SUMMARY
Entomopathogenic bacteria produce insecticidal proteins that accumulate in inclusion bodies or parasporal crystals (such as the Cry and Cyt proteins) as well as insecticidal proteins that are secreted into the culture medium. Among the latter are the Vip proteins, which are divided into four families according to their amino acid identity. The Vip1 and Vip2 proteins act as binary toxins and are toxic to some members of the Coleoptera and Hemiptera. The Vip1 component is thought to bind to receptors in the membrane of the insect midgut, and the Vip2 component enters the cell, where it displays its ADP-ribosyltransferase activity against actin, preventing microfilament formation. Vip3 has no sequence similarity to Vip1 or Vip2 and is toxic to a wide variety of members of the Lepidoptera. Its mode of action has been shown to resemble that of the Cry proteins in terms of proteolytic activation, binding to the midgut epithelial membrane, and pore formation, although Vip3A proteins do not share binding sites with Cry proteins. The latter property makes them good candidates to be combined with Cry proteins in transgenic plants (Bacillus thuringiensis-treated crops [Bt crops]) to prevent or delay insect resistance and to broaden the insecticidal spectrum. There are commercially grown varieties of Bt cotton and Bt maize that express the Vip3Aa protein in combination with Cry proteins. For the most recently reported Vip4 family, no target insects have been found yet.
INTRODUCTION
Entomopathogenic bacteria have enormous potential for insect control, and they can provide us with an arsenal of insecticidal compounds (1). By far, the most widely used and best-known insecticidal proteins are the Cry proteins from Bacillus thuringiensis. These proteins accumulate in the parasporal crystal at the time of sporulation and are released into the culture medium only after the cell wall disintegrates. Formulations based on B. thuringiensis crystals and spores have been successfully used to control a wide range of lepidopteran pests as well as some coleopteran, blackfly, and mosquito species (2, 3). The insecticidal potency of some Cry proteins is such that their respective cry genes have been transferred to plants, conferring total or very-high-level protection against the most damaging pests (4–6).
Despite the wide success of Cry proteins in insect control, some important pests were found to be highly tolerant to Cry proteins, such as Agrotis ipsilon (Lepidoptera: Noctuidae) and Diabrotica spp. (Coleoptera: Chrysomelidae), which cause significant damage to corn. Screening programs that aimed to evaluate active insecticidal components in culture supernatants from Bacillus isolates identified a culture supernatant from Bacillus cereus AB78 that induced 100% mortality of Diabrotica virgifera virgifera and Diabrotica longicornis barberi larvae (7). The active component of this supernatant was found to be proteinaceous. Anion exchange chromatography followed by SDS-PAGE showed that the insecticidal activity was due to two different proteins of 80 and 45 kDa, which were named Vip1Aa and Vip2Aa, respectively (for vegetative insecticidal protein). Sequences with homology to the respective vip1Aa and vip2Aa genes were found in ∼12% of the 463 B. thuringiensis strains tested (7). In that same study, the vegetative culture supernatant from B. thuringiensis strain AB88 contained an 88.5-kDa protein that was highly toxic to A. ipsilon and other lepidopteran larvae, which was named Vip3Aa. More recently, Vip4Aa was reported (NCBI GenBank accession number AEB52299). In silico analysis predicted a molecular mass of ∼108 kDa for Vip4Aa (8).
Alternative names for Vip proteins were also given before standardization by the Bt Toxin Nomenclature Committee (9) (Fig. 1), such as insecticidal secreted proteins (Isp), with the classes Isp1, Isp2, and Isp3 (NCBI GenBank accession numbers AJ871923, AJ871924, and AJ872070, respectively), which are homologous to Vip1, Vip2, and Vip3, respectively. It should be mentioned that another secreted insecticidal protein from B. thuringiensis, named Sip, has been reported. This protein shares no homology with the Vip proteins and should not be mistaken for one of them (10).
FIG 1.
Nomenclature system for Vip proteins. The system consists of four ranks based on amino acid sequence identity (9). The primary, secondary, and tertiary ranks distinguish proteins with less than ∼45, 78, and 95% sequence identities, respectively. The quaternary rank distinguishes proteins sharing >95% sequence identity, which can be considered products of “allelic” forms of the same gene but can also have the same sequence that originated from different isolates.
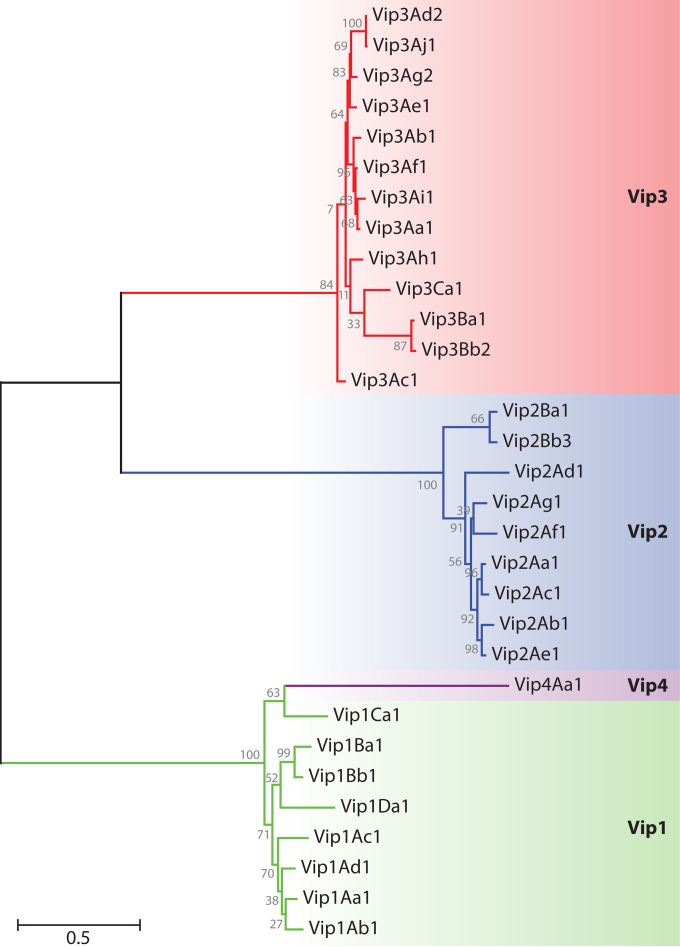
To date, 15 Vip1 proteins, 20 Vip2 proteins, 101 Vip3 proteins, and 1 Vip4 protein have been reported (9). Figure 2 shows a dendrogram with the hierarchy of the Vip proteins based on their degree of amino acid identity. Vip1 and Vip2 act as binary toxins for some members of the Coleoptera and Hemiptera (7, 11–14), and Vip3 is active against a wide range of species of Lepidoptera (15, 16). No target insects have as yet been found for Vip4. Vip1, Vip2, Vip3, and Vip4 share almost no sequence homology with each other, with Vip1 and Vip4 being the most similar (34% amino acid identity).
FIG 2.
Dendrogram showing the relationships among Vip proteins based on their degree of amino acid identity. Amino acid sequences were aligned by using the Clustal X interface (120). The evolutionary distance was calculated by maximum likelihood analysis, and the tree was constructed by using the MEGA5 program (121). The proteins used in this analysis are as follows: Vip1Aa1 (sequence identification number [Seq. ID no.] 5 in reference 28), Vip1Ab1 (Seq. ID no. 21 in reference 28), Vip1Ac1 (GenBank accession number HM439098), Vip1Ad1 (accession number JQ855505), Vip1Ba1 (accession number AAR40886), Vip1Bb1 (accession number AAR40282), Vip1Ca1 (accession number AAO86514), Vip1Da1 (accession number CAI40767), Vip2Aa1 (RCSB Protein Data Bank accession number 1QS1_A), Vip2Ab1 (Seq. ID no. 20 in reference 28), Vip2Ac1 (accession number AAO86513), Vip2Ad1 (accession number CAI40768), Vip2Ae1 (accession number EF442245), Vip2Af1 (accession number ACH42759), Vip2Ag1 (accession number JQ855506), Vip2Ba1 (accession number AAR40887), Vip2Bb3 (accession number AIA96500), Vip3Aa1 (accession number AAC37036), Vip3Ab1 (accession number AAR40284), Vip3Ac1 (named PS49C; Seq. ID no. 7 in K. Narva and D. Merlo, U.S. patent application 20,040,128,716), Vip3Ad2 (accession number CAI43276), Vip3Ae1 (accession number CAI43277), Vip3Af1 (accession number CAI43275), Vip3Ag2 (accession number ACL97352), Vip3Ah1 (accession number ABH10614), Vip3Ai1 (accession number KC156693), Vip3Aj1 (accession number KF826717), Vip3Ba1 (accession number AAV70653), Vip3Bb2 (accession number ABO30520), Vip3Ca1 (accession number ADZ46178), and Vip4Aa1 (accession number HM044666).
THE BINARY Vip1/Vip2 TOXIN
In addition to B. cereus and B. thuringiensis, vip1 and vip2 genes have also been found in other bacterial species, such as Lysinibacillus sphaericus (formerly Bacillus sphaericus) and Brevibacillus laterosporus (17, 18). Studies on the distribution of vip1 and vip2 genes have shown that they are found in ∼10% of B. thuringiensis strains (7, 19–22). These two genes have been found in the same operon and with two different open reading frames separated by an intergenic spacer of 4 to 16 bp within a 4- to 5-kb genomic sequence (7, 23, 24) and in a megaplasmid (∼328 kb in length) in B. thuringiensis strain IS5056 (25). At the time of writing of this review, the Bacillus thuringiensis Toxin Nomenclature database lists the following vip1 and vip2 genes: 3 vip1Aa, 1 vip1Ab, 1 vip1Ac, 1 vip1Ad, 2 vip1Ba, 3 vip1Bb, 1 vip1Bc, 2 vip1Ca, and 1 vip1Da, and 3 vip2Aa, 1 vip2Ab, 2 vip2Ac, 1 vip2Ad, 3 vip2Ae, 2 vip2Af, 2 vip2Ag, 2 vip2Ba, and 4 vip2Bb genes (9).
Vip1 and Vip2 proteins are expressed concomitantly, and translation from the same transcript appears to be essential to ensure high levels of both proteins. They are produced during the vegetative growth phase of B. thuringiensis, and their levels remain high until after the sporulation stage. Gene transcripts are detected at the start of the logarithmic phase, reaching their maximum expression levels in the stationary phase and remaining at high levels in the sporulation stage (15, 23, 24, 26).
Protein Structure and Function
Classical binary bacterial toxins of the “A-B” type, such as cholera toxin, interact with cells as a complex composed of one or several polypeptides associated in solution (in the case of cholera toxin, the A component is surrounded by 5 B polypeptides). Alternatively, Gram-positive bacilli from the genera Clostridium and Bacillus produce proteins with a synergistic binary mode of action in which the two proteins do not form an aggregate before binding to the cell surface (binary toxins of the “A+B” type) (27). The Vip1/Vip2 toxin is an example of an A+B toxin related to mammalian toxins from Clostridium spp. (C. botulinum, C. difficile, C. perfringens, and C. spiroforme) and Bacillus anthracis. Sequence homology with the mammalian toxins, the lack of toxicity of the individual proteins, data from translational frameshift mutation experiments with the vip1 gene, along with data from toxicity bioassays against susceptible insects confirmed the binary mode of action of these proteins (7).
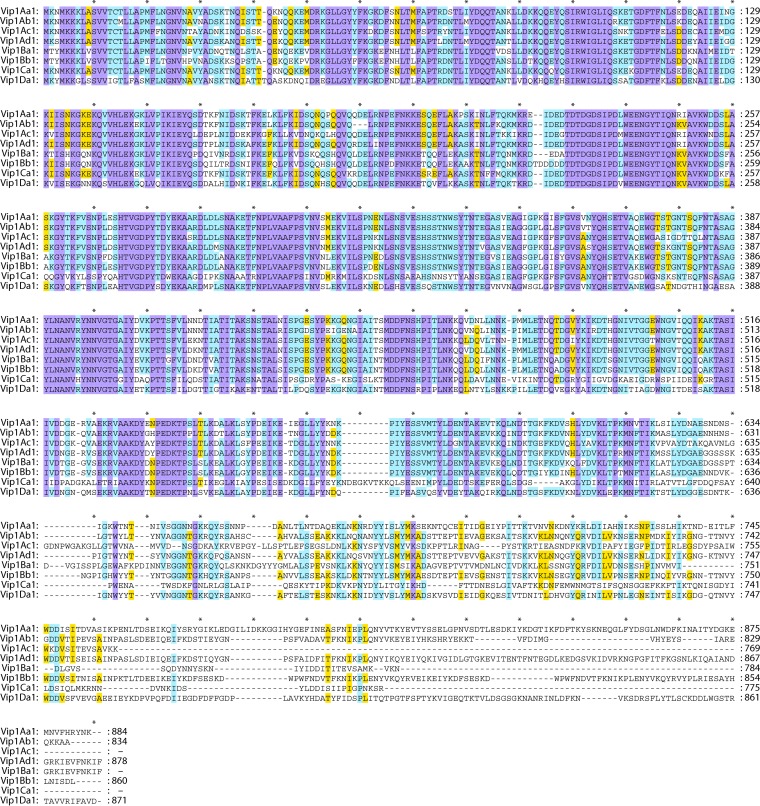
Sequence analysis of the Vip1Aa and Vip2Aa proteins revealed the presence of N-terminal signal peptides of ∼30 and 50 amino acids, respectively (23, 26, 28). The signal peptide was shown to be cleaved during secretion, rendering mature proteins of ∼82 kDa (for Vip1Aa) and 45 kDa (for Vip2Aa) (7, 24). Sequence alignment revealed that the N terminus of Vip1 is highly conserved (75 to 91% identity) (Fig. 3). In contrast, the C terminus of Vip1 is much less conserved (23 to 35% identity) (7, 23, 26).
FIG 3.
Multiple-sequence alignment of the Vip1 proteins. Sequence identity is indicated by shading, where violet is 100% sequence identity, pale blue is 80 to 100%, yellow is 60 to 80%, and white is <60%. Intervals of 10 amino acids are marked with “*.” SP, signal peptide. Proteins used in this analysis are as follows: Vip1Aa1 (Seq. ID no. 5 in reference 28), Vip1Ab1 (Seq. ID no. 21 in reference 28), Vip1Ac1 (GenBank accession number HM439098), Vip1Ad1 (accession number JQ855505), Vip1Ba1 (accession number AAR40886), Vip1Bb1 (accession number AAR40282), Vip1Ca1 (accession number AAO86514), and Vip1Da1 (accession number CAI40767).
Vip1 has moderate sequence identity with the binding component C2-II of the C2 C. botulinum toxin (29%) and the Ib component of iota-toxin from C. perfringens (31%). It also shares 33 to 38% identity with the C. spiroforme toxin, the B. anthracis protective antigen, and toxin B of C. difficile at amino acids 142 to 569 (23, 26, 29). Vip2 shares >30% sequence identity with the clostridial Rho-ADP-ribosylating exotoxin C3 (30). These similarities suggested that the Vip1 protein is the “B” component and that the Vip2 protein is the “A” component of the binary toxin (27). Thus, Vip1 acts as the binding and translocation component (channel-forming protein) (31–33), and Vip2 enters the cell and exerts its toxic effect.
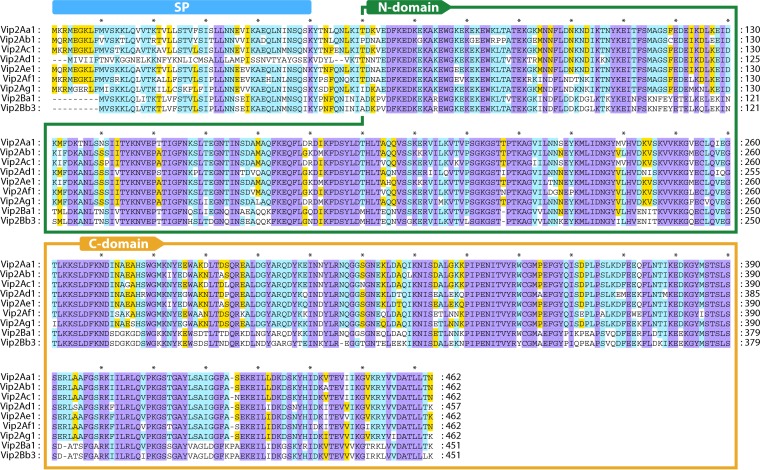
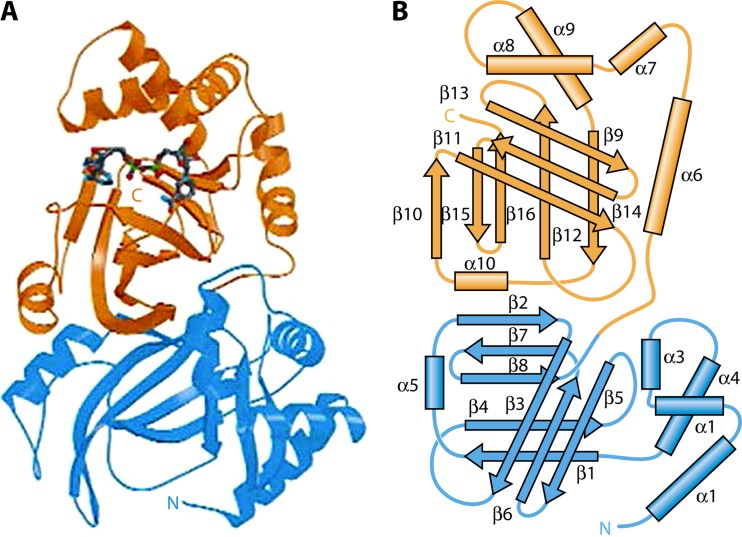
Vip2 is a NAD-dependent actin-ADP-ribosylating toxin (34) that has two distinctive domains: the N-terminal domain, composed of amino acids 60 to 265, and the C-terminal domain, composed of amino acids 266 to 461, which is the NAD-binding domain (Fig. 4) (30). Despite their limited sequence homology to each other, crystallography structure analysis of the Vip2 N- and C-terminal domains showed homology in their structures (Fig. 5). Each domain core is formed mainly by the perpendicular packing of a five-stranded mixed β-sheet with a three-stranded antiparallel β-sheet. The three-stranded sheet is flanked by four consecutive α-helices, and the five-stranded sheet is flanked by an additional α-helix (30). The overall fold of each domain resembles the catalytic domains of classical A-B toxins. In fact, crystal structure superposition of Vip2 and the clostridial toxin C3, along with sequence alignment, suggests that the class of Vip2 toxins has arisen by a single gene duplication of an ancestral ADP-ribosyltransferase. This duplication event would have been followed by further divergence by which the N-terminal domain would have lost catalytic function and evolved into a binding component, to finally give rise to a new protein family with the ability to bind to other carrier proteins (e.g., Vip1) and thereby act as binary toxins (30, 35, 36).
FIG 4.
Multiple-sequence alignment of the Vip2 proteins. Sequence identity is indicated by shading, where violet is 100% sequence identity, pale blue is 80 to 100%, yellow is 60 to 80%, and white is <60%. Intervals of 10 amino acids are marked with “*.” SP, signal peptide. The N-terminal domain (N-domain) and C-terminal domain (C-domain) are framed within boxes. The protein sequences used in this analysis are as follows: Vip2Aa1 (RCSB Protein Data Bank accession number 1QS1_A), Vip2Ab1 (Seq. ID no. 20 in reference 28), Vip2Ac1 (GenBank accession number AAO86513), Vip2Ad1 (accession number CAI40768), Vip2Ae1 (accession number EF442245), Vip2Af1 (accession number ACH42759), Vip2Ag1 (accession number JQ855506), Vip2Ba1 (accession number AAR40887), and Vip2Bb3 (accession number AIA96500).
FIG 5.
Tridimensional structure of Vip2 showing the two domains in different colors (N-terminal domain in blue and C-terminal domain in orange). (A) Schematic ribbon representation showing the NAD molecule (in blue) bound to the C-terminal domain. (B) Schematic drawing with secondary structure nomenclature. (Reprinted from reference 30 by permission from Macmillan Publishers Ltd.)
Insecticidal Activity
The toxicity of Vip1, Vip2, and their combination has been tested against a number of insect species belonging to the orders Coleoptera, Lepidoptera, Diptera, and Hemiptera as well as nematodes (Table 1). So far, toxicity against 10 coleopteran species (7, 11, 12, 24, 37, 38) and the hemipteran species Aphis gossypii (13, 14) has been found.
TABLE 1.
Spectrum of activity of individual Vip1 and Vip2 protoxins and their combinations as binary toxins
| Protein | Insect order | Insect species | Activitya (LC50) | Reference(s) |
|---|---|---|---|---|
| Vip1Aa | Coleoptera | D. virgifera virgifera | NA | 7 |
| Vip1Ac | Coleoptera | C. suppressalis, Holotrichia oblita | NA | 13 |
| Tenebrio molitor | NA | 26 | ||
| Lepidoptera | H. armigera, S. litura | NA | 13 | |
| S. exigua | NA | 26 | ||
| Diptera | C. quinquefasciatus | NA | 26 | |
| Hemiptera | A. gossypii | NA | 13, 26 | |
| Vip1Ad | Coleoptera | Anomala corpulenta, H. oblita, Holotrichia parallela | NA | 24 |
| Vip1Ae | Hemiptera | A. gossypii | NA | 14 |
| Vip1Da | Coleoptera | D. virgifera virgifera | NA | 37 |
| Vip2Aa | Coleoptera | D. virgifera virgifera | NA | 7 |
| Vip2Ac | Coleoptera | T. molitor | NA | 26 |
| Lepidoptera | H. armigera, S. exigua, S. litura | NA | 26 | |
| Vip2Ad | Coleoptera | D. virgifera virgifera | NA | 37 |
| Vip2Ae | Coleoptera | H. oblita, T. molitor | NA | 13 |
| Lepidoptera | C. suppressalis, H. armigera, S. exigua | NA | 13 | |
| Diptera | C. quinquefasciatus | NA | 13 | |
| Hemiptera | A. gossypii | NA | 13, 14 | |
| Vip2Ag | Coleoptera | A. corpulenta, H. oblita, H. parallela | NA | 24 |
| Vip1Aa + Vip2Aa | Coleoptera | Diabrotica longicornis barberi | +++ (NI) | 7 |
| Diabrotica undecimpunctata howardi | + (NI) | 7 | ||
| D. virgifera virgifera | +++ (40/20b ng/g diet) | 7 | ||
| Leptinotarsa decemlineata, T. molitor | NA | 7 | ||
| Lepidoptera | A. ipsilon, H. virescens, H. zea, M. sexta, O. nubilalis, S. exigua, S. frugiperda | NA | 7 | |
| Diptera | Culex pipiens | NA | 7 | |
| Vip1Aa + Vip2Ab | Coleoptera | D. virgifera virgifera | +++ (NI) | 7 |
| Vip1Ab + Vip2Aa | Coleoptera | D. virgifera virgifera | NA | 7 |
| Vip1Ab + Vip2Ab | Coleoptera | D. virgifera virgifera | NA | 7 |
| Vip1Ac + Vip2Ac | Coleoptera | T. molitor | NA | 26 |
| Lepidoptera | H. armigera, S. exigua, S. litura | NA | 26 | |
| Vip1Ac + Vip2Ae | Coleoptera | H. oblita, T. molitor | NA | 13 |
| Lepidoptera | C. suppressalis, H. armigera, S. exigua | NA | 13 | |
| Diptera | C. quinquefasciatus | NA | 13 | |
| Hemiptera | A. gossypii | +++ (87.5 ng/ml) | 13 | |
| Vip1Ad + Vip2Ag | Coleoptera | A. corpulenta | +++ (220 ng/g soil) | 24 |
| H. oblita | +++ (120 ng/g soil) | 24 | ||
| H. parallela | +++ (80 ng/g soil) | 24 | ||
| Vip1Ae + Vip2Ae | Hemiptera | A. gossypii | ++ (96/481b ng/ml) | 14 |
| Vip1Ca + Vip2Aa | Coleoptera | T. molitor | NA | 23 |
| Lepidoptera | H. armigera, S. exigua, S. litura | NA | 23 | |
| Diptera | C. quinquefasciatus | NA | 23 | |
| Vip1Da + Vip2Ad | Coleoptera | Anthonomus grandis | + (207 μg/ml) | 37 |
| D. longicornis barberi | +++ (213 ng/ml) | 37 | ||
| D. undecimpunctata howardi | ++ (4.91 μg/ml) | 37 | ||
| D. virgifera virgifera | +++ (437 ng/ml) | 37 | ||
| L. decemlineata | +++ (37 ng/ml) | 37 | ||
| Lepidoptera | H. virescens, H. zea, M. sexta, O. nubilalis, Sesamia nonagrioides, S. littoralis, S. frugiperda | NA | 37 | |
| Vip1Ac-like/Vip2Ac-like | Coleoptera | Sitophilus zeamais | ++ (NI) | 38 |
| Vip1/Vip2 | Nematoda | Caenorhabditis elegans, Pristionchus pacificus | NA | 122 |
| Vip1Ba1-Vip2Ba1 | Coleoptera | D. virgifera virgifera | +++ (NI) | 11 |
| Vip1Aa2-Vip2Aa2 | Coleoptera | D. virgifera virgifera | +++ (NI) | 12 |
| Lepidoptera | H. virescens, H. zea | NA | 12 | |
| Vip1Bb1-Vip2Bb1 | Coleoptera | D. virgifera virgifera | +++ (NI) | 12 |
| Lepidoptera | H. virescens, H. zea | NA | 12 |
The number of “+” symbols reflects the activity level. NA, not active; NI, no information on the LC50.
Proportion of Vip1/Vip2 that gives 50% mortality.
Testing of individual Vip1 or Vip2 proteins against a number of insect species from different orders confirmed the fact that these proteins must act together to be toxic, since neither protein alone displayed any toxic activity against the species tested (Table 1). Another interesting feature of these toxins comes from experiments combining different pairs of proteins. The Vip1Aa/Vip2Aa binary toxin (carried on and expressed from the same operon) is active against D. virgifera virgifera, but Vip1Ab/Vip2Ab (carried on and expressed from the same operon) has no activity against this insect. Interestingly, the Vip1Aa/Vip2Ab combination is active, whereas its counterpart Vip1Ab/Vip2Aa is not, suggesting that the lack of toxicity of the Vip1Ab/Vip2Ab pair to D. virgifera virgifera is due to the Vip1Ab component (7).
Mode of Action
The molecular mechanism of the insecticidal activity of the Vip1/Vip2 toxin is not totally understood (Fig. 6). The multistep process begins with the ingestion of the toxin by the larva, followed by proteolytic activation in the midgut by trypsin-like proteases. The activated monomer of Vip1Ac has been shown to form oligomers containing seven Vip1 molecules (29). These oligomers recognize specific receptors in the midgut brush border membrane, where the toxin is then inserted into the membrane. Evidence that the Vip1 component is involved in receptor recognition was in part provided by the finding that Vip1Aa cannot be replaced by Vip1Ab without losing toxicity to D. virgifera virgifera (7). The first Vip1-binding protein described was identified in A. gossypii by ligand blot analysis and was ∼50 kDa; concomitantly, no binding of Vip1 to brush border membrane vesicles (BBMVs) from nonsusceptible insect species was observed (14).
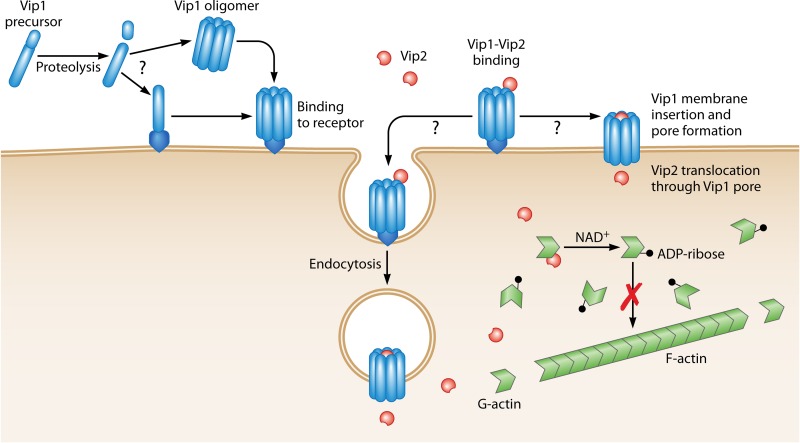
FIG 6.
Proposed mode of action of the binary Vip1/Vip2 toxin. The Vip1 protoxin is proteolytically processed by midgut proteases. The activated toxin binds to specific receptors either as a monomeric form or after oligomerization. Vip2 then binds to the oligomeric Vip1 protein and enters the cell either by endocytosis of the whole complex or directly through the pore formed by Vip1. Once inside the cytosol, Vip2 catalyzes the transfer of the ADP-ribose group from NAD to the actin monomers, preventing their polymerization.
In vitro experiments showed that Vip1 formed membrane pores in artificial lipid bilayers (29). The pores had two different conductance states, suggesting the simultaneous formation of two different channels. Vip1Ac channels are asymmetric and moderately anion selective. The putative channel-forming domain of Vip1 contains two negatively charged (E340 and E345) and two positively charged (K351 and H363) amino acids, which are hypothesized to contribute to the selectivity of the channel (29).
The mechanism by which Vip2 enters the cell is still unknown, but based on its homology with the C2-I component of the C2 clostridial binary toxin, it seems likely that Vip2 enters the cell via receptor-mediated endocytosis (27). Leuber et al. (29) proposed a second possibility, in which the strong outward proton gradient across the midgut brush border membrane of insect cells (maintained by the highly alkaline midgut fluids of the larvae) could favor Vip2Ac being “directly” delivered into the cytoplasm of the midgut cells via the channel formed by Vip1Ac. Experimental evidence favoring either one of these mechanisms is lacking. Once inside the cytosol, the catalytic Vip2 domain would catalyze the transfer of the ADP-ribose group from NAD to actin, preventing its polymerization and thus inhibiting microfilament network formation (30, 34).
Expression in Plants
Despite the economic importance of Vip1 and Vip2 as effective toxins against the major corn pest D. virgifera virgifera, expression of the binary toxin in planta has not been possible due to the cytotoxic activity of the Vip2 protein. In fact, Vip2 expression in yeast resulted in serious developmental pathology and phenotypic alterations (34). To overcome this problem, Jucovic et al. (34) designed a new zymogene strategy that consisted of the expression of a zymogenic form of Vip2 called “ProVip2.” The Vip2 proenzyme was obtained by extension of the C-terminal portion of the protein in such a way that it masked the enzymatic activity. The additional C-terminal peptide was effectively eliminated by the proteolytic action of D. virgifera virgifera midgut enzymes, and insects on a diet containing ProVip2 transgenic corn and Vip1 were all killed. Transformed plants had a phenotype unrecognizable from that of controls.
THE Vip3 LEPIDOPTERAN-ACTIVE PROTEIN
Similarly to the Vip1 and Vip2 proteins, Vip3 proteins are produced during the vegetative growth phase of B. thuringiensis and can be detected in culture supernatants from 15 h postinoculation to beyond sporulation, which reflects their high stability (15, 39). A study of the vip3Aa16 gene reported that the transcription start point was located 101 bp upstream of the start codon and that the −35 and −10 promoter regions were very similar to the B. subtilis promoters that are under the control of the σE holoenzyme. These results strongly suggested that the vip3Aa16 gene is transcribed by a σ35 holoenzyme, the B. thuringiensis homolog of σE (39).
Genes coding for Vip3 proteins are commonly found among B. thuringiensis strains, and hence, some studies have even found them in 50% and up to 87% of the strains tested and in >90% of strains carrying cry1 and cry2 genes (20, 21, 40–44). vip3 genes are ∼2.4 kb in length, and they are normally carried on large plasmids (43, 45), although in some cases, they have been proposed to be located in the bacterial chromosome (46). Many strategies for screening of B. thuringiensis isolates have been performed with the aim of isolating new vip3 genes (19, 20, 25, 42, 46–52). At the time of writing of this review, there have been 54 vip3Aa, 2 vip3Ab, 1 vip3Ac, 4 vip3Ad, 1 vip3Ae, 3 vip3Af, 15 vip3Ag, 1 vip3Ah, 1 vip3Ai, 2 vip3Ba, 3 vip3Bb, and 4 vip3Ca genes reported (9). It is not surprising that most studies on the Vip3 proteins have been carried out with the most abundant Vip3Aa proteins, and hence, very little information is available on the Vip3B and Vip3C proteins and other less common proteins of the Vip3A family (Vip3Ab and Vip3Ac, etc.). Unfortunately, early papers omitted the tertiary rank for the Vip3 proteins, referring just to Vip3A. Although these studies were most likely carried out on Vip3Aa, in this review, we follow the nomenclature provided by the authors whenever we found that it was not possible to identify the protein by accession number or by any other means.
Protein Structure and Function
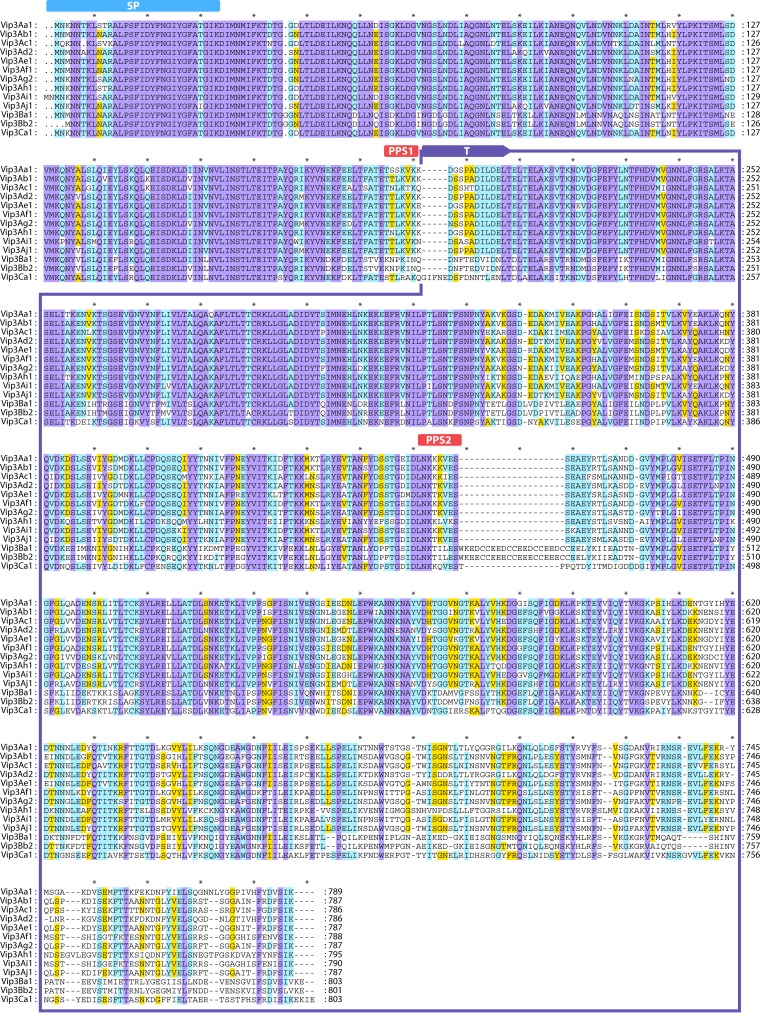
The number of amino acids in any particular Vip3 protein is ∼787, and the protein has an average molecular mass of ∼89 kDa. The N terminus of Vip3 is highly conserved, while the C-terminal region is highly variable (16, 50, 53) (Fig. 7); thus, the C-terminal region was proposed to be involved in target specificity (53).
FIG 7.
Multiple-sequence alignment of the Vip3A proteins. Sequence identity is indicated by shading, where violet is 100% sequence identity, pale blue is 80 to 100%, yellow is 60 to 80%, and white is <60%. SP, signal peptide (50); “T,” 65-kDa fragment after proteolysis; “PPS1” and “PPS2,” first and second processing sites, respectively (50). Intervals of 10 amino acids are marked with “*.” The protein sequences used in this analysis are as follows: Vip3Aa1 (GenBank accession number AAC37036), Vip3Ab1 (accession number AAR40284), Vip3Ac1 (named PS49C; Seq. ID no. 7 in Narva and Merlo, U.S. patent application 20,040,128,716), Vip3Ad2 (accession number CAI43276), Vip3Ae1 (accession number CAI43277), Vip3Af1 (accession number CAI43275), Vip3Ag2 (accession number ACL97352), Vip3Ah1 (accession number ABH10614), Vip3Ai1 (accession number KC156693), Vip3Aj1 (accession number KF826717), Vip3Ba1 (accession number AAV70653), Vip3Bb2 (accession number ABO30520), and Vip3Ca1 (accession number ADZ46178).
Vip3A proteins contain three cysteine residues. Point mutations in each of these three residues resulted in a loss of activity. However, this loss of activity was related to trypsin sensitivity rather than to the disruption of potential disulfide bonds (54).
The N terminus of Vip3 proteins contains a signal peptide that is responsible for the translocation of the protein across the cell membrane. It consists of a few positively charged amino acids, followed by a hydrophobic region, which are not removed after secretion from the bacterial cell (15, 55, 56). Without a clear putative cleavage site, the size of the signal peptide varies depending on the protein sequence itself and on the program used for prediction and ranges from 11 to 28 amino acids (15, 55, 56). Since the secretion of proteins commonly implies the excision of the signal peptide, the secretion mechanism for the Vip3 proteins is still unclear.
The highly conserved amino acid sequence of the N-terminal region of Vip3A proteins suggests that this region likely plays an important role in protein structure and insecticidal activity. However, contradictory results have been obtained in experiments testing the insecticidal activity of mutant Vip3A proteins with deletions at the N-terminal end. Deletion of the first 198 amino acids (which corresponds to the 22-kDa proteolytic fragment described by Estruch and Yu [57]) abolished toxicity to Helicoverpa armigera (Lepidoptera: Noctuidae) and Spodoptera exigua (Lepidoptera: Noctuidae) (58). Deletion of the first 27 N-terminal amino acids from Vip3Aa rendered the protein inactive due to a total loss of solubility (56). The deletion of the first 39 N-terminal amino acids from Vip3Aa differentially affected the toxicity of this protein toward the two susceptible insect species Spodoptera litura (Lepidoptera: Noctuidae) and Chilo partellus (Lepidoptera: Crambidae) (59). Contrary to the above-described results, Gayen et al. (60) found that a deletion of the first 200 N-terminal amino acids enhanced the insecticidal potency of the core active toxin ∼2 to 3-fold against H. armigera, A. ipsilon, Spodoptera littoralis (Lepidoptera: Noctuidae), and Scirpophaga incertulas (Lepidoptera: Pyralidae). Similarly, in another study (49), a deletion of 33 amino acids from the Vip3Aa N terminus caused no loss of toxicity against S. litura, Plutella xylostella (Lepidoptera: Plutellidae), and Earias vitella (Lepidoptera: Noctuidae).
The function of some C-terminal modifications has also been studied, without leading to a general conclusion. Usually, the effect of the same change varies among different insect species, preventing a consensus about the contribution of certain regions or amino acid positions to the toxicity of Vip3A proteins (49, 56, 58–60). There is generally agreement in that the last amino acids of the C terminus are critical for the activity and stability of Vip3 proteins, since their deletion, their replacement by nonconservative residues, or the addition of amino acids to the end of the protein completely abolishes protein activity (59, 60) and increases susceptibility to proteases (57, 58). A triple mutation at the C terminus of Vip3Aa1 resulted in an unstable protein that was completely hydrolyzed by the midgut juice of A. ipsilon larvae but retained toxicity against Sf9 cells (57).
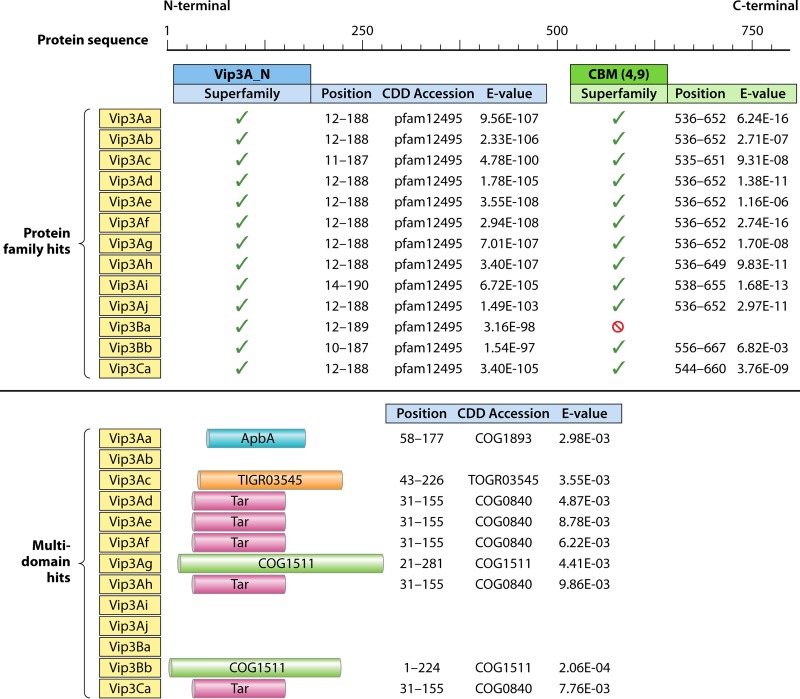
An analysis of Vip3 protein sequences using the NCBI CDD database (61) which we conducted revealed the presence of a carbohydrate-binding motif (CBM) (CBM_4_9 superfamily; pfam02018) in all Vip3 proteins with the exception of Vip3Ba (Fig. 8). The CBM spans from position 536 to a position near amino acid 652, with a consistent E value of between 10 e−4 and 10 e−17, depending on the Vip3 protein being considered. Analysis of Vip3 sequences also revealed positive hits with different multidomains in the N-terminal region, with lower E values of ∼10 e−4 and with differences depending on the Vip3 protein being considered. We did not detect any hydrophobic region susceptible to forming a transmembrane domain other than the short succession of hydrophobic amino acids in the signal peptide (15, 55).
FIG 8.
Conserved Domain Database (CDD) analysis of representative Vip3 proteins. The same sequences as those shown in Fig. 4 were used. CBM, carbohydrate-binding motif; ApbA, ketopantoate reductase motif; Tar, methyl-accepting chemotaxis protein motif; COG1511, motif of a predicted protein membrane of unknown function. TIGR03545 represents a relatively rare but broadly distributed uncharacterized family of proteins, distributed in 1 to 2% of bacterial genomes.
Comparison of the Vip3Aa1 sequence with those of the Vip3B- and Vip3C-type proteins reveals differences distributed throughout the length of the protein (Fig. 7). Nevertheless, maximum divergence was found at the C terminus, as occurs among Vip3A family proteins. The N termini of the putative signal sequences of Vip3B and Vip3C are almost identical to those of all Vip3A proteins. The proteolytic processing sites are less conserved among the three Vip3 proteins, but major differences in the middle of the protein sequence are found: an insertion of 5 amino acids downstream of the first processing site for Vip3Ca1 and an insertion of 17 amino acids downstream of the second processing site for Vip3Ba1 are responsible for the change in the expected size of the toxin “active form” from 66 kDa to 69 kDa. The inserted Vip3B sequence consists of three repetitions of the pattern DCCEE, which is characterized by its high content of negatively charged amino acids (D and E) and cysteine residues. Of a total of 11 cysteine residues found in Vip3B proteins, 8 (78%) are located in this inserted sequence (50, 62). Whether the insertion of this repetitive pattern contributes to the limited insecticidal activity of the Vip3B proteins is not known.
The conformational three-dimensional (3D) structure of Vip3 proteins has not yet been elucidated. Secondary structure prediction suggests that the N terminus is composed mainly of α-helix structures, whereas the essential components of the C terminus are β-helix structures and coils, which would be consistent with its proposed role in insect specificity (50, 53). The fact that Vip3 proteins do not show homology to any protein outside their group prevents in silico modeling based on structure homology. Only a partial tertiary structure of the Vip3 protein corresponding to the last 200 amino acids has been modeled by homology to domain II of the Cry proteins (53).
Insecticidal Activity
Most of the information on the insecticidal activity of Vip3 proteins has been obtained with the most abundant variants of the Vip3Aa subfamily, and there are very few data on the toxicity of Vip3B, Vip3C, and other Vip3A proteins outside the Vip3Aa subfamily.
Insecticidal spectrum of Vip3 proteins.
Vip3A proteins are toxic to a large number of lepidopteran species. It is worth mentioning that Vip3A proteins are very active against insect species of the genus Agrotis, which are known to be tolerant to Cry proteins, and also against species of the genus Spodoptera, which display low susceptibility to Cry proteins (63). In this regard, it has been shown that deletion of the vip3A gene from the B. thuringiensis HD1 strain significantly decreased this strain's toxicity to A. ipsilon and S. exigua (64). On the other hand, other species susceptible to Cry proteins, such as Ostrinia nubilalis (Lepidoptera: Crambidae), Culex quinquefasciatus (Diptera: Culicidae), and Chironomus tepperi (Diptera: Chironomidae), are marginally or not susceptible to any Vip3A protein tested (15, 55, 65, 66). With Vip3 proteins, depending on the Vip3 protein and the insect species being considered, it is not uncommon to find that while mortality is reached with a high concentration of Vip3 protein, strong growth inhibition (or even complete growth arrest) is observed with lower concentrations (16, 62, 67–69). Therefore, “functional mortality” (dead insects plus those remaining at larval instar L1) better represents the effectiveness of the Vip3 protein in these cases (16, 70, 71).
Table 2 summarizes the results that have been reported on the insecticidal activity of proteins of the Vip3Aa subfamily. Only the values for the protoxin form are given, since there are no reports indicating relevant differences in the insecticidal activities of the protoxin and the activated forms (16), with the exceptions of the activities of Vip3Aa16 against S. exigua and Vip3Af1 against Spodoptera frugiperda (Lepidoptera: Noctuidae) (71, 72). Despite the very small differences among Vip3Aa sequences, some proteins may exhibit significant differences in toxicity to the same insect species (16, 59, 73). For example, among all Vip3Aa proteins tested, only Vip3Aa1 and Vip3Aa14 have been described to have low or no activity against H. armigera (Table 2). Nonetheless, considering that most of the data in Table 2 were obtained in different laboratories, insecticidal differences are likely to come from factors other than slight differences in protein sequence, such as the protocol used for protein preparation, purity of the sample, method of quantification, bioassay conditions, or variability among insect populations. Independent laboratories have observed a decrease in the toxicity of some Vip3A proteins after purification with metal chelate chromatography (47, 72). The effect of the method of purification on toxicity depends on both the type of protein and the insect species tested. The duration of the bioassay can also drastically affect the final outcome for some proteins, as has been shown for Vip3Aa16 with S. exigua and S. frugiperda, for which the 50% lethal concentrations (LC50s) decreased by a factor of 10 when mortality was scored at 10 days instead of 7 days (71). Ali and Luttrell (70) found that the insecticidal responses of Helicoverpa zea and Heliothis virescens (Lepidoptera: Noctuidae) to Vip3Aa varied greatly among different batches of the same protein as well as with the buffer used.
TABLE 2.
Spectrum of activity and toxicity of the Vip3Aa subfamily proteins
| Protein | Insect species | Larval instarc | Assay type | LC50 (ng/cm2)a | Scoring time (days) | Reference(s) |
|---|---|---|---|---|---|---|
| Vip3Aa | A. ipsilon | 2nd–3rd | Diet incorporation | <200.0 | 2 | 65 |
| O. nubilalis | 2nd–3rd | Diet incorporation | NA | 2 | 65 | |
| S. frugiperda | 2nd–3rd | Diet incorporation | <200.0 | 2 | 65 | |
| H. armigera | Neonate | Surface contamination | 155 | 7 | 123 | |
| Helicoverpa punctigera | Neonate | Surface contamination | 22 | 7 | 123 | |
| H. virescens | Neonate | Diet incorporation | NI | 7 | 123 | |
| H. zea | Neonate | Diet incorporation | NI | 7 | 123 | |
| A. ipsilon | 1st | Surface contamination | 17.1 | 5 | 87 | |
| Danaus plexippus | 1st | Surface contamination | NA | 5 | 87 | |
| H. zea | 1st | Surface contamination | 112.5 | 5 | 87 | |
| M. sexta | 1st | Surface contamination | 176.3 | 5 | 87 | |
| O. nubilalis | 1st | Surface contamination | NA | 5 | 87 | |
| S. frugiperda | 1st | Surface contamination | 55.9 | 5 | 87 | |
| H. armigera | Neonate | Diet incorporation | 89 | 5 | 60, 118 | |
| A. ipsilon | Neonate | Diet incorporation | 63 | 5 | 60, 118 | |
| S. littoralis | Neonate | Diet incorporation | 36 | 5 | 60, 118 | |
| S. incertulas | Neonate | Diet incorporation | 60 | 5 | 60 | |
| Vip3Aa1 | A. ipsilon | Neonate | Diet incorporation | <28 | 6 | 15 |
| H. virescens | Neonate | Diet incorporation | <420 | 6 | 15 | |
| H. zea | Neonate | Diet incorporation | ≥420 | 6 | 15 | |
| O. nubilalis | Neonate | Diet incorporation | >420 | 6 | 15 | |
| S. exigua | Neonate | Diet incorporation | <28 | 6 | 15 | |
| S. frugiperda | Neonate | Diet incorporation | <70 | 6 | 15 | |
| B. mori | Neonate | Surface contamination | 1,986 | 7 | 77 | |
| H. zea | Neonate | Surface contamination | 27.7 | 7 | 77 | |
| S. frugiperda | Neonate | Surface contamination | 6.9 | 7 | 77 | |
| S. frugiperda | Neonate | Surface contamination | 49.3 | 7 | 95 | |
| A. ipsilon | Neonate | Surface contamination | 14 | 7 | 72 | |
| S. frugiperda | Neonate | Surface contamination | 620 | 7 | 72 | |
| H. armigera | Neonate | Surface contamination | 1,660 | 7 | 16 | |
| Lobesia botrana | Neonate | Diet incorporation | 1.3 μg/ml | 7 | 16 | |
| Mamestra brassicae | Neonate | Surface contamination | 14.4 | 7 | 16 | |
| S. littoralis | Neonate | Surface contamination | 4.0 | 7 | 16 | |
| Vip3Aa7 | H. armigera | Neonate | Leaf dip | 35.6 ng/ml | 3 | 80 |
| P. xylostella | 3rd | Diet incorporation | 28.9 ng/ml | 3 | 80 | |
| S. exigua | Neonate | Diet incorporation | 46.1 ng/ml | 7 | 80 | |
| P xylostella | 3rd | Leaf dip | 4.9 | 3 | 54, 79 | |
| Vip3Aa9 | A. ipsilon | 1st | Leaf dip | 2,165 | 1 | 59 |
| C. partellus | 1st | Leaf dip | 8 | 1 | 59 | |
| Phthorimaea operculella | 1st | Leaf dip | 370 | 1 | 59 | |
| P xylostella | 1st | Leaf dip | 36 | 1 | 59 | |
| S. litura | 1st | Leaf dip | 5 | 1 | 59 | |
| Vip3Aa10 | A. ipsilon | Neonate/1st | Surface contamination | 80.7 | 6 | 55 |
| B. mori | Neonate/1st | Surface contamination | NA | 6 | 55 | |
| C. quinquefasciatus | Neonate/1st | In water | NA | 6 | 55 | |
| H. armigera | Neonate/1st | Surface contamination | 325.2 | 6 | 55 | |
| P. xylostella | Neonate/1st | Leaf dip | 220.7 | 6 | 55 | |
| S. litura | Neonate/1st | Surface contamination | 45.4 | 6 | 55 | |
| Vip3Aa11 | H. armigera | 1st | Diet incorporation | 25.7 ng/mg | 7 | 42 |
| Ostrinia furnacalis | 1st | Diet incorporation | 720 μg/ml | 7 | 42 | |
| P. xylostella | 1st | Leaf dip | 4.2 mg/ml | 4 | 42 | |
| S. exigua | 1st | Diet incorporation | 1.3 ng/mg | 7 | 42 | |
| Vip3Aa13 | H. armigera | Neonate | Diet incorporation | 160 ng/ml | 2 | 56 |
| S. exigua | Neonate | Diet incorporation | 740 ng/ml | 2 | 56 | |
| S. litura | Neonate | Diet incorporation | 270 ng/ml | 2 | 56 | |
| Vip3Aa14 | Earias vitella | Neonate | Leaf dip | 794 | 3 | 49 |
| H. armigera | Neonate | Leaf dip | NA | 3 | 49 | |
| Pieris brassicae | Neonate | Leaf dip | NA | 3 | 49 | |
| P. xylostella | Neonate | Leaf dip | 120 | 3 | 49 | |
| S. litura | Neonate | Leaf dip | 12 | 3 | 49 | |
| H. armigera | Neonate | Diet incorporation | NA | 3 | 78 | |
| P. xylostella | Neonate | Leaf dip | NA | 3 | 78 | |
| S. litura | Neonate | Leaf dip | 0.1 | 3 | 78 | |
| Vip3Aa16 | P. oleae | 3rd | Leaf dip | NI | 5 | 97 |
| S. littoralis | 1st | Surface contamination | 305 | 6 | 68 | |
| E. kuehniella | 1st | Diet incorporation | 36 | 6 | 88 | |
| S. exigua | Neonate | Surface contamination | 2,600 | 7 | 71 | |
| 290 | 10 | 71 | ||||
| S. frugiperda | Neonate | Surface contamination | 340 | 7 | 71 | |
| 24 | 10 | 71 | ||||
| A. segetum | 1st | Surface contamination | 86 | 6 | 69 | |
| Tuta absoluta | 3rd | Leaf dip | 335 | 3 | 90 | |
| Ectomyelois ceratoniae | Neonate | Diet incorporation | 40b | 5 | 91 | |
| Vip3Aa19 | H. armigera | 1st | Diet incorporation | 24.1 ng/mg | 7 | 42 |
| O. furnacalis | 1st | Diet incorporation | >100 μg/ml | 7 | 42 | |
| P. xylostella | 1st | Leaf dip | 59.8 μg/ml | 4 | 42 | |
| S. exigua | 1st | Diet incorporation | 1.4 ng/mg | 7 | 42 | |
| H. virescens | 1st | Diet incorporation | 1.35 μg/ml | 7 | 124 | |
| P. xylostella | 1st | Leaf dip | 2236 μg/ml | 5 | 124 | |
| H. zea | Neonate | Surface contamination | 500 | 7 | 109 | |
| Vip3Aa29 | C. quinquefasciatus | — | In water | NA | 2 | 66 |
| C. suppressalis | — | Diet incorporation | 24.0 μg/ml | 5 | 66 | |
| C. tepperi | — | In water | NA | 2 | 66 | |
| H. armigera | — | Diet incorporation | 22.6 μg/mld | 5 | 66 | |
| S. exigua | — | Diet incorporation | 36.6 μg/ml | 5 | 66 | |
| Vip3Aa43 | S. albula | Neonate | Surface contamination | 3.9 | 7 | 74 |
| S. cosmioides | Neonate | Surface contamination | 2.8 | 7 | 74 | |
| S. eridania | Neonate | Surface contamination | 3.4 | 7 | 74 | |
| S. frugiperda | Neonate | Surface contamination | 24.7 | 7 | 74 | |
| Vip3Aa45 | Chrysodeixis chalcites | Neonate | Surface contamination | 1,044.6 | 7 | 73 |
| L. botrana | Neonate | Diet incorporation | 1.96 μg/ml | 7 | 73 | |
| M. brassicae | Neonate | Surface contamination | 39.7 | 7 | 73 | |
| S. exigua | Neonate | Surface contamination | 119.7 | 7 | 73 | |
| S. littoralis | Neonate | Surface contamination | 18.7 | 7 | 73 | |
| Vip3Aa50 | Anticarsia gemmatalis | Neonate | Surface contamination | 20.3 | 7 | 125 |
| S. frugiperda | Neonate | Surface contamination | 79.6 | 7 | 125 | |
| Vip3Aa58 | S. exigua | Neonate | Surface contamination | 160 | 10 | 47 |
| Cydia pomonella | Neonate | Surface contamination | 2,380 | 10 | 47 | |
| Dendrolimus pini | 2nd | Leaf dip | 23,550 | 10 | 47 | |
| Vip3Aa59 | S. exigua | Neonate | Surface contamination | 190 | 10 | 47 |
| C. pomonella | Neonate | Surface contamination | 2,750 | 10 | 47 | |
| D. pini | 2nd | Leaf dip | 16,260 | 10 | 47 |
Unless otherwise stated, LC50s are given in nanograms per square centimeter and refer to the protoxin form of the proteins. NA, not active; NI, no information on the LC50 is available, although the protein was active.
Although the LC50 is given in nanograms per square centimeter, the bioassay was performed by using diet incorporation.
—, not specified.
The 50% inhibitory concentration is shown instead of the LC50.
Table 3 summarizes the bioassay data on Vip3A proteins other than those of the Vip3Aa subfamily, and Table 4 summarizes the bioassay data on Vip3 proteins other than those of the Vip3A family.
TABLE 3.
Spectrum of activity and toxicity of Vip3A proteins other than those of the Vip3Aa subfamily
| Protein | Insect species | Larval instar | Assay type | LC50 (ng/cm2)a | Reference |
|---|---|---|---|---|---|
| Vip3Ab1 | A. ipsilon | Neonate | Surface contamination | 62 | 16 |
| S. exigua | Neonate | Surface contamination | 597 | 16 | |
| S. frugiperda | Neonate | Surface contamination | 2,020 | 16 | |
| S. littoralis | Neonate | Surface contamination | 163 | 16 | |
| Vip3Ac1 | Anopheles gambiae | —b | — | NA | 77 |
| B. mori | Neonate | Surface contamination | 44.8 | 77 | |
| D. virgifera virgifera | — | — | NA | 77 | |
| H. zea | Neonate | Surface contamination | 133.7 | 77 | |
| O. nubilalis | Neonate | Surface contamination | NA | 77 | |
| S. frugiperda | Neonate | Surface contamination | 11.6 | 77 | |
| Vip3Ad2 | A. ipsilon | Neonate | Surface contamination | >4,000 | 72 |
| S. frugiperda | Neonate | Surface contamination | >4,000 | 72 | |
| Vip3Ae1 | A. ipsilon | Neonate | Surface contamination | 4 | 72 |
| S. frugiperda | Neonate | Surface contamination | 28 | 72 | |
| S. exigua | Neonate | Surface contamination | 11.1 | 94 | |
| S. frugiperda | Neonate | Surface contamination | 20 | 94 | |
| H. armigera | Neonate | Surface contamination | 4,460 | 16 | |
| L. botrana | Neonate | Diet incorporation | 0.2 μg/ml | 16 | |
| M. brassicae | Neonate | Surface contamination | 258 | 16 | |
| S. littoralis | Neonate | Surface contamination | 8 | 16 | |
| Vip3Af1 | S. frugiperda | Neonate | Surface contamination | 49.3 | 95 |
| A. ipsilon | Neonate | Surface contamination | 18 | 72 | |
| S. frugiperda | Neonate | Surface contamination | 60 | 72 | |
| H. armigera | Neonate | Surface contamination | 840 | 16 | |
| L. botrana | Neonate | Diet incorporation | 0.8 μg/ml | 16 | |
| M. brassicae | Neonate | Surface contamination | 6 | 16 | |
| S. littoralis | Neonate | Surface contamination | 43.2 | 16 | |
| Vip3Ag4 | C. chalcites | Neonate | Surface contamination | 45.5 | 73 |
| L. botrana | Neonate | Diet incorporation | 1.1 | 73 | |
| M. brassicae | Neonate | Surface contamination | >2,500 | 73 | |
| S. exigua | Neonate | Surface contamination | 265.2 | 73 | |
| S. littoralis | Neonate | Surface contamination | 34.9 | 73 |
LC50s refer to mortality at 7 days for the protoxin form of the proteins and are given in nanograms per square centimeter unless otherwise stated; NA, not active.
—, information not available.
TABLE 4.
Spectrum of activity and toxicity of Vip3B and Vip3C protein families
| Protein | Insect species | Larval instarb | Assay type | LC50 (ng/cm2)a | Scoring time (days) | Reference |
|---|---|---|---|---|---|---|
| Vip3Ba1 | O. nubilalis | Neonate | Surface contamination | NA | 7 | 50 |
| P. xylostella | 2nd | Leaf dip | NA | 7 | 50 | |
| Vip3Bb2 | A. gossypii | Nymph | Diet incorporation | NA | 7 | 40 |
| C. tepperi | 4th | Liquid solution | NA | 4 | 40 | |
| H. armigera | Neonate | Surface contamination | NI | 7 | 40 | |
| H. punctigera | Neonate | Surface contamination | NI | 7 | 40 | |
| Tribolium castaneum | — | Diet incorporation | NA | 10 | 40 | |
| Vip3Ca3 | A. ipsilon | Neonate | Surface contamination | >4,000 | 10 | 62 |
| C. chalcites | Neonate | Surface contamination | <400 | 10 | 62 | |
| H. armigera | Neonate | Surface contamination | <4,000 | 10 | 62 | |
| L. botrana | Neonate | Diet incorporation | >100 μg/ml | 10 | 62 | |
| M. brassicae | Neonate | Surface contamination | <4,000 | 10 | 62 | |
| O. nubilalis | Neonate | Surface contamination | >4,000 | 10 | 62 | |
| S. exigua | Neonate | Surface contamination | >4,000 | 10 | 62 | |
| S. frugiperda | Neonate | Surface contamination | >4,000 | 10 | 62 | |
| S. littoralis | Neonate | Surface contamination | <4,000 | 10 | 62 | |
| T. ni | Neonate | Surface contamination | <4,000 | 10 | 62 |
Unless otherwise stated, LC50s are given in nanograms per square centimeter and refer to the protoxin form of the proteins. NA, not active; NI, no information on the LC50 is available, although the protein was active.
—, not specified.
Interactions with other insecticidal proteins.
Synergism has been observed between the Vip3Aa and Cyt2Aa proteins against Chilo suppressalis (Lepidoptera: Crambidae) and S. exigua after their coexpression in Escherichia coli; contrarily, this protein combination was slightly antagonistic against C. quinquefasciatus (66). Bergamasco et al. (74) reported synergism between Vip3A and Cry1Ia in three Spodoptera species (S. frugiperda, S. albula, and S. cosmioides [Lepidoptera: Noctuidae]) but slight antagonism in Spodoptera eridania (Lepidoptera: Noctuidae). Antagonism between the Vip3A and Cry1A or Cry1Ca proteins in H. virescens was described (75): antagonism was found for the combination of Cry1Ca and Vip3Aa, Vip3Ae, or Vip3Af and for the combination of Vip3Af and either Cry1Aa or Cry1Ac. In that same study, Vip3Aa and Cry1Ca showed antagonism in S. frugiperda, whereas the same combination was synergistic in Diatraea saccharalis (Lepidoptera: Crambidae).
The mechanisms underlying synergism and antagonism are unknown. For the antagonism between the Vip3A and Cry1C proteins, Lemes et al. (75) hypothesized that a physical interaction of the two proteins impairs the access of the binding epitopes to the membrane receptor. Hetero-oligomer formation with an increased ability for membrane insertion or pore formation was proposed to explain the synergism between Cry1Ac and Cry1Aa (76). However, this possibility for the Vip3 and Cry1 proteins seems very unlikely because of their lack of homology.
Genetically engineered vip3A genes.
Genetic engineering allows the construction of chimeric genes that code for parts of different proteins to obtain new proteins with novel or improved properties. Knowledge of the domains of a protein is of great advantage in the design of chimeric proteins. Despite the lack of information on the tertiary structure of Vip3A proteins, two chimeras have been created by sequence swapping between the vip3Aa and vip3Ac genes with the aim of increasing host specificity (77). These chimeras were created by combining ∼600 amino acids from the N terminus of one protein and ∼180 amino acids of the C terminus of the other (Table 5). The two chimeric proteins exhibited new toxicity properties: Vip3AcAa (with the N terminus of Vip3Ac) was more toxic to all the insects tested than the two original proteins, and it even caused growth inhibition of Vip3A-tolerant O. nubilalis. In contrast, Vip3AaAc was less toxic than its counterpart and the original proteins, and it even completely lost activity against Bombyx mori (Lepidoptera: Bombycidae) (77) (Table 5). Li et al. (58) achieved an 18-fold increase in toxicity against S. exigua by changing the last two amino acids of the chimeric Vip3AcAa protein (from IK to LR).
TABLE 5.
Genetically engineered Vip3A proteins and effects on insect toxicity
| Protein | Modification type(s) | Descriptionb | Effect(s) of modificationa | Reference |
|---|---|---|---|---|
| Vip3AcAa | Domain swapping | Chimera of Vip3Ac N terminus (600 aa) and Vip3Aa C terminus (189 aa) | Gain of toxicity against O. nubilalis; IA against S. frugiperda, H. zea, and B. mori | 77 |
| Vip3AaAc | Domain swapping | Chimera of Vip3Aa1 N terminus (610 aa) and Vip3Ac C terminus (179 aa) | DA against S. frugiperda and H. zea; LA against B. mori | 77 |
| Vip3Aa14 | Protein fusion | Chimera of Vip3Aa14 and Cry1Ac | As effective as Cry1Ac against H. armigera and P. xylostella but DA compared to Vip3Aa against S. litura | 78 |
| Vip3Aa7 | Gene promoter change and protein fusion | Chimera of Cry1C promoter with truncated Vip3Aa7 (39 aa deleted at N terminus) and Cry1C C-terminal region | Higher yield of Vip3Aa7, Vip relocation in Bt inclusion bodies but DA against P. xylostella, H. armigera, and S. exigua | 80 |
| Vip3Aa7 | Protein fusion | Chimera of Vip3Aa7 and Cry9Ca N terminus | IA against P. xylostella | 79 |
DA, decrease of activity; IA, increase of activity; LA, loss of activity.
aa, amino acids.
Similar attempts have been conducted by combining the vip and cry genes. Fusion of the vip3Aa gene with cry1A rendered a fusion protein that retained the toxicity of Cry1Ac but partially lost that of Vip3Aa, possibly due to incorrect Vip3A folding (78). In another study, the vip3Aa gene was fused with the 5′ region of cry9Ca, and the resultant chimeric protein was more toxic than the individual proteins and the mixture of them, probably because Vip3Aa increased the solubility of the Cry9Ca protein (79). In an attempt to improve the Vip3Aa yield, a mutant vip3Aa gene (with the signal peptide deleted) was fused with the promoter and the 3′-terminal half of cry1C, with the result of a 9-fold increase in the expression of the recombinant protein, which was concentrated in inclusion bodies. Unfortunately, this protein showed lower insecticidal activity against the insects tested than the original Vip3Aa protein, probably due to low solubilization or improper folding of the protein (80) (Table 5).
Another type of approach has been the introduction and expression of vip3A genes in B. thuringiensis strains expressing different cry genes, to create new strains to be used in insecticidal formulations with a broader spectrum of action. Commercial formulations of B. thuringiensis strains contain small amounts of Vip proteins, since these proteins are secreted into the growth medium, which is mostly discarded during the processing of the formulation (81). This problem can be alleviated by directing the expression of the vip3A gene to the sporulation stage by using sporulation-dependent promoters and specific transcription sequences from different cry genes (82–85). The engineered strains in all these cases showed improved production of Vip3A proteins and higher toxicity to the insects tested. Cloning and expression of the vip3Aa gene in Pseudomonas fluorescens have also been accomplished with the aim of producing spray insecticides based on the Vip3A protein, either combined with Cry proteins or not (86). The heterologously expressed Vip3Aa protein, which was not secreted into the medium and remained “encapsulated” within the bacterial cell, retained full toxicity.
Mode of Action
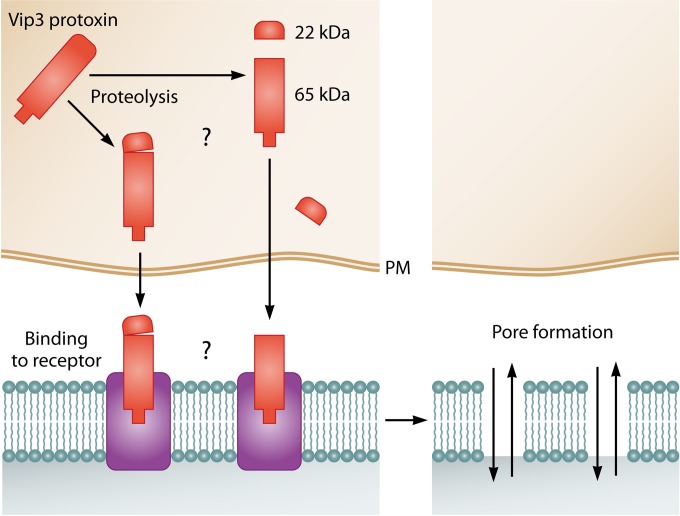
Study of the mode of action of the Vip3 proteins started soon after their discovery in 1996 by Estruch et al. (15), who proposed that Vip3 proteins would exert their toxicity via a process different from that of the Cry proteins, based on the lack of structural homology of these two types of proteins. Despite being so different, both types of toxins exert their toxic action through apparently the same sequence of events: activation by midgut proteases, crossing the peritrophic membrane, binding to specific proteins in the apical membrane of the epithelial midgut cells, and pore formation (87) (Fig. 9). So far, all reported studies on the mode of action of Vip3 proteins have been carried out with those of the Vip3A family, mostly those of the Vip3Aa subfamily. Ongoing studies on the Vip3Ca protein indicate that this protein has a mode of action similar to that of the Vip3A proteins (J. Gomis-Cebolla, I. Ruiz de Escudero, M. Chakroun, N. M. Vera-Velasco, P. Hernández-Martínez, C. S. Hernández-Rodríguez, Y. Bel, B. Escriche, P. Caballero, and J. Ferré, presented at the 48th Annual Meeting of the Society for Invertebrate Pathology, Vancouver, BC, Canada, 8 to 13 August 2015).
FIG 9.
Proposed mode of action of the Vip3 proteins. The full-length protoxin is proteolytically processed by midgut proteases. The 65-kDa fragment binds to specific receptors (with the 22-kDa fragment still bound or not). Pores are then formed, which leads to the death of the cell.
Behavioral and histopathological effects.
The behavioral symptoms observed in susceptible insects after ingestion of the Vip3Aa protein resemble the ones observed after Cry intoxication: feeding cessation, loss of gut peristalsis, and overall paralysis of the insect (65). Analysis of gut cross sections of susceptible insects after ingestion of the Vip3Aa protein shows extensive damage in the midgut, with disrupted, swollen, and/or lysed epithelial cells and leakage of cellular material into the lumen (65, 68, 69, 88–91). No damage was observed in either the foregut or the hindgut, nor was damage observed in the midgut of nonsusceptible insects (65).
Proteolytic processing.
In vitro proteolysis of full-length Vip3Aa proteins using insect midgut juice showed that they are processed to several major proteolytic products, generally of ∼62 to 66, 45, 33, and 22 kDa (65, 68, 69, 88, 89). The 22-kDa fragment corresponds to the N terminus of the protein (amino acids 1 to 198 in Vip3Aa1), the 66-kDa fragment corresponds to the remainder of the protein (from amino acid 199 to the end of Vip3Aa1), and the 45- and 33-kDa fragments are thought to be derived from the 66-kDa portion (57).
The minimal toxic fragment of the Vip3Aa protein has also been studied. Although an early study claimed that the minimal fragment that retained insecticidal activity after proteolysis was the 33-kDa fragment (57), all subsequent studies are in favor of the 62- to 66-kDa fragment being the Vip3Aa toxic core (58, 60, 68, 69, 71, 72, 87–89, 92–94). Interestingly, the 20-kDa fragment produced upon proteolytic processing of the Vip3Aa16 protoxin copurifies with the 62-kDa fragment, suggesting that after activation of the full-length protein, the two fragments remain together (89).
Compared to Cry proteins, the 62- to 66-kDa toxic core of the Vip3A proteins is more susceptible to the action of proteases. Incubation of either Vip3Aa or Vip3Ae with commercial serine proteases or insect midgut juice showed the unstable nature of the 62-kDa fragment, which started to break down even before all the protoxin was processed (65, 69, 88, 89, 94). Partial purification of peptidases from S. frugiperda midgut juice showed that cationic trypsin-like and anionic chymotrypsin-like peptidases are involved in the formation of the Vip3A 62-kDa fragment, whereas cationic chymotrypsin-like peptidases participated in its further processing (94).
In general, proteolytic activation does not seem to be a critical step in determining Vip3A insect toxicity and specificity. It has been shown that the midgut juice of a nonsusceptible insect (O. nubilalis) could process Vip3A in vitro into a 65-kDa fragment that was fully toxic when fed to susceptible insects (65). However, in some cases, the rate of processing of the full-length protein was proposed to account for differences in the toxicity of a given Vip3A protein to different insect species (71, 88, 94). Indeed, some studies have shown that differences in mortality disappeared when the trypsin-activated protein was used instead of the full-length protein (71, 72).
Binding to the larval midgut epithelium.
In vivo immunolocalization studies have shown that Vip3A binds to the apical microvilli of midgut epithelial cells (65, 89) (Fig. 10). Specific binding to brush border membrane vesicles (BBMVs) prepared from susceptible insects was first shown by using biotin-labeled Vip3Aa and Vip3Af (68, 69, 74, 87, 92, 93, 95). Interestingly, Vip3Aa also binds specifically to BBMVs of the nonsusceptible insect O. nubilalis (87), which indicates that specific binding is not sufficient to produce toxicity.
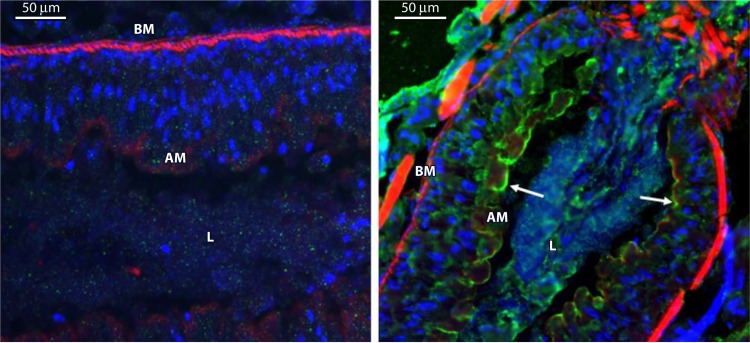
FIG 10.
Immunolocalization of Vip3Aa in midgut tissue sections after ingestion by S. frugiperda larvae. (Left) Control larvae. (Right) Larvae that ingested Vip3Aa. Nuclei were stained blue, and the apical and basal membranes were stained red. Binding of Vip3Aa to the apical membrane is shown in green. BM, basal membrane; AM, apical membrane; L, gut lumen. (Reprinted from reference 89.)
Quantitative binding parameters were obtained by using 125I-labeled Vip3Aa and S. frugiperda BBMVs. This binding was found to be saturable, mostly irreversible, and differentially affected by the presence of divalent cations (89). Vip3A proteins were also found to have lower affinities but higher numbers of binding sites than the Cry1A and Cry2A proteins. Interestingly, homologous competition showed that both the 62-kDa and the 20-kDa fragments of trypsin-activated 125I-labeled Vip3Aa bound to BBMVs, and both fragments were displaced by the addition of nonlabeled Vip3Aa. In contrast, using biotin-labeled Vip3Aa, Liu et al. (93) found that only the 62-kDa fragment was able to bind to H. armigera BBMVs and also that the 20-kDa fragment was found exclusively in the supernatant of the binding reaction mixture.
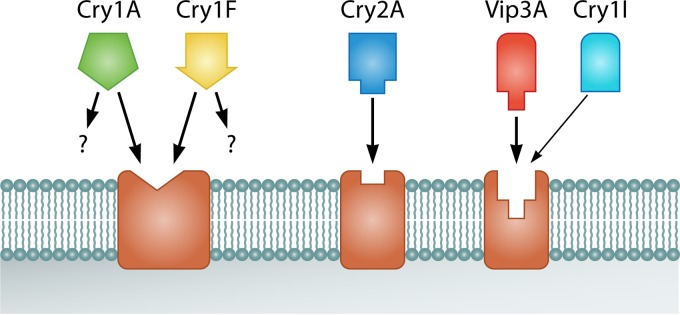
Competition binding assays showed the absence of shared binding sites between Vip3A and Cry proteins. This has been shown for Vip3Aa with Cry1Ac, Cry1Ab, Cry1Fa, Cry2Ae, and Cry2Ab in all insect species tested and for Vip3Af with Cry1Ab and Cry1F in S. frugiperda (69, 89, 92, 93, 95, 96). However, Bergamasco et al. (74) reported partial competition of Cry1Ia for the Vip3Aa-binding sites in S. eridania BBMVs but not in S. frugiperda, S. albula, and S. cosmioides BBMVs. Competition among proteins of the Vip3A family has been tested only with S. frugiperda (89). Vip3Ae, Vip3Af, and even the nonactive Vip3Ad protein competed for the Vip3Aa-binding sites, with no significant differences in their binding parameters. A general model of the binding sites of Vip3A proteins in relation to Cry proteins is shown in Fig. 11.
FIG 11.
General binding site model for the Cry and Vip proteins in the midgut epithelial membrane of lepidopteran larvae. Cry1Fa and Cry1A proteins, in addition to the shared binding site, may have other sites depending on the insect species considered. Recognition of Vip3Aa sites by Cry1Ia has been found only in S. eridania (of four Spodoptera species tested).
The interaction of Vip3Aa with BBMVs of susceptible insects involves specific binding molecules different from the ones recognized by Cry1A proteins. Ligand blot analyses revealed that Vip3Aa recognized 80- and 110-kDa proteins in Manduca sexta (Lepidoptera: Sphingidae), while Cry1Ab bound to proteins of 120 and 210 kDa (87). That same study showed that Vip3Aa was unable to bind to purified aminopeptidase N (APN) and the cadherin ectodomain toxin-binding region (TBR) from M. sexta, both membrane proteins known to bind Cry proteins (87). In Prays oleae (Lepidoptera: Yponomeutidae) and Agrotis segetum (Lepidoptera: Noctuidae), Vip3Aa bound to a 65-kDa protein, while Cry1Ac bound to a 210-kDa band in P. oleae and to a 120-kDa band in A. segetum (69, 97). In S. littoralis, Vip3Aa bound proteins of 55 and 100 kDa (68), and in Ephestia kuehniella (Lepidoptera: Pyralidae), S. frugiperda, S. albula, S. cosmioides, and S. eridania, Vip3Aa bound to a protein of 65 kDa (74, 88), to which Cry1Ia also bound in the four Spodoptera species (74).
Very few studies have addressed the identity of the Vip3A-binding molecules in the insect midgut. Two Vip3Aa-binding molecules have been identified so far by using the yeast two-hybrid system. The first one was a 48-kDa protein from A. ipsilon with homology to a family of extracellular glycoproteins called tenascins, which could be associated with apoptotic processes (57). The second binding molecule was the S2 ribosomal protein from S. litura, identified as a Vip3A receptor in Sf21 cells (98). Silencing of the S2 gene reduced the toxicity of Vip3A to both Sf21 cells and fifth-instar S. litura larvae. Both S2 and Vip3Aa colocalized on the surface and in the cytoplasm of Sf21 cells, suggesting that the interaction takes place on the cell surface and, once pores are produced, that the Vip3-S2 complex internalizes (98). How this S2-Vip3A protein interaction could trigger the lysis of cells was not explained and remains unknown. In H. armigera, the molecules that bind to Vip3Aa were found to be slightly associated with lipid rafts (93).
In an attempt to understand how midgut cells respond to intoxication by Vip3 proteins, gene expression profiles of S. exigua larvae treated with a sublethal dose of Vip3Aa were obtained by using a genome-wide microarray that included >29,000 unigenes (unique assembled sequences obtained from a transcriptome) (99). No alteration in the expression levels of the two Vip3A-binding proteins described above (S2 and the tenascin of the X-tox type) was found, nor were there alterations in the transcription levels of genes related to the mode of action of the Cry proteins. It was concluded that the lack of significant changes in the transcription levels of the above-mentioned genes was most likely either due to the fact that they were not involved in the Vip3 mode of action or because the mechanisms of defense against Vip3A toxins do not rely on the regulation of the members involved in the mode of action.
Pore formation.
Despite the absence of any predicted pore-forming structure in the Vip3 proteins, the pore formation activity of the Vip3Aa protein activated with trypsin or midgut juice has been demonstrated by voltage clamping assays with dissected midguts of M. sexta (Lepidoptera: Sphingidae) and also in planar lipid bilayers (87). In contrast, the full-length Vip3Aa protein was unable to form pores. The ion channels were able to destroy the transmembrane potential, and they were voltage independent and cation selective (87). The pore-forming ability of activated Vip3Aa was also demonstrated by fluorescence quenching using H. armigera BBMVs (93). The formation of Vip3Aa ion channels was restricted to susceptible insects, and they have been found to have biophysical properties that differ from those of Cry1Ab in M. sexta (87).
Resistance and Cross-Resistance
Very few cases of resistance to Vip3 proteins have been reported. Laboratory selection of a H. virescens colony led to 2,040-fold resistance to Vip3Aa compared to the unselected population (100). Resistance was found to be polygenic, with possible paternal influence, and ranged from almost completely recessive to incompletely dominant; fitness costs were temperature dependent, with reduced mating success, fecundity, and fertility (101). After 12 generations of selection with Vip3A, a freshly established laboratory colony of S. litura reached a resistance level of 285-fold compared to a susceptible colony (102). The resistant insects were found to lack two casein-degrading bands in nondenaturing electrophoretic gels and to have reduced proteolytic activity (∼2-fold) toward several protease substrates.
The presence of Vip3Aa resistance alleles in field populations of H. armigera and Helicoverpa punctigera (Lepidoptera: Noctuidae) was studied in Australia by using the F2 screening method (103). The results showed that resistance alleles in both insect species existed as natural polymorphisms at a relatively high frequency (0.027 and 0.008, respectively), above mutation rates normally encountered (103). Interestingly, within each species, the resistance of two different F2 families was due to alleles at the same locus, and resistance was found to be essentially recessive, most probably conferred by a single gene, and did not result in cross-resistance to Cry1Ac or Cry2Ab (103). The frequency of resistant alleles in H. armigera did not increase over the following four seasons (until 2014/2015), and resistant insects were found to activate the Vip3Aa protoxin more slowly than susceptible insects, although no significant differences in binding to membrane receptors were found (M. Chakroun, N. Banyuls, T. Walsh, S. Downes, B. James, and J. Ferré, submitted for publication). Further studies on the resistant H. punctigera strain confirmed that there was no linkage between the Vip3A and the Cry2Ab resistance loci (104). A study on the presence of Vip3Aa resistance alleles in field populations of S. frugiperda from different states of Brazil, using the F2 method, estimated an overall frequency of 0.0009, which is relatively low (105).
The increased use of Vip3 toxins in pyramided B. thuringiensis-treated crops (Bt crops) to improve both pest control and resistance management sparked interest in the evaluation of cross-resistance between Cry and Vip3A proteins (106). So far, no significant cross-resistance between these two classes of proteins has been described. Vip3Aa was found to be equally toxic to one susceptible and three Cry-resistant H. virescens strains (YHD2, resistant to Cry1Ac and Cry1F and slightly cross-resistant to Cry2A, and CXC and KCBhyb, both resistant to Cry1Ac, Cry1Aa, Cry1Ab, Cry1F, and Cry2Aa2) (107). Two studies on Cry1Ac-resistant strains of H. zea showed no significant cross-resistance to Vip3A or Cry2Ab (108, 109). A study on two H. armigera populations from Cry1Ac-cotton planting regions in China showed a lack of significant correlation between the responses to Vip3Aa and those to Cry1Ac, suggesting little or no cross-resistance between these two toxins (110). Cross-resistance to Vip3A has also been studied in two S. frugiperda Cry1F-resistant populations, one collected from Bt maize fields in Puerto Rico and the other collected from the southeast United States. Both populations were very susceptible to Vip3Aa, indicating the absence of cross-resistance between the Vip3Aa and Cry1F proteins (111, 112). A study using a different Vip3A protein, Vip3Ac, showed that it was equally toxic to susceptible and Cry1Ac-resistant Trichoplusia ni (Lepidoptera: Noctuidae) strains (77). However, in this case, the resistant strain was slightly less susceptible to Vip3Aa (resistance ratio of 2.1) and to two Vip3A chimeric proteins (resistance ratios of 1.8 and 3.2) (77).
Expression in Plants
The vip3Aa gene has been successfully introduced into cotton and corn and was later combined with other cry genes to confer higher-level protection and delay insect resistance (http://www.epa.gov/ingredients-used-pesticide-products/current-previously-registered-section-3-plant-incorporated). VipCot and Agrisure Viptera were registered in the United States in 2008 and 2009, respectively (Syngenta Seeds, Inc.). The former is the result of the transformation event COT102 in cotton, which produces the Vip3Aa19 protein [see http://www.isaaa.org/gmapprovaldatabase/gene/default.asp?GeneID=24&Gene=vip3A(a) and http://en.biosafetyscanner.org/schedaevento.php?evento=208], whereas the latter is the result of event MIR162 in corn, which produces the Vip3Aa20 protein (http://iaspub.epa.gov/apex/pesticides/f?p=CHEMICALSEARCH:30#p). Both events were pyramided with cry1Ab (VipCot Vip3Aa plus mCry1Ab and Agrisure Viptera Vip3Aa plus Cry1Ab) and later with cry1Fa (VipCot Vip3Aa plus Cry1Ac plus Cry1Fa and Agrisure Viptera Vip3Aa plus Cry1Ab plus Cry1Fa) to confer wider and more robust protection against Lepidoptera (113–115). Furthermore, corn event MIR162 has been stacked with other cry genes expressing proteins that are active against Coleoptera (Cry3A and eCry3.1Ab) to confer protection against these two insect orders (116). A 3-year study on the field performance of VipCot expressing just the Vip3Aa protein indicated that the plants were highly efficacious against H. armigera early in the season but that efficacy declined as the season progressed although not so drastically as Cry1Ac in Bollgard or Ingard cotton (117). In 2015, the first modified Vip3A protein, with improved toxicity, was introduced into tobacco, conferring almost total protection against H. armigera, A. ipsilon, and S. littoralis (118).
Cotton has also been transformed with a synthetic vip3A gene fused to a chloroplast transit peptide coding sequence (119). The Vip3A protein accumulated in chloroplasts, and its concentration in plants was higher than that in plants transformed with just the synthetic gene. Transformed plants provoked 100% mortality in larvae of S. frugiperda, S. exigua, and H. zea.
ACKNOWLEDGMENTS
We thank C. S. Hernández-Rodríguez for her critical comments on the manuscript and for designing and drawing Fig. 6.
This research was supported by the Spanish Ministry of Science and Innovation (grant AGL2012-39946-C02-01), by grants ACOMP/2011/094 and GVPROMETEOII-2015-001 from the Generalitat Valenciana, and by European FEDER funds. N.B. was a recipient of a Ph.D. grant from the Spanish Ministry of Science and Innovation (grant BES-2010-039487).
Biographies

Maissa Chakroun obtained her Ph.D. in Biology from the University of Valencia (UV) (Spain) in October 2015, with the work Biochemical and Molecular Study of the Bacillus thuringiensis Vegetative Insecticidal Protein (Vip3A) Mode of Action in Spodoptera Species (thesis). This work was carried out at the Interdisciplinary Research Structure in Biotechnology and Biomedicine of the UV (ERI Biotecmed) at the Department of Genetics. She received an M.Sc. in Molecular and Cellular Biology from the University of Sfax/Centre of Biotechnology of Sfax (Tunisia) in 2009 and a B.Sc. in Life Sciences from the University of Sfax in 2007. She plans to continue her work on the mode of action of microbial toxins.

Núria Banyuls is a Ph.D. student at the University of Valencia (Spain) in the Interdisciplinary Research Structure in Biotechnology and Biomedicine of the UV (ERI Biotecmed) at the Department of Genetics. She received an M.Sc. in Integrated Pest Management from the University of Lleida (Spain) in 2009 and a B.Sc. in Biology from the University of Valencia in 2007. She is currently writing her thesis under the supervision of Prof. Ferré. Her thesis is focused on the Vip3A proteins, including biochemical properties, protein engineering, and function diversity and its relation with protein structure.

Yolanda Bel studied Biology (major in Biochemistry) and received her Ph.D. in Biology from the University of Valencia (UV), Spain, in 1991. Her Ph.D. was carried out at both the Department of Genetics of the UV and the Biology Division of the Oak Ridge National Laboratory, Oak Ridge, TN. She did her postdoctoral studies (1992 to 1993) in Sandoz, Basel, Switzerland, working on DNA adducts. After a contract at the University of Valencia (1993 to 1996), when she started working on Bacillus thuringiensis (Bt), she completed a master's in Industrial Wastewater Treatment and worked for six years on the microbiology of water and food in the private industry. In 2003, she returned to the University of Valencia, where she still works as a Research Associate. Her current research interests include the study of the binding of Bt toxins to insect midguts, the study of their mode of action, and the biochemical and genetic bases of insect resistance.

Baltasar Escriche is Associate Professor of Genetics at the University of Valencia (Spain), and he currently serves as Head of the Department of Genetics and as Director of the Master in Research in Genetics and Molecular and Cellular Biology of the Faculty of Biology. He studied Biology at the University of Valencia and completed his postdoctoral work at the University of Limburg (Belgium), funded by a European Union grant. He obtained a tenure track at the University of Valencia funded with the prestigious Spanish program Ramon y Cajal in 2002. He has worked on different aspects of Bacillus thuringiensis toxins and their production and application because of their relevance as an environmentally friendly pesticide, starting with his Ph.D. in 1990. He is especially interested in technology transfer to developing countries. Recently, he finished a collaborative project with several African research groups to control sweet potato pests, participating in a project funded by the Bill and Melinda Gates Foundation.

Juan Ferré received his Ph.D. in Chemistry at the University of Valencia (UV), Spain, with the work Study of the Pteridines and Quinolines from Drosophila melanogaster Eyes (thesis), which he carried out at both the Department of Genetics of the UV and the Biology Division of the Oak Ridge National Laboratory, Oak Ridge, TN. He did his postdoctoral studies at the Department of Reproductive Genetics of Magee Women's Hospital (Pittsburgh, PA, USA). He became Professor of Genetics in 2000 and served as Head of the Department of Genetics of the UV for 7 years. He is currently Director of the Interdisciplinary Research Structure in Biotechnology and Biomedicine of the UV (ERI Biotecmed). His current research interests, starting in 1990, are (i) to understand the biochemical and genetic bases of insect resistance to Bacillus thuringiensis (Bt) toxins, (ii) to study the mode of action of Bt toxins, and (iii) to find novel Bt strains and insecticidal protein genes for the development of Bt-based insecticides to control agricultural insect pests.
REFERENCES
- 1.De Maagd RA, Bravo A, Berry C, Crickmore N, Schnepf HE. 2003. Structure, diversity, and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu Rev Genet 37:409–433. doi: 10.1146/annurev.genet.37.110801.143042. [DOI] [PubMed] [Google Scholar]
- 2.Sanchis V. 2011. From microbial sprays to insect-resistant transgenic plants: history of the biopesticide Bacillus thuringiensis. A review. Agron Sustain Dev 31:217–231. doi: 10.1051/agro/2010027. [DOI] [Google Scholar]
- 3.Sanahuja G, Banakar R, Twyman RM, Capell T, Christou P. 2011. Bacillus thuringiensis: a century of research, development and commercial applications. Plant Biotechnol J 9:283–300. doi: 10.1111/j.1467-7652.2011.00595.x. [DOI] [PubMed] [Google Scholar]
- 4.Estruch JJ, Carozzi NB, Desai N, Duck NB, Warren GW, Koziel MG. 1997. Transgenic plants: an emerging approach to pest control. Nat Biotechnol 15:137–141. doi: 10.1038/nbt0297-137. [DOI] [PubMed] [Google Scholar]
- 5.Shelton AM. 2012. Genetically engineered vegetables expressing proteins from Bacillus thuringiensis for insect resistance: successes, disappointments, challenges and ways to move forward. GM Crops Food 3:175–183. doi: 10.4161/gmcr.19762. [DOI] [PubMed] [Google Scholar]
- 6.James C. 2014. Global status of commercialized biotech/GM crops: 2014. ISAAA brief no. 49 ISAAA, Ithaca, NY. [Google Scholar]
- 7.Warren GW. 1997. Vegetative insecticidal proteins: novel proteins for control of corn pests, p 109–121. In Carozzi NB, Koziel M (ed), Advances in insect control, the role of transgenic plants. Taylor & Francis Ltd, London, United Kingdom. [Google Scholar]
- 8.Palma L, Muñoz D, Berry C, Murillo J, Caballero P. 2014. Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins (Basel) 6:3296–3325. doi: 10.3390/toxins6123296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crickmore N, Baum J, Bravo A, Lereclus D, Narva K, Sampson K, Schnepf E, Sun M, Zeigler DR. 2014. Bacillus thuringiensis toxin nomenclature. http://www.btnomenclature.info/ Accessed 5 July 2015.
- 10.Donovan WP, Engleman JT, Donovan JC, Baum JA, Bunkers GJ, Chi DJ, Clinton WP, English L, Heck GR, Ilagan OM, Krasomil-Osterfeld KC, Pitkin JW, Roberts JK, Walters MR. 2006. Discovery and characterization of Sip1A: a novel secreted protein from Bacillus thuringiensis with activity against coleopteran larvae. Appl Microbiol Biotechnol 72:713–719. doi: 10.1007/s00253-006-0332-7. [DOI] [PubMed] [Google Scholar]
- 11.Schnepf HE, Narva KE, Stockhoff BA, Lee SF, Waltz M, Sturgis B. August 2003. Pesticidal toxins and genes from Bacillus laterosporus strains. US patent 6,605,701 B2.
- 12.Feitelson JS, Schnepf HE, Narva KE, Stockhoff BA, Schmeits J, Loewer D, Dullum CJ, Muller-Cohn J, Stamp L, Morrill G, Finstad-Lee S. December 2003. Pesticidal toxins and nucleotide sequences which encode these toxins. US patent no. 6,656,908 B2.
- 13.Yu X, Liu T, Liang X, Tang C, Zhu J, Wang S, Li S, Deng Q, Wang L, Zheng A, Li P. 2011. Rapid detection of vip1-type genes from Bacillus cereus and characterization of a novel vip binary toxin gene. FEMS Microbiol Lett 325:30–36. doi: 10.1111/j.1574-6968.2011.02409.x. [DOI] [PubMed] [Google Scholar]
- 14.Sattar S, Maiti MK. 2011. Molecular characterization of a novel vegetative insecticidal protein from Bacillus thuringiensis effective against sap-sucking insect pest. J Microbiol Biotechnol 21:937–946. doi: 10.4014/jmb.1105.05030. [DOI] [PubMed] [Google Scholar]
- 15.Estruch JJ, Warren GW, Mullins MA, Nye GJ, Craig JA, Koziel MG. 1996. Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc Natl Acad Sci U S A 93:5389–5394. doi: 10.1073/pnas.93.11.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz de Escudero I, Banyuls N, Bel Y, Maeztu M, Escriche B, Muñoz D, Caballero P, Ferré J. 2014. A screening of five Bacillus thuringiensis Vip3A proteins for their activity against lepidopteran pests. J Invertebr Pathol 117:51–55. doi: 10.1016/j.jip.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Ruiu L. 2013. Brevibacillus laterosporus, a pathogen of invertebrates and a broad-spectrum antimicrobial species. Insects 4:476–492. doi: 10.3390/insects4030476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnepf HE, Narva KE, Stockhoff BA, Lee SF, Walz M, Sturgis B. October 2005. Pesticidal toxins and genes from Bacillus laterosporus strains. US patent 6,956,116.
- 19.Sattar S, Biswas PK, Hossain MA, Maiti MK, Sen SK, Basu A. 2008. Search for vegetative insecticidal proteins (VIPs) from local isolates of Bacillus thuringiensis effective against lepidopteran and homopteran insect pests. J Biopest 1:216–222. [Google Scholar]
- 20.Hernández-Rodríguez CS, Boets A, Van Rie J, Ferré J. 2009. Screening and identification of vip genes in Bacillus thuringiensis strains. J Appl Microbiol 107:219–225. doi: 10.1111/j.1365-2672.2009.04199.x. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Zheng A, Zhu J, Wang S, Wang L, Deng Q, Li S, Liu H, Li P. 2011. Characterization of vegetative insecticidal protein vip genes of Bacillus thuringiensis from Sichuan Basin in China. Curr Microbiol 62:752–757. doi: 10.1007/s00284-010-9782-3. [DOI] [PubMed] [Google Scholar]
- 22.Shingote PR, Moharil MP, Dhumale DR, Deshmukh AG, Jadhav PV, Dudhare MS, Satpute NS. 2013. Distribution of vip genes, protein profiling and determination of entomopathogenic potential of local isolates of Bacillus thuringiensis. Bt Res 4:14–20. [Google Scholar]
- 23.Shi Y, Ma W, Yuan M, Sun F, Pang Y. 2007. Cloning of vip1/vip2 genes and expression of Vip1Ca/Vip2Ac proteins in Bacillus thuringiensis. World J Microbiol Biotechnol 23:501–507. doi: 10.1007/s11274-006-9252-z. [DOI] [Google Scholar]
- 24.Bi Y, Zhang Y, Shu C, Crickmore N, Wang Q, Du L, Song F, Zhang J. 2015. Genomic sequencing identifies novel Bacillus thuringiensis Vip1/Vip2 binary and Cry8 toxins that have high toxicity to Scarabaeoidea larvae. Appl Microbiol Biotechnol 99:753–760. doi: 10.1007/s00253-014-5966-2. [DOI] [PubMed] [Google Scholar]
- 25.Murawska E, Fiedoruk K, Bideshi DK, Swiecicka I. 2013. Complete genome sequence of Bacillus thuringiensis subsp. thuringiensis strain IS5056, an isolate highly toxic to Trichoplusia ni. Genome Announc 1(2):e00108-13. doi: 10.1128/genomeA.00108-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y, Xu W, Yuan M, Tang M, Chen J, Pang Y. 2004. Expression of vip1/vip2 genes in Escherichia coli and Bacillus thuringiensis and the analysis of their signal peptides. J Appl Microbiol 97:757–765. doi: 10.1111/j.1365-2672.2004.02365.x. [DOI] [PubMed] [Google Scholar]
- 27.Barth H, Aktories K, Popoff MR, Stiles BG. 2004. Binary bacterial toxins: biochemistry, biology, and applications of common Clostridium and Bacillus proteins. Microbiol Mol Biol Rev 68:373–402. doi: 10.1128/MMBR.68.3.373-402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warren GW, Koziel MG, Mullin MA, Nye GJ, Carr B, Desai NM, Kostichka K, Duck NB, Estruch JJ. June 1998. Auxiliary proteins for enhancing the insecticidal activity of pesticidal proteins. US patent 5,770,696.
- 29.Leuber M, Orlik F, Schiffler B, Sickmann A, Benz R. 2006. Vegetative insecticidal protein (Vip1Ac) of Bacillus thuringiensis HD201: evidence for oligomer and channel formation. Biochemistry 45:283–288. doi: 10.1021/bi051351z. [DOI] [PubMed] [Google Scholar]
- 30.Han S, Craig JA, Putnam CD, Carozzi NB, Tainer JA. 1999. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat Struct Biol 6:932–936. doi: 10.1038/13300. [DOI] [PubMed] [Google Scholar]
- 31.Knapp O, Benz R, Gibert M, Marvaud JC, Popoff MR. 2002. Interaction of Clostridium perfringens iota-toxin with lipid bilayer membranes. Demonstration of channel formation by the activated binding component Ib and channel block by the enzyme component Ia. J Biol Chem 277:6143–6152. [DOI] [PubMed] [Google Scholar]
- 32.Schmid A, Benz R, Just I, Aktories K. 1994. Interaction of Clostridium botulinum C2 toxin with lipid bilayer membranes; formation of cation-selective channels and inhibition of channel function by chloroquine. J Biol Chem 269:16706–16711. [PubMed] [Google Scholar]
- 33.Blaustein RO, Koehler TM, Collier RJ, Finkelstein A. 1989. Anthrax toxin: channel-forming activity of protective antigen in planar phospholipid bilayers. Proc Natl Acad Sci U S A 86:2209–2213. doi: 10.1073/pnas.86.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jucovic M, Walters FS, Warren GW, Palekar NV, Chen JS. 2008. From enzyme to zymogen: engineering Vip2, an ADP-ribosyltransferase from Bacillus cereus, for conditional toxicity. Protein Eng Des Sel 21:631–638. doi: 10.1093/protein/gzn038. [DOI] [PubMed] [Google Scholar]
- 35.Han S, Arvai AS, Clancy SB, Tainer JA. 2001. Crystal structure and novel recognition motif of rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: structural insights for recognition specificity and catalysis. J Mol Biol 305:95–107. doi: 10.1006/jmbi.2000.4292. [DOI] [PubMed] [Google Scholar]
- 36.Barth H, Hofmann F, Olenik C, Just I, Aktories K. 1998. The N-terminal part of the enzyme component (C2I) of the binary Clostridium botulinum C2 toxin interacts with the binding component C2II and functions as a carrier system for a Rho ADP-ribosylating C3-like fusion toxin. Infect Immun 66:1364–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boets A, Arnaut G, Van Rie J, Damme N. April 2011. Toxins. US patent 7,919,609 B2.
- 38.Shingote PR, Moharil MP, Dhumale DR, Jadhav PV, Satpute NS, Dudhare MS. 2013. Screening of vip1/vip2 binary toxin gene and its isolation and cloning from local Bacillus thuringiensis isolates. Sci Asia 39:620–624. doi: 10.2306/scienceasia1513-1874.2013.39.620. [DOI] [Google Scholar]
- 39.Mesrati LA, Tounsi S, Jaoua S. 2005. Characterization of a novel vip3-type gene from Bacillus thuringiensis and evidence of its presence on a large plasmid. FEMS Microbiol Lett 244:353–358. doi: 10.1016/j.femsle.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Beard CE, Court L, Boets A, Mourant R, Van Rie J, Akhurst RJ. 2008. Unusually high frequency of genes encoding vegetative insecticidal proteins in an Australian Bacillus thuringiensis collection. Curr Microbiol 57:195–199. doi: 10.1007/s00284-008-9173-1. [DOI] [PubMed] [Google Scholar]
- 41.Espinasse S, Chaufaux J, Buisson C, Perchat S, Gohar M, Bourguet D, Sanchis V. 2003. Occurrence and linkage between secreted insecticidal toxins in natural isolates of Bacillus thuringiensis. Curr Microbiol 47:501–507. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Song F, Zhang J, Liu R, He K, Tan J, Huang D. 2007. Identification of vip3A-type genes from Bacillus thuringiensis strains and characterization of a novel vip3A-type gene. Lett Appl Microbiol 45:432–438. doi: 10.1111/j.1472-765X.2007.02217.x. [DOI] [PubMed] [Google Scholar]
- 43.Mesrati LA, Tounsi S, Kamoun F, Jaoua S. 2005. Identification of a promoter for the vegetative insecticidal protein-encoding gene vip3LB from Bacillus thuringiensis. FEMS Microbiol Lett 247:101–104. doi: 10.1016/j.femsle.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 44.Rice WC. 1999. Specific primers for the detection of vip3A insecticidal gene. Lett Appl Microbiol 28:378–382. doi: 10.1046/j.1365-2672.1999.00536.x. [DOI] [Google Scholar]
- 45.Wu ZL, Guo WY, Qiu JZ, Huang TP, Li XB, Guan X. 2004. Cloning and localization of vip3A gene of Bacillus thuringiensis. Biotechnol Lett 26:1425–1428. doi: 10.1023/B:BILE.0000045645.45536.3f. [DOI] [PubMed] [Google Scholar]
- 46.Franco-Rivera A, Benintende G, Cozzi J, Baizabal-Aguirre VM, Valdez-Alarcón JJ, López-Meza JE. 2004. Molecular characterization of Bacillus thuringiensis strains from Argentina. Antoine Van Leeuwenhoek 86:87–92. doi: 10.1023/B:ANTO.0000024913.94410.05. [DOI] [PubMed] [Google Scholar]
- 47.Baranek J, Kaznowski A, Konecka E, Naimov S. 2015. Activity of vegetative insecticidal proteins Vip3Aa58 and Vip3Aa59 of Bacillus thuringiensis against lepidopteran pests. J Invertebr Pathol 130:72–81. doi: 10.1016/j.jip.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Loguercio LL, Barreto ML, Rocha TL, Santos CG, Teixeira FF, Paiva E. 2002. Combined analysis of supernatant-based feeding bioassays and PCR as a first-tier screening strategy for Vip-derived activities in Bacillus thuringiensis strains effective against tropical fall armyworm. J Appl Microbiol 93:269–277. doi: 10.1046/j.1365-2672.2002.01694.x. [DOI] [PubMed] [Google Scholar]
- 49.Bhalla R, Dalal M, Panguluri SK, Jagadish B, Mandaokar AD, Singh AK, Kumar PA. 2005. Isolation, characterization and expression of a novel vegetative insecticidal protein gene of Bacillus thuringiensis. FEMS Microbiol Lett 243:467–472. doi: 10.1016/j.femsle.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Rang C, Gil P, Neisner N, Van Rie J, Frutos R. 2005. Novel Vip3-related protein from Bacillus thuringiensis. Appl Environ Microbiol 71:6276–6281. doi: 10.1128/AEM.71.10.6276-6281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abulreesh HH, Osman GEH, Assaeedi ASA. 2012. Characterization of insecticidal genes of Bacillus thuringiensis strains isolated from arid environments. Indian J Microbiol 52:500–503. doi: 10.1007/s12088-012-0257-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asokan R, Swamy HM, Arora DK. 2012. Screening, diversity and partial sequence comparison of vegetative insecticidal protein (vip3A) genes in the local isolates of Bacillus thuringiensis Berliner. Curr Microbiol 64:365–370. doi: 10.1007/s00284-011-0078-z. [DOI] [PubMed] [Google Scholar]
- 53.Wu J, Zhao F, Bai J, Deng G, Qin S, Bao Q. 2007. Evidence for positive Darwinian selection of vip gene in Bacillus thuringiensis. J Genet Genomics 34:649–660. doi: 10.1016/S1673-8527(07)60074-5. [DOI] [PubMed] [Google Scholar]
- 54.Dong F, Zhang S, Shi R, Yi S, Xu F, Liu Z. 2012. Ser-substituted mutations of Cys residues in Bacillus thuringiensis Vip3Aa7 exert a negative effect on its insecticidal activity. Curr Microbiol 65:583–588. doi: 10.1007/s00284-012-0201-9. [DOI] [PubMed] [Google Scholar]
- 55.Doss VA, Kumar KA, Jayakumar R, Sekar V. 2002. Cloning and expression of the vegetative insecticidal protein (vip3V) gene of Bacillus thuringiensis in Escherichia coli. Protein Expr Purif 26:82–88. doi: 10.1016/S1046-5928(02)00515-6. [DOI] [PubMed] [Google Scholar]
- 56.Chen J, Yu J, Tang L, Tang M, Shi Y, Pang Y. 2003. Comparison of the expression of Bacillus thuringiensis full-length and N-terminally truncated vip3A gene in Escherichia coli. J Appl Microbiol 95:310–316. doi: 10.1046/j.1365-2672.2003.01977.x. [DOI] [PubMed] [Google Scholar]
- 57.Estruch JJ, Yu CG. September 2001. Plant pest control. US patent 6,291,156 B1.
- 58.Li C, Xua N, Huanga X, Wanga W, Chenga J, Wuc K, Shen Z. 2007. Bacillus thuringiensis Vip3 mutant proteins: insecticidal activity and trypsin sensitivity. Biocontrol Sci Technol 17:699–708. doi: 10.1080/09583150701527177. [DOI] [Google Scholar]
- 59.Selvapandiyan A, Arora N, Rajagopal R, Jalali SK, Venkatesan T, Singh SP, Bhatnagar RK. 2001. Toxicity analysis of N- and C-terminus-deleted vegetative insecticidal protein from Bacillus thuringiensis. Appl Environ Microbiol 67:5855–5858. doi: 10.1128/AEM.67.12.5855-5858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gayen S, Hossain MA, Sen SK. 2012. Identification of the bioactive core component of the insecticidal Vip3A toxin peptide of Bacillus thuringiensis. J Plant Biochem Biotechnol 21:S128–S135. [Google Scholar]
- 61.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palma L, Hernández-Rodríguez CS, Maeztu M, Hernández-Martínez P, Ruiz de Escudero I, Escriche B, Muñoz D, Van Rie J, Ferré J, Caballero P. 2012. Vip3C, a novel class of vegetative insecticidal proteins from Bacillus thuringiensis. Appl Environ Microbiol 78:7163–7165. doi: 10.1128/AEM.01360-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Frankenhuyzen K, Nystrom C. 2009. The Bacillus thuringiensis toxin specificity database. http://www.glfc.forestry.ca/bacillus/BtSearch.cfm Accessed 4 February 2016.
- 64.Donovan WP, Donovan JC, Engleman JT. 2001. Gene knockout demonstrates that vip3A contributes to the pathogenesis of Bacillus thuringiensis toward Agrotis ipsilon and Spodoptera exigua. J Invertebr Pathol 78:45–51. doi: 10.1006/jipa.2001.5037. [DOI] [PubMed] [Google Scholar]
- 65.Yu CG, Mullins MA, Warren GW, Koziel MG, Estruch JJ. 1997. The Bacillus thuringiensis vegetative insecticidal protein Vip3Aa lyses midgut epithelium cells of susceptible insects. Appl Environ Microbiol 63:532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu X, Liu T, Sun Z, Guan P, Zhu J, Wang S, Li S, Deng Q, Wang L, Zheng A, Li P. 2012. Co-expression and synergism analysis of Vip3Aa29 and Cyt2Aa3 insecticidal proteins from Bacillus thuringiensis. Curr Microbiol 64:326–331. doi: 10.1007/s00284-011-0070-7. [DOI] [PubMed] [Google Scholar]
- 67.Jamoussi K, Sellami S, Abdelkefi-Mesrati L, Givaudan A, Jaoua S. 2009. Heterologous expression of Bacillus thuringiensis vegetative insecticidal protein-encoding gene vip3LB in Photorhabdus temperata strain K122 and oral toxicity against the Lepidoptera Ephestia kuehniella and Spodoptera littoralis. Mol Biotechnol 43:97–103. doi: 10.1007/s12033-009-9179-3. [DOI] [PubMed] [Google Scholar]
- 68.Abdelkefi-Mesrati L, Boukedi H, Dammak-Karray M, Sellami-Boudawara T, Jaoua S, Tounsi S. 2011. Study of the Bacillus thuringiensis Vip3Aa16 histopathological effects and determination of its putative binding proteins in the midgut of Spodoptera littoralis. J Invertebr Pathol 106:250–254. doi: 10.1016/j.jip.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 69.Ben Hamadou-Charfi D, Boukedi H, Abdelkefi-Mesrati L, Tounsi S, Jaoua S. 2013. Agrotis segetum midgut putative receptor of Bacillus thuringiensis vegetative insecticidal protein Vip3Aa16 differs from that of Cry1Ac toxin. J Invertebr Pathol 114:139–143. doi: 10.1016/j.jip.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 70.Ali MI, Luttrell RG. 2011. Susceptibility of Helicoverpa zea and Heliothis virescens (Lepidoptera: Noctuidae) to Vip3A insecticidal protein expressed in VipCot cotton. J Invertebr Pathol 108:76–84. doi: 10.1016/j.jip.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 71.Chakroun M, Bel Y, Caccia S, Abdelkefi-Mesrati L, Escriche B, Ferré J. 2012. Susceptibility of Spodoptera frugiperda and S. exigua to Bacillus thuringiensis Vip3Aa insecticidal protein. J Invertebr Pathol 110:334–339. doi: 10.1016/j.jip.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 72.Hernández-Martínez P, Hernández-Rodríguez CS, Van Rie J, Escriche B, Ferré J. 2013. Insecticidal activity of Vip3Aa, Vip3Ad, Vip3Ae, and Vip3Af from Bacillus thuringiensis against lepidopteran corn pests. J Invertebr Pathol 113:78–81. doi: 10.1016/j.jip.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 73.Palma L, Ruiz de Escudero I, Maeztu M, Caballero P, Muñoz D. 2013. Screening of vip genes from a Spanish Bacillus thuringiensis collection and characterization of two Vip3 proteins highly toxic to five lepidopteran crop pests. Biol Control 66:141–149. doi: 10.1016/j.biocontrol.2013.05.003. [DOI] [Google Scholar]
- 74.Bergamasco VB, Mendes DR, Fernandes OA, Desidério JA, Lemos MV. 2013. Bacillus thuringiensis Cry1Ia10 and Vip3Aa protein interactions and their toxicity in Spodoptera spp. (Lepidoptera). J Invertebr Pathol 112:152–158. doi: 10.1016/j.jip.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 75.Lemes ARN, Davolos CC, Legori PCBC, Fernandes OA, Ferré J, Lemos MVF, Desiderio JA. 2014. Synergism and antagonism between Bacillus thuringiensis Vip3A and Cry1 proteins in Heliothis virescens, Diatraea saccharalis and Spodoptera frugiperda. PLoS One 9:e107196. doi: 10.1371/journal.pone.0107196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee MK, Curtiss A, Alcantara E, Dean DH. 1996. Synergistic effect of the Bacillus thuringiensis toxins CryIAa and CryIAc on the gypsy moth, Lymantria dispar. Appl Environ Microbiol 62:583–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fang J, Xu X, Wang P, Zhao JZ, Shelton AM, Cheng J, Feng MG, Shen Z. 2007. Characterization of chimeric Bacillus thuringiensis Vip3 toxins. Appl Environ Microbiol 73:956–996. doi: 10.1128/AEM.02079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saraswathy N, Nain V, Sushmita K, Kumar PA. 2008. A fusion gene encoding two different insecticidal proteins of Bacillus thuringiensis. Indian J Biotechnol 7:204–209. [Google Scholar]
- 79.Dong F, Shi R, Zhang S, Zhan T, Wu G, Shen J, Liu Z. 2012. Fusing the vegetative insecticidal protein Vip3Aa7 and the N terminus of Cry9Ca improves toxicity against Plutella xylostella larvae. Appl Microbiol Biotechnol 96:921–929. doi: 10.1007/s00253-012-4213-y. [DOI] [PubMed] [Google Scholar]
- 80.Song R, Peng D, Yu Z, Sun M. 2008. Carboxy-terminal half of Cry1C can help vegetative insecticidal protein to form inclusion bodies in the mother cell of Bacillus thuringiensis. Appl Microbiol Biotechnol 80:647–654. doi: 10.1007/s002253-008-1613-0. [DOI] [PubMed] [Google Scholar]
- 81.Lisansky SC, Quinlan R, Tassoni G. 1993. The Bacillus thuringiensis production handbook. CPL Scientific Ltd, Newbury, United Kingdom. [Google Scholar]
- 82.Arora N, Selvapandiyan A, Agrawal N, Bhatnagar RK. 2003. Relocating expression of vegetative insecticidal protein into mother cell of Bacillus thuringiensis. Biochem Biophys Res Commun 310:158–162. doi: 10.1016/j.bbrc.2003.08.137. [DOI] [PubMed] [Google Scholar]
- 83.Zhu C, Ruan L, Peng D, Yu Z, Sun M. 2006. Vegetative insecticidal protein enhancing the toxicity of Bacillus thuringiensis subsp kurstaki against Spodoptera exigua. Lett Appl Microbiol 42:109–114. doi: 10.1111/j.1472-765X.2005.01817.x. [DOI] [PubMed] [Google Scholar]
- 84.Thamthiankul Chankhamhaengdecha S, Tantichodok A, Panbangred W. 2008. Spore stage expression of a vegetative insecticidal gene increase toxicity of Bacillus thuringiensis subsp. aizawai SP41 against Spodoptera exigua. J Biotechnol 136:122–128. doi: 10.1016/j.jbiotec.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 85.Sellami S, Jamoussi K, Dabbeche E, Jaoua S. 2011. Increase of the Bacillus thuringiensis secreted toxicity against lepidopteron larvae by homologous expression of the vip3LB gene during sporulation stage. Curr Microbiol 63:289–294. doi: 10.1007/s00284-011-9976-3. [DOI] [PubMed] [Google Scholar]
- 86.Hernández-Rodríguez CS, Ruiz de Escudero I, Asensio AC, Ferré J, Caballero P. 2013. Encapsulation of the Bacillus thuringiensis secretable toxins Vip3Aa and Cry1Ia in Pseudomonas fluorescens. Biol Control 66:159–165. doi: 10.1016/j.biocontrol.2013.05.002. [DOI] [Google Scholar]
- 87.Lee MK, Walters FS, Hart H, Palekar N, Chen JS. 2003. The mode of action of the Bacillus thuringiensis vegetative insecticidal protein Vip3Aa differs from that of Cry1Ab delta-endotoxin. Appl Environ Microbiol 69:4648–4657. doi: 10.1128/AEM.69.8.4648-4657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abdelkefi-Mesrati L, Boukedi H, Chakroun M, Kamoun F, Azzouz H, Tounsi S, Rouis S, Jaoua S. 2011. Investigation of the steps involved in the difference of susceptibility of Ephestia kuehniella and Spodoptera littoralis to the Bacillus thuringiensis Vip3Aa16 toxin. J Invertebr Pathol 107:198–201. doi: 10.1016/j.jip.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 89.Chakroun M, Ferré J. 2014. In vivo and in vitro binding of Vip3Aa to Spodoptera frugiperda midgut and characterization of binding sites by 125I radiolabeling. Appl Environ Microbiol 80:6258–6265. doi: 10.1128/AEM.01521-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sellami S, Cherif M, Abdelkefi-Mesrati L, Tounsi S, Jamoussi K. 2015. Toxicity, activation process, and histopathological effect of Bacillus thuringiensis vegetative insecticidal protein Vip3Aa16 on Tuta absoluta. Appl Biochem Biotechnol 175:1992–1999. doi: 10.1007/s12010-014-1393-1. [DOI] [PubMed] [Google Scholar]
- 91.Boukedi H, Ben Khedher S, Triki N, Kamoun F, Saadaoui I, Chakroun M, Tounsi S, Abdelkefi-Mesrati L. 2015. Overproduction of the Bacillus thuringiensis Vip3Aa16 toxin and study of its insecticidal activity against the carob moth Ectomyelois ceratoniae. J Invertebr Pathol 127:127–129. doi: 10.1016/j.jip.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 92.Lee MK, Miles P, Chen JS. 2006. Brush border membrane binding properties of Bacillus thuringiensis Vip3A toxin to Heliothis virescens and Helicoverpa zea midguts. Biochem Biophys Res Commun 339:1043–1047. doi: 10.1016/j.bbrc.2005.11.112. [DOI] [PubMed] [Google Scholar]
- 93.Liu JG, Yang AZ, Shen XH, Hua BG, Shi GL. 2011. Specific binding of activated Vip3Aa10 to Helicoverpa armigera brush border membrane vesicles results in pore formation. J Invertebr Pathol 108:92–97. doi: 10.1016/j.jip.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 94.Caccia S, Chakroun M, Vinokurov K, Ferré J. 2014. Proteolytic processing of Bacillus thuringiensis Vip3A proteins by two Spodoptera species. J Insect Physiol 67:76–84. doi: 10.1016/j.jinsphys.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 95.Sena JAD, Hernández-Rodríguez CS, Ferré J. 2009. Interaction of Bacillus thuringiensis Cry1 and Vip3Aa proteins with Spodoptera frugiperda midgut binding sites. Appl Environ Microbiol 75:2236–2237. doi: 10.1128/AEM.02342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gouffon C, Van Vliet A, Van Rie J, Jansens S, Jurat-Fuentes JL. 2011. Binding sites for Bacillus thuringiensis Cry2Ae toxin on heliothine brush border membrane vesicles are not shared with Cry1A, Cry1F, or Vip3A toxin. Appl Environ Microbiol 77:3182–3188. doi: 10.1128/AEM.02791-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abdelkefi-Mesrati L, Rouis S, Sellami S, Jaoua S. 2009. Prays oleae midgut putative receptor of Bacillus thuringiensis vegetative insecticidal protein Vip3LB differs from that of Cry1Ac toxin. Mol Biotechnol 43:15–19. doi: 10.1007/s12033-009-9178-4. [DOI] [PubMed] [Google Scholar]
- 98.Singh G, Sachdev B, Sharma N, Seth R, Bhatnagar RK. 2010. Interaction of Bacillus thuringiensis vegetative insecticidal protein with ribosomal S2 protein triggers larvicidal activity in Spodoptera frugiperda. Appl Environ Microbiol 76:7202–7209. doi: 10.1128/AEM.01552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bel Y, Jakubowska AK, Costa J, Herrero S, Escriche B. 2013. Comprehensive analysis of gene expression profiles of the beet armyworm Spodoptera exigua larvae challenged with Bacillus thuringiensis Vip3Aa toxin. PLoS One 8:e81927. doi: 10.1371/journal.pone.0081927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pickett BR. 2009. Studies on resistance to vegetative (Vip3A) and crystal (Cry1A) insecticidal toxins of Bacillus thuringiensis in Heliothis virescens (Fabricius). PhD thesis. Imperial College London, London, United Kingdom. [Google Scholar]
- 101.Gulzar A, Pickett B, Sayyed AH, Wright DJ. 2012. Effect of temperature on the fitness of a Vip3A resistant population of Heliothis virescens (Lepidoptera: Noctuidae). J Econ Entomol 105:964–970. doi: 10.1603/EC11110. [DOI] [PubMed] [Google Scholar]
- 102.Barkhade UP, Thakare AS. 2010. Protease mediated resistance mechanism to Cry1C and Vip3A in Spodoptera litura. Egypt Acad J Biol Sci 3:43–50. [Google Scholar]
- 103.Mahon RJ, Downes SJ, James B. 2012. Vip3A resistance alleles exist at high levels in Australian targets before release of cotton expressing this toxin. PLoS One 7:e39192. doi: 10.1371/journal.pone.0039192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Walsh TK, Downes SJ, Gascoyne J, James W, Parker T, Armstrong J, Mahon RJ. 2014. Dual Cry2Ab and Vip3A resistant strains of Helicoverpa armigera and Helicoverpa punctigera (Lepidoptera: Noctuidae); testing linkage between loci and monitoring of allele frequencies. J Econ Entomol 107:1610–1617. doi: 10.1603/EC13558. [DOI] [PubMed] [Google Scholar]
- 105.Bernardi O, Bernardi D, Ribeiro RS, Okuma DM, Salmeron E, Fatoretto J, Medeiros FCL, Burd T, Omoto C. 2015. Frequency of resistance to Vip3Aa20 toxin from Bacillus thuringiensis in Spodoptera frugiperda (Lepidoptera: Noctuidae) populations in Brazil. Crop Prot 76:7–14. doi: 10.1016/j.cropro.2015.06.006. [DOI] [Google Scholar]
- 106.Kurtz RW. 2010. A review of Vip3A mode of action and effects on Bt Cry protein-resistant colonies of lepidopteran larvae. Southwest Entomol 35:391–394. doi: 10.3958/059.035.0321. [DOI] [Google Scholar]
- 107.Jackson RE, Marcus MA, Gould F, Bradley JR Jr, Van Duyn JW. 2007. Cross-resistance responses of Cry1Ac-selected Heliothis virescens (Lepidoptera: Noctuidae) to the Bacillus thuringiensis protein Vip3A. J Econ Entomol 100:180–186. doi: 10.1093/jee/100.1.180. [DOI] [PubMed] [Google Scholar]
- 108.Anilkumar KJ, Rodrigo-Simón A, Ferré J, Pusztai-Carey M, Sivasupramaniam S, Moar WJ. 2008. Production and characterization of Bacillus thuringiensis Cry1Ac-resistant cotton bollworm Helicoverpa zea (Boddie). Appl Environ Microbiol 74:462–469. doi: 10.1128/AEM.01612-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Welch KL, Unnithan GC, Degain BA, Wei J, Zhang J, Li X, Tabashnik BE, Carrière Y. 2015. Cross-resistance to toxins used in pyramided Bt crops and resistance to Bt sprays in Helicoverpa zea. J Invertebr Pathol 132:149–156. doi: 10.1016/j.jip.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 110.An J, Gao Y, Wu K, Gould F, Gao J, Shen Z, Lei C. 2010. Vip3Aa tolerance response of Helicoverpa armigera populations from a Cry1Ac cotton planting region. J Econ Entomol 103:2169–2173. doi: 10.1603/EC10105. [DOI] [PubMed] [Google Scholar]
- 111.Vélez AM, Spencer TA, Alves AP, Moellenbeck D, Meagher RL, Chirakkal H, Siegfried BD. 2013. Inheritance of Cry1F resistance, cross-resistance and frequency of resistant alleles in Spodoptera frugiperda (Lepidoptera: Noctuidae). Bull Entomol Res 103:700–713. doi: 10.1017/S0007485313000448. [DOI] [PubMed] [Google Scholar]
- 112.Huang F, Qureshi JA, Meagher RL Jr, Reisig DD, Head GP, Andow DA, Ni X, Kerns D, Buntin GD, Niu Y, Yang F, Dangal V. 2014. Cry1F resistance in fall armyworm Spodoptera frugiperda: single gene versus pyramided Bt maize. PLoS One 9:e112958. doi: 10.1371/journal.pone.0112958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kurtz RW, McCaffery A, O'Reilly D. 2007. Insect resistance management for Syngenta's VipCot transgenic cotton. J Invertebr Pathol 95:227–230. doi: 10.1016/j.jip.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 114.Adamczyk JJ Jr, Mahaffey JS. 2008. Efficacy of Vip3A and Cry1Ab transgenic traits in cotton against various lepidopteran pests. Fla Entomol 91:570–575. [Google Scholar]
- 115.Burkness EC, Dively G, Patton T, Morey AC, Hutchison WD. 2010. Novel Vip3A Bacillus thuringiensis (Bt) maize approaches high-dose efficacy against Helicoverpa zea (Lepidoptera: Noctuidae) under field conditions: implications for resistance management. GM Crops 1:337–343. doi: 10.4161/gmcr.1.5.14765. [DOI] [PubMed] [Google Scholar]
- 116.Carrière Y, Crickmore N, Tabashnik BE. 2015. Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat Biotechnol 33:161–168. doi: 10.1038/nbt.3099. [DOI] [PubMed] [Google Scholar]
- 117.Llewellyn DJ, Mares CL, Fitt GP. 2007. Field performance and seasonal changes in the efficacy against Helicoverpa armigera (Hübner) of transgenic cotton expressing the insecticidal protein Vip3A. Agric For Entomol 9:93–101. doi: 10.1111/j.1461-9563.2007.00332.x. [DOI] [Google Scholar]
- 118.Gayen S, Samanta MK, Hossain MA, Mandal CC, Sen SK. 2015. A deletion mutant ndv200 of the Bacillus thuringiensis vip3BR insecticidal toxin gene is a prospective candidate for the next generation of genetically modified crop plants resistant to lepidopteran insect damage. Planta 242:269–281. doi: 10.1007/s00425-015-2309-1. [DOI] [PubMed] [Google Scholar]
- 119.Wu J, Luo X, Zhang X, Shi Y, Tian Y. 2011. Development of insect-resistant transgenic cotton with chimeric TVip3A* accumulating in chloroplasts. Transgenic Res 20:963–973. doi: 10.1007/s11248-011-9483-0. [DOI] [PubMed] [Google Scholar]
- 120.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Iatsenko I, Nikolov A, Sommer RJ. 2014. Identification of distinct Bacillus thuringiensis 4A4 nematicidal factors using the model nematodes Pristionchus pacificus and Caenorhabditis elegans. Toxins 6:2050–2063. doi: 10.3390/toxins6072050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liao C, Heckel DG, Akhurst R. 2002. Toxicity of Bacillus thuringiensis insecticidal proteins for Helicoverpa armigera and Helicoverpa punctigera (Lepidoptera: Noctuidae), major pests of cotton. J Invertebr Pathol 80:55–63. doi: 10.1016/S0022-2011(02)00035-6. [DOI] [PubMed] [Google Scholar]
- 124.Gulzar A, Wright DJ. 2015. Sub-lethal effects of Vip3A toxin on survival, development and fecundity of Heliothis virescens and Plutella xylostella. Ecotoxicology 24:1815–1822. doi: 10.1007/s10646-015-1517-6. [DOI] [PubMed] [Google Scholar]
- 125.Figueiredo CS, Marucci SC, Tezza RID, Lemos MVF, Desidério JA. 2013. Caracterização do gene vip3A e toxicidade da proteína Vip3Aa50 à lagarta-do-cartucho e à lagarta-da-soja. Pesq Agropec Bras 48:1220–1227. doi: 10.1590/S0100-204X2013000900005. [DOI] [Google Scholar]