Abstract
N'-substituted 1,2-diaminoethylphosphonic acids and 1,2-diaminoethylphosphinic dipeptides were explored to unveil the structural context of the unexpected selectivity of these inhibitors of M1 alanine aminopeptidases (APNs) versus M17 leucine aminopeptidase (LAP). The diaminophosphonic acids were obtained via aziridines in an improved synthetic procedure that was further expanded for the phosphinic pseudodipeptide system. The inhibitory activity, measured for three M1 and one M17 metalloaminopeptidases of different sources (bacterial, human and porcine), revealed several potent compounds (e.g., Ki = 65 nM of 1u for HsAPN). Two structures of an M1 representative (APN from Neisseria meningitidis) in complex with N-benzyl-1,2-diaminoethylphosphonic acid and N-cyclohexyl-1,2-diaminoethylphosphonic acid were determined by the X-ray crystallography. The analysis of these structures and the models of the phosphonic acid complexes of the human ortholog provided an insight into the role of the additional amino group and the hydrophobic substituents of the ligands within the S1 active site region.
Keywords: metalloaminopeptidases, aminopeptidase N, Neisseria meningitidis, phosphonic and phosphinic acids, APN-inhibitor complex structures, S1 binding mode
Graphical Abstract
Introduction
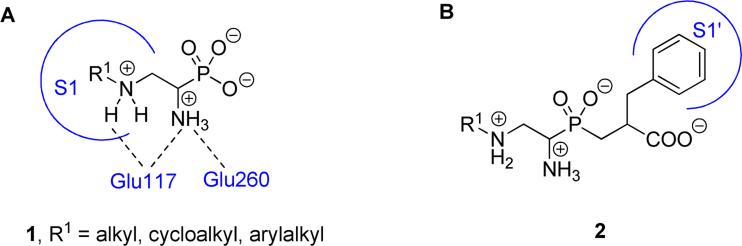
A detailed substrate fingerprint study [1] has recently revealed a broad substrate tolerance of the S1 pocket of the M1 alanine metalloaminopeptidase from Neisseria meningitidis (NmAPN) – a gram-negative diplococcus bacterium that is considered the major causative agent of meningitis and other meningococcal diseases [2-4]. Fluorogenic substrates, derived from unnatural α-amino acids that comprise bulky hydrophobic P1 side-chain residues bearing a fragment of a basic character, were ranked among the most favored ligands. A similar hydrophobic and base-oriented P1 specificity profile was observed for mammalian (human and porcine) [5] and protozoan (Plasmodium falciparum [6] and Eimeria tenella [7]) alanine aminopeptidases, the abundantly spread metallohydrolases of multiple functions and medical implications [8-10]. The privileged features of the amino acid substrates can be readily translated into the structure of potential inhibitors. The α-aminoalkylphosphonic acids provide an opportunity to install the preferred P1 substituents on the N-C-P scaffold and are commonly recognized as transition state analogue inhibitors of zinc metalloaminopeptidases [11]. However, the insertion of an additional heteroatom-based group into the substituent structure is a separate and not trivial task, in particular to be performed in a parallel manner. One such convenient modification is aziridinephosphonate ring opening to yield N'-substituted 1,2-diaminoethylphosphonic acids, which was originally proposed to provide inhibitors of metalloaminopeptidases from the porcine kidney [12]. The compounds contain an extra β-amino group that modifies the character of the P1 substituent to basic. Indeed, several compounds were found to be good inhibitors of mono-zinc alanyl aminopeptidase and discriminate versus two zinc atom-containing leucine aminopeptidase (LAP), for which they exhibited poor or no inhibition [12]. This was a quite unique observation, as the structural fragment H2N-C-PO2 typically provides much more effective complexation systems for the two zinc ions in LAP than for the single one in APNs [11,13]. Apparently, the additional β-amino group does not allow convenient P1-S1 side-chain docking (hydrophobic residues are strongly preferred) and distorts the overall binding mode to this particular aminopeptidase.The precise reasons for the good affinity to the porcine APN remain elusive. For NmAPN, we recently postulated that an additional heteroatom group located in the proximity of the side chain is able to enhance interactions with the glutamate-rich anionic site that is formed by Glu117, Glu260 and Glu316 and typically dedicated to the binding the N-terminus of peptidic substrates (Fig. 1, panel A) [1].
Figure 1.
Structure of N'-substituted 1,2-diaminoethylphosphonic acids (1) and 1,2-diaminoethylphosphinic pseudodipeptides (2) and the presumed role of the amino groups in binding to NmAPN.
As N'-substituted 1,2-diaminoethylphosphonic acids (1) are also promising lead compounds to obtain specific and potent inhibitors of alanyl aminopeptidase by the P1' elongation to phosphinic dipeptides (2, Fig. 1, panel B), we have decided to revisit the issue. An extensive series of novel compounds was obtained in an improved synthetic approach. Their inhibitory activity was tested towards four enzymes: NmAPN, human (HsAPN) and porcine APN (SsAPN), and LAP. Finally, the crystal structures of two inhibitor complexes (NmAPN-1h, and NmAPN-1n) were determined by X-ray crystallography and compared against modeled NmAPN-inhibitor and HsAPN-inhibitor complexes, providing structural insight of the ligand-protein interactions and inhibition modes. The potential of the P1' extension was evaluated for selected N' substituents.
Results and discussion
Synthesis of 1,2-diaminoethylphosphonic acids
The α,β-unsaturated system of dialkyl vinylphosphonates 3 (Scheme 1) is the usual substrate for the synthesis of 1,2-diaminoethyl derivatives 1 in a pathway via aziridinephosphonates. In the classical Gabriel-Cromwell method, bromine is readily added to the unsaturated bond to produce 1,2-dibromophosphonate esters 4 [14]. Then, consecutive eliminations of HBr with gaseous and liquid ammonia yield the heterocyclic system 5 in the historical literature method (Scheme 1, pathway A) [14]. Nucleophilic substitution with a primary or secondary amine enables the aziridine ring opening [15]. However, the approach is characterized by at least two shortcomings. First, ammonia is the reagent of choice to obtain the non-substituted aziridine, but the elimination is capricious and not pure [14,16]. Second, for not fully clear reasons, esters of aziridinephosphonate 5 are very resistant to the nucleophilic attack of an amine and ring opening. As a consequence, they demand hydrolysis to free acids 6 prior to substitution [15,16]. These complications were associated with problematic purification, as a careful ion-exchange chromatography was applied after each of those steps.
Scheme 1.
Reagents and conditions: a) Br2, CH2Cl2; b) gaseous NH3 then liquid NH3 (or NH3/H2O followed by NaOH/ H2O); c) HCl/H2O; d) R1(R2)NH, EtOH; e) TsNH2, PhI=O, CuCl, MS 4 Å, MeCN/MeNO2 (4:1, vv); f) BzlNH2; g) Et3N then Ph(R)CHNH2; h) H2, 10% Pd/C, MeOH; i) R1(R2)NH, H2O/dioxan; j) H+/RP-HPLC. For the expanded structures of R1 and R2, see Table 1.
N-Tosylaziridines are considered much more susceptible to nucleophilic substitution than their non-modified counterparts. Accordingly, our first alternative approach involved the verification of N-tosylaziridinephosphonates 7 as convenient intermediates (Scheme 1, pathway B). Indeed, we managed to open a tosylated phosphonate diester with benzylamine in a simple and clean manner. However, obtaining such a material (7) by nitrene-mediated aziridination appears to be far more problematic. Contrary to the literature data, different versions of both metal and non-metal assisted oxidative additions of Chloramine T (N-chlorotosylamide sodium salt) to the double bond of 3 are ineffective [17-19]. Extensive optimization of the reaction conditions with other tosyl precursors and hypervalent iodobenzene-based reagents [20-22] finally led to the desired products under copper(I) catalysis [21], but in a very poor yield (< 10% after column chromatography). Although the subsequent substitution to compounds 8 looked promising, we decided to abandon the approach.
In such circumstances, the elimination option was reconsidered, but ammonia was replaced by Et3N (the first stage) and subsequently by a benzylamine derivative (the second step) [23]. The use of benzyl or α-methylbenzylamine gave N-substituted aziridinephosphones 9 in a reasonable yield (Scheme 1, pathway C). The benzylamine derivatives were chosen as they could be readily hydrogenated to free aziridine 5. Diphenylmethylamine (benzhydrylamine) did not work to the same extent, apparently because of the decreased nucleophilicity and steric hindrance of the amino group. Products 9 were easily purified by flash chromatography on silica. The subsequent catalytic hydrogenation was performed in standard conditions prior to the ring opening with selected amines. The two-step procedure appeared more practical to obtain 5 than the direct use of ammonia.
An excess of the reacting amine, harsh reaction conditions and the use of a water medium in place of an organic solvent ensured the substitution without the prerequisite hydrolysis of the ester groups. As a matter of fact, both transformations proceeded simultaneously, and the 1,2-diaminoethylphosphonate products were in the form of ammonium salts of phosphonate monoesters 10 and preparatively separated after HPLC chromatography to afford 11. Apparently, the monohydrolysis occurring in basic aqueous conditions sufficiently increased the susceptibility of the aziridine to substitution for it to be opened without requiring exhaustive hydrolysis. Different primary and secondary structurally diversified (linear, branched, cyclic and benzyl, including heteroatom-substituted, listed in Scheme 1 and Table 1) amines reacted readily. In the cases of difunctional amines, the high excess of the nucleophilic component ensured mono-substitution, including evident regioselectivity. For example, the aromatic amino function of 4-aminobenzylamine remained non-modified, as evidenced in products 11r.
Table 1.
Structure-activity relationship for 1,2-diaminoethylphosphonic acid inhibitors of N. meningitides APN and mammalian aminopeptidases: porcine and human APNs and porcine LAP (NI – no inhibition up to 0.8 mM inhibitor concentration). In the cases of compounds previously tested toward SsAPN and LAP [12] the originally measured activity is presented, unless the difference exceeds 25%.
| No. |

|
Ki [μM] | ||||
|---|---|---|---|---|---|---|
| R1 | R2 | NmAPN | SsAPN | HsAPN | LAP | |
| 1a | H | H | 0.589 ± 0.17 | 2.88 ± 0.43 (19.0 [12]) | 10.6 ± 2.2 | NI [12] |
| 1b | Me | H | 29.6 ± 4.4 | 46.0 [12] | 7.42 ± 0.96 | NI [12] |
| 1c |
|
H | 6.72 ± 1.1 | 1.91 [12] | 1.66 ± 0.13 | NI [12] |
| 1d |

|
H | 23.3 ± 0.6 | 1.71 [12] | 2.51 ± 0.29 | NI [12] |
| 1e |
|
H | 1.11 ± 0.11 | 1.21 ± 0.22 | 0.297 ± 0.093 | 119 ± 14 |
| 1f |
|
H | 43.1 ± 5.4 | 32.4 ± 3.9 | 41.1 ± 2.5 | 289 ± 23 |
| 1g |

|
H | 26.0 ± 1.4 | 19.3 ± 0.46 | 5.74 ± 0.26 | 72.6 ± 6.0 |
| 1h |

|
H | 2.38 ± 0.28 | 0.87 [12] | 0.691 ± 0.046 | NI [12] |
| 1i |

|
H | 1.68 ± 0.03 | 0.520 ± 0.040 | 0.397 ± 0.077 | 20.8 ± 2.7 |
| 1j |

|
H | 26.5 ± 3.7 | 27.7 ± 1.6 | 13.5 ± 0.70 | 240 ± 8.6 |
| 1k |

|
H | 9.67 ± 0.49 | 3.64 ± 0.38 | 0.782 ± 0.015 | 69.1 ± 6.8 |
| 11 |

|
H | 11.4 ± 1.7 | 9.39 ± 0.80 | 3.29 ± 0.52 | 68.4 ± 6.8 |
| 1m |

|
H | 27.7 ± 0.46 | 7.00 ± 0.89 | 3.22 ± 0.15 | 55.8 ± 7.6 |
| 1n |

|
H | 16.6 ± 0.71 | 2.30 [12] | 0.341 ± 0.091 | 350 [12] |
| 1o |

|
H | 15.4 ± 0.97 | 14.5 ± 2.4 (2.80 [12]) | 0.391 ± 0.11 | 600 [12] |
| 1p |

|
H | 1.42 ± 0.050 | 6.58 ± 0.25 | 2.55 ± 0.37 | 71.1 ± 1.2 |
| 1q |

|
H | 3.00 ± 0.035 | 6.04 ± 0.81 | 1.22 ± 0.18 | 108 ± 7.8 |
| 1r |

|
H | 4.79 ± 0.18 | 2.49 ± 0.057 | 1.49 ± 0.19 | 7.02 ± 0.42 |
| 1s |

|
H | 4.32 ± 0.52 | 2.29 ± 0.046 | 0.357 ± 0.036 | 26.2 ± 2.0 |
| 1t |

|
H | 4.19 ± 1.2 | 5.42 ± 0.28 | 2.89 ± 0.43 | 26.5 ± 2.1 |
| 1u |

|
H | 2.33 ± 0.14 | 0.723 ± 0.040 | 0.065 ± 0.009 | 19.5 ± 2.4 |
| 1v |

|
12.5 ± 2.1 | 14.7 [12] | 6.04 ± 0.38 | 1410 [12] | |
| 1w |

|
197 ± 8.9 | 94.2 ± 11 | 37.7 ± 1.6 | NI | |
Acidic conditions, applied to remove the remaining alkyl ester group, completed the reaction sequence. Hydrogenation, ring opening/monohydrolysis and the final hydrolysis could be also performed in a convenient one-pot manner. Each step proceeded efficiently and can be easily controlled by 31P NMR, so that the removal of solvents is the only operation demanded before the following stage. The separation of the target compounds 1 involves standard treatment with propylene oxide. Analytical samples could be conveniently recrystallized from water. In summary, even though the overall procedure is extended by one step, it seems to be advantageous to the approach described previously. The detailed synthetic procedure together with the spectroscopic and analytical data for novel compounds is given in the Supporting Materials.
SAR studies and binding modes of 1,2-diaminoethylphosphonic acids
Amines of a different structure were used to explore the efficiency of binding of the N' substituent of 1,2-diaminoethylphosphonic acids to the NmAPN and the reference aminopeptidases (Table 1). The collection included the unsubstituted compound 1a, being the product of the reaction of aziridine with ammonia, alkyl (1b-e), cycloalkyl (1h and 1i) and phenylalkyl derivatives (1n-q). The three series of compounds were further diversified with representatives containing an additional functionalization: 1f and 1g for alkyls, 1j-m for cycloalkyls and 1r-u for arylalkyls. The functionalization mainly involved an extra terminal amino group or an ether function, which were dedicated to forming interactions with the distal part lining the S1 binding region of NmAPN. Finally, piperazine (1v) and morpholine analogues (1w) were derived from cyclic secondary amines.
The studied compounds were found to be good, generally low-micromolar-range competitive and fast-binding inhibitors of NmAPN. Unsubstituted 1,2-diaminoethylphosphonic acids (1a) appeared to be the most potent inhibitor, with a submicromolar Ki value (0.6 μM). The binding mode of 1a with NmAPN obtained by molecular modeling (Fig. S1) principally confirmed the assumed interactions of the amino groups with the active site glutamate residues. Despite unfavorable electrostatic repulsions both positively charged nitrogen atoms are aimed at Glu117 and Glu260 (compare also Fig. 1, panel A). Compounds derived with bulky, extended alkyl, cycloalkyl and arylalkyl fragments (1e, 1h, 1i, 1p and 1q) were only slightly less potent (Ki = 1.1-3.0 μM). The presence of a smaller alkyl chain (1b-d), a heterocyclic ring (k-m), a benzyl portion (1n and 1o) and cyclic piperazine or morpholine (1v and 1w) diminished the activity, typically by 1-2 orders of magnitude. Interestingly, an additional terminal amino or methoxy group located at the benzyl residue (compounds 1r and 1s) improved the affinity by 3-to-4-fold compared with the non-modified analogue 1n (4.2 and 4.8 μM versus 16.6 μM). A similar effect was achieved for a more flexible and basic 4-aminomethyl functionalization of benzylic R1 (1t, Ki = 4.2 μM). Twice as potent as the heteroatom-modified benzyls was the elongated 4-methoxy-functionalized phenylethyl fragment of compound 1u (Ki = 2.3 μM).
The actual binding mode of selected 1,2-diaminoalkylphosphonic acids to the active site of NmAPN was revealed by crystallographic studies (Fig. 2, panel A for N'-cyclohexyl and panel B for N'-benzyl derivative). The crystals of the recombinant enzyme soaked with an inhibitor do not show significant changes in the overall binding site architecture compared with the ligand-unbound protein [24], nor to those visualized for NmAPN complexed with 1-aminoalkylphosphonic acids [25]. Two negatively charged phosphonic oxygen atoms coordinate the catalytic zinc ion (2.41 and 2.23 Å for 1h and 2.07 and 2.20 Å for 1n), mimicking the gem-diolate transition state of the scissile peptide bond. One of them also interacts with the hydroxyl group of Tyr377 (2.55 Å for 1h and 2.52 Å for 1n), the key residue for the hydrolytic mechanism. Nevertheless, the phosphonate of 1,2-diaminophosphonic acids seems to be buried somewhat shallower than in the 1-amino analogues for which the corresponding distances are slightly shorter, typically, O-Zn below 2.1 Å and O-Tyr377 below 2.5 Å [25]. The fundamental contacts (the salt bridges) of the α-amino group with the glutamates responsible for the binding of the N-terminus are also well reproduced, in particular for the N'-benzyl derivative 1n, with distances 2.65 Å to Glu117 and Glu260. In general, the NCP fragment of compounds 1 binds to the NmAPN in a fairly predictable way with some minor distortions, apparently caused by the bulkiness of the N' substituent.
Figure 2.
Close-up views showing interactions of N'-cyclohexyl- (1h, panel A) and N'-benzyl-1,2-diaminoalkylphosphonic acid (1n, panel B) ligands with NmAPN as observed in the crystal structures. The hydrogen bonds and ligand-metal interactions are marked as green lines, distances are shown in Angstroms.
Large-size P1 residues fill the hydrophobic S1 cleft well. To our surprise, the β-amino group is indiscriminately positioned and plays a variable role in binding. For the N'-cyclohexyl residue of 1h, it is aimed at the interior of the pocket, and its hydrogen-bonding potential is somewhat shielded by the intramolecular alkyl and cycloalkyl surroundings. Nevertheless, it is directed straight to the negatively charged surface (compare also Fig. S1), although beyond the hydrogen bond distance (3.43 Å to carboxylate oxygen atom of Glu117). For the N'-benzyl, the β-amino functionality is located in the opposite conformation. It is also at the limit of the interatomic distances of hydrogen bonding to C=O of Ala258 and to the third, solvent-exposed, phosphonate oxygen atom (both distances 3.14 Å). Most probably these plausible interactions are not essential effects, as 1-amino-3-phenylpropylphosphonic acid (the homophenylalanine analogue), the compound missing the β-amino group, is bound in a quite similar manner (Fig. S2) but exhibits an improved activity (Ki = 1.26 μM [1]).
The range of inhibition obtained for porcine kidney SsAPN is quite similar to that measured for the N. meningitidis ortholog. In Table 1, the results obtained for novel compounds 1e, 1g, 1j-l and 1n are compiled with the data acquired previously (if currently measured Ki values are consistent with the literature data for compounds 1a-d, 1f, 1h, 1i and 1m [12] and do not differ by more than 25%, the original number is given; otherwise, both are included). For SsAPN, a much broader tolerance to different substitutions is visible. The majority of the studied inhibitors show a dissociation constant below 10 μM, with the newly synthesized cycloheptyl derivative 1g being the most potent (Ki = 0.5 μM).
Far more impressive data in terms of the inhibition constant value were collected for human alanine aminopeptidase. Several aliphatic compounds display Ki below 1 μM, being equally (N'-n-propyl, 1c, N'-cyclohexyl, 1h, and N'-cycloheptyl, 1i) or more active (N'-n-hexyl, 1e, Ki = 0.3 μM) than for the highly homologic porcine APN. Significantly, arylalkyl residues are much better accepted for human than for pig APN. N'-Benzyl, 1n, N'-(α-methylbenzyl), 1o, and N'-(4-methoxybenzyl), 1s, displayed strong inhibition, with Ki values falling in a submicromolar range (all values below 0.4 μM). The higher homolog of the last compound (N'-2-(4-methoxyphenyl)ethyl, 1u) appeared to be the most potent compound found in this study (Ki = 65 nM). To the best of our knowledge, it is the most active amino acid analogue inhibitor of HsAPN reported to date. To discuss the presumed binding mode and the reasons for the potency of selected compounds, molecular modeling studies were performed. The crystal structure of alanyl aminopeptidase from H. sapiens [26] was used to dock the ligand and analyze the interactions. The HsAPN-1u complex (Fig. 3) shows a typical pattern of contacts of the ligand N-C-P fragment with the enzyme, similar to those described above for NmAPN, namely, the involvement of two P-O oxygen atoms in Zn complexation and the ion pairing / hydrogen bonding of the α-amino with glutamates / glutamine (Glu355, Glu414 and Gln213). Interestingly, the β-amino group adapt a conformation similar to that evidenced in the crystal structure of the N'-benzyl-1,2-diaminoalkylphosphonic acid-NmAPN complex. The group is exposed externally out of the S1 enzyme cavity and tentatively linked with the solved-aimed oxygen atom via an intramolecular hydrogen bond. The (4-methoxyphenyl)ethyl fragment fits particularly well to the S1 binding site, filling it very tightly (Fig. 3 and Graphical Abstract). The aromatic ring is surrounded by the phenyl of Phe348 (edge to face) and the amide groups of Gln211 and Asn350. The electron-rich character of the aromatic ring definitely improves the contacts with the neighboring residues. The ether oxygen atom is in proximity to the N-terminal amide N-H of Asn350, but the potential hydrogen bonding can be a vague suggestion because of a not favored geometry. Suggestion of the interaction between the inhibitor oxygen atom of OMe and the side-chain amide NH2 group of Asn350 seems to be more justified for compound 1s, a methylene group shorter homologue of 1u. The high activity of inhibitor 1s (Ki = 357 nM) can be attributed to this specific bond (Fig. S3) as the overall P1-S1 fit is not so perfectly tight as modeled for 1u. The latter statement is also valid for the N'-2-phenylethyl substituent of 1q, lacking OMe group compared to 1u (Fig. S4), which gives rise to a moderate inhibitory potency. To conclude, the length and character of the N'-2-(4-methoxyphenyl)ethyl portion can be considered optimal to bind to the S1 pocket of HsAPN.
Figure 3.
A model of the complex of N'-[2-(4-methoxyphenyl)ethyl]-1,2-diaminoalkylphosphonic acid (1u) with HsAPN showing P1-S1 interactions. Intra- and intermolecular hydrogen bonds and ligand-metal interactions are marked in green.
Discussing the selectivity issues between APNs from different sources, several interesting discrimination cases can be observed. For example, unsubstituted derivative 1a clearly favors binding to NmAPN. The difference in inhibition constants is approximately 5-fold to porcine and 20-fold to human enzyme. The opposite cases (mammalian APNs versus the bacterial one) are even more pronounced. For example, compound 1d (N'-isopropyl) exhibits over an order of magnitude higher potency in favor of SsAPN and HsAPN than NmAPN. Certain inhibitors preferentially bind to only the human ortholog. The N'-α-methylbenzyl (1o) derivative distinguishes human aminopeptidase, with Ki = 0.4 μM versus Ki = 15 μM for the other two APNs. The much more potent N'-[2-(4-methoxyphenyl)ethyl]-1,2-diaminoalkylphosphonic acid (1u) is also at least 10-fold more active for HsAPN than for the alanyl aminopeptidase of other organisms.
The selectivity ratio between the APNs and LAP is indeed significant. In general, N'-alkyl-and N'-cycloalkyldiaminoethylphosphonic acids do not inhibit (up to 800 μM inhibitor concentration) or poorly inhibit porcine kidney leucine aminopeptidase, whereas they appear to be quite active for APNs (Ki ~ 1 μM). The cyclohexyl derivative 1h is the most significant example. It is a low micromolar inhibitor of three alanyl aminopeptidases and does not influence the LAP activity at all. In this and similar instances, the selectivity ratio can be estimated to be three orders of magnitude. When the effect of LAP inhibition is measurable, the ratio is typically 1.5-2 orders of magnitude (compare, for example, the Ki values found for compound 1e: 1.1 μM for NmAPN, 1.2 μM for SsAPN and 0.3 μM for HsAPN versus 119 μM for LAP). Only individual compounds (arylalkyl) show somewhat significant affinity toward LAP. Among them, the N'-4-aminobenzyl analogue (1k) can be considered a universal inhibitor of aminopeptidases, with all the Ki values between 2.5 and 7.0 μM.
Synthesis and activity of 1,2-diaminoethylphosphinic-derived dipeptides
In continuation, we undertook an effort to adapt the optimized protocols of obtaining and opening the corresponding aziridine system for a suitable phosphinic dipeptide precursor. Such a building block should contain an appropriate P1' fragment. The aromatic ring of the benzyl-derived portion is favorably bound in a shallow and hydrophobic S1' cleft [27], so we selected unsaturated compound 16 and aziridine 18 (Scheme 2) as the target scaffolds. The synthetic challenge to prepare vinylphosphinate 16 involves a multistep approach starting from methyl α-benzylacrylate 12 [28,29]. Diethyl phosphite was readily added to the acrylate in a phospha-Michael reaction under strong base catalysis [30] to obtain 13. The triester was selectively monodealkylated with NaI in acetone [31] to the mixed diester 14. Subsequently, the phosphonic monoacid function was converted into the corresponding chloridate 15 by thionyl chloride [32]. Without preparative separation, phosphonochloridate was reacted with vinylmagnesium chloride [33], the Grignard compound, to form the demanded vinyl-to-phosphorus portion. The following steps of the procedure were analogous to those elaborated for the amino acid analogs. They involved the addition of bromine to yield 17 and ring closure to aziridine 18. The last compound was heated in parallel with selected amines (or ammonia) to obtain the N'-substituted diaminoethyl fragment of 19. Catalytic hydrogenation and a final hydrolysis led to the target products 2.
Scheme 2.
Reagents and conditions: a) t-BuOK, HP(O)(OEt)2, MeOH; b) NaI, acetone; c) SOCl2, CH2Cl2; d) CH2=CHMgCl, CH2Cl2; e) Br2, CH2Cl2; f) Et3N then Ph(R)CHNH2; g) R1NH2, H2O/dioxan; h) H2, 10% Pd/C, MeOH; i) HCl/H2O.
As evidenced by previous studies on organophosphorus inhibitors, more potent inhibitors of aminopeptidases are those extended to the P1' fragment rather than simple amino acid analogues designed to bind only within the S1 region [11,25,27,34]. This is not the case in the current study. The phosphinic dipeptides 2, although containing selected favorable N'-substituents, appeared to be less potent inhibitors of alanyl aminopeptidases than the corresponding phosphonic amino acids 1 (Table 2). A particular drop in activity was evidenced for NmAPN. The Ki values increased by 1-2 orders of magnitude and exceeded 100 μM for the majority of compounds. A minor loss of activity was evidenced for the porcine APN, typically a 2-5-fold decrease. For the human enzyme, the results obtained for dipeptides are comparable with those acquired for the amino acid analogues. N'-Alkyl and cycloalkyl derivatives 2b-e showed good affinity, with Ki values between 0.2 and 1.1 μM (the bulkier the better). Each of these results was slightly improved when compared with corresponding compounds 1c, 1d, 1i and 1j comprising only the P1 substituent (Ki = 0.4-2.5 μM). The affinity of the most potent amino acid compound 1u did not change (within the experimental error) upon the structure elongation and remained at a very good nanomolar level for 2f.
Table 2.
Structure-activity relationship for 1,2-diaminoethylphosphinic dipeptide inhibitors of N. meningitidis APN and mammalian aminopeptidases.
| No. |

|
Ki [μM] |
|||
|---|---|---|---|---|---|
| NmAPN | SsAPN | HsAPN | LAP | ||
| 2a | R1 = H | 171 ± 5.5 | 29.0 ± 2.3 | 4.45 ± 0.50 | 64.4 ± 2.6 |
| 2b |
|
131 ± 16 | 6.74 ± 0.54 | 1.11 ± 0.057 | 21.0 ± 2.5 |
| 2c |
|
101 ± 13 | 4.54 ± 0.26 | 0.499 ± 0.031 | 15.2 ± 2.6 |
| 2d |

|
114 ± 14 | 3.83 ± 0.43 | 0.275 ± 0.040 | 28.3 ± 2.5 |
| 2e |

|
109 ± 13 | 3.24 ± 0.41 | 0.207 ± 0.019 | 23.8 ± 2.4 |
| 2f |
|
39.5 ± 9.6 | 1.48 ± 0.18 | 0.078 ± 0.009 | 21.1 ± 2.6 |
The affinity of pseudodipeptidic compounds 2 towards LAP flattened. Those amino acids that exhibited a moderate potency (Ki ~ 20 μM, 1i and 1u) retained the activity upon the structure elongation (2e and 2f, respectively). Those that were inactive (1c, 1d and 1h) gained affinity after the P1' addition (2b-d) to the aforementioned level. Only the N'-unsubstituted derivative 2a displayed three-fold worse inhibition parameters than the other dipeptides. This improvement is definitely associated with benzyl incorporation, a favorable residue accommodated in the S1' binding sub-site. Participation of the P1' part of the molecules in interactions with LAP seems to be decisive in determining the compound activity, which does not depend on the P1 portion structure.
Discussion and conclusions
A careful and thorough optimization of the P1 and P1' substituent structure (modification/extension) in phosphorus-based transition state inhibitors and the examination of their interactions with the corresponding sub-sites has led to the discovery of very potent and selective inhibitors of metallo-dependent aminopeptidases, including APNs [25,27,34]. Although synthetically challenging, this method seems to be a rational and attractive approach for the construction of new ligands and an alternative to the elongation of pseudopeptidic sequences [35]. The selectivity issues are not trivial aspects, as the amino acid content and architecture of the APN active sites are well conserved between species. Nevertheless, certain structural nuances can be explored by systematic guest-host matching, which in turn demands access to extensive libraries of compounds. The parallel opening of aziridine phosphonates/phosphinates with amines fulfills these requirements. We have presented here an improved and versatile version of the reaction, validated for both amino acid and dipeptide analogues. It enables the preparation of a vast collection of diaminoethyl compounds, substituted at the β-amino group. The substitution is defined by the structure of the amine substrates, which include alkyls, cycloalkyls and arylalkyls, also variously modified. Very potent inhibitors were identified among the N'-derived 1,2-diaminoethylphosphonic acids, and what is equally important, the compounds showed satisfactory selectivity. In particular, the studied bacterial ortholog, NmAPN, could be exclusively inhibited by the non-substituted compound 1a. N'-Arylalkyls (e.g., α-methylbenzyl in compound 1o) are specific residues that give rise to Ki constants at least one order of magnitude lower for human APN than for the remaining two enzymes. One member of this group (N'-[2-(4-methoxyphenyl)ethyl]-substituted phosphonic acid 1u) appeared to be the most potent inhibitor revealed in the study (Ki = 65 nM). N'-substitution with an extended alkyl and cycloalkyl (e.g., 1e and 1i) produced universal ligands with good affinity to all three bacterial and mammalian APNs. Less bulky derivatives (1c, 1d and 1h) of an aliphatic character retain their activity towards APNs and do not inhibit LAP at all, even though both aminopeptidases show a quite similar substrate specificity [36,37]. The SAR results of analogous compounds, alkyl and cycloalkyl-substituted α-aminophosphonic acids (missing the β-amino group), are mirror-reflected: they display a low micromolar and submicromolar inhibition of LAP and poor inhibition of APN (porcine kidney) (Fig. 4) [12,38].
Figure 4.
Preference of the P1 substituents in aminophosphonic acid inhibitors of neutral aminopeptidases, a heteroatom-modified cycloalkyl of 1h favored by APNs and a typically lipophilic cycloalkyl favored by LAP [12,38].
Accordingly, the motivation of our study on diaminoethylphosphonic/phosphinic compounds was also focused on addressing the basic question of the structural reasons for such an unusual behavior. The crystal structures of a few ligands complexed with NmAPN did not provide a definite answer on the role of the β-amino group in binding. Instead, two possible conformations were envisaged (Fig. 2 and S1). For the N'-cyclohexyl derivative 1h (Fig. 2A), the β-amino group is buried in the S1 pocket without any close contacts, yet it is clearly pointed at the negatively polarized glutamate region, in particular at Glu117. This is in general accordance with our hypothesis depicted in Fig. 1A, confirmed by molecular modelling for unsubstituted analogue 1a (Fig. S1), and interactions displayed by certain disubstituted ligands, including diaminocarboxylic compounds complexed with Escherichia coli APN [39].
In the structure of N'-benzyl compound 1n (Fig. 2B), the β-amino group is exposed outside of the S1 cavity of NmAPN. In this case, the ligand conformation is the most likely to be stabilized by an intramolecular interaction with the phosphonic acid and a hydrogen bond with a neighboring residue (Ala258). Similar arrangements could be observed in three modeled inhibitor-HsAPN complexes (Fig. 3, S3, and S4). These interactions might produce a certain energetic gain upon complexation. However, taking into account the number of hydrogen bonds with water that need to be released upon the inhibitor entry to the active site, the formation of new weak contacts is only moderately profitable.
The lack of potency of diaminoalkylphosponates towards LAP is easier to explain. The overall binding mode of the N-C-P ligands to the two-zinc containing aminopeptidase is quite different. LAP does not contain such a precisely defined acidic region. Instead, Asp273 and one of the metal ions are involved in the binding of the N-terminal amino group. This is the supplementary role of the zinc apart from participation in the complexation of the gem-diolate transition state product of the scissile bond hydrolysis. This was confirmed by the crystal structure of transition state inhibitor, the phosphonic analogues of leucine bound to this aminopeptidase [40]. An additional β-amino functionality would definitely alter the favorable and fragile interaction network.
The decrease in potency, in particular towards NmAPN, observed for phosphinic dipeptides 2 upon extension of the diaminoethylphosphonate structure 1 by the P1' fragment seems to be the most unexpected result revealed in this study. Typically, tightening organophosphorus ligand-APN interactions by P1'-S1' contacts provides at least a one to two order of magnitude gain in the binding constants [11,25]. Molecular modeling has somewhat clarified this issue. The steric fit of diaminoalkylphosphinic dipeptide possessing the additional β-amino group within the S1' subsite has to be virtually identical to that evidenced for the phosphinic analogue of hPhePhe, a potent inhibitor of NmAPN (Ki = 302 nM) [25]. Most probably the difference concerns the S1 region. Superimposition of two enzyme-inhibitor structures containing either N'-benzyl-1,2-diaminoalkylphosphonic acids (Fig. 2B) or the phosphinic analogue hPhePhe revealed that the P1 benzyl fragments of the two compounds adopt quite comparable conformations (Fig. 5). However, for 1n, an intramolecular interaction P-O...HN that has been proposed for the amino acids is not allowed in the case of the dipeptide – the phosphonic oxygen involved in the bonding is replaced with a methylene linking the P1' portion. Typically, this oxygen atom is not involved in any specific contact, but in our case, its removal visibly alters the profitability. In general, this discussion adequately illustrates the subtlety of the design and development of bioactive ligands at the molecular level. Despite the rational approaches, the actual behavior of the molecules is governed by nuances that are difficult to predict.
Figure 5.
Superimposition of N'-benzyl-1,2-diaminoalkylphosphonic acid (1n) structure with phosphinic hPhePhe analogue complexed with NmAPN ([25], PDB code 4QME). Intermolecular hydrogen bonds and ligand-metal interactions are marked in green.
Supplementary Material
Highlights.
Convenient approach to 1,2-diaminoethylphosphonates and phosphinates was elaborated.
The SAR studies confirmed an unexpected inhibition profile of APNs versus LAP.
Inhibitor complexes with N. meningitides APN showed two alternative binding modes.
Acknowledgments
The work was financed by a statutory activity subsidy from the Polish Ministry of Science and Higher Education for the Faculty of Chemistry of Wrocław University of Technology. Ewelina Węglarz-Tomczak was supported by a grant from the Polish National Science Centre (Grant UMO-2012/05/N/ST5/01145). The Biovia Discovery Studio package was used under a Polish country-wide license. The use of software resources (Biovia Discovery Studio program package) of the Wrocław Centre for Networking and Supercomputing is also kindly acknowledged. The Structural Biology Center beamlines at APS are supported by the U.S. Department of Energy Office of Biological and Environmental Research program under Contract DE-AC02-06CH11357. The structural studies were performed at the Midwest Center for Structural Genomics supported by the National Institutes of Health Grant GM094585. We gratefully acknowledge Dr. M. Soroka for samples of N'-substituted diaminoethylphosphonic acids from MSJZ87 collection (compounds 1a, 1b, 1e, 1f, 1o, 1v and 1w).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information
Details regarding preparation, purification and characterization of the compounds (experimental procedures and NMR, MS, and HPLC data; purity of the final compounds assessed at >95% by analytical reverse-phase HPLC using gradient elution) as well as the enzyme preparation, the kinetic data with the methodology used to calculate the inhibition constants, and crystallographic data collection and structural determination. This material is available free of charge via the Internet at http://.
Accession Codes. PDB codes for Neisseria meningitidis alanyl aminopeptidase complexed with organophosphorus inhibitors are as follows: 4QPE (compound 1h), 5DYF (compound1n).
References
- 1.Węglarz-Tomczak E, Poręba M, Byzia A, Berlicki Ł, Nocek B, Mulligan, Joachimiak A, Drąg M, Mucha A. An integrated approach to the ligand binding specificity of Neisseria meningitidis M1 alanine aminopeptidase by fluorogenic substrate profiling, inhibitory studies and molecular modeling. Biochimie. 2013;95:419–428. doi: 10.1016/j.biochi.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mace SE. Acute bacterial meningitis. Emerg. Med. Clin. North Am. 2008;26:281–317. doi: 10.1016/j.emc.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Harrison LH, Trotter CL, Ramsey ME. Global epidemiology of meningococcal disease. Vaccine. 2009;275:B51–B63. doi: 10.1016/j.vaccine.2009.04.063. [DOI] [PubMed] [Google Scholar]
- 4.Leimkugel J, Racloz V, Jacintho da Silva L, Pluschke G. Global review of meningococcal disease. A shifting etiology. J. Bacteriol. Res. 2009;1:6–18. [Google Scholar]
- 5.Drąg M, Bogyo M, Ellman JA, Salvesen GS. Aminopeptidase fingerprints. An integrated approach for identification of good substrates and optimal inhibitors. J. Biol. Chem. 2010;285:3310–3318. doi: 10.1074/jbc.M109.060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poręba M, McGowan S, Skinner-Adams TS, Trenholme KR, Gardiner DL, Whisstock JC, G J. To, Salvesen S, Dalton JP, Drąg M. Fingerprinting the Substrate Specificity of M1 and M17 Aminopeptidases of Human Malaria, Plasmodium Falciparum. PLoS One. 2012;7:e31938. doi: 10.1371/journal.pone.0031938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gras S, Byzia A, Gilbert FB, McGowan S, Drąg M, Silvestre A, Niepceron A, Lecaille F, Lalmanach G, Brossier F. Aminopeptidase N1 (EtAPN1), an M1 metalloprotease of the apicomplexan parasite Eimeria tenella, participates in parasite development. Eukaryotic Cell. 2014;13:884–895. doi: 10.1128/EC.00062-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mina-Osorio P. The moonlighting enzyme CD13: Old and new functions to target. Trends Mol. Med. 2008;14:361–371. doi: 10.1016/j.molmed.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wickström M, Larsson R, Nygren P, Gullbo J. Aminopeptidase N (CD13) as a target for cancer chemotherapy. Cancer Sci. 2011;102:501–508. doi: 10.1111/j.1349-7006.2010.01826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skinner-Adams TS, Stack CM, Trenholme KR, Brown CL, Grembecka J, Lowther J, Mucha A, Drąg M, Kafarski P, McGowan S, Whisstock JC, Gardiner DL, Dalton JP. Plasmodium falciparum neutral nminopeptidases: New targets for anti-malarials. Trends Biochem. Sci. 2010;35:53–61. doi: 10.1016/j.tibs.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Mucha A, Kafarski P, Berlicki L. Remarkable potential of the α-aminophosphonate/phosphinate structural motif in medicinal chemistry. J. Med. Chem. 2011;54:5955–5980. doi: 10.1021/jm200587f. [DOI] [PubMed] [Google Scholar]
- 12.Lejczak B, Kafarski P, Zygmunt J. Inhibition of aminopeptidases by aminophosphonates. Biochemistry. 1989;28:3549–3555. doi: 10.1021/bi00434a060. [DOI] [PubMed] [Google Scholar]
- 13.Mucha A, Drag M, Dalton JP, Kafarski P. Metalloaminopeptidase inhibitors. Biochimie. 2010;92:1509–1529. doi: 10.1016/j.biochi.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rengaraju S, Berlin KD. Slow nitrogen inversion due to intramolecular hydrogen bonding. Slow nitrogen inversion in diethyl 2-aziridinylphosphonate from the paramagnetic-induced shifts in the PMR spectra using tris(dipivalomethanato)europium (III), and solvent shifts. J. Org. Chem. 1972;37:3304–3310. [Google Scholar]
- 15.Zygmunt J. Aziridine-2-phosphonic acid, the valuable synthon for synthesis of 1-amino-2-functionalized ethanephosphonic acids. Tetrahedron. 1985;41:4979–4982. [Google Scholar]
- 16.Zygmunt J, Mastalerz P. Synthesis of 1-amino-2-hydroxyethanephosphonic acid, the phosphonic analog of serine. Pol. J. Chem. 1981;55:411–414. [Google Scholar]
- 17.Ando T, Kano D, Minakata S, Ryu I, Komatsu M. Iodine-catalyzed aziridination of alkenes using Chloramine-T as a nitrogen source. Tetrahedron. 1998;54:13485–13494. [Google Scholar]
- 18.Thakur VV, Sudalai A. N-Bromoamides as versatile catalysts for aziridination of olefins using Chloramine-T. Tetrahedron Letters. 2003;44:989–992. [Google Scholar]
- 19.Jain SL, Sharma VB, Sain B. An efficient transition metal-free aziridination of alkenes with Chloramine-T using aqueous H2O2/HBr. Tetrahedron Letters. 2004;45:8731–8732. [Google Scholar]
- 20.Kim DY, Rhie DY. Synthesis of α-aminoalkylphosphonates from vinylphosphonates via aziridinylphosphonates. Tetrahedron. 1997;53:13603–13608. [Google Scholar]
- 21.Inoue S, Okauchi T, Minami T. New synthesis of gem-bis(phosphono)ethylenes and their applications. Synthesis. 2003:1971–1976. [Google Scholar]
- 22.Liu Y, Che CM. [Fe(III)(F(20)-tpp)Cl] is an effective catalyst for nitrene transfer reactions and amination of saturated hydrocarbons with sulfonyl and aryl azides as nitrogen source under thermal and microwave-assisted conditions. Chemistry. 2010;16:10494–10501. doi: 10.1002/chem.201000581. [DOI] [PubMed] [Google Scholar]
- 23.Doğan O, Babiz H, Gözen G, Budak S. Synthesis of 2-aziridinyl phosphonates by modified Gabriel-Cromwell reaction and their antibacterial activities. Eur. J. Med. Chem. 2011;46:2485–2489. doi: 10.1016/j.ejmech.2011.03.037. [DOI] [PubMed] [Google Scholar]
- 24.Nocek B, Mulligan R, Bargassa M, Collart F, Joachimiak A. Crystal structure of aminopeptidase N from human pathogen Neisseria meningitides. Proteins. 2008;70:273–279. doi: 10.1002/prot.21276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vassiliou S, Węglarz-Tomczak E, Berlicki Ł, Pawełczak M, Nocek B, Mulligan R, Joachimiak A, Mucha A. Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases. J. Med. Chem. 2014;57:8140–8151. doi: 10.1021/jm501071f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong AHM, Zhou D, M Rini J. The X-ray crystal structure of human aminopeptidase N reveals a novel dimer and the basis for peptide processing. J. Biol. Chem. 2012;287:36804–36813. doi: 10.1074/jbc.M112.398842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grembecka J, Mucha A, Cierpicki T, Kafarski P. The most potent organophosphorus inhibitors of leucine aminopeptidase. Structure-based design, chemistry, and activity. J. Med. Chem. 2003;46:2641–2655. doi: 10.1021/jm030795v. [DOI] [PubMed] [Google Scholar]
- 28.Stetter H, Kuhlmann H. Eine einfache Herstellung von α-Alkylacrylsaure-estern. Synthesis. 1979:29–30. [Google Scholar]
- 29.Vassiliou S, Mucha A, Cuniasse P, Georgiadis D, Lucet-Levannier K, Beau F, Kannan R, Murphy G, Knäuper V, Rio M-C, Basset P, Yiotakis A, Dive V. Phosphinic pseudo-tripeptides as potent inhibitors of matrix metalloproteinases: a structure-activity study. J. Med. Chem. 1999;42:2610–2620. doi: 10.1021/jm9900164. [DOI] [PubMed] [Google Scholar]
- 30.Cristau H-J, Coulombeau A, Genevois-Borella A, Sanchez F, Pirat J-L. Preparation of phosphinodipeptide analogs as building blocks for pseudopeptides synthesis. J. Organomet. Chem. 2002:643–644. 381–391. [Google Scholar]
- 31.Hoffmann M. An efficient synthesis of dibenzyl and di-p-nitrobenzyl N-protected 1-aminoalkanephosphonates using isoureas. J. Prakt. Chem. 1988;330:820–824. [Google Scholar]
- 32.Mucha A, Kafarski P, Plenat F, Cristau H-J. The preparation of phosphono peptides containing a phosphonamidate bond. Tetrahedron. 1994;50:12743–12754. [Google Scholar]
- 33.Dunne KS, Bisaro F, Odell B, Paris J-M, Gouverneur V. Diastereoselective ring-closing metathesis: synthesis of P-stereogenic phosphinates from prochiral phosphinic acid derivatives. J. Org. Chem. 2005;70:10803–10809. doi: 10.1021/jo0518708. [DOI] [PubMed] [Google Scholar]
- 34.Mucha A, Drąg M, Dalton JP, Kafarski P. Metallo-aminopeptidase inhibitors. Biochimie. 2010;92:1509–1529. doi: 10.1016/j.biochi.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Noble F, Mothé A, Meudal H, Coric P, Danascimento S, Roques BP, George P, Fournié-Zaluski M-C. J. Med. Chem. 2000;43:1398–1408. doi: 10.1021/jm990483l. [DOI] [PubMed] [Google Scholar]
- 36.Turner AJ. Aminopeptidase N. In: Rawlings ND, Salvesen G, editors. Handbook of Proteolytic Enzymes. 3rd ed. Academic Press; Amsterdam: 2013. pp. 397–403. [Google Scholar]
- 37.Sträter N, Lipscomb WN. Leucyl Aminopeptidase (Animal). In: Rawlings ND, Salvesen G, editors. Handbook of Proteolytic Enzymes. 3rd ed. Academic Press; Amsterdam: 2013. pp. 1465–1470. [Google Scholar]
- 38.Drąg M, Grembecka J, Pawełczak M, Kafarski P. α-Aminoalkylphosphonates as a tool in experimental optimisation of P1 side chain shape of potential inhibitors in S1 pocket of leucine- and neutral aminopeptidases. Eur. J. Med. Chem. 2005;40(8):764–771. doi: 10.1016/j.ejmech.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 39.Gumpena R, Kishor C, Ganji RJ, Addlagatta A. Discovery of α,β- and α,γ-diamino acid scaffolds for the inhibition of M1 family aminopeptidases. ChemMedChem. 2011;6:1971–1976. doi: 10.1002/cmdc.201100298. [DOI] [PubMed] [Google Scholar]
- 40.Sträter N, Lipscomb WN. Transition state analogue L-leucinephosphonic acid bound to bovine lens leucine aminopeptidase: X-ray structure at 1.65 Å resolution in a new crystal form. Biochemistry. 1995;34:9200–9210. doi: 10.1021/bi00028a033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.