Abstract
Flow cytometry is a powerful tool capable of simultaneously analyzing multiple parameters on a cell-by-cell basis. Lung tissue preparation for flow cytometry requires creation of a single-cell suspension, which often employs enzymatic and mechanical dissociation techniques. These practices may damage cells and cause cell death that is unrelated to the experimental conditions under study. We tested methods of lung tissue dissociation and sought to minimize cell death in the epithelial, endothelial, and hematopoietic lineage cellular compartments. A protocol that involved flushing the pulmonary circulation and inflating the lung with Dispase, a bacillus-derived neutral metalloprotease, at the time of tissue harvest followed by mincing, digestion in a DNase and collagenase solution, and filtration before staining with fluorescent reagents concurrently maximized viable yields of epithelial, endothelial, and hematopoietic lineage cells compared with a standard method that did not use enzymes at the time of tissue harvest. Flow cytometry identified each population—epithelial (CD326+CD31−CD45−), endothelial (CD326−CD31+CD45−), and hematopoietic lineage (CD326−CD31−CD45+)—and measured cellular viability by 7-aminoactinomycin D (7-AAD) staining. The Dispase method permitted discrimination of epithelial vs. endothelial cell death in a systemic lipopolysaccharide model of increased pulmonary vascular permeability. We conclude that application of a dissociative enzyme solution directly to the cellular compartments of interest at the time of tissue harvest maximized viable cellular yields of those compartments. Investigators could employ this dissociation method to simultaneously harvest epithelial, endothelial, and hematopoietic lineage and other lineage-negative cells for flow-cytometric analysis.
Keywords: flow cytometry, lung, tissue digestion, tissue processing, cell death
flow cytometry is a sophisticated technique that permits investigators to simultaneously analyze multiple parameters on a cell-by-cell basis thousands of times per second. Tissue preparation for flow cytometry requires generation of a single-cell suspension, and digestion protocols typically employ a combination of enzymatic and mechanical tissue disruption to achieve a single-cell suspension amenable to flow-cytometric analysis. However, tissue processing can injure cells and cause cell death that is due not directly to the biological system under study but rather to the technique used to prepare the sample. Lungs are composed of numerous cellular compartments, for example, epithelial, endothelial, and hematopoietic lineage cells, organized in a specific and intricate manner to form functional tissue structures. Lung tissue processing for flow-cytometric analysis is complicated in part because both epithelial and endothelial cells are adherent to their respective basement membranes and are also separated from each other by an interstitial layer of cells and matrix proteins.
Recent publications have reported methods to harvest pulmonary cells for flow-cytometric investigation but have largely focused on a single cellular compartment for analysis (2, 3, 8, 11). For example, a combination of enzymatic and mechanical dissociation (3) or solely mechanical dissociation (11) yielded a single-cell suspension amenable to immune cell analysis. The bulk of these published methods use a combination of DNase and collagenase to digest harvested lung tissue prior to flow-cytometric analysis, but enzymatic dissociation with the enzyme Dispase facilitated endothelial cell isolation and viability (8). However, it is unclear how this methodology performed when assessing epithelial cell viability. Bantikassegn et al. (2) were able to examine multiple compartments in a diffuse lung adenocarcinoma model; however, their protocol required enzymatic digestion followed by magnetic bead or fluorescence-activated cell sorting (FACS) separations. Despite this involved protocol, viability varied dramatically: from 57 to 96% between various cellular compartments and separation methods.
Given the complex organization of the various cellular compartments within the lung, a focus on harvesting one cellular compartment for analysis may lead to increased, and variable, damage and cell death within other cellular compartments. To that end, we sought to optimize a method to prepare lung tissue for flow-cytometric assessment that curtails processing-related cell death of epithelial, endothelial, and hematopoietic lineage cellular compartments and therefore maximizes yield of viable cells. Here we describe a method to digest mouse lung tissue that minimizes cell death related to sample preparation and generates viable populations of epithelial, endothelial, and hematopoietic lineage cells from a single sample of mouse lung tissue. These cell lineages represent major compartments that investigators may wish to examine simultaneously using flow cytometry, and we apply our method to a model of endotoxin-induced pulmonary vascular permeability.
METHODS
The Johns Hopkins Institutional Animal Care and Use Committee approved all animal protocols. Male C57BL/6 wild-type mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and euthanized at 8–12 wk of age. Euthanasia was performed by pentobarbital overdose followed by transection of the caudal vena cava. A 3-mm skin incision was made overlying the larynx; subcutaneous tissues were gently retracted to allow direct visualization of the proximal-mid trachea. A suture was then passed behind the trachea. The trachea was transorally intubated with an opaque 1-in. 20-gauge catheter, and correct placement was confirmed by direct visualization. The endotracheal catheter was secured in place by tying the suture around the trachea.
The chest cavity was then exposed by midline sternotomy and retraction of the chest wall. A small incision was made into each ventricle, and the pulmonary circulation was flushed with ∼2 ml of phosphate-buffered saline (PBS) followed by instillation of 1 ml of fluid (see Table 1) delivered via a 20-gauge catheter inserted into the main pulmonary artery under direct visualization. Flushed lungs blanched and appeared white in color.
Table 1.
Description of methods
| Method | Pulmonary Circulation Flush Fluid | Lung Inflation Fluid | Digestion Dish Fluid |
|---|---|---|---|
| Control | PBS | DNase/collagenase | |
| Dispase | Dispase | Dispase | DNase/collagenase |
Enzyme solutions were prepared fresh in PBS/0.5% BSA and kept on ice until use. Collagenase I (Worthington Biochemical Corporation, Lakewood, NJ) was prepared at 5 mg/ml, and DNase I (Sigma-Aldrich) was prepared at 1 mg/ml. Dispase was used in undiluted form directly from the manufacturer (Corning Life Sciences, Tewksbury, MA) after freezing into aliquots and thawing on ice immediately before use.
The left lung was clamped at the hilum and two sequential volumes of 0.7 ml of PBS were gently instilled and withdrawn [bronchoalveolar lavage (BAL)]. For the Dispase method, a volume of 0.7 ml of Dispase (see Table 1) was delivered via the endotracheal catheter followed by 0.5 ml of 1% low-melting-point agarose (Sigma-Aldrich, St. Louis, MO) heated to 50°C, which when cooled served as a semisolid plug to keep enzyme-containing fluid apposed to the lung tissue. The hilum was ligated with suture, and both lungs were then removed separately and placed in PBS on ice for immediate processing.
Each lung was removed from PBS and placed into a 30-mm dish, referred to herein as the digestion dish, containing 2 ml of DNase and collagenase solution (see Table 1). The tissue was minced with scissors into pieces no larger than 2–3 mm and then incubated at 37°C with an orbital speed of 125 revolutions per minute for 30 min.
After incubation, remaining tissue and all fluid in the digestion dish were drawn via an 18-gauge needle into a 5-ml syringe and then discharged back into the digestion dish two to three times to facilitate tissue disruption. On the final aspiration, the fluid was discharged onto a 70-μm filter (BD Biosciences, San Jose, CA) that had been prewetted with 1 ml of PBS containing 0.5% bovine serum albumin (BSA, Sigma-Aldrich) suspended over a 50-ml conical tube. The digestion dish was washed with 1 ml of PBS/0.5% BSA to ensure maximal cellular retrieval and then this fluid was aspirated into the syringe and discharged onto the filter. The syringe plunger was then used to gently disrupt any remaining intact tissue before washing the filter with 2 ml of cold PBS/0.5% BSA.
Cell pellets were obtained via centrifugation at 300 g for 5 min at 4°C (all spins). The supernatant was discarded by aspiration and 2 ml of ACK lysis solution (Invitrogen, Carlsbad, CA) was added to the pellet to lyse remaining erythrocytes. The pellet was then resuspended by gentle vortex and allowed to incubate in the ACK lysis solution for 4 min at room temperature before addition of 4 ml PBS to each sample (to normalize tonicity) and passing the cell suspension through a second prewetted 70-μm filter. Samples were recentrifuged and the pellet was finally resuspended by gentle vortex in 1 ml of cold PBS/2.5% BSA. Cell counts were determined manually in a hemocytometer by an investigator blinded to the experimental condition (M. Damarla).
An aliquot of each sample containing 1–1.5 × 106 cells was loaded into a well of a 96-well plate, which was centrifuged, and the supernatant was removed by decanting and blotting the plate on a paper towel. Then 50 μl of PBS/0.5% BSA containing a 1:100 dilution of Fc Block (BD Pharmingen) was added to the cells. Cells were resuspended by tapping the plate and then incubated for 10 min at 4°C, and 50 μl of PBS/0.5% BSA containing fluorochrome-conjugated antibodies (see Table 2) was then added to each well followed by a 20-min incubation in the dark at 4°C. Each well was washed with 150 μl of PBS/0.5% BSA and then the plate was centrifuged. After removal of the supernatant as above, the cells in each well were resuspended in 250 μl of PBS/0.5% BSA and added to 750 μl of PBS/0.5% BSA in a 12 × 75-mm polystyrene tube for staining with 7-aminoactinomycin D (7-AAD; cell death marker) as per the manufacturer's recommendations (SYTOX AADvanced Dead Cell Stain, Life Technologies, Carlsbad, CA).
Table 2.
Reagents and cytometer setup
| Marker | Fluorochrome | Company, catalog number (location) | Volume (μl) Added to Stain 1–1.5 × 106 Cells in 100 μl | Laser Lines (color) | Emission Filters |
|---|---|---|---|---|---|
| CD326 (epithelial marker) | BV421* | BioLegend, no. 118225 (San Diego, CA) | 1 | 405 nm (violet) | 450/50 |
| CD31 (endothelial marker) | PE-CF594 | BD Horizon, no. 563616 (San Jose, CA) | 0.1 | 488 nm (blue) | 610/20 |
| CD45 (hematopoietic lineage marker) | PE-Cy7 | BioLegend, no. 103114 (San Diego, CA) | 0.25 | 488 nm (blue) | 780/60 |
| 7-AAD [SYTOX AADvanced] (cell permeability/death marker) | PerCP-Cy5.5 | Life Technologies, no. C10427 (Carlsbad, CA) | 1 (in 1 ml) | 488 nm (blue) | 695/40 |
7-AAD, 7-aminoactinomycin D; BV421, Brilliant Violet 421; PE, phycoerythrin; PerCP, peridinin chlorophyll protein complex.
Some experiments were performed using an allophycocyanin-Cy7-conjugated anti-CD326 antibody (BioLegend, no. 118218 [San Diego, CA]) excited by a 633-nm (red) laser and detected with a 780/60 emission filter.
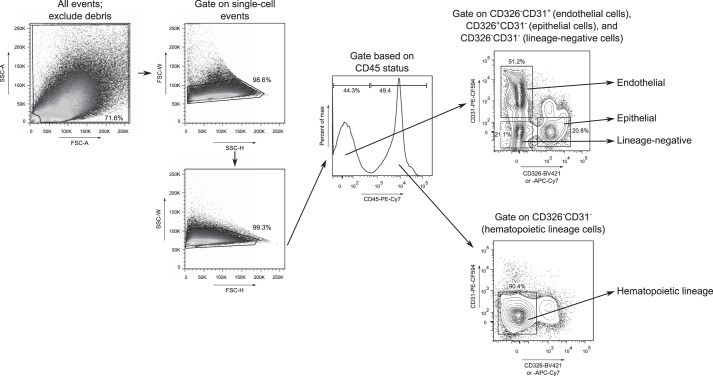
Data acquisition was performed on a custom FACSAria II instrument running FACSDiva acquisition software (BD Biosciences; see Table 2 for cytometer setup). Compensation was performed as previously described (15) with UltraComp eBeads (eBioscience, San Diego, CA) for the surface antibody fluorochromes, a singly stained aliquot of lung cells for the 7-AAD fluorochrome, and unstained lung cells to compensate for background autofluorescence; 5 × 105 events were obtained per sample and analyzed by using FlowJo 7.6.5 (Tree Star, Ashland, OR) as outlined in Fig. 1. We defined cell compartments as follows: epithelial, CD326+CD31−CD45−; endothelial, CD326−CD31+CD45−; hematopoietic lineage, CD326−CD31−CD45+; lineage-negative, CD326−CD31−CD45− (2, 6, 8–10). Dead cells were identified by positive 7-AAD (SYTOX AADvanced) staining (12).
Fig. 1.
Gating strategy to identify epithelial, endothelial, hematopoietic lineage, and lineage-negative cells. Forward and side scatter (FSC and SSC) gating on area (A), height (H), and width (W) excluded debris and non-single cell events. We then interrogated CD45 status and subsequently assessed CD326 and CD31 status within the CD45− and CD45+ populations to identify epithelial (CD326+CD31−CD45−), endothelial (CD326−CD31+CD45−), lineage-negative (CD326−CD31−CD45−), and hematopoietic lineage (CD326−CD31−CD45+) cells. The single-cell suspension was generated by the Dispase method. max, Maximum.
We sought to validate the ability of our Dispase method to measure compartment-specific cell death. To that end, we utilized a previously published model of increased pulmonary vascular permeability and harvested lung tissue 6 h after intravenous administration of lipopolysaccharide (LPS) 7.5 mg/kg or PBS, as described by our group (5). At the time of harvest, right lungs were subjected to our Dispase method and flow cytometry as above.
Differences between groups were compared by two-tailed Mann-Whitney U-tests. Multiple group comparisons were performed by two-way ANOVA with the Sidak correction for multiple comparisons. Nonparametric data were log-transformed where indicated. Significance was determined at alpha values less than 0.05.
RESULTS
Previous reports have minced the lung following harvest and used enzymatic digestion, either a proprietary blend (2) or permutations of DNase, collagenase, and Dispase II (8), for preparation of single-cell suspensions and have noted variable rates of cellular viability depending on the cell type of interest. Similar to prior work from our group examining lung lymphocytes (15) and a common method found in the literature (3), we used enzymatic digestion with DNase and collagenase prior to filtration as a control method. We compared this control method to a method meant to maximize epithelial and endothelial cell dissociation using a Dispase pulmonary vascular flush and inflation, respectively, prior to DNase and collagenase digestion. Compared with the control method, the Dispase method generated a higher total cellular yield (Fig. 2A) and yield of epithelial, endothelial, and lineage-negative cells (Fig. 2B). Hematopoietic lineage cellular yield was similar and the frequency of each compartment as a function of total lung cells was also similar (Table 3) between methods.
Fig. 2.
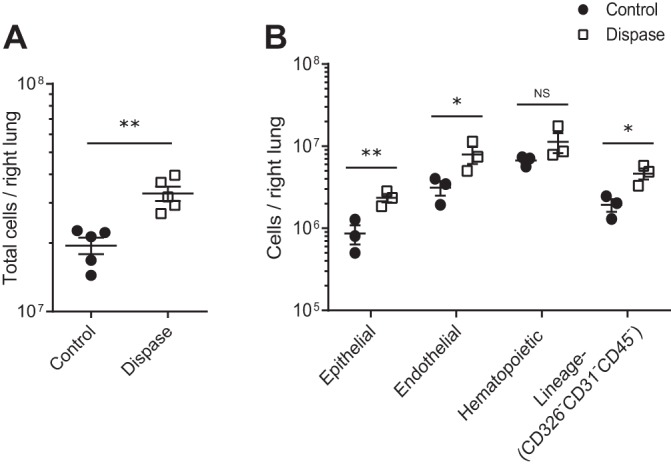
Total cellular yields and yields of epithelial (CD326+CD31−CD45−), endothelial (CD326−CD31+CD45−), hematopoietic lineage (CD326−CD31−CD45+), and lineage-negative (CD326−CD31−CD45−) cells per right lung after control and Dispase dissociation methods. A: total cellular yield per right lung is shown on a log scale; n = 5 per group. B: compartment cellular yields within each population including the lineage-negative (CD326−CD31−CD45−) population are shown on a log scale; ANOVA P < 0.0001 for control vs. Dispase using log-transformed data; n = 3 per group. *P < 0.05, **P < 0.01. NS, not significant. Values reported are means ± SE.
Table 3.
Cellular frequencies for control and Dispase methods
| Cellular Frequency (Percent of Total Lung Cells) |
||||
|---|---|---|---|---|
| Method | Epithelial | Endothelial | Hematopoietic lineage | Lineage-negative |
| Control (IV PBS, DNase/collagenase in digestion dish) | 3.9 ± 0.6 | 18.4 ± 2.8 | 29.1 ± 6.1 | 17.4 ± 7.6 |
| Dispase (IV and IT Dispase, DNase/collagenase in digestion dish) | 7.5 ± 0.6 | 18.2 ± 1.9 | 31.4 ± 3.4 | 10.9 ± 0.6 |
Values are percentage of total lung cells (singlets as per the third step of Fig. 1) represented by each cell type ± SE, n = 4–6, ANOVA P for control vs. Dispase > 0.8. IV, intravenous (referring to flushing the pulmonary circulation); IT, intratracheal (referring to inflating the lung).
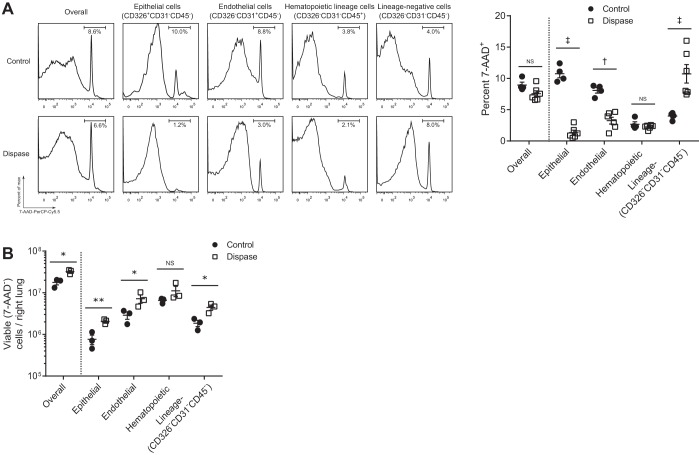
We next sought to compare viability between the control and Dispase methods. 7-AAD positivity (representing dead cells) was lower among epithelial and endothelial cells with use of the Dispase method (Fig. 3A). The Dispase method also generated a higher yield of viable cells overall as well as epithelial, endothelial, and lineage-negative cells compared with the control method (Fig. 3B).
Fig. 3.
Viability of all cells, epithelial (CD326+CD31−CD45−), endothelial (CD326−CD31+CD45−), hematopoietic lineage (CD326−CD31−CD45+), and lineage-negative (CD326−CD31−CD45−) cells after processing by the control vs. Dispase method. A: percentage of 7-aminoactinomycin D (7-AAD)+ (dead) cells is shown within each population; ANOVA P < 0.0001 for control vs. Dispase; n = 4–6 per group. B: viable cells overall and within each compartment including the lineage-negative (CD326−CD31−CD45−) population are shown on a log scale; ANOVA P < 0.001 for control vs. Dispase using log-transformed data; n = 3 per group. *P < 0.05, **P < 0.01, †P < 0.001, ‡P < 0.0001. Values reported are means ± SE.
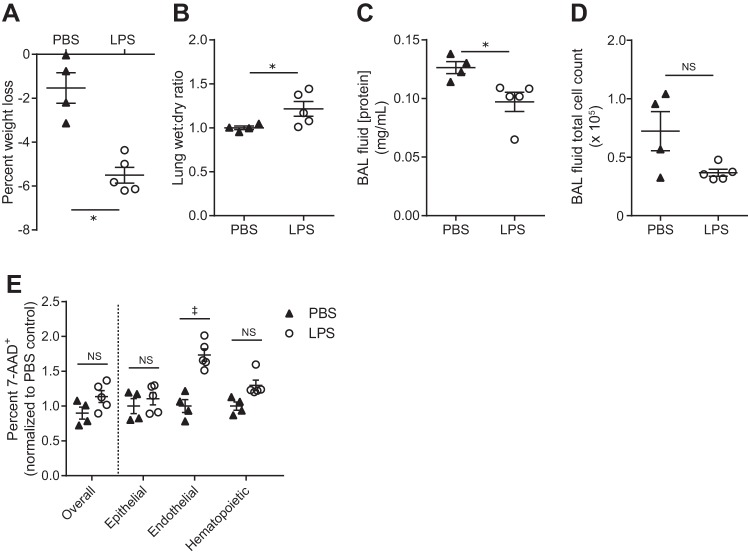
Since the Dispase method was able to generate a high yield of viable cells, we examined whether the method could be used to detect differential rates of cell death among epithelial, endothelial, and hematopoietic lineage cells following systemic LPS-induced pulmonary vascular injury. Compared with a PBS injection, mice that received systemic LPS by intravenous (IV) injection lost more weight and displayed an increased lung wet-to-dry ratio, demonstrating a significant increase in pulmonary vascular permeability in response to IV LPS (Fig. 4, A and B). However, there was no evidence of alveolitis (epithelial barrier injury) as evidenced by a decreased BAL fluid protein concentration and similar BAL fluid total cell counts 6 h after IV LPS (Fig. 4, C and D), similar to our previously published results (5). Right lungs of these mice were subjected to our Dispase method of cellular dissociation and flow-cytometric analysis. Epithelial cell death rates were not affected by LPS administration whereas endothelial cell death significantly increased (Fig. 4E).
Fig. 4.
Dispase dissociation method applied to a systemic lipopolysaccharide (LPS) model of increased pulmonary vascular permeability. A: percent weight loss shown 6 h after injection. B–D: lung wet-to-dry ratios (B), bronchoalveolar lavage (BAL) fluid protein concentrations (C), and BAL fluid total cell counts (D). E: percent cell death (7-AAD+) overall and within each population assessed by flow cytometry following the Dispase dissociation method and normalized to PBS control; ANOVA P < 0.0001 for PBS vs. LPS; n = 4–5 per group. *P < 0.05, ‡P < 0.0001. Values reported are means ± SE.
DISCUSSION
Flow cytometry is a powerful modality to concurrently assess multiple parameters, but processing-related cellular damage can limit its usefulness by introducing a survival bias especially when cell death itself is a readout of interest. In this study, we explored a method to prepare a single-cell suspension from mouse lung tissue that simultaneously maximized viable yields of epithelial, endothelial, and hematopoietic lineage cells. Compared with a standard control method that did not use enzymes at the time of tissue harvest, we observed an increase in compartment-specific viable cellular yield using a method that flushed the pulmonary circulation and inflated the lungs with Dispase. With this method, epithelial and endothelial viable cellular yield increased by ∼170 and 150%, respectively, without diminishing hematopoietic viable cellular yield compared with the control method. Thus our Dispase method represents an effective strategy to generate viable cell populations from multiple compartments.
Our Dispase method resulted in similar endothelial cell viability to Kawasaki et al. (8) in their protocol to isolate endothelial cells while preserving cellular viability in other examined cellular compartments. Our method also generated similar or higher structural cellular frequencies to those observed by Bantikassegn et al. (2) in preseparation fractions of their diffuse lung adenocarcinoma model. Although our Dispase method requires the additional steps of vascular flush and lung inflation, after a brief run-in period we were able to reduce our harvest time to less than 10 min per mouse and obtain flow cytometry results less than 3 h later. Our short processing time prevented the need for fixation and allowed for analysis of live cells. However, our method could be modified to incorporate fixation, and subsequent permeabilization would allow for analysis of numerous intracellular markers.
An advantage of our Dispase method is the high and reproducible rate of cellular viability observed. All cell lineages analyzed had a high viability rate (∼95% or higher) with epithelial and hematopoietic lineage cells displaying ∼98% viability. The ∼90% viability of the lineage-negative (CD326−CD31−CD45−) cellular compartment, which includes fibroblasts, smooth muscle cells, and other mesenchymal cells, could enable analysis of additional cell types. Our flow-cytometric analysis likely overestimates cell death in specific cell populations, as our markers defined lung cellular compartments in a coarse fashion. For example, whereas we used CD326 as a pan-epithelial marker, subgrouping into type 1 and 2 alveolar epithelial cells is possible by adding additional markers and employing a protocol similar to the method of Mock et al. (10). Application of subtype markers to the epithelial, endothelial, and hematopoietic lineage and lineage-negative compartments would permit more granular analysis, and the low background cell death rate of our Dispase method could facilitate detection of rare cell types. Notably, if only one compartment is of interest, a simpler method could be employed. Our control method, which does not require enzymes at the time of tissue harvest, is adequate to analyze hematopoietic lineage cells, and our group has employed this method to characterize lung lymphocytes (15). A similar protocol can assess lung macrophages (3). Solely mechanical dissociation techniques may also create single-cell suspensions that permit characterization of lung leukocytes (11), although viability should be ensured following mechanical processing.
The low processing-related cell death imparted by our Dispase method facilitated flow-cytometric analysis of lung epithelial and endothelial cell death at an early time point following systemic administration of LPS. LPS caused increased pulmonary vascular permeability (increased lung edema); however, the changes at this time point appear to be a principally interstitial, rather than alveolar, injury as evidenced by the lack of cellular alveolitis and a paradoxically decreased BAL fluid protein concentration in response to LPS, as shown in these studies and previously noted by our group (5). This constellation of findings suggests a primarily endothelial rather than epithelial injury, and our flow cytometry assessment of cell death supports this hypothesis with an ∼70% increase in endothelial cell death with no change in epithelial cell death. Going forward, our Dispase method could allow for identification of subtle differences in cell death due to varied experimental conditions in multiple cellular compartments and permit specific interrogation of the apoptotic pathway. Validated flow cytometry reagents to measure externalization of phosphatidylserine (fluorescent annexin V conjugates), membrane asymmetry (fluorescent ratiometric probe F2N12S), and activation of specific caspases (fluorescent substrates activated by the cysteine proteases that regulate the apoptotic pathway) by flow cytometry (4, 7, 12–14, 16) increase the detail to which investigators could assess apoptosis in the lung using our method.
Several limitations accompany our Dispase method. Investigators that use our protocol must be aware that enzymatic dissociation, while critical in promoting cellular viability, may cleave surface markers of interest and give the appearance of a low cellular yield. Dispase can cleave some immunologically relevant surface molecules and may need to be avoided in applications such as T cell analysis (1). Although it is possible that our digestion protocol cleaves some surface markers, we do not believe cleavage completely explains our results. For example, our data show that incorporating Dispase at the time of tissue harvest increases viable endothelial cellular yield compared with not using Dispase. Processing methods that generate a low frequency or yield of a given cellular compartment because of true cellular loss or cleavage of a surface marker should be reevaluated and optimized by trying a different enzyme solution and applying it to the cellular compartment of interest. Additionally, we show comparison data from basal conditions, and investigators will need to ensure that a chosen dissociation protocol generates acceptable cellular recovery and viability in their model system.
In summary, our Dispase method (Dispase to flush the pulmonary circulation and inflate the lung before digestion in a DNase and collagenase solution) to prepare mouse lung tissue provides a low rate of processing-associated cell death that allows for simultaneous analysis of epithelial, endothelial, and hematopoietic lineage cells by flow cytometry. Our results suggest that beginning enzymatic tissue digestion at the time of harvest limits damage inflicted during later processing steps.
GRANTS
Our work was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under F32 HL120400 and K08 HL128867 (B. D. Singer) and K08 HL097024 (M. Damarla) as well as the Parker B. Francis Research Opportunity Award (B. D. Singer), the American Heart Association under 11FTF7280014 (N. R. Aggarwal), and the Flight Attendant Medical Research Institute Young Clinical Scientist Award (N. R. Aggarwal).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
B.D.S., J.R.M., F.R.D., N.R.A., and M.D. conception and design of research; B.D.S., N.R.A., P.M., L.J., and M.D. performed experiments; B.D.S. and M.D. analyzed data; B.D.S., J.R.M., F.R.D., N.R.A., and M.D. interpreted results of experiments; B.D.S. and M.D. prepared figures; B.D.S. and M.D. drafted manuscript; B.D.S., J.R.M., F.R.D., N.R.A., L.J., and M.D. edited and revised manuscript; B.D.S., J.R.M., F.R.D., N.R.A., P.M., L.J., and M.D. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Raffaello Cimbro for assistance in the Johns Hopkins Bayview Flow Cytometry Core.
B. D. Singer is now an Assistant Professor in the Division of Pulmonary and Critical Care at the Northwestern University Feinberg School of Medicine, Chicago, IL. J. R. Mock is now a Clinical Instructor in the Division of Pulmonary Diseases and Critical Care Medicine at the University of North Carolina, Chapel Hill, NC.
REFERENCES
- 1.Autengruber A, Gereke M, Hansen G, Hennig C, Bruder D. Impact of enzymatic tissue disintegration on the level of surface molecule expression and immune cell function. Eur J Microbiol Immunol (Bp) 2: 112–120, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bantikassegn A, Song X, Politi K. Isolation of epithelial, endothelial, and immune cells from lungs of transgenic mice with oncogene-induced lung adenocarcinomas. Am J Respir Cell Mol Biol 52: 409–417, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bharat A, Bhorade SM, Morales-Nebreda L, McQuattie-Pimentel AC, Soberanes S, Ridge K, DeCamp MM, Mestan KK, Perlman H, Budinger GRS, Misharin AV. Flow cytometry reveals similarities between lung macrophages in humans and mice. Am J Respir Cell Mol Biol 54: 147–149, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breu A, Rosenmeier K, Kujat R, Angele P, Zink W. The cytotoxicity of bupivacaine, ropivacaine, and mepivacaine on human chondrocytes and cartilage. Anesth Analg 117: 514–522, 2013. [DOI] [PubMed] [Google Scholar]
- 5.Damarla M, Parniani AR, Johnston L, Maredia H, Serebreni L, Hamdan O, Sidhaye VK, Shimoda LA, Myers AC, Crow MT, Schmidt EP, Machamer CE, Gaestel M, Rane MJ, Kolb TM, Kim BS, Damico RL, Hassoun PM. Mitogen-activated protein kinase-activated protein kinase 2 mediates apoptosis during lung vascular permeability by regulating movement of cleaved caspase 3. Am J Respir Cell Mol Biol 50: 932–941, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujino N, Kubo H, Ota C, Suzuki T, Suzuki S, Yamada M, Takahashi T, He M, Suzuki T, Kondo T, Yamaya M. A novel method for isolating individual cellular components from the adult human distal lung. Am J Respir Cell Mol Biol 46: 422–430, 2012. [DOI] [PubMed] [Google Scholar]
- 7.Huang TC, Chen JY. Proteomic analysis reveals that pardaxin triggers apoptotic signaling pathways in human cervical carcinoma HeLa cells: cross talk among the UPR, c-Jun and ROS. Carcinogenesis 34: 1833–1842, 2013. [DOI] [PubMed] [Google Scholar]
- 8.Kawasaki T, Nishiwaki T, Sekine A, Nishimura R, Suda R, Urushibara T, Suzuki T, Takayanagi S, Terada J, Sakao S, Tatsumi K. Vascular repair by tissue-resident endothelial progenitor cells in endotoxin-induced lung injury. Am J Respir Cell Mol Biol 53: 500–512, 2015. [DOI] [PubMed] [Google Scholar]
- 9.Messier EM, Mason RJ, Kosmider B. Efficient and rapid isolation and purification of mouse alveolar type II epithelial cells. Exp Lung Res 38: 363–373, 2012. [DOI] [PubMed] [Google Scholar]
- 10.Mock JR, Garibaldi BT, Aggarwal NR, Jenkins J, Limjunyawong N, Singer BD, Chau E, Rabold R, Files DC, Sidhaye V, Mitzner W, Wagner EM, King LS, D'Alessio FR. Foxp3(+) regulatory T cells promote lung epithelial proliferation. Mucosal Immunol 7: 1440–1451, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel BV, Tatham KC, Wilson MR, O'Dea KP, Takata M. In vivo compartmental analysis of leukocytes in mouse lungs. Am J Physiol Lung Cell Mol Physiol 309: L639–L652, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petit JM, Denis-Gay M, Ratinaud MH. Assessment of fluorochromes for cellular structure and function studies by flow cytometry. Biol Cell 78: 1–13, 1993. [DOI] [PubMed] [Google Scholar]
- 13.Rosich L, Xargay-Torrent S, López-Guerra M, Campo E, Colomer D, Roué G. Counteracting autophagy overcomes resistance to everolimus in mantle cell lymphoma. Clin Cancer Res 18: 5278–5289, 2012. [DOI] [PubMed] [Google Scholar]
- 14.Shynkar VV, Klymchenko AS, Kunzelmann C, Duportail G, Muller CD, Demchenko AP, Freyssinet JM, Mely Y. Fluorescent biomembrane probe for ratiometric detection of apoptosis. J Am Chem Soc 129: 2187–2193, 2007. [DOI] [PubMed] [Google Scholar]
- 15.Singer BD, Mock JR, Aggarwal NR, Garibaldi BT, Sidhaye VK, Florez MA, Chau E, Gibbs KW, Mandke P, Tripathi A, Yegnasubramanian S, King LS, D'Alessio FR. Regulatory T cell DNA methyltransferase inhibition accelerates resolution of lung inflammation. Am J Respir Cell Mol Biol 52: 641–652, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe M, Hitomi M, van der Wee K, Rothenberg F, Fisher SA, Zucker R, Svoboda KKH, Goldsmith EC, Heiskanen KM, Nieminen AL. The pros and cons of apoptosis assays for use in the study of cells, tissues, and organs. Microsc Microanal 8: 375–391, 2002. [DOI] [PubMed] [Google Scholar]