Abstract
Background
Epidemiological data on antibiotic susceptibility of Staphylococcus strains isolated from nasal carriers in each region can be helpful to select appropriate drugs to eradicate carriage states, control nosocomial infections and also treat patients.
Objectives
The current study aimed to investigate the antibiotic resistance profile and the molecular prevalence of the ermA, ermB, ermC and msrA genes among Staphylococcus strains isolated from the anterior nares of hospital employees.
Patients and Methods
In this cross-sectional study, a total of 100 Staphylococcus isolates, 51 Staphylococcus aureus, 49 coagulase-negative staphylococci (CoNS) were isolated from the anterior nares of hospital employees in Khorramabad, Iran. Susceptibility pattern to macrolide antibiotics were determined using the disk diffusion method. The polymerase chain reaction (PCR) assay was applied to determine the major erythromycin-resistant genes (ermA, ermB, ermC and msrA).
Results
Fifty-three (53%) isolates were simultaneously resistant to erythromycin, azithromycin and clarithromycin (cross-resistance); while 8 (8%) isolates had variable macrolide susceptibility pattern. Among the S. aureus isolates, the difference in prevalence of resistance to erythromycin between males and females was significant (P = 0.011). The frequency of ermA, ermB, ermC, and msrA genes were 3%, 5%, 33% and 20%, respectively. It was also found that out of 53 isolates resistant to erythromycin, 44 (83%) isolates (eight S. aureus and thirty-six CoNS strains) carried at least one of the four tested genes. Eight (8%) isolates had intermediate phenotype to erythromycin, in which 4 (50%) isolates carried ermB or ermC genes. In addition, out of 39 erythromycin-susceptible isolates, 3 (7.7%) isolates were positive for ermB or ermC genes.
Conclusions
No entire association was found between genotype and phenotype methods to detect macrolides-resistant isolates. In addition, distribution of genetically erythromycin-resistant isolates is geographically different among staphylococci. It is recommend removing S. aureus from nasal carriers by proved approaches such as local or systemic administration of effective antibiotics or bacterial interference.
Keywords: Coagulase-Negative Staphylococci, Erythromycin, Nasal, PCR, erm, Staphylococcus aureus
1. Background
Staphylococcus aureus and coagulase-negative staphylococci (CoNS) are well known to most clinicians as a cause of nosocomial and community-associated infections worldwide (1, 2). Staphylococci are able to develop resistance to many routinely used antibiotics to which they were susceptible first (1). Moreover, this condition has become more complicated because of various factors such as horizontal acquisition of foreign genetic elements and ability to establish persistent infections (e.g. biofilm formation) in the host. Nowadays, due to wide spread use of indwelling and prosthetic medical devices, CoNS have gained much attention as considerable agents involved in nosocomial infections (3). A major part of these infections are caused by the transmission of the bacteria from hospital employees as reservoir to patients. In particular, this situation can lead to serious infections in immunocompromised patients who are hospitalized with underlying diseases (4-8). Macrolides including erythromycin are the antibiotics used against Gram-positive and some Gram-negative bacteria. Three mechanisms in Gram-positive bacteria that result in resistance to erythromycin are as follows: (I) modification in the ribosomal target site, mediated by the methyltransferases encoded by erm (erythromycin resistance methylase) genes, (II) efflux pump encoded by msrA/B (macrolide specific resistance genes) and (III) ereA/B (erythromycin esterases) genes (9-12). Among these mechanisms, the erm encoded methylases are the major factor of resistance to macrolides. Among many reported and sequenced erm genes, three major genes of ermA, ermB and ermC are present in staphylococci (12, 13). Due to the development of resistant strains, epidemiological data on antibiotic susceptibility of Staphylococcus strains isolated from nasal carriers in each region can be helpful to select appropriate drugs to eradicate carriage states, control the nosocomial infections and also treat the patients (14, 15).
2. Objectives
The current study aimed to investigate the antibiotic susceptibility profile and the prevalence of the ermA, ermB, ermC and msrA genes among staphylococcal strains isolated from the nares of hospital personnel in Khorramabad, Iran.
3. Patients and Methods
3.1. Isolation and Identification of Nasal Staphylococci
During this cross-sectional study, a total of 340 volunteers from hospital staff (males and females), employed in various wards of four referral and state hospitals (Shohaday-e-ashayer, Asalian, Shahid Madani and Taemin Ejtemai) in Khorramabad city (west of Iran) were included and screened for nasal carriage of S. aureus from July 2011 to January 2012.The subjects with a history of any antibiotic consumption for at least ten days at the time of sampling were excluded. The sample size was estimated via formula to calculate the ratio with an accuracy of 0.05 (d = 0.05).
| Equation 1. |
To collect samples, the convenience sampling method was used. Three hospitals under study were general (400, 350 and 150 beds) and one hospital was specialized (100 beds). Finally 100 adults of both genders (40 males and 60 females) were enrolled. Information about variables including gender, work experience and the ward which hospital staff worked was collected by a questionnaire. The samples were collected from both anterior nares of the subjects by rotating a sterile cotton wool swab. The swabs were placed and transported in 3 mL tryptic soy broth (Merck, Germany) and incubated at 37°C. After overnight incubation, a loop full of inoculated broth medium was subcultured on 10% sheep blood agar, mannitol salt agar and nutrient agar (Merck, Germany) (16). Presumptive staphylococcal colonies (growth on mannitol salt agar, Gram-positive and catalase-positive cocci) were tested further for the production of coagulase and DNase enzymes according to the standard bacteriological procedures (17). Isolates with positive reactions (coagulase-positive and DNase-positive) were identified as S. aureus; other staphylococcal species with negative test results designated as CoNS. Duplicate isolates from the same subject were excluded.
3.2. Bacterial Susceptibility Testing
Susceptibility of Staphylococcus isolates to antibiotics (Mast, Merseyside, United Kingdom) including erythromycin (15 µg), clarithromycin (15 µg), azithromycin (15 µg), cefoxitin (30 µg), penicillin G (10 U), vancomycin (30 µg), tetracycline (30 µg), clindamycin (2 µg), trimethoprim/sulfamethoxazole (1.25/23.75 µg) and rifampin (5 µg) was determined by the Kirby-Bauer disk diffusion method on muller-hinton agar, according to the procedures described by the clinical and laboratory standards institute (CLSI) (18).
3.3. DNA Extraction
Total DNA (consisting of the chromosomal and the plasmid) of all the isolates was extracted and collected using AccuPrep® Genomic DNA extraction kit (Bioneer, Korea) with some modifications. Briefly, a 1 ml of Luria-Bertani broth (LB) bacterial colony of each strain was suspended in 1 mL of lysogeny broth (LB) medium, and incubated at 37°C for 18 hours. The bacterial cells were harvested by centrifugation at 7,500 g for 10 minutes and resuspended in 200 μL of phosphate-buffered saline (PBS) and mixed well. To ensure adequate cell breakage, 10 μL lysostaphin (100 μg/ml in sterile deionized water; Sigma) was added to the bacterial mixture and incubated at 37°C for 30 minutes (19). Then, 10 μL of proteinase K (20 mg/mL) was added to the sample and after incubating at 56°C for 30 minutes, the process was continued according to the manufacturer's instructions. Finally, DNA was extracted and preserved at -20°C.
3.4. Primers and PCR
The four primer pairs used to amplify the ermA, ermB, ermC and msrA genes are listed in Table 1 (12). In the current study, multiplex polymerase chain reaction (PCR) amplification was performed for ermA, ermC and msrA genes as previously described by Zmantar et al. (12) in a 25 µL reaction mixture consisting of: 1X PCR reaction buffer, 2.5 mM MgCl2, 0.2 mM dNTP, 25 pmol of each of the three primer pairs, 1.2 U of Taq DNA polymerase (Fermentas, Lithuania) and 200 ng DNA template. For ermB gene amplification, the ermB primers (25 pmol) were added in a separate PCR reaction under the same above-mentioned conditions. Amplification was carried out using a gradient thermal cycler (Bio-Rad, My cycler, US) as the following program: five minutes at 94°C, 30 cycles of amplification, consisting one minute at 95°C, 0.5 minute at 55°C and two minutes at 72°C, ending with 10 minutes at 72°C (final extension). To verify amplification, 5 μL of PCR products were subjected to agarose gel electrophoresis (1.5% agarose, 1X TAE, 100V). Ethidium bromide-stained DNA fragments were then visualized on a UV transilluminator at 300 nm in comparison with a 50-bp molecular size standard ladder (Fermentas, Lithuania).
Table 1. Oligonucleotide Sequences Used for Erythromycin-Resistant Genes Amplification.
| Resistance Gene | Primers Sequences (5’ → 3’) | Product Size, bp |
|---|---|---|
| ermA | 139 | |
| Forward | TATCTTATCGTTGAGAAGGGATT | |
| Reverse | CTACACTTGGCTTAGGATGAAA | |
| ermB | 142 | |
| Forward | CTATCTGATTGTTGAAGAAGGATT | |
| Reverse | GTTTACTCTTGGTTTAGGATGAAA | |
| ermC | 190 | |
| Forward | CTTGTTGATCACGATAATTTCC | |
| Reverse | ATCTTTTAGCAAACCCGTATTC | |
| msrA | 163 | |
| Forward | TCCAATCATAGCACAAAATC | |
| Reverse | AATTCCCTCTATTTGGTGGT |
To verify the authenticity of the amplicons, PCR products of three positive samples containing one of the genes of ermB, ermC or msrA as representative were subjected to sequencing (Macrogen, Korea). The resulting sequences were aligned with corresponding sequences in the GenBank database using at the national center for biotechnology information (NCBI) BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi? program = blastn and PAGE-TYPE = blast search and LINK-LOC = blasthome). To minimize the random errors, all of the devices were calibrated before use. All observations were carried out by one microbiologist.
3.5. Statistical Analysis
Descriptive data were presented as frequencies and percentages via SPSS software version 16. Chi-square test and T-test were used to determine any significant differences between prevalence of the tested genes among S. aureus and CoNS strains. P values less than 0.05 were considered statistically significant.
3.6. Ethical Issues
The study was approved by ethical committee of Lorestan University of Medical Sciences, Khorramabad, Iran (No. 200/49017). All nasal samples were taken from hospital employees volunteered for the study. Informed consent was obtained from all precipitants before sampling. Privacy and confidentiality of personal information were saved and protected.
4. Results
4.1. Staphylococcal Isolates and its Susceptibility Patterns
A total of 51 S. aureus strains were isolated from anterior nares of the 340 screened subjects (i.e. the nasal carriage rate for S. aureus was 15%). Of the 51 S. aureus-carriers, 22 (43.1%) were males and 29 (56.9%) were females. On the other hand, 49 randomly selected CoNS strains, isolated from nares of non-S. aureus carriers [18 males (36.7%), 31 females (63.3%)] were also included and tested in the study. There was no statistical difference between the two groups (S. aureus nasal carriers and CoNS) in terms of gender (P = 0.72). Average work experience for CoNS obtained subjects was 6.4 ± 0.89 years, and within S. aureus-nasal carriers were 4.8 ± 0.68 years. Based on the T-test result, difference in the mean of work experience between the two groups was not statistically significant (P = 0.22). The resistance profile for all isolates to macrolides as well as other tested antibiotics is listed in Table 2. The majority of isolates expressed resistance to penicillin (96%). No resistance to vancomycin was noted. Twenty-nine (29%) isolates were resistant to cefoxitin (eight S. aureus and twenty-one CoNS). Fifty-three (53%) isolates were simultaneously resistant to erythromycin, azithromycin and clarithromycin (cross-resistance); while 8 (8%) isolates had various macrolide susceptibility patterns. For example, two isolates were intermediate to erythromycin, while they were resistant to azithromycin and clarithromycin. In addition, two isolates were found susceptible to erythromycin, while they were resistant to clarithromycin. The frequency of resistance to all tested antibiotics between CoNS and S. aureus isolates were statistically significant (P < 0.001). Distribution of susceptibility phenotypes to erythromycin among the isolates of S. aureus and CoNS according to the gender is shown in Table 3. Statistical analyses showed that among the S. aureus isolates difference in prevalence of resistance to erythromycin was significant between males and females (P = 0.011); while such difference was not significant within CoNS isolates (P = 0.771).
Table 2. Prevalence of Resistance to the Tested Antibiotics Among the Isolates of Staphylococcus aureus and CoNS Using the Disk Diffusion Method a,b.
| Antibiotics | S. aureus, n = 51 | CoNS, n = 49 | Total, n = 100 |
|---|---|---|---|
| Penicillin | 49 (96.1) | 47 (96.1) | 96 (96) |
| Clarithromycin | 13 (25.5) | 44 (89.8) | 57 (57) |
| Azithromycin | 11 (21.6) | 44 (89.8) | 55 (55) |
| Erythromycin | 10 (19.6) | 43 (87.7) | 53 (53) |
| Tetracycline | 12 (23.5) | 47 (96) | 49 (49) |
| Clindamycin | 9 (17.6) | 37 (75.5) | 46 (46) |
| TS | 5 (9.8) | 38 (77.5) | 43 (43) |
| Cefoxitin | 8 (15.7) | 21 (42.8) | 29 (29) |
| Kanamycin | 7 (13.7) | 18 (36.7) | 25 (25) |
| Rifampin | 3 (5.9) | 4 (8.2) | 7 (7) |
Abbreviations: CoNS, coagulase-negative staphylococci; TS, Trimethoprim/ sulfamethoxazole.
aP value according to the Chi-square test < 0.001.
bValues are expressed as No. (%).
Table 3. Erythromycin Susceptibility Among the Isolates of Staphylococcus aureus and CoNS Based on Gender.
| Isolatesa | N | Erythromycin Resistant, No. (%) | Erythromycin Susceptible, No. (%) | Erythromycin Intermediate, No. (%) | P Valueb |
|---|---|---|---|---|---|
| S. aureus | 51 | 0.011 | |||
| Male | 22 | 1 (4.5) | 20 (90.9) | 1 (4.5) | |
| Female | 29 | 9 (31) | 15 (51.7) | 5 (17.3) | |
| CoNS | 49 | 0.771 | |||
| Male | 18 | 15 (83.3) | 2 (11.1) | 1 (5.6) | |
| Female | 31 | 28 (90.3) | 2 (6.5) | 1 (3.2) |
Abbreviation: CoNS, coagulase-negative staphylococci.
aComparison between isolates performed by using Chi-square test, P value < 0.001.
bChi-square test.
4.2. Distribution of Macrolides Resistance Genes Using PCR
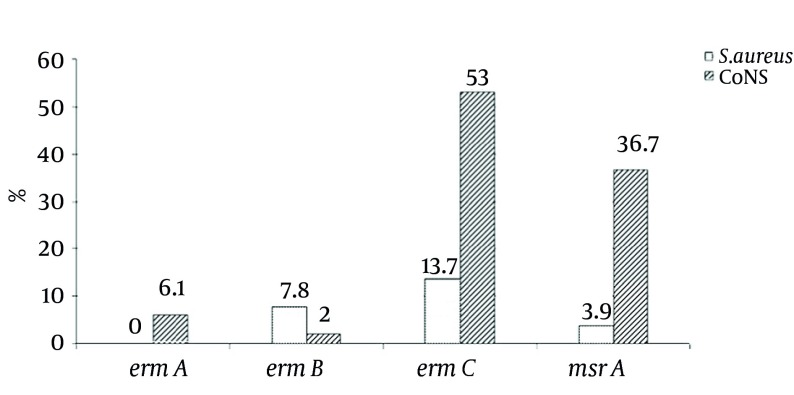
All of the isolates were screened for the presence of the four genes ermA, ermB, ermC and msrA as the main causative agents of resistance to macrolides. As predicted, the positive isolates for the ermA, ermB, ermC and msrA genes showed 139, 142, 190 and 163 bp bands, respectively (Figure 1). The frequency rate of ermA, ermB, ermC and msrA genes were 3%, 5%, 33% and 20%, respectively; separately illustrated for S. aureus and CoNS isolates in Figure 2. It was found that among the 53 isolates resistant to erythromycin, 44 (83%) isolates (eight S. aureus and thirty-six CoNS) carried at least one of the four tested genes. Of these erythromycin-resistant isolates, nine (approximately 17%) isolates (one S. aureus and eight CoNS) harbored genes of ermC and msrA, and one (%1.8) CoNS isolate also harbored ermA and ermC genes simultaneously. However, combination of three or four genes was not found among the tested isolates (Table 4). Eight (8%) isolates had intermediate phenotype to erythromycin, in which 4 (50%) isolates carried ermB or ermC genes (Table 4). In addition, it was found that out of 39 erythromycin-susceptible isolates, 36 (92.3%) isolates did not harbor any of the four antibiotic resistant genes, but the remaining isolates (three isolates) were positive for ermB or ermC genes, while they were still susceptible to erythromycin (Table 4). Interestingly, of the 29 methicillin-resistant staphylococci, 22 (75.9%) isolates were resistant to erythromycin and 19 (65.5%) isolates harbored at least one of the tested genes.
Figure 1. Gel Electrophoresis of DNA Fragments generated by PCR of erm and msrA genes in Erythromycin-Resistant Staphylococci Isolated From the Nares of Hospital Employees in Khorramabad, Iran.
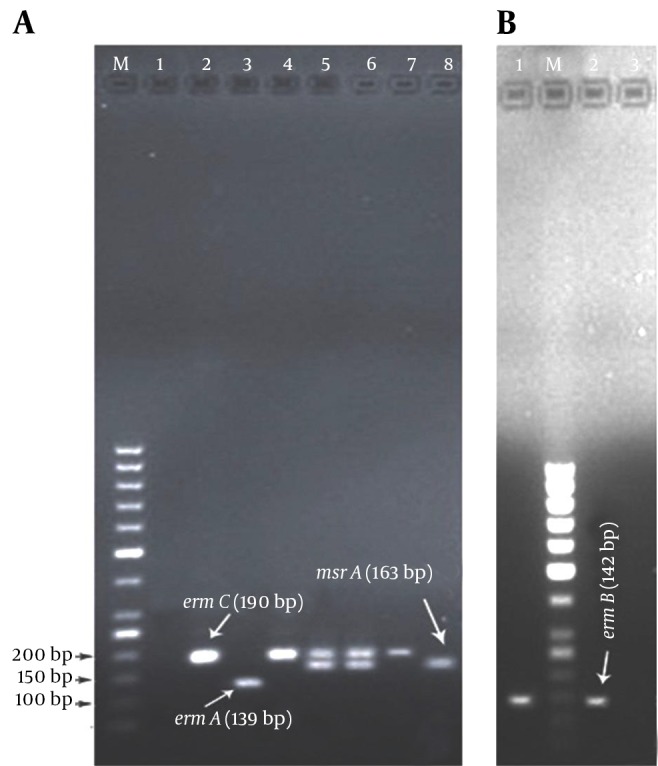
A, Multiplex PCR amplification of ermA, ermC and msrA genes; source of DNA for lanes: 1, negative control strain of erythromycin-sensitive Staphylococcus aureus ATCC 25923; lanes 2, 4 and 7, three different S. aureus isolates (ermC); lane 3, a coagulase-negative staphylococci (CoNS) isolate (ermA); lanes 5 and 6, two different CoNS isolates contain ermC and msrA genes simultaneously; lane 8, a CoNS isolate contains msrA gene. B, PCR products resulted from amplification of ermB gene; lanes 1 and 2, two different S. aureus isolates contain ermB gene; lane 3, S. aureus ATCC 29213 (negative control); lanes M, DNA molecular size marker (50-bp ladder; Fermentas, Lithonia); the electrophoresis was run on 1.5% agarose gel, stained with ethidium bromide.
Figure 2. The Prevalence of Erythromycin-Resistant Genes in Staphylococcus aureus (n = 51) and Coagulase-Negative Staphylococci (n = 49) Isolates as Determined by PCR Assay.
Table 4. The Prevalence of Resistant Genes in Staphylococci with Different Erythromycin Susceptibility.
| N | Resistant Genes, No. (%) | P Valuea | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ermA | ermB | ermC | msrA | ermA and ermC | ermC and msrA | Total | |||
| Erythromycin resistant | 53 | 0.581 | |||||||
| S. aureus | 10 | 0 | 1 (10) | 5 (50) | 1 (10) | 0 | 1 (10) | 8 (80) | |
| CoNS | 43 | 2 (4.6) | 1 (2.3) | 14 (32.5) | 10 (23.2) | 1 (2.3) | 8 (18.6) | 36 (83.7) | |
| Erythromycin susceptible | 39 | 0.386 | |||||||
| S. aureus | 35 | 0 | 1 (2.8) | 1 (2.8) | 0 | 0 | 0 | 2 (5.7) | |
| CoNS | 4 | 0 | 0 | 1 (25) | 0 | 0 | 0 | 1 (25) | |
| Erythromycin intermediate | 8 | 0.999 | |||||||
| S. aureus | 6 | 0 | 2 (33.3) | 0 | 0 | 0 | 0 | 2 (33.3) | |
| CoNS | 2 | 0 | 0 | 2 (100) | 0 | 0 | 0 | 2 (100) | |
Abbreviation: CoNS, coagulase-negative staphylococci.
aChi-square test.
4.3. Nucleotide Sequence Accession Numbers
Nucleotide sequences were submitted to GenBank under the accession numbers AB937975, AB937976 and AB937977 for the ermB, ermC and msrA genes, respectively.
5. Discussion
Staphylococcus aureus carrier status can lead to nosocomial infections in hospitalized patients (6, 8). On the other hand, CoNs are leading cause agents of opportunistic and device-related infections (3). In the current study, nasal carriage rate for S. aureus was 15%. In several studies performed in various regions of the world, this carriage status in healthcare workers have been reported ranging ranging from 18.6% to 50% (20). Moreover, in Iran, the frequency of nasal carriage among hospital employees is reported approximately 12.7% - 45% (21). Unfortunately, the results of disk diffusion testing showed that all the evaluated isolates were multidrug-resistant. So that, 29% of strains were methicillin-resistant (Table 2). It is a serious threat when a methicillin-resistant stain is transferred and spread among hospitalized patients (therapeutic failure) (22). It was observed that the frequency of resistance among CoNS isolates was significantly higher than those among S. aureus isolates; and this finding was also attributable to resistance to erythromycin and cefoxitin (Table 2). While the majority of the erythromycin-resistant strains had cross-resistance to other macrolides; however eight strains exhibited heterogeneous susceptibility pattern toward macrolide antibiotics (Table 2). It is presumed that this discrepancy is due to alterations in the chemical structure of various generations of macrolides. The association between resistance to methicillin and the presence of tested genes was significant (P = 0.031). The simple explanation for this phenomenon is carrying and transmission of the various adjacent resistance genes on genetic elements such as plasmids. Several studies concerning the distribution of erm genes and efflux pumps are conducted using southern hybridization and PCR techniques (13, 23, 24). Despite previous studies in which have primarily focused on clinical strains of staphylococci (1, 11, 23-28), the current study presents the first report on the occurrence of main genes implicated in macrolides- resistance in Staphylococcus strains obtained from nares of hospital employees. The obtained PCR results demonstrated that the most prevalent gene in both S. aureus and CoNS strains was ermC (33%). Also, the frequency of this gene (ermC) in erythromycin-resistant S. aureus and CoNS were 13.7% and 53%, respectively. These results are consistent with those of the study by Cetin et al. (25) performed with CoNS in Turkey in which a frequency of 30% for ermC was yielded. Martineau et al. (24) reported an occurrence of 66% for S. epidermidis strains. Fluit et al. (9) found that the ermC gene is located on a small plasmid; therefore, horizontal transmission of this gene from resistant strains to susceptible strains is unavoidable. Regarding ermA, some studies showed that this gene, as the most prevalent gene among clinical strains of S. aureus, has a frequency of 21% to 67% (1, 24-27). Similar findings were also published in Denmark, where ermA was responsible for 80.6% resistance to erythromycin among 428 tested clinical S. aureus strains from 1959 to 1988 (23). In contrast, the current study PCR results demonstrated that ermA was not observed in the tested S. aureus strains. It was also found that ermB gene was carried in a minority of S. aureus (7.8%) and CoNS (2%) strains; while previous studies revealed that this gene was primarily originated from animal strains and generally its frequency was low (24-27). However, Zmantar et al. in two different studies (12, 28) reported that ermB was the most common gene when they evaluated 81 S. aureus strains isolated from human auricular. Finally, msrA gene, encoding an ATP-dependent efflux pump (24), was more prevalent in CoNS (41.8%) than S. aureus strains (20%). These observations are in line with the findings of Lina et al. (26) who reported the prevalence of msrA gene as 14.6% and 2.1% among CoNS and S. aureus strains, respectively. Similar data were also obtained (41%) from a study conducted by Zmantar et al. (28) when 77 clinical CoNS strains were investigated. Some staphylococcal strains in the current study showed discrepancies between the genotype and phenotype. So that, three strains (two S. aureus and one CoNS) harbored ermB or ermC gene, while they were susceptible to erythromycin. Zmantar et al. (28) and Coutinho et al. (1) also hypothesized that this phenomenon may be caused by down-regulation or mutation in promoter region of erm gens. On the other hand, Martineau et al. (24) demonstrated that such strains carrying the resistant genes would exhibit phonotypical resistance, upon subculturing in media with increasing gradients of the corresponding antibiotic. Moreover, nine Staphylococcus isolates were resistant to erythromycin, but did not carry any of the tested erythromycin-resistant genes. Authors presume that other variants of erm genes or efflux pump (msrB) involve in this subject, which were not evaluated in the study (1, 24, 29, 30). Although the current study is the first report on the occurrence of genetic determinants responsible for macrolide resistance among Staphylococcus strains originated from hospital employees; however, lack of further in vitro investigations, such as determining the minimum inhibitory concentration (MIC) for erythromycin, detection of other genes involved in macrolide resistance and limited sample size, were the weakness of the current study. In conclusion, no entire association was found between genotype and phenotype methods to detect macrolides resistance. In addition, distribution of genetically erythromycin-resistant isolates is geographically different among staphylococci. It is recommend controlling S. aureus in nasal carriers by proved approaches such as local or systemic administration of effective antibiotics or bacterial interference. In addition, to prevent CoNS-associated nosocomial and indwelling medical device infections, it is recommended that healthcare workers wash hands and avoid touching their noses during the patient examination, use sterile masks and gloves and finally, insert prosthetic and catheter devices aseptically.
Acknowledgments
Authors are grateful to the personnel of Razi herbal medicines research center for their cooperation. Authors also thank Mr. Babak Bashiri for native revision of the manuscript.
Footnotes
Authors’ Contribution:The concept, design, study supervision, PCR set up, manuscript preparation and critical revision of the manuscript: Gholam Reza Goudarzi; sample collection, culture and performance of PCR: Farzad Tahmasbi; statistical analysis of data: Khatereh Anbari; sample collection in women’s hospital: Masoumeh Ghafarzadeh; all authors have read and approved the final manuscript.
Funding/Support:This study was supported by a research grant (No.1220) from Lorestan University of Medical Sciences, Khorramabad, Iran.
References
- 1.Coutinho VLS, Paiva RM, Reiter KC, de-Paris F, Barth AL, Machado ABMP. Distribution of erm genes and low prevalence of inducible resistance to clindamycin among staphylococci isolates. Braz J Infect Dis. 2010;14(6):564–8. doi: 10.1016/S1413-8670(10)70113-6. [DOI] [PubMed] [Google Scholar]
- 2.Pfaller MA, Herwaldt LA. Laboratory, clinical, and epidemiological aspects of coagulase-negative staphylococci. Clin Microbiol Rev. 1988;1(3):281–99. doi: 10.1128/cmr.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otto M. Staphylococcus epidermidis--the 'accidental' pathogen. Nat Rev Microbiol. 2009;7(8):555–67. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen KD, Anson JJ, Parsons LA, Frost NG. Staff carriage of methicillin-resistant Staphylococcus aureus (EMRSA 15) and the home environment: a case report. J Hosp Infec. 1997;35(4):307–11. doi: 10.1016/S0195-6701(97)90225-5. [DOI] [PubMed] [Google Scholar]
- 5.Mitsuda T, Arai K, Ibe M, Imagawa T, Tomono N, Yokota S. The influence of methicillin-resistant Staphylococcus aureus (MRSA) carriers in a nursery and transmission of MRSA to their households. J Hosp Infect. 1999;42(1):45–51. doi: 10.1053/jhin.1998.0551. [DOI] [PubMed] [Google Scholar]
- 6.Eveillard M, Martin Y, Hidri N, Boussougant Y, Joly-Guillou ML. Carriage of methicillin-resistant Staphylococcus aureus among hospital employees: prevalence, duration, and transmission to households. Infect Control Hosp Epidemiol. 2004;25(2):114–20. doi: 10.1086/502360. [DOI] [PubMed] [Google Scholar]
- 7.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10(3):505–20. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandenbergh MF, Verbrugh HA. Carriage of Staphylococcus aureus: epidemiology and clinical relevance. J Lab Clin Med. 1999;133(6):525–34. doi: 10.1016/s0022-2143(99)90181-6. [DOI] [PubMed] [Google Scholar]
- 9.Fluit AC, Visser MR, Schmitz FJ. Molecular detection of antimicrobial resistance. Clin Microbiol Rev. 2001;14(4):836–71. doi: 10.1128/CMR.14.4.836-871.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ardic N, Ozyurt M, Sareyyupoglu B, Haznedaroglu T. Investigation of erythromycin and tetracycline resistance genes in methicillin-resistant staphylococci. Int J Antimicrob Agents. 2005;26(3):213–8. doi: 10.1016/j.ijantimicag.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Khan SA, Nawaz MS, Khan AA, Cerniglia CE. Simultaneous detection of erythromycin-resistant methylase genes ermA and ermC from Staphylococcus spp. by multiplex-PCR. Mol Cell Probes. 1999;13(5):381–7. doi: 10.1006/mcpr.1999.0265. [DOI] [PubMed] [Google Scholar]
- 12.Zmantar T, Chaieb K, Ben Abdallah F, Ben Kahla-Nakbi A, Ben Hassen A, Mahdouani K, et al. Multiplex PCR detection of the antibiotic resistance genes in Staphylococcus aureus strains isolated from auricular infections. Folia Microbiol (Praha). 2008;53(4):357–62. doi: 10.1007/s12223-008-0055-5. [DOI] [PubMed] [Google Scholar]
- 13.Maravic G. Macrolide resistance based on the Erm-mediated rRNA methylation. Curr Drug Targets Infect Disord. 2004;4(3):193–202. doi: 10.2174/1568005043340777. [DOI] [PubMed] [Google Scholar]
- 14.Erami M, Soltani B, Taghavi Ardakani A, Moravveji A, Haji Rezaei M, Soltani S, et al. Nasal Carriage and Resistance Pattern of Multidrug Resistant Staphylococcus aureus Among Healthy Children in Kashan, Iran. Iran Red Crescent Med J. 2014;16(9) doi: 10.5812/ircmj.21346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghadiri K. Nasal carriage rate of community-and hospital-acquired methicillin-resistant staphylococcus aureus in children, Kermanshah, Iran. Arch Clin Infect Dis. 2012;6(3):117–20. [Google Scholar]
- 16.Khorvash F, Abdi F, Ataei B, Fattahi Neisiani H, Hassanzadeh Kashani H, Narimani T. Nasal carriage of Staphylococcus aureus: Frequency and antibiotic resistance in healthy adults. J Res Med Sci. 2012;17 [Google Scholar]
- 17.Monson LS. Staphylococci. In: Mahon CR, Lehman DC, Manuselis G, editors. Textbook of diagnostic microbiology. Maryland Heights: Saunders Elsevier Company; 2011. [Google Scholar]
- 18.CLSI . Performance Standards for antimicrobial Susceptibility Testing. 21 ed. Wayne, USA: Clinical and laboratory Standards Institute; 2011. p. informational supplement. [Google Scholar]
- 19.Yadegar A, Sattari M, Mozafari NA, Goudarzi GR. Prevalence of the genes encoding aminoglycoside-modifying enzymes and methicillin resistance among clinical isolates of Staphylococcus aureus in Tehran, Iran. Microb Drug Resist. 2009;15(2):109–13. doi: 10.1089/mdr.2009.0897. [DOI] [PubMed] [Google Scholar]
- 20.Vinodhkumaradithyaa A, Uma A, Shirivasan M, Ananthalakshmi I, Nallasivam P, Thirumalaikolundusubramanian P. Nasal carriage of methicillin-resistant Staphylococcus aureus among surgical unit staff. Jpn J Infect Dis. 2009;62(3):228–9. [PubMed] [Google Scholar]
- 21.Rahimi-Alang S, Asmar M, Cheraghali F, Yazarlou S, Amini A, Shakeri F, et al. Frequency of methicillin resistant Staphylococcus aureus in health care. Zahedan J Res Med Sci. 2011;13(1):17–22. [Google Scholar]
- 22.Hoseini Alfatemi SM, Motamedifar M, Hadi N, Sedigh Ebrahim Saraie H. Analysis of Virulence Genes Among Methicillin Resistant Staphylococcus aureus (MRSA) Strains. Jundishapur J Microbiol. 2014;7(6):e25701. doi: 10.5812/jjm.10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westh H, Hougaard DM, Vuust J, Rosdahl VT. Prevalence of erm gene classes in erythromycin-resistant Staphylococcus aureus strains isolated between 1959 and 1988. Antimicrob Agents Chemother. 1995;39(2):369–73. doi: 10.1128/aac.39.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martineau F, Picard FJ, Lansac N, Menard C, Roy PH, Ouellette M, et al. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44(2):231–8. doi: 10.1128/aac.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cetin ES, Gunes H, Kaya S, Aridogan BC, Demirci M. Distribution of genes encoding resistance to macrolides, lincosamides and streptogramins among clinical staphylococcal isolates in a Turkish university hospital. J Microbiol Immunol Infect. 2010;43(6):524–9. doi: 10.1016/S1684-1182(10)60081-3. [DOI] [PubMed] [Google Scholar]
- 26.Lina G, Quaglia A, Reverdy ME, Leclercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother. 1999;43(5):1062–6. doi: 10.1128/aac.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitz FJ, Sadurski R, Kray A, Boos M, Geisel R, Kohrer K, et al. Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J Antimicrob Chemother. 2000;45(6):891–4. doi: 10.1093/jac/45.6.891. [DOI] [PubMed] [Google Scholar]
- 28.Zmantar T, Kouidhi B, Miladi H, Bakhrouf A. Detection of macrolide and disinfectant resistance genes in clinical Staphylococcus aureus and coagulase-negative staphylococci. BMC Res Notes. 2011;4:453. doi: 10.1186/1756-0500-4-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdollahi S, Ramazanzadeh R, Delami Khiabani Z, Kalantar E, Menbari S. Molecular detection of inducible clindamycin resistance among Staphylococcal strains isolated from hospital patients, [in Persian]. J Ardabil Uni Med Sci. 2013;13(1):59–68. [Google Scholar]
- 30.Park AK, Kim H, Jin HJ. Phylogenetic analysis of rRNA methyltransferases, Erm and KsgA, as related to antibiotic resistance. FEMS Microbiol Lett. 2010;309(2):151–62. doi: 10.1111/j.1574-6968.2010.02031. [DOI] [PubMed] [Google Scholar]