold versus warm temperatures, respectively (Kapun et al., 2014). However, an important caveat is that in the same experiment In(3R)P itself was rapidly lost, from an initial frequency of ~20%, in both cold and warm environments. Thus, together with the findings mentioned above, unknown selective factors other than – or in addition to –temperature must play a major role in maintaining this inversion. It will clearly be of great interest – as well as a major challenge – to determine the selective factors affecting In(3R)P in future work.
Conclusions
Here, we have demonstrated that the chromosomal inversion In(3R)P affects several size-related traits in North American populations of D. melanogaster. Remarkably, these effects go in the same direction – and are of similar magnitude (e.g., see Table S2) – as those that have been previously reported for the Australian cline (Rako et al., 2006). In conjunction with the Australian data, our results thus suggest a major role of In(3R)P in shaping clinal size variation across both continents, thereby considerably strengthening the case for spatially varying selection acting on body size via genetic variants contained within this inversion. However, the effects we have identified here remain correlational; future efforts will be required to dissect the functional links between size and the causative genetic variants harbored by this inversion.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers and Kathrin Garschall for helpful comments; David Houle, Brian Hollis and Fiona Hollis for advice on morphometrics; Tad Kawecki, Jérôme Goudet and Tanja Schwander for statistical advice; and Daria Martynow for help in the laboratory. Our research was supported by the Swiss National Science Foundation (SNSF) (grant PP00P3_133641 to T.F.), the National Institutes of Health (NIH) (grant 5R01GM100366 to P.S.S.), the National Science Foundation (NSF) (grant DEB 0921307 to P.S.S.), and the Department of Ecology and Evolution at the University of Lausanne.
Footnotes
Data accessibility
Raw data have been deposited at Dryad (doi:10.5061/dryad.8ns67).
References
- Anderson AR, Collinge JE, Hoffmann AA, Kellett M, McKechnie SW. Thermal tolerance trade-offs associated with the right arm of chromosome 3 and marked by the hsr-omega gene in Drosophila melanogaster. Heredity. 2003;90:195–202. doi: 10.1038/sj.hdy.6800220. [DOI] [PubMed] [Google Scholar]
- Anderson AR, Hoffmann AA, McKechnie SW, Umina PA, Weeks AR. The latitudinal cline in the In(3R)Payne inversion polymorphism has shifted in the last 20 years in Australian Drosophila melanogaster populations. Mol Ecol. 2005;14:851–858. doi: 10.1111/j.1365-294X.2005.02445.x. [DOI] [PubMed] [Google Scholar]
- Aulard S, David JR, Leumeunier F. Chromosomal inversion polymorphism in Afrotropical populations of Drosophila melanogaster. Genet Res. 2002;79:49–63. doi: 10.1017/s0016672301005407. [DOI] [PubMed] [Google Scholar]
- Ayrinhac A, Debat V, Gibert P, Kister AG, Legout H, Moreteau B, et al. Cold adaptation in geographical populations of Drosophila melanogaster: phenotypic plasticity is more important than genetic variability. Funct Ecol. 2004;18:700–706. [Google Scholar]
- Azevedo RBR, James AC, McCabe J, Partridge L. Latitudinal Variation of Wing:Thorax Size Ratio and Wing-Aspect Ratio in Drosophila melanogaster. Evolution. 1998;52:1353–1362. doi: 10.1111/j.1558-5646.1998.tb02017.x. [DOI] [PubMed] [Google Scholar]
- Bergland AO, Behrman EL, O’Brien KR, Schmidt PS, Petrov DA. Genomic Evidence of Rapid and Stable Adaptive Oscillations over Seasonal Time Scales in Drosophila. PLoS Genet. 2014;10:e1004775. doi: 10.1371/journal.pgen.1004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland AO, Tobler R, González J, Schmidt P, Petrov D. Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster. Mol Ecol. 2015 doi: 10.1111/mec.13455. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair SS. Wing Vein Patterning in Drosophila and the Analysis of Intercellular Signaling. Annu Rev Cell Dev Biol. 2007;23:293–319. doi: 10.1146/annurev.cellbio.23.090506.123606. [DOI] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Calboli FCF, Kennington WJ, Partridge L. Qtl Mapping Reveals a Striking Coincidence in the Positions of Genomic Regions Associated with Adaptive Variation in Body Size in Parallel Clines of Drosophila melanogaster on Different Continents. Evolution. 2003;57:2653–2658. doi: 10.1111/j.0014-3820.2003.tb01509.x. [DOI] [PubMed] [Google Scholar]
- Capy P, Pla E, David JR. Phenotypic and genetic variability of morphometrical traits in natural populations of Drosophila melanogaster and D. simulans I Geographic variations. Genet Sel Evol. 1993;25:517–536. [Google Scholar]
- Charlesworth B, Charlesworth D. Elements of Evolutionary Genetics. Roberts and Company Publishers; Greenwood Village: 2010. [Google Scholar]
- Cohen J. Statistical power analysis for the behavioral sciences. Lawrence Erlbaum Associates; Hillsdale: 1988. [Google Scholar]
- Corbett-Detig RB, Cardeno C, Langley CH. Sequence-based detection and breakpoint assembly of polymorphic inversions. Genetics. 2012;192:131–137. doi: 10.1534/genetics.112.141622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Beecham E. Heritability of two morphological characters within and among natural populations of Drosophila melanogaster. Genetics. 1987;117:727–737. doi: 10.1093/genetics/117.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Singh BN. Genetic differentiation and inversion clines in Indian natural populations of Drosophila melanogaster. Genome. 1991;34:618–625. doi: 10.1139/g91-094. [DOI] [PubMed] [Google Scholar]
- David JR, Bocquet C. Evolution in a cosmopolitan species: Genetic latitudinal clines in Drosophila melanogaster wild populations. Experientia. 1975;31:164–166. doi: 10.1007/BF01990682. [DOI] [PubMed] [Google Scholar]
- David JR, Capy P. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 1988;4:106–111. doi: 10.1016/0168-9525(88)90098-4. [DOI] [PubMed] [Google Scholar]
- David JR, Capy P, Payant V, Tsakas S. Thoracic trident pigmentation in Drosophila melanogaster: differentiation of geographical populations. Genet Sel Evol. 1985;17:211–224. doi: 10.1186/1297-9686-17-2-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debat V, Bloyer S, Faradji F, Gidaszewski N, Navarro N, Orozco-Terwengel P, et al. Developmental stability: a major role for cyclin G in Drosophila melanogaster. PLoS Genet. 2011;7:e1002314. doi: 10.1371/journal.pgen.1002314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong G, Bochdanovits Z. Latitudinal clines in Drosophila melanogaster: body size, allozyme frequencies, inversion frequencies, and the insulin-signalling pathway. J Genet. 2003;82:207–223. doi: 10.1007/BF02715819. [DOI] [PubMed] [Google Scholar]
- Dickinson MH, Lehmann FO, Sane SP. Wing rotation and the aerodynamic basis of insect flight. Science. 1999;284:1954–1960. doi: 10.1126/science.284.5422.1954. [DOI] [PubMed] [Google Scholar]
- Edgar B. How flies get their size: genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- Endler JA. Geographic Variation, Speciation, and Clines. Princeton University Press; Princeton: 1977. [PubMed] [Google Scholar]
- Fabian DK, Kapun M, Nolte V, Kofler R, Schmidt PS, Schlötterer C, et al. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol Ecol. 2012;21:4748–4769. doi: 10.1111/j.1365-294X.2012.05731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian DK, Lack JB, Mathur V, Schlötterer C, Schmidt PS, Pool JE, et al. Spatially varying selection shapes life history clines among populations of Drosophila melanogaster from sub-Saharan Africa. J Evol Biol. 2015;28:826–840. doi: 10.1111/jeb.12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T. The evolutionary genetics of canalization. Quart Rev Biol. 2005;80:287–316. doi: 10.1086/432265. [DOI] [PubMed] [Google Scholar]
- Flatt T. Genomics of clinal variation in Drosophila: disentangling the interactions of selection and demography. Mol Ecol. 2016 doi: 10.1111/mec.13534. in press. [DOI] [PubMed] [Google Scholar]
- Gibert P, Capy P, Imasheva A, Moreteau B, Morin JP, Pétavy G, et al. Comparative analysis of morphological traits among Drosophila melanogaster and D. simulans: genetic variability, clines and phenotypic plasticity. Genetica. 2004;120:165–179. doi: 10.1023/b:gene.0000017639.62427.8b. [DOI] [PubMed] [Google Scholar]
- Gibert P, Moreteau B, Pétavy G, Karan D, David JR. Chill-coma tolerance, a major climatic adaptation among Drosophila species. Evolution. 2001;55:1063–1068. doi: 10.1554/0014-3820(2001)055[1063:cctamc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Gilchrist AS, Azevedo RBR, Partridge L, O’Higgins P. Adaptation and constraint in the evolution of Drosophila melanogaster wing shape. Evol Dev. 2000;2:114–124. doi: 10.1046/j.1525-142x.2000.00041.x. [DOI] [PubMed] [Google Scholar]
- Gilchrist AS, Partridge L. The contrasting genetic architecture of wing size and shape in Drosophila melanogaster. Heredity. 2001;86:144–152. doi: 10.1046/j.1365-2540.2001.00779.x. [DOI] [PubMed] [Google Scholar]
- Glinka S, Stephan W, Das A. Homogeneity of common cosmopolitan inversion frequencies in Southeast Asian Drosophila melanogaster. J Genet. 2005;84:173–178. doi: 10.1007/BF02715842. [DOI] [PubMed] [Google Scholar]
- Gockel J, Kennington WJ, Hoffmann AA, Goldstein DB, Partridge L. Nonclinality of molecular variation implicates selection in maintaining a morphological cline of Drosophila melanogaster. Genetics. 2001;158:319–323. doi: 10.1093/genetics/158.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gockel J, Robinson SJW, Kennington WJ, Goldstein DB, Partridge L. Quantitative genetic analysis of natural variation in body size in Drosophila melanogaster. Heredity. 2002;89:145–153. doi: 10.1038/sj.hdy.6800121. [DOI] [PubMed] [Google Scholar]
- Goenaga J, Fanarar JJ, Hasson E. Latitudinal Variation in Starvation Resistance is Explained by Lipid Content in Natural Populations of Drosophila melanogaster. Evol Biol. 2013;40:601–612. [Google Scholar]
- Hoffmann AA, Anderson A, Hallas R. Opposing clines for high and low temperature resistance in Drosophila melanogaster. Ecol Lett. 2002;5:614–618. [Google Scholar]
- Hoffmann AA, Harshman LG. Desiccation and starvation resistance in Drosophila: patterns of variation at the species, population and intrapopulation levels. Heredity. 1999;83:637–643. doi: 10.1046/j.1365-2540.1999.00649.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Parsons PA. An integrated approach to environmental stress tolerance and life-history variation: desiccation tolerance in Drosophila. Biol J Linn Soc. 2009;37:117–136. [Google Scholar]
- Hoffmann AA, Rieseberg LH. Revisiting the Impact of Inversions in Evolution: From Population Genetic Markers to Drivers of Adaptive Shifts and Speciation? Annu Rev Ecol Evol Syst. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Sgrò CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–485. doi: 10.1038/nature09670. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Sgrò CM, Weeks AR. Chromosomal inversion polymorphisms and adaptation. Trends Ecol Evol. 2004;19:482–488. doi: 10.1016/j.tree.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Shirriffs J. Geographic variation for wing shape in Drosophila serrata. Evolution. 2002;56:1068–1073. doi: 10.1111/j.0014-3820.2002.tb01418.x. [DOI] [PubMed] [Google Scholar]
- Imasheva AG, Bubli OA, Lazebny OE. Variation in wing length in Eurasian natural populations of Drosophila melanogaster. Heredity. 1994;72:508–514. doi: 10.1038/hdy.1994.68. [DOI] [PubMed] [Google Scholar]
- Inoue Y, Watanabe TK. Inversion polymorphisms in Japanese natural populations of D. melanogaster. Jpn J Genet. 1979;54:69–82. [Google Scholar]
- James AC, Partridge L. Thermal evolution of rate of larval development in Drosophila melanogaster in laboratory and field populations. J Evol Biol. 1995;8:315–330. [Google Scholar]
- James AC, Azevedo RBR, Partridge L. Cellular basis and developmental timing in a size cline of Drosophila melanogaster. Genetics. 1995;140:659–666. doi: 10.1093/genetics/140.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AC, Azevedo RBR, Partridge L. Genetic and environmental responses to temperature of Drosophila melanogaster from a latitudinal cline. Genetics. 1997;146:881–890. doi: 10.1093/genetics/146.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao JY, Zubair A, Salomon MP, Nuzhdin SV, Campo D. Population genomic analysis uncovers African and European admixture in Drosophila melanogaster populations from the south-eastern United States and Caribbean Islands. Mol Ecol. 2015;24:1499–1509. doi: 10.1111/mec.13137. [DOI] [PubMed] [Google Scholar]
- Kapun M, Fabian DK, Goudet J, Flatt T. Genomic Evidence for Adaptive Inversion Clines in Drosophila melanogaster. Mol Biol Evol. 2016 doi: 10.1093/molbev/msw016. in press. [DOI] [PubMed] [Google Scholar]
- Kapun M, van Schalkwyk H, McAllister B, Flatt T, Schlötterer C. Inference of chromosomal inversion dynamics from Pool-Seq data in natural and laboratory populations of Drosophila melanogaster. Mol Ecol. 2014;23:1813–1827. doi: 10.1111/mec.12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A. Drosophila melanogaster’s history as a human commensal. Curr Biol. 2007;17:R77–R81. doi: 10.1016/j.cub.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Kennington WJ, Hoffmann AA, Partridge L. Mapping Regions Within Cosmopolitan Inversion In(3R)Payne Associated With Natural Variation in Body Size in Drosophila melanogaster. Genetics. 2007;177:549–556. doi: 10.1534/genetics.107.074336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepsatel P, Gáliková M, Huber CD, Flatt T. Similarities and differences in altitudinal versus latitudinal variation for morphological traits in Drosophila melanogaster. Evolution. 2014;68:1385–1398. doi: 10.1111/evo.12351. [DOI] [PubMed] [Google Scholar]
- Knibb WR. Chromosome inversion polymorphisms in Drosophila melanogaster II. Geographic clines and climatic associations in Australasia, North America and Asia. Genetica. 1982;58:213–221. [Google Scholar]
- Knibb WR, Oakeshott JG, Gibson JB. Chromosome Inversion Polymorphisms in Drosophila melanogaster I Latitudinal Clines and Associations between Inversions in Australasian Populations. Genetics. 1981;98:833–847. doi: 10.1093/genetics/98.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B, Kern AD, Holloway AK, Begun DJ. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics. 2011;187:245–260. doi: 10.1534/genetics.110.123059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SF, Chen Y, Varan AK, Wee CW, Rako L, Axford JK, et al. Molecular basis of adaptive shift in body size in Drosophila melanogaster: functional and sequence analyses of the Dca gene. Mol Biol Evol. 2011;28:2393–2402. doi: 10.1093/molbev/msr064. [DOI] [PubMed] [Google Scholar]
- Lemeunier F, Aulard S. Inversion polymorphism in Drosophila melanogaster. In: Krimbas CB, Powell JR, editors. Drosophila Inversion Polymorphism. CRC Press; Boca Raton: 1992. pp. 339–405. [Google Scholar]
- Luckinbill LS, Reddy S, Dudekonda V, Curtsinger JW. Analysis of two components of flight using recombinant inbred lines of Drosophila melanogaster. Genetica. 2005;124:235–245. doi: 10.1007/s10709-005-2375-6. [DOI] [PubMed] [Google Scholar]
- Matamoro-Vidal A, Salazar-Ciudad I, Houle D. Making quantitative morphological variation from basic developmental processes: Where are we? The case of the Drosophila wing. Dev Dyn. 2015;244:1058–1073. doi: 10.1002/dvdy.24255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin LM, Merritt TJS, Zhu CT, Eanes WF. The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics. 2005;170:1143–1152. doi: 10.1534/genetics.104.038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez EJ, Cabeen R, Woods RP, Houle D. The Measurement of Local Variation in Shape. Evol Biol. 2012;39:419–439. doi: 10.1007/s11692-012-9159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Animal Species and Evolution. The Belknap Press of Harvard University Press; Cambridge, Mass: 1963. [Google Scholar]
- McColl G, Hoffmann AA, McKechnie SW. Response of two heat shock genes to selection for knockdown heat resistance in Drosophila melanogaster. Genetics. 1996;143:1615–1627. doi: 10.1093/genetics/143.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan MW, Artiss JD, Strandbergh DR, Zak B. A peroxidase-coupled method for the colorimetric determination of serum triglycerides. Clin Chem. 1983;29:538–542. [PubMed] [Google Scholar]
- McKechnie SW, Blacket MJ, Song SV, Rako L, Carroll X, Johnson TK, et al. A clinally varying promoter polymorphism associated with adaptive variation in wing size in Drosophila. Mol Ecol. 2010;19:775–784. doi: 10.1111/j.1365-294X.2009.04509.x. [DOI] [PubMed] [Google Scholar]
- Mettler LE, Voelker RA, Mukai T. Inversion Clines in Populations of Drosophila melanogaster. Genetics. 1977;87:169–176. doi: 10.1093/genetics/87.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth CK, Shingleton AW. Integrating Body and Organ Size in Drosophila: Recent Advances and Outstanding Problems. Front Endocrin. 2012;3:1–13. doi: 10.3389/fendo.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth CK, Shingleton AW. The roles of juvenile hormone, insulin/target of rapamycin, and ecydsone signaling in regulating body size in Drosophila. Comm Integr Biol. 2014;7:e971568. doi: 10.4161/cib.29240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munjal AK, Karan D, Gibert P, Moreteau B, Parkash R, David JR. Thoracic trident pigmentation in Drosophila melanogaster: latitudinal and altitudinal clines in Indian populations. Genet Sel Evol. 1997;29:601–610. [Google Scholar]
- Nässel DR, Liu Y, Luo J. Insulin/IGF signaling and its regulation in Drosophila. Gen Comp Endocrinol. 2015;221:255–266. doi: 10.1016/j.ygcen.2014.11.021. [DOI] [PubMed] [Google Scholar]
- Oldham S, Hafen E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 2003;13:79–85. doi: 10.1016/s0962-8924(02)00042-9. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Schmidt PS. Functional Significance of Allelic Variation at methuselah, an Aging Gene in Drosophila. PLoS ONE. 2008;3:e1987. doi: 10.1371/journal.pone.0001987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, Bergland AO, Behrman EL, Schmidt PS. A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation. Evolution. 2014;68:3395–3409. doi: 10.1111/evo.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Coyne JA. Bergmann’s rule in ectotherms: is it adaptive? Evolution. 1997;51:632–635. doi: 10.1111/j.1558-5646.1997.tb02454.x. [DOI] [PubMed] [Google Scholar]
- Partridge L, Barrie B, Fowler K, French V. Evolution and development of body size and cell size in Drosophila melanogaster in response to temperature. Evolution. 1994;48:1269–1276. doi: 10.1111/j.1558-5646.1994.tb05311.x. [DOI] [PubMed] [Google Scholar]
- Rako L, Anderson AR, Sgrò CM, Stocker AJ, Hoffmann AA. The association between inversion In(3R)Payne and clinally varying traits in Drosophila melanogaster. Genetica. 2006;128:373–384. doi: 10.1007/s10709-006-7375-7. [DOI] [PubMed] [Google Scholar]
- Rane RV, Rako L, Kapun M, Lee SF, Hoffmann AA. Genomic evidence for role of inversion 3RP of Drosophila melanogaster in facilitating climate change adaptation. Mol Ecol. 2015;24:2423–2432. doi: 10.1111/mec.13161. [DOI] [PubMed] [Google Scholar]
- Reinhardt JA, Kolaczkowski B, Jones CD, Begun DJ, Kern AD. Parallel Geographic Variation in Drosophila melanogaster. Genetics. 2014;197:361–373. doi: 10.1534/genetics.114.161463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SJW, Zwaan B, Partridge L. Starvation resistance and adult body composition in a latitudinal cline of Drosophila melanogaster. Evolution. 2000;54:1819–1824. doi: 10.1111/j.0014-3820.2000.tb00726.x. [DOI] [PubMed] [Google Scholar]
- Rohatgi A. WebPlotDigitizer. 2015 http://arohatgi.info/WebPlotDigitizer.
- Savolainen O, Lascoux M, Merilä J. Ecological genomics of local adaptation. Nat Rev Genet. 2013;14:807–820. doi: 10.1038/nrg3522. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Matzkin L, Ippolito M, Eanes WF. Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution. 2005;59:1721–1732. [PubMed] [Google Scholar]
- Schmidt PS, Paaby AB. Reproductive Diapause and Life-History Clines in North American Populations of Drosophila melanogaster. Evolution. 2008;62:1204–1215. doi: 10.1111/j.1558-5646.2008.00351.x. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Zhu CT, Das J, Batavia M, Yang L, Eanes WF. An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc Natl Acad Sci USA. 2008;105:16207–16211. doi: 10.1073/pnas.0805485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingleton AW, Frankino WA, Flatt T, Nijhout HF, Emlen DJ. Size and shape: the developmental regulation of static allometry in insects. BioEssays. 2007;29:536–548. doi: 10.1002/bies.20584. [DOI] [PubMed] [Google Scholar]
- Sperlich D, Pfriem P. Chromosomal polymorphism in natural and experimental populations. In: Ashburner M, Carson HL, Thompson JR, editors. The Genetics and Biology of Drosophila. 3e. Academic Press; New York: 1986. pp. 257–309. [Google Scholar]
- Stalker HD. Chromosome Studies in Wild Populations of Drosophila melanogaster II Relationship of Inversion Frequencies to Latitude, Season, Wing-Loading and Flight Activity. Genetics. 1980;95:211–223. doi: 10.1093/genetics/95.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Takano-Shimizu T. Divergent enhancer haplotype of ebony on inversion In(3R)Payne associated with pigmentation variation in a tropical population of Drosophila melanogaster. Mol Ecol. 2011;20:4277–4287. doi: 10.1111/j.1365-294X.2011.05260.x. [DOI] [PubMed] [Google Scholar]
- Telonis-Scott M, Hoffmann AA, Sgrò CM. The molecular genetics of clinal variation: a case study of ebony and thoracic trident pigmentation in Drosophila melanogaster from eastern Australia. Mol Ecol. 2011;20:2100–2110. doi: 10.1111/j.1365-294X.2011.05089.x. [DOI] [PubMed] [Google Scholar]
- Tennessen JM, Barry WE, Cox J, Thummel CS. Methods for studying metabolism in Drosophila. Methods. 2014;68:105–115. doi: 10.1016/j.ymeth.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau T. Package “coxme”: Mixed effects Cox models. R package version 2.2–5. 2012 Available at: https://cran.r-project.org/web/packages/coxme/coxme.pdf.
- Tsuneizumi K, Nakayama T, Kamoshida Y, Kornberg TB, Christian JL, Tabata T. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature. 1997;389:627–631. doi: 10.1038/39362. [DOI] [PubMed] [Google Scholar]
- Umina PA, Weeks AR, Kearney MR, McKechnie SW, Hoffmann AA. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science. 2005;308:691–693. doi: 10.1126/science.1109523. [DOI] [PubMed] [Google Scholar]
- van der Linde K, Houle D. Inferring the Nature of Allometry from Geometric Data. Evol Biol. 2009;36:311–322. [Google Scholar]
- van’t Land J, Van Putten P, Zwaan B, Kamping A, van Delden W. Latitudinal variation in wild populations of Drosophila melanogaster: heritabilities and reaction norms. J Evol Biol. 1999;12:222–232. [Google Scholar]
- Weeks AR, McKechnie SW, Hoffmann AA. Dissecting adaptive clinal variation: markers, inversions and size/stress associations in Drosophila melanogaster from a central field population. Ecol Lett. 2002;5:756–763. [Google Scholar]
- Zwaan BJ, Azevedo R, James AC, Land JV. Cellular basis of wing size variation in Drosophila melanogaster: a comparison of latitudinal clines on two continents. Heredity. 2000;84:338. doi: 10.1046/j.1365-2540.2000.00677.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.