SUMMARY
Chlamydia species infect millions of individuals worldwide and are important etiological agents of sexually transmitted disease, infertility, and blinding trachoma. Historically, the genetic intractability of this intracellular pathogen has hindered the molecular dissection of virulence factors contributing to its pathogenesis. The obligate intracellular life cycle of Chlamydia and restrictions on the use of antibiotics as selectable markers have impeded the development of molecular tools to genetically manipulate these pathogens. However, recent developments in the field have resulted in significant gains in our ability to alter the genome of Chlamydia, which will expedite the elucidation of virulence mechanisms. In this review, we discuss the challenges affecting the development of molecular genetic tools for Chlamydia and the work that laid the foundation for recent advancements in the genetic analysis of this recalcitrant pathogen.
INTRODUCTION
The phylum Chlamydiae is composed of obligate intracellular pathogens grouped into the single class Chlamydiae and the order Chlamydiales. Members of the Chlamydiales are classified into one of the following eight families: Parachlamydiaceae, Criblamydiaceae, Waddliaceae, Simkaniaceae, Rhabdochlamydiaceae, Clavichlamydiaceae, Piscichlamydiaceae, and Chlamydiaceae (1). With the exception of the Chlamydiaceae, most members of this order infect various hosts in the environment and are collectively referred to as “environmental” Chlamydiae. In contrast, the members of Chlamydiaceae, which contains the single genus Chlamydia, are considered pathogenic and contribute to disease burdens in humans and animal species of commercial importance. The genus Chlamydia comprises nine species: Chlamydia trachomatis, C. muridarum, C. pneumoniae, C. pecorum, C. suis, C. abortus, C. felis, C. caviae, and C. psittaci (2, 3). Undoubtedly, the best-characterized species in this genus is C. trachomatis, which infects humans and is a major cause of ocular and urogenital diseases. This species is classified into serovars (serological variants) and two human biovars (trachoma and lymphogranuloma venereum [LGV]), dictated by the nature of the diseases that they cause. Based on high-resolution genome-wide single-nucleotide polymorphism (SNP) analysis, the population structure of the trachoma biovar is further subdivided into two lineages (4): lineage 1 comprises clinically prevalent urogenital serovars (D, E, and F), whereas lineage 2 includes uncommon urogenital serovars (G, Ia, J, and K). These serovars are the primary cause of sexually transmitted diseases, such as cervicitis and urethritis, which can further progress into pelvic inflammatory disease in women and epididymitis in men (5). The ocular serotypes (A, B, and C) cause trachoma, a chronic inflammatory disease resulting in infectious blindness (6), and cluster into a lineage that likely evolved from a urogenital ancestor (4). The LGV biovar contains invasive serovars L1 to L3, including the epidemic L2b strain, which can disseminate to regional lymph nodes and cause invasive disease, including ulcer formation, inguinal lymphadenopathy, and hemorrhagic proctitis (7).
Given the impact of C. trachomatis on human health, most of the work discussed below focuses on this species. There is great interest in understanding the molecular mechanisms underlying its pathogenesis for the improvement of vaccine development. A critical step in this direction is the identification of chlamydial factors that contribute to virulence. Several experimental approaches have been employed to identify these factors, such as ectopic expression in heterologous hosts and cell lines, gene expression analysis, proteomic analysis, and comparative genomics; however, approaches that utilize genetic strategies, particularly those that satisfy Koch's molecular postulates (8), have been unavailable for all Chlamydiae. This lack of genetic tools remains frustrating, especially because some Chlamydia species have been propagated in vitro since the 1950s (9).
However, in the last decade, significant inroads have been made toward developing systems for genetic analysis of this recalcitrant obligate intracellular pathogen. A landmark event was the sequencing of the first Chlamydiae genome (10), which revealed that C. trachomatis is capable of inserting exogenous DNA into its genome because it encodes an intact DNA recombination machinery. Subsequent studies demonstrated that lateral gene transfer (LGT) events in Chlamydia are frequent and robust and occur both in nature and in the laboratory (4, 11–13). Transformation of exogenous DNA into Chlamydia has been achieved via several methods, including electroporation, dendrimer-based delivery, and a calcium chloride-based treatment (14–19). Successful and stable transformation of C. trachomatis LGV L2 with an Escherichia coli-C. trachomatis L2 shuttle plasmid conferring β-lactam resistance led to the development of expression vectors for expressing Chlamydia open reading frames (ORFs), fluorescent proteins (green fluorescent protein [GFP], cyan fluorescent protein [CFP], mCherry, and mKate2), and reporter proteins (β-galactosidase, adenylate cyclase, and glycogen synthase kinase [GSK]-tagged proteins) (19–26). A conditional expression vector was also developed using a tetracycline-inducible system as well as vectors conferring chloramphenicol and blasticidin resistance (21, 26–29). In contrast, the development of genome engineering methods has lagged behind. Consequently, the use of chemical mutagenesis coupled with whole-genome sequencing and mismatch-specific endonucleases for mapping mutant alleles led to the emergence of experimental platforms to perform forward and reverse genetic screens in Chlamydiae (30–32). More recently, targeted mutagenesis was achieved using a group II intron-based gene knockout system (TargeTron) (33). These advances, although recent, promise to revolutionize our understanding of the molecular basis of Chlamydia pathogenesis. In this review, we discuss the challenges associated with developing genetic techniques for Chlamydia that follow Koch's molecular postulates, and we review the efforts that led to the development of the molecular tools currently available for genetic analyses in Chlamydia.
CHALLENGES IN THE GENETIC MANIPULATION OF CHLAMYDIA
The Chlamydia Life Cycle
One major hurdle for delivering exogenous DNA into Chlamydia is likely its unique life cycle. Chlamydia species alternate between two morphologically distinct forms: a spore-like form called the elementary body (EB) and a vegetative form termed the reticulate body (RB). EBs are small (∼0.2 μm) and are characterized by the presence of a dense nucleoid structure that consists of DNA tightly packed by bacterial histone-like proteins (34–41). EBs are infectious, and their rigid cell walls aid in their dissemination throughout the extracellular environment. In contrast, RBs are larger (∼1 μm), their chromosomal DNA is uncondensed, and they are osmotically fragile. The infectious cycle begins when EBs adhere to host epithelial cells and induce their internalization to form a membrane-bound vacuole called the inclusion. Upon internalization and throughout the life cycle, Chlamydia remodels the inclusion membrane to escape host lysosomal fusion and to establish a replicative niche. Within the confines of the inclusion, EBs transition into RBs at 6 to 8 h postinfection. Newly differentiated RBs divide by binary fission and populate the inclusion; between 12 and 30 h postinfection, depending on the Chlamydia species, RBs differentiate back into EBs in an asynchronous manner (42). At 46 to 72 h postinfection, EBs are released from the inclusion by promoting host cell and inclusion lysis or by extrusion of the inclusion from its host cell (43).
The various features of the Chlamydia developmental forms pose formidable challenges to the delivery of exogenous DNA. For instance, the EB cell wall is an array of tightly cross-linked proteins that provides rigidity to EB cell walls and protects them from osmotic and shear stress during dissemination (44). A rigid cell wall might render EBs refractory to the uptake of large macromolecules, thereby limiting the acquisition of foreign DNA during extracellular sojourns. Indeed, this barrier might explain the relatively low frequencies of insertion elements, phage remnants, and pathogenicity islands or the lack of genes coding for restriction enzymes in the genomes of chlamydial species. Nonetheless, DNA has successfully been introduced into Chlamydia following the electroporation and chemical transformation of EBs (14, 18, 19), although such transformation events require large amounts of DNA (5 to 10 μg) and occur at a very low frequency.
RB cell walls lack the latticework of cross-linked proteins found in EB cell walls (45, 46) and have low and constricted levels of peptidoglycan (47), potentially facilitating DNA uptake. RBs also undergo cell division and express DNA repair enzymes that mediate the chromosomal integration of DNA by homologous recombination during division, and thus RBs are likely to be naturally competent for transformation. However, targeting RBs for transformation within infected cells requires exogenous DNA to traverse through four lipid bilayers (the host plasma membrane, the inclusion membrane, and the RB outer and inner membranes) before encountering the RB chromosome. RBs can be isolated and potentially transformed in axenic media (48). However, because RBs are noninfectious, their utility in pathogenesis studies is limited unless methods are developed to permit their transition to EBs.
Selection with Antibiotics
Another impediment in establishing robust systems for genetic manipulation in Chlamydia is the lack of selectable markers available to identify transformed bacterial cells. Restrictions on the use of antibiotics due to their clinical use for the treatment of infected patients limit available antibiotic resistance markers. According to the 2010 sexually transmitted disease (STD) treatment guidelines established by the U.S. Centers for Disease Control and Prevention (49), tetracycline, azithromycin, doxycycline, erythromycin, levofloxacin, ofloxacin, and amoxicillin are to be used exclusively for clinical treatment regimens and are prohibited, without special dispensation and approval, from use in generating recombinant strains. Furthermore, the introduction of β-lactamases into urogenital serovars is also restricted due to the use of β-lactams for the treatment of infected women who are pregnant (49).
Because Chlamydia resides in an inclusion within the host, antibiotics must penetrate at least two lipid bilayers (four if the target resides within the bacterium), making some antibiotics (i.e., kanamycin, gentamicin, and streptomycin) unsuitable. Furthermore, the higher MICs required for the delivery of these antibiotics into the inclusion can lead to toxicity in the host cell, which imposes further limits on the use of antibiotics for selection. Several antibiotics have been used successfully in genetic selections, including chloramphenicol, kasugamycin, nalidixic acid, rifampin, spectinomycin, trimethoprim, tetracycline (only for naturally resistant veterinary strains), β-lactams (only for LGV serovars), and blasticidin S (12, 14, 18, 19, 27, 31, 50–53). However, the use of mutant versions of chlamydial factors, such as 16S rRNA, RpoB, and GyrA, that render them resistant to antibiotics as selectable markers is limited because the gene mutations that confer resistance to these antibiotics are often recessive. Exogenous drug resistance cassettes conferring chloramphenicol (cat), β-lactam (bla), and blasticidin (Shble) resistance are currently used as markers to select transformed Chlamydia (18, 19, 27, 28). However, the bla cassette cannot be used in urogenital strains; chloramphenicol can cause mitochondrial stress (54), limiting its use during continuous passaging; and blasticidin S exhibits antibiotic activity toward both prokaryotic and eukaryotic cells and thus can be toxic to host cells. In addition, not all Chlamydiae species are susceptible to these antibiotics, such as Parachlamydia acanthamoeba, which is naturally resistant to β-lactams (55).
Clonal Isolation of DNA Transformants
After incubation with recombinant DNA, transformants can be enriched in the presence of antibiotics; however, it is imperative to eventually isolate clonal populations to minimize the potential carryover of untransformed bacteria that are protected by bystander effects. In contrast to many free-living bacteria, for which clonal populations can be isolated on an agar plate, plaque formation on a monolayer of cells is often employed for Chlamydia. This plaque method consists of laying a culture medium containing agarose over a monolayer of cells infected with bacteria. The overlay restricts the dispersal of bacterial to only neighboring cells. Chlamydia exits its host by promoting host cell lysis, and after several rounds of infection, a zone of clearance becomes visible to the naked eye. The bacteria present within the plaque can be picked from the agarose overlay and further expanded. Although this method is amenable for the clonal isolation of several Chlamydia species (56, 57), most clinical isolates form plaques poorly, which limits the recovery of clonal populations (58). Isolating clonal populations by plaque assay has not been reported for environmental Chlamydiae strains that can only be propagated in amoebal hosts. In these situations, alternative approaches, such as limiting dilution, flow cytometry (59, 60), laser microdissection, or use of micromanipulators, have been employed to isolate bacterial cells directly from infected cells in a monolayer, as reported for Chlamydia and other intracellular pathogens (61, 62).
Chlamydia Shuttle Plasmids
The transformation of chlamydial strains by use of shuttle plasmids is complicated by the presence of native plasmids in some Chlamydia species. The C. trachomatis plasmid is a highly conserved 7.5-kb plasmid that is nonconjugative, nonintegrative, and maintained at up to 8 copies per cell (63, 64). The plasmid carries eight ORFs and is required for the production of glycogen within the inclusion (57, 65). In C. trachomatis, C. muridarum, and C. caviae, a plasmid is required for the expression of several virulence-associated chromosomal genes (23, 66–68). Plasmid loss is associated with reduced activation of Toll-like receptor 2 (TLR2)-dependent inflammatory responses, and plasmid-deficient C. trachomatis and C. muridarum strains have been used as live attenuated vaccine strains against genital and ocular infections (69–71).
Because plasmids containing the same origin of replication are generally incompatible and rarely coexist together in a cell, the maintenance of a native plasmid limits the introduction of an exogenous recombinant plasmid (72). Competition for plasmid replication factors leads to competition between transformed recombinant plasmids and native plasmids, and native plasmids presumably replicate more efficiently due to their smaller size. Plasmid incompatibility therefore decreases the transformation efficiencies of exogenous plasmids, though this can potentially be circumvented by using plasmid-free recipient strains (65, 73–75). Plasmid-deficient strains have been isolated following treatment with novobiocin (65) or other curing agents, such as ethidium bromide or acridine orange, although these agents can be mutagenic and occasionally foment an increase in plasmid copy number (64).
DNA EXCHANGE IN CHLAMYDIA
Evidence from Chlamydia Genomic Signatures
Despite the barriers to Chlamydia transformation by recombinant DNA, there is ample evidence indicating that Chlamydia can import and integrate exogenous DNA into its genome. Homologous DNA recombination is required to repair double-stranded DNA (dsDNA) breaks, which can lead to stalled replication forks (reviewed in reference 76). In E. coli, initiation of DNA recombination involves the recognition and processing of dsDNA breaks into linear single-stranded DNA by RecBCD protein complexes (77), and homologs of all three proteins are carried by Chlamydia. Following the recognition and processing of dsDNA breaks, single-stranded DNA pairs with homologous DNA in a process mediated by a multimeric complex containing RecA, single-stranded DNA binding protein (SSB), RecF, RecO, and RecR (77), all of which are encoded by chlamydial genomes (10, 78–82). Holliday junction formation and branch migration mediated by the RuvAB complex follow, with subsequent resolution of junctions by the endonuclease RuvC (77). Branch migration and junction resolution can also be performed by RecG (77). The ruvA, ruvB, ruvC, and recG genes are carried in Chlamydia genomes, indicating that Holliday junction formation, branch migration, and resolution might occur in a manner similar to that in E. coli.
HGT Events in Chlamydia
Because a number of C. trachomatis genes seem to have been acquired from eukaryotes via horizontal gene transfer (HGT) events, ancestral chlamydial species must have been capable of acquiring foreign DNA (10). Some examples are the C. trachomatis genes encoding SET (nuE) and SWIB domain-containing proteins, as well as the Swi/Snf2 family of helicases (CT555 and CT708), all of which are otherwise exclusively found in eukaryotes (10, 83). Moreover, phylogenetic analyses have revealed that several C. trachomatis protein-encoding sequences are most closely related to genes carried by chloroplasts in photosynthetic cyanobacteria (84). Genome sequencing and phylogenetic reconstructions suggest that progenitor Chlamydiae organisms contributed genetic material to other species, as observed in the genomes of microsporidians (85) and in more than 60 archaeplastidal genes in photosynthetic eukaryotes (1, 86–91). The latter ancestral exchange is hypothesized to have contributed to the evolution of algae and plants (90).
The genomes of most sequenced Chlamydiae organisms are devoid of phages, transposons, and genes encoding DNA restriction and modification systems, suggesting that in contrast to ancestral species, extant Chlamydiae rarely engage in horizontal gene transfer events. An example of this trend is the C. trachomatis type III secretion system (T3SS). Rather than the components of the machinery being encoded in pathogenicity islands or plasmids acquired by HGT events, as commonly observed in other bacteria, C. trachomatis T3SS components are encoded by 10 distinct operons dispersed throughout the genome (92). Another example is the average G/C ratio of open reading frames, which varies among microbial genomes; regions exhibiting high or low G/C ratios are thought to reflect HGT events from organisms with different G/C ratios. Among Chlamydia strains that are pathogenic to humans, the variability in G/C ratios among open reading frames is notably low and, in fact, is among the lowest observed among microbial genome sequences (84). In contrast, high variability is observed in the genomes of Neisseria species, which undergo frequent horizontal gene transfer events (84).
In contrast to human-adapted Chlamydia species, other Chlamydia species have been exposed to HGT events, as indicated by the discovery of bacteriophages in several Chlamydia species that are zoonotic pathogens. The first bacteriophage discovered, chlamydiaphage 1 (Chp1), was initially observed by electron microscopy in thin sections of C. psittaci EBs (93). Other chlamydiaphages include ϕCPG1 from C. caviae (94, 95), Chp2 and Chp4 from C. abortus (96–98), ϕCPAR39 from C. pneumoniae (79), and Chp3 from C. pecorum (99). Chlamydiaphages are icosahedral single-stranded DNA (ssDNA) phages belonging to a subfamily of Microviridae termed Gokushovirinae (100). Chlamydiaphages are lytic to their hosts and might affect the infectivity and virulence of C. caviae (101) and delay RB replication and EB transitioning in C. abortus (102). The discovery of chlamydiaphages is exciting, and future investigation into the prevalence of chlamydiaphages among human chlamydial pathogens, their influence on disease outcomes, and their potential application to molecular genetic manipulation warrants further investigation. Mobile insertion elements have also contributed to HGT events in Chlamydia, such as in tetracycline-resistant chlamydial pathogens of swine (C. suis) that harbor mobile insertion elements encoding tetracycline efflux pumps (103–106).
These observations indicate that Chlamydiae organisms are capable of acquiring nonchlamydial DNA, although this appears to be rare and restricted to a few species. This apparent lack of horizontal gene transfer events is likely the result of ecological isolation from other microbial species due to the obligate intracellular lifestyle of Chlamydia.
Evidence of LGT Events among Chlamydia Clinical Isolates
Although Chlamydiae might undergo limited horizontal acquisition of foreign DNA, there is strong evidence of active gene transfer events between closely related chlamydial serovars and species (lateral gene transfer [LGT]). Preliminary evidence of LGT in Chlamydia was observed based on phylogenetic studies of the genetic structure and diversity of C. trachomatis strains between infected humans. Initial classifications were made by serotyping clinical isolates with panels of monoclonal antibodies raised against the highly immunoreactive C. trachomatis major outer membrane protein (MOMP). This membrane protein, encoded by ompA, consists of four highly polymorphic domains called variable domains (VD1 to VD4). Antibody responses are directed primarily against the variable domains of MOMP, and each protein variant has been used to subclassify C. trachomatis serological variants into serovars. Interestingly, MOMP serotyping uncovered a surprisingly large number of clinical isolates that failed to react to MOMP antibodies (107–110). Detailed sequence analysis of ompA alleles in these variants revealed the presence of ompA mosaic alleles, many of which are hybrids composed of ompA sequences from different serovars. Hybrid ompA alleles likely originated from recombination events between variable domains of ompA alleles from different chlamydial urogenital and trachoma serovars (110–115). The emergence of MOMP variants is thought to be driven by selective pressures leading to antigenic variation, because MOMP is the most abundant surface protein in EBs and RBs and is therefore a prominent target of immune responses. The widespread recombination observed between ompA alleles also explains why ompA-based strain typing in some cases correlates poorly with the clinical phenotypes and infection site tropisms of C. trachomatis serovars. Furthermore, typing of loci from highly variable regions of the genome, such as the plasticity zone and the polymorphic membrane protein (pmp) genes, as well as multilocus sequencing typing (MLST), further supports the notion that recombination extends beyond the ompA locus and is pervasive throughout the C. trachomatis genome (116–121).
Whole-genome sequencing provided the most compelling evidence for DNA interchange between C. trachomatis strains. This was first demonstrated by Jeffrey and colleagues, who determined that large regions of the genome of a cervical nonfusogenic isolate (C. trachomatis Ds/2923) were homologous to the genomes of serovar E and F isolates (urogenital isolates), whereas the ompA and flanking sequences were most closely related to serotype D (C. trachomatis D/UW3) (13) sequences. Further scrutiny of the ompA recombinant region uncovered a crossover event within the rs2 gene (CT680) and a site within ompA that generated an ompA allele encoding a chimeric MOMP with variable domains from different serovars. Interestingly, crossover events in the same genomic region (a 3.7-kb region encompassing the rs2, ompA, and pbpB genes) have been identified in 13 clinical isolates (C/CL-1, E/CL-3, G/CL-5, H/CL6, I/CL-8, Ja/CL-10, C/CL-1, Da/CL-2, E/CL-3, G/CL-4, G/CL-5, H/CL-6, and I/CL-8) (119), indicating that this region is under strong selective pressure to diversify. Finer-scale mapping revealed other regions with telltale signs of recombination, supporting the conclusion that multiple regions in the Ds/2923 genome are products of recombination events between the chromosomes of multiple serovars (13). Somboonna and colleagues also identified intrabiovar lateral gene transfer events (122) based on the whole-genome sequencing of a hypervirulent clinical strain (L2C) isolated from a male patient diagnosed with hemorrhagic proctitis. In this strain, genetic exchange between an LGV L2 strain and a serotype D strain gave rise to a novel hybrid strain. The L2C genome inherited a serotype D region encoding a partial yet functional toxin that might contribute to the hypervirulent phenotype associated with this strain and that is notably absent from other sequenced LGV strains. An increasing number of LGV-causing serovars (L1, L2, L3, L2a, L2b, and L2C) have been identified (122–124). These strains predominantly infect monocytes and macrophages and disseminate to inguinal lymph nodes, leading to their classification as a biovar (LGV) distinct from the noninvasive urogenital and ocular serovars (trachoma biovars) (125). Importantly, the data provided by Somboonna et al. were the first evidence of the emergence of a hypervirulent C. trachomatis strain resulting from natural lateral DNA exchange between Chlamydia biovars.
Comparative genomic analyses of more than 65 clinical strains have provided evidence that LGT events within and between C. trachomatis biovars are natural and common occurrences (4, 13, 126–128). Such analyses also indicated that genetic exchange is more likely between strains with tropism for the same site of infection (4, 126–128). However, there are now multiple examples of interbiovar DNA exchange (4, 122, 127) in which strains with tropisms for different infection sites exchange DNA, suggesting that there are no absolute barriers to genetic exchange. The evidence for DNA exchange inferred from comparative genomic studies correlates well with reports of mixed chlamydial infections. For instance, in addition to ocular strains found in patients with trachoma, urogenital strains have also been observed in single or mixed infections of the conjunctiva (129). Mixed infections are also prevalent in populations with a high propensity for acquiring STDs (108, 111, 130–133). Moreover, opportunities for intrabiovar recombination events are abundant in coinfections of the rectal mucosa with LGV and noninvasive urogenital strains, such as those from serotypes D, G, J, E, F, and K, that also infect the rectum (134–139). Alternatively, several reports suggest that chlamydial species might persist in humans as commensals of the lower gastrointestinal tract (reviewed in reference 140), which could provide fertile grounds for inter- and intrabiovar exchange of DNA.
Although it is clear that widespread exchange of DNA occurs between C. trachomatis biovars and serovars, the mechanism for this phenomenon remains unclear. Such events might occur within the confines of infected epithelial cells, and several studies have demonstrated that more than one C. trachomatis strain can simultaneously infect the same cell and form a single mixed intracellular inclusion (20, 141, 142). Once inside the host cell, coinfecting strains may exchange DNA, presumably while coinhabiting the same inclusion, although inclusion fusion might not be an absolute requirement for DNA exchange, because C. trachomatis isolates with nonfusogenic inclusions can exchange DNA in vitro (143). Moreover, many C. trachomatis strains (such as serotype G, D, K, F, and E strains) form fibers extending from their inclusion to neighboring infected cells (144) that could function as conduits for DNA trafficking. In this model, replicating RBs receive and incorporate linear fragments of DNA via homologous recombination. The transport of linear DNA could be mediated by the C. trachomatis CT339 ORF (CTL0593 [LGV L2 434/Bu]), which resembles the Bacillus subtilis porin ComEC (48% amino acid similarity), a multiple-spanning membrane protein required for linear DNA uptake through the inner and outer membranes (145, 146). Interestingly, the C. trachomatis ComEC homolog appears to be transcribed predominantly in RBs (147), suggesting that RBs might be naturally competent. Thus, DNA exchange between RBs may occur within the confines of an inclusion, thereby limiting exposure of their genomes to foreign DNA. This mode of exchange seems the most likely given the absence of any identifiable DNA conjugation machinery. It is also possible that during chemical transformation of EBs (see the sections below), extracellular DNA deposited on the surface of EBs is incorporated by Chlamydia after the EBs transition to the RB form.
Reproducing LGT Events in the Laboratory
Elegant studies undertaken by Robert Demars and colleagues demonstrated that the process of LGT can be replicated in the laboratory. These authors isolated doubly antibiotic-resistant recombinants from coinfections of HeLa cells with C. trachomatis (serovar LGV L1) strains resistant to ofloxacin (gyrA T249G/A247C), lincomycin (23S rRNA gene A2039C mutant), rifampin (rpoB T1383G), or trimethoprim. Recombinant emergence was detected at a frequency of 10−4 to 10−3 (1 event in 10,000 to 1 event in 1,000), which is several orders of magnitude higher than spontaneous mutation rates, strongly suggesting that the emergence of these doubly resistant recombinants in a coinfection setting was the result of DNA transfer events (12).
Subsequent studies in which an ofloxacin-resistant C. trachomatis serovar L1 strain was “crossed” with a rifampin-resistant C. trachomatis serovar D strain further confirmed that DNA exchange can occur between strains of distinct biovars (11), as has been observed among clinical isolates (122). All the crossover events in 14 recombinant genomes resulting from this cross were mapped, and the lengths of the exchanged DNAs ranged from 336 to 790 kb (11). Similar studies performed with 12 C. trachomatis recombinants generated in vitro revealed 190 homologous recombination events that occurred in these strains, without any evidence of crossover hot spots (143). These results are consistent with comparative genomic analyses of clinical isolates in which crossover events were found to be unbiased, although the recombined regions spanned segments ranging from 3 to 50,141 bp (4). In addition, LGT events are not restricted to intraspecies exchange but also occur between Chlamydia species, since an acquired tetracycline-resistant marker from the C. suis R19 strain was transferred to several C. trachomatis and C. muridarum strains in vitro (50). The transfer of tetracycline resistance involves the insertion of fragments of C. suis DNA ranging from 40 to 100 kb into recipient strains (50), again highlighting the apparent absence of barriers to genetic exchange within and between Chlamydia species in both in vivo and in vitro settings.
MOLECULAR GENETIC MANIPULATION OF CHLAMYDIA
Transient Plasmid Transformation
The first successful transformation of Chlamydiae was reported in 1994 (18). In that work, a chimeric shuttle plasmid (pPBW100) was constructed by ligating an E. coli plasmid (a version of pUC9 encoding kanamycin resistance) to the linearized C. trachomatis serotype E endogenous plasmid pCTE1. A promoterless cat gene, encoding chloramphenicol acetyltransferase, was placed under the control of the pCTE1-based promoter P7248 (near DNA position 7248 in pCTE1). The resulting plasmid (pPBW100) was electroporated into EBs, which were then used to infect McCoy cells. Inclusions containing chloramphenicol-resistant Chlamydia were initially detected; however, these transformants were lost upon further passaging. The P7248::cat cassette was expressed transiently during the early stages of RB development (18), likely explaining the inability to recover stable resistant bacteria. This study was the first to demonstrate that exogenous DNA could be delivered into Chlamydia EBs via electroporation and that transformants could be selected in cell culture, although alternative promoters for driving the expression of heterologous selectable markers were clearly needed.
Allelic Exchange
Surprisingly, 14 years passed from the initial report of the first transformation event in Chlamydiae (18) until the next successful transformation of a chlamydial species was reported (14). Both circular and linearized plasmids containing an allele of the 16S rRNA gene from C. psittaci harboring two single-nucleotide substitutions conferring resistance to both kasugamycin and spectinomycin were electroporated into C. psittaci EBs. The use of the 16S rRNA gene variant (present as a single copy in the C. psittaci genome) ensured that any doubly drug-resistant recombinants resulted from an allelic exchange event that eliminated the wild-type copy, because both mutations in the 16S rRNA gene variant are recessive in a merodiploid strain and because spontaneous resistance to both antibiotics is exceptionally rare (14).
This constraint also precluded the selection of plasmid integration through single-crossover homologous recombination events. Antibiotic-resistant strains were isolated with both linear and circular DNA substrates, and gene conversion events were rare. Maximum recombination frequencies were obtained with 10 or 20 μg of circular plasmid DNA prepared from E. coli strains deficient in DNA methylation (HsdS-, Dcm-, and Dam-defective strains), and recombination frequencies decreased when the flanking homologous DNA sequence (less than 2 kb) and rRNA locus length (from 8.1 to 2.5 kb) were reduced. Although the use of kasugamycin and spectinomycin resistances as selectable markers limits their use as markers for allelic exchange at loci outside the rRNA gene locus, this elegant study provided the proof of principle that recombinant DNA can be stably introduced into the chromosome of a Chlamydia strain.
Stable Plasmid Transformation
Stable transformation of Chlamydia with recombinant DNA was a landmark event in Chlamydia biology (19). By utilizing a chimeric plasmid (pBR325::L2) generated by ligating a C. trachomatis serovar LGV L2 (434/Bu) plasmid and an E. coli plasmid (pBR325) carrying β-lactamase (bla) and chloramphenicol acetyltransferase (cat) genes, Ian Clarke and colleagues successfully transformed C. trachomatis (LGV L2) by using chemical transformation rather than electroporation of EBs (19). Transformants were generated by incubating a mixture of EBs and plasmid DNA in a buffer containing calcium chloride and were isolated by selecting for penicillin G-resistant bacteria in McCoy cells. Stable transformants lost their endogenous plasmid and expressed both the β-lactamase and chloramphenicol acetyltransferase genes, indicating that the standard E. coli promoters driving the expression of both drug resistance cassettes are functional in C. trachomatis. Subsequently, EBs were transformed with the hybrid shuttle plasmid pGFP::SW2, which consists of a variant L2 plasmid (pSW2) with a 377-bp deletion in CDS1, isolated from a Swedish LGV L2 clinical isolate (148), and an E. coli plasmid expressing β-lactamase and GFP fused to chloramphenicol acetyltransferase under the control of a Neisseria promoter. GFP-expressing bacteria within inclusions were readily detectable (19). Given that the use of molecular genetic tools and recombinant DNA has provided the basis for most bacterial pathogenesis studies, these results represent a major turning point that will expedite our understanding of the biology of these important pathogens.
Ectopic Gene Expression
Following the development of a transformation system for C. trachomatis, the repertoire of molecular genetic tools available for Chlamydia expanded rapidly as a series of shuttle vectors with versatile multiple-cloning sites (MCS), fluorescent protein reporters, inducible promoters, and new selectable markers were generated (Table 1; Fig. 1 and 2). For example, the plasmid p2TK2-SW2 (20) and the pBOMB4 series of plasmids (21) combined a β-lactamase-encoding gene (bla) and a multiple-cloning site into the L2 pSW2 and pL2 (L2/434/Bu) plasmids, respectively. The pBOMB4 series offers the additional benefit of utilizing an intact L2 plasmid (pL2) rather than the SW2 variant plasmid, which harbors a 377-bp deletion in CDS1 (148). These vectors are well suited for expressing epitope-tagged or untagged gene products under the control of native promoters. pBOMB4 and its derivative pBOMB4-MCI also express GFP and mCherry, respectively, from the Neisseria promoter used in pSW2:GFP (19), providing a convenient marker to confirm the presence and maintenance of recombinant plasmids in transformants (21). Genes can also be expressed by using constitutive promoters, such as the rpoB promoter, located upstream of the MCS in pBOMB4R (GFP+) and pBOMB4R-MCI (mCherry+) (21). If precise control of gene expression at different times in the Chlamydia developmental cycle is required, several vectors in which gene expression is controlled by anhydrotetracycline via the tetracycline repressor are available. These include pASK-GFP-L2/mKate 2 (GFP+ mCherry+), in which GFP expression is controlled by the tetA promoter (the gene of choice can be swapped with the gfp ORF), and pBOMB4-Tet-mCherry (mCherry+), which carries a multiple-cloning site under the control of a tetA promoter (21, 26) (Table 1; Fig. 1). The use of a tetracycline-inducible system is advantageous with Chlamydia because tetracycline derivatives, such as anhydrotetracycline, can cross biological membranes and activate gene expression at concentrations that are not toxic to Chlamydia. The development of these plasmids has expanded the repertoire of genetic tools available for ectopic expression in Chlamydia. These plasmids are also well suited for use in mutant complementation, for gene overexpression or conditional expression, and for expressing secreted effectors. In the latter case, C-terminal tags, such as FLAG, CyaA (adenylate cyclase), GSK (glycogen synthase kinase), and TEM-1 β-lactamase, can be used to monitor effector secretion and translocation (20, 21, 29, 149–152).
TABLE 1.
The Chlamydia molecular genetic toolbox
| Plasmid category and name | Parental plasmid | Selectable marker(s) | Fluorophore(s)a | Application(s) | Feature | Reference |
|---|---|---|---|---|---|---|
| Plasmids for ectopic gene expression | ||||||
| p2TK2-SW2 plasmid series | ||||||
| p2TK2-SW2 | pGFP-SW2 | bla | None | Ectopic expression from native promoter | Multiple-cloning site | 20 |
| p2TK2-SW2 IncDProm-RSGFP-IncDTerm | p2TK2-SW2 | bla | RSGFP | Fluorescent bacteria; ectopic gene expression from incD promoter | Gene expression from incD promoter | 20 |
| p2TK2-SW2 IncDProm-mCherry-IncDTerm | p2TK2-SW2 | bla | mCherry | Fluorescent bacteria; ectopic gene expression from incD promoter | Gene expression from incD promoter | 20 |
| p2TK2-SW2 IncDProm-CFP-IncDTerm | p2TK2-SW2 | bla | CFP | Fluorescent bacteria; ectopic gene expression from incD promoter | Gene expression from incD promoter | 20 |
| pBOMB4 vector series | ||||||
| pBOMB4 | pL2 [C. trachomatis LGV L2 (434/Bu)] | bla | GFP | Fluorescent bacteria; ectopic gene expression from native promoters | Multiple-cloning site | 21 |
| pBOMB4-MCI | pL2 [C. trachomatis LGV L2 (434/Bu)] | bla | mCherry | Fluorescent bacteria; ectopic gene expression from native promoters | Multiple-cloning site | 21 |
| pBOMB4R | pL2 [C. trachomatis LGV L2 (434/Bu)] | bla | GFP | Fluorescent bacteria; ectopic gene expression from rpoB promoter | rpoB promoter upstream of multiple-cloning site | 21 |
| pBOMB4R-MCI | pL2 [C. trachomatis LGV L2 (434/Bu)] | bla | mCherry | Fluorescent bacteria; ectopic gene expression from rpoB promoter | rpoB promoter upstream of multiple-cloning site | 21 |
| Tetracycline-inducible vectors | ||||||
| pBOMB4-Tet-mCherry | pL2 [C. trachomatis LGV L2 (434/Bu)] | bla | GFP, mCherry | Fluorescent bacteria; inducible ectopic gene expression | Tetracycline-inducible promoter | 21 |
| pASK-GFP/mKate2-L2 | pL2 [C. trachomatis LGV L2 (434/Bu)] | bla | RSGFP, mKate2 | Fluorescent bacteria; inducible ectopic gene expression | Tetracycline-inducible promoter | 26 |
| Plasmids for blasticidin/chloramphenicol selection | ||||||
| pGFPBSD/Z:SW2 | pGFP-SW2 | bsd, ble | RSGFP | Fluorescent bacteria; ectopic gene expression from native promoters | Blasticidin resistance selectable marker | 27 |
| pGFP-CAT::SW2 | pGFP-SW2 | cat | RSGFP | Fluorescent bacteria; ectopic gene expression from native promoters | Chloramphenicol resistance selectable marker | 28 |
| Plasmids for targeted gene disruption | ||||||
| pDFTT3-bla (TargeTron) | pACD4K-C | bla (GII intron), cat | None | Gene disruption in C. trachomatis LGV L2 | incA (C. trachomatis LGV L2) gene disruption | 33 |
| pDFTT3-aadA (TargeTron) | pDFTT3-bla | aadA (GII intron), cat | None | Gene disruption in C. trachomatis LGV L2 | Spectinomycin resistance | 53 |
RSGFP, red-shifted GFP.
FIG 1.
C. trachomatis LGV L2 expression vectors encoding a β-lactamase resistance marker for use in LGV L2 strains. (A) The p2TK2-SW2 plasmids (20) are derivatives of the pGFP-SW2 plasmid (19). pSW2 is a C. trachomatis LGV L2 plasmid isolated from the Swedish SW2 strain, which contains a 377-bp deletion in CDS1 (148). The p2TK2-SW2 vector features a versatile multiple-cloning site for ectopic gene expression under the control of native promoters. p2TK2-SW2 plasmids expressing rsgfp, mCherry, and cfp fluorophores from the incD promoter are ideal for generating fluorescently labeled bacteria. Fluorophore-encoding genes can be substituted with a gene of interest for expression under the control of the incD promoter. (B) pBOMB4 vectors (21) are derived from an intact C. trachomatis LGV L2 (434/Bu) plasmid. These vectors include a multiple-cloning site in addition to fluorescent markers to confirm the presence of recombinant plasmids in transformed bacteria. pBOMB4 (GenBank accession no. KF790906) and pBOMB4-MCI (GenBank accession no. KF790907) are ideal for expressing proteins from native promoters, and pBOMB4R (GenBank accession no. KF790908) and pBOMB4R-MCI (GenBank accession no. KF790909) promote constitutive protein expression from the rpoB promoter. (C) Both pBOMB4-Tet-mCherry (GenBank accession no. KF790910) (21) and pASK-GFP/mKate2-L2 (26) are derived from an intact C. trachomatis LGV L2 (434/Bu) plasmid. Protein expression is controlled by the inducible tetA promoter in both plasmids. pBOMB4-Tet-mCherry features a multiple-cloning site and encodes GFP as a fluorescent marker. pASK-GFP/mKate2-L2 encodes mKate2 as a far-red fluorescent marker for transformed strains. (D) Multiple-cloning sites in each vector. All of the unique restriction sites are labeled in red. bla, β-lactamase-encoding gene; MCS, multiple-cloning site; pSW2, plasmid from C. trachomatis LGV L2 strain SW2; pL2, plasmid from C. trachomatis LGV L2 (434/Bu) strain. (The vector maps in panel A are adapted from reference 20 with permission, the vector maps in panel B and the pBOMB4-Tet-mCherry map in panel C are adapted from reference 21 with permission, and the pASK-GFP/mKate2-L2 map in panel C is adapted from reference 26 with permission.)
FIG 2.
C. trachomatis expression vectors for use in non-LGV L2 strains. Plasmid pGFPBSD/Z::SW2 (27) is a derivative of pGFP:SW2 (19) in which the chloramphenicol acetyltransferase gene (cat) has been replaced by a blasticidin S deaminase gene (bsd) and the β-lactamase-encoding gene (bla) has been replaced by the Shble (zeocin resistance cassette) gene. pGFP-CAT::SW2 (28) is another derivative of pGFP:SW2, in which the β-lactamase-encoding gene (bla) has been removed. pGFPBSD/Z::SW2- and pGFP-CAT::SW2-transformed cells can be selected by using blasticidin and chloramphenicol, respectively. (The pGFPBSD/Z:SW2 map is adapted from reference 27 with permission, and the pGFP-CAT::SW2 map is adapted from reference 28 with permission.)
The majority of ectopic expression plasmids use β-lactamase as a selectable marker. Because amoxicillin, a β-lactam, is used to treat pregnant women with cervicitis, the use of β-lactamases for the selection of transformants in non-C. trachomatis LGV strains (trachoma and urogenital serovars) is not permitted by the guidelines for research involving recombinant DNA established by the National Institutes of Health. Two plasmids have been generated with alternative antibiotic resistance markers. The plasmid pGFPBSD/Z::SW2 (27) is a derivative of pGFP:SW2 (19) in which the chloramphenicol acetyltransferase gene (cat) is replaced by a blasticidin S deaminase gene (bsd). This gene replacement results in a vector that encodes a GFP-BSD fusion protein, and transformants carrying this plasmid can be selected using blasticidin. In addition, the gene encoding β-lactamase (bla) has been replaced with the Shble gene to generate a β-lactamase-free plasmid (27) (Table 1; Fig. 2). In pGFP-CAT::SW2 (28), which is another derivative of pGFP:SW2, the bla cassette has been removed to generate a shuttle plasmid that can be selected using chloramphenicol because the parental pGFP::SW2 vector expresses a GFP-CAT (chloramphenicol acetyltransferase) fusion (Table 1; Fig. 2).
Gene Inactivation by Targeted Mutagenesis
The establishment of a plasmid transformation protocol enabled the development of a plasmid-based approach for targeted gene disruption in chlamydial chromosomal DNA. This approach is based on the TargeTron technology marketed by Sigma. The TargeTron system relies on mobile group II introns that target prokaryotic genomes. Type II introns are ribozymes that insert into target sites by using a retrotransposition mechanism called retrohoming (153), which requires the activity of the intron-encoded protein LtrA, which has endogenous RNA maturase, endonuclease, and reverse transcriptase functions. After chromosomal integration, the type II intron is mobilized by a posttranscriptional RNA splicing mechanism also mediated by LtrA (154). Chromosomal target recognition is mediated by base pairing between the intron target recognition domain and the target gene itself. Successful integration requires proper selection of potential target sites and intron retargeting by modification of the intron (155). In the TargeTron system, the ltrA gene has been removed from the intron and replaced with a selectable marker; both the intron and ltrA are carried on a suicide plasmid. Expression of the intron and LtrA in transformed bacterial cells leads to insertion of the intron into the targeted gene. Because the TargeTron plasmid cannot replicate in Chlamydia, the intron cannot be spliced out, leading to stable and heritable insertional gene inactivation.
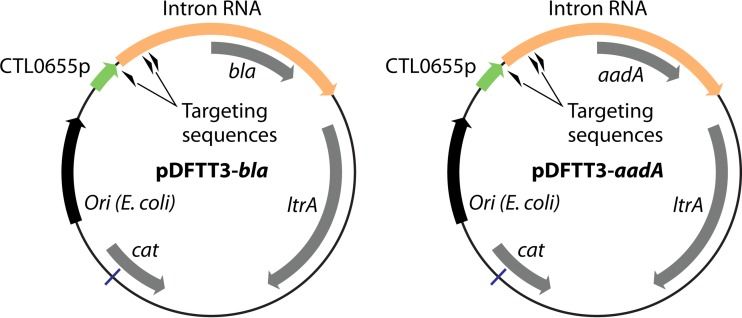
This system was used to engineer a modified TargeTron vector (pDFTT3-bla) (Fig. 3) for use in C. trachomatis by placing the group II intron under the control of a C. trachomatis LGV L2-specific promoter (CTL0655p) and inserting bla for selection in C. trachomatis LGV L2 biovars (33). In proof-of-principle experiments, the resulting vector was retargeted for homing into the incA locus, which encodes an inclusion membrane protein that mediates homotypic inclusion fusion (156, 157). The selection of stable penicillin-resistant transformants generated strains with inactivated incA and fragmented inclusions, as has been observed in naturally occurring IncA-negative C. trachomatis strains (157). That study was the first report of targeted gene inactivation in Chlamydiae. A second TargeTron vector (pDFTT3-aadA) (Fig. 3), carrying a spectinomycin resistance cassette (aadA), was also generated (53) and successfully adapted to generate a C. trachomatis strain bearing an inactivated rsbV1 gene (rsbV1::GII[aadA]), encoding the anti-anti-sigma factor RsbV1 (52). A similar approach also enabled the generation of a double mutant strain bearing loss-of-function alleles of incA and rsbV1 (incA::GII[aadA] and rsbV1::GII[bla]) (53). The pDFTT3-aadA TargeTron vector provides an additional selectable marker for use in inactivation of multiple genes in Chlamydia strains, especially for non-LGV serovars, and allows for the use of the aadA cassette for complementation studies. Importantly, this technique now allows the use of this system for targeted gene inactivation, genome editing (via Cre-lox systems [158]), delivery of genetic material, and mutant strain complementation with single-copy constructs that are chromosomally integrated.
FIG 3.
TargeTron vectors adapted for targeted mutagenesis in C. trachomatis. The suicide plasmid pDFTT3-bla features a group II intron carrying a bla marker that is targeted for integration into the incA locus. Intron RNA expression is driven by the CTL0655 promoter from C. trachomatis LGV L2/434/Bu. pDFTT3-aadA is a reengineered version of pDFTT3-bla in which the bla marker has been replaced with the spectinomycin resistance marker aadA. (The pDFTT3-bla map is adapted from reference 33 with permission, and the pDFTT3-aadA map is based on data from reference 53.)
Forward and Reverse Genetic Approaches
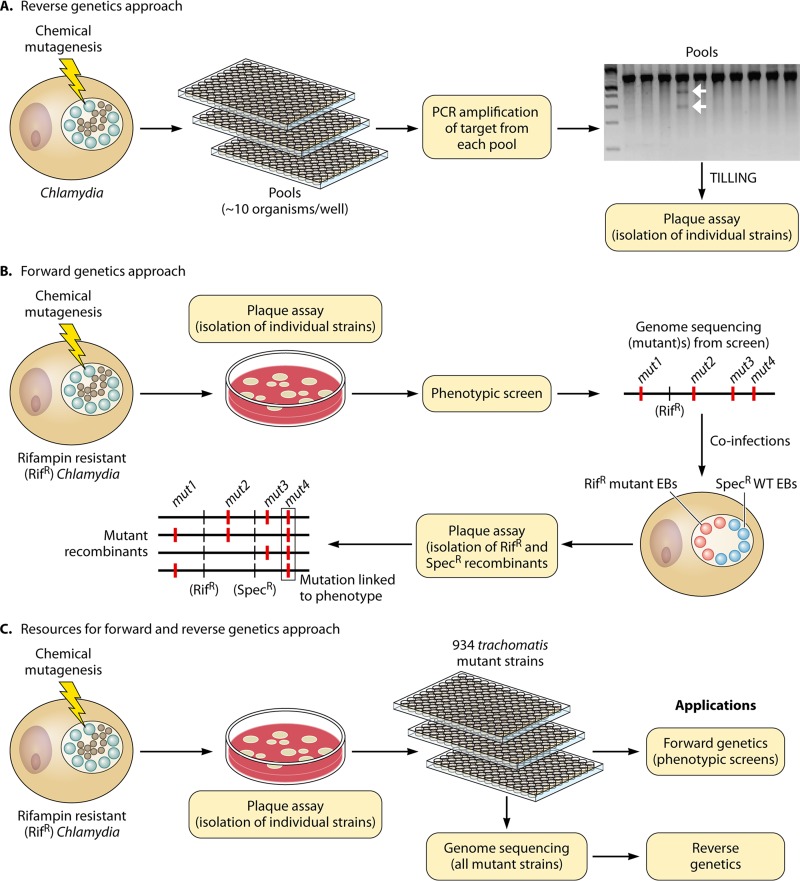
Although the TargeTron system permits site-specific gene disruption, it depends on the inefficient delivery of the TargeTron plasmid into chlamydial cells as well as the selection of an appropriate targeting sequence, and not all constructs are effective at driving GII intron insertion. An alternative approach for the generation of mutants relies on the use of chemical mutagens. Despite the random nature of these lesions, two approaches have been described that enable the identification of mutant strains with desired genetic lesions. In one approach (32) (Fig. 4A), pools of mutagenized C. trachomatis strains were used to infect host cells and then screened for mutants bearing mutations in the trpBA operon by TILLING (targeted induced local lesions in genomes) (159). In this approach, mutants of interest were identified by screening for mismatches between a wild-type gene and alleles generated by chemical mutagenesis. In brief, PCR products spanning the trpBA locus from pools of mutants were hybridized against wild-type trpBA and digested with the CEL1 endonuclease, which targets mismatches in heteroduplex DNA (32). Using this approach, a strain bearing a nonsense mutation in trpB was identified.
FIG 4.
Strategies for genetic analyses of C. trachomatis. (A) Reverse genetic approach for selecting mutant strains harboring mutations in a gene of interest. Pools of ∼10 organisms are generated and arrayed in 96-well plates. A target of choice, such as an ORF, operon, or promoter region, is amplified from each pool, and mutated targets are identified by CEL1 digestion (TILLING). The gel image depicts a representative CEL1 digest. Sanger sequencing is then used to determine the genotypes of mutant targets in the positive pools. Individual strains carrying the mutant target of interest are isolated from each pool by a standard plaque assay. (B) Strategy for forward genetic analysis of Chlamydia. A rifampin-resistant C. trachomatis strain is mutagenized by chemical mutagenesis. Individual mutant strains are isolated by a standard plaque assay. Mutant strains are selected from phenotypic screens of plaque-purified strains, and their genomes are sequenced to identify genetic lesions. To establish linkage between a gene lesion and a phenotype, recombinant strains are selected in the presence of rifampin and spectinomycin after coinfection of host cells with a wild-type strain (Specr) and a mutant (Rifr) strain. TILLING can be utilized to follow the segregation of mutant alleles in recombinants displaying the phenotype. (C) A library of 934 plaque-purified C. trachomatis (LGV L2 434/Bu) mutants has been generated in which all single-nucleotide substitutions have been mapped by whole-genome sequencing. This collection can be utilized for phenotypic screens (forward genetic approach) or to isolate mutant strains harboring a mutant allele of interest (reverse genetic approach). Linkage between a mutant allele and a phenotype of interest can be determined as described for panel B.
In a similar approach (Fig. 4B), chemical mutagenesis was employed to generate pools of mutants in a rifampin-resistant variant of a C. trachomatis LGV L2 strain, which were screened for mutants with aberrant plaque morphologies in a standard plaque assay (31). From this screen, several mutant strains accumulating intrainclusion glycogen aggregates were isolated for further analysis. Whole-genome sequencing revealed that these strains harbored between 3 and 20 single-nucleotide variants (SNVs) in their genomes. Mutations linked to glycogen aggregates were then identified by coinfecting host cells with each rifampin-resistant mutant strain and a spectinomycin-resistant wild-type strain. Recombinant strains generated by LGT were genotyped, and mutations in glgB were found to exclusively segregate with strains containing glycogen deposits and to be absent in recombinant strains lacking glycogen aggregates. Thus, whole-genome sequencing combined with Chlamydia's propensity for exchanging DNA allows the genetic links between causal mutations and phenotypes to be established quickly for mutagenized strains.
Resources for Genome-Wide Genetic Analysis
Screening for strains carrying a mutant allele of interest by forward or reverse genetic approaches can be cumbersome and laborious. To streamline this process, a collection of 934 chemically mutagenized strains was generated in which all SNVs present in each strain were identified by whole-genome sequencing (30). This collection offers over 5,000 nonsynonymous mutations dispersed across the entire C. trachomatis LGV L2 434-Bu genome for use in genome-wide genetic surveys (Fig. 4C). Among these, 99 nonsense mutations in 84 open reading frames comprise a collection of putative loss-of-function mutations in a variety of alleles that function in central metabolism, amino acid metabolism, DNA processing, transcription, and membrane transport and stability (Fig. 5). This collection of mutants also serves as a platform for reverse genetics and has facilitated the identification of several strains carrying recently characterized mutations of interest (160–162). Furthermore, the remaining point mutations offer a comprehensive source of point mutations that can be screened for suppressors, adaptive mutations, conditional alleles, and partial loss- and gain-of-function mutations. Additionally, point mutations can disrupt gene function in operons without incurring polar effects.
FIG 5.
C. trachomatis LGV L2 434/Bu alleles harboring nonsense single-nucleotide substitutions.
The mutant collection also functions as a platform for forward genetic screens, as exemplified by a recent report (30) in which a phenotypic screen using this collection identified a mutant strain that failed to polymerize actin filaments at the periphery of the bacterial inclusion. The mutant strain harbored 12 nonsynonymous SNVs within its genome, and a genetic link between a nonsense mutation in CTL0184 (InaC [inclusion membrane protein for actin assembly]) and loss of actin recruitment was established by LGT. This genetic link was confirmed by transcomplementation with a plasmid expressing wild-type inaC.
Because transposon mutagenesis and the use of mobile introns for gene inactivation at a genomic scale are not yet feasible, integrating chemical mutagenesis and mapping of mutations by whole-genome sequencing or TILLING currently remains the sole strategy for genome-wide analyses of Chlamydiae. In addition, because this approach is not limited by DNA transformation efficiencies and as the costs of whole-genome sequencing continue to plummet, this strategy is readily applicable to all Chlamydiae.
FUTURE DIRECTIONS
Stable transformation with recombinant DNA has been the most formidable barrier to molecular genetic manipulation of Chlamydiae, and the development of a transformation procedure for C. trachomatis was a major breakthrough (19). Nevertheless, significant challenges remain. First and foremost, the process of DNA transformation remains highly inefficient and must be improved. Electroporation protocols should be revisited, as they are highly efficient in promoting the delivery of heterologous DNA into cells. Similarly, high transformation efficiencies might be achievable by targeting chlamydial RBs, which are likely to be naturally competent. However, delivering DNA into RBs remains technically challenging. Lipid-encapsulated nanoparticles (nanosomes/liposomes) might be effective as vehicles for DNA delivery, as has been reported for Plasmodium falciparum (163). Dendrimers, which have been utilized to transform Chlamydia (15–17), can also serve as vehicles for trafficking DNA to RBs. Second, a wider variety of selectable markers, particularly markers that are not used in clinical settings, is required.
The improvement of transformation efficiencies will also enable the application of other genetic tools, such as transposon mutagenesis and associated technology, for a genome-scale assessment of gene function, e.g., via transposon insertion sequencing (164). Higher transformation efficiencies should also facilitate targeted gene disruption and complementation via allelic exchange. Because allelic exchange requires rare double-crossover events between donor and recipient DNAs, counterselectable markers will need to be developed to facilitate the isolation of gene knockout mutant strains. Counterselectable markers can also be used to recycle selectable markers and to generate strains with multiple gene knockouts, which is desirable due to the limited number of selectable markers available for use in Chlamydia. In addition, knockout strains might eventually be constructed by recombineering using bacteriophage λ Red recombinase (165). With this system, targeted gene disruption can be achieved in vivo by cotransforming a linear donor sequence together with a suicide plasmid expressing the λ Red recombinase.
Although tools for targeted gene inactivation are clearly needed, the inactivation of many genes might be problematic because the small size of chlamydial genomes suggests that many genes are essential. In this scenario, inducible systems for gene ablation would be crucial. A system based on FRT/FLP recombination would permit in vivo gene ablation by introducing flanking FRT sites into a locus of interest and promoting gene excision by inducing trans-expression of the FLP recombinase. Plasmids in which expression is controlled by the Tet-inducible operon have already been developed and can easily be coopted for such approaches (21, 26). An inducible FRT/FLP system could also be harnessed for genome editing and selectable marker recycling during the engineering of strains with multiple gene knockouts. In addition, an inducible expression system would be beneficial for epigenetic gene silencing approaches, such as the CRISPR/Cas9 system (166) and TALENS (167, 168), or for the expression of programmable repressors and dominant negative protein variants. These procedures involve the ectopic expression of specialized proteins (e.g., endonucleases and zinc finger DNA binding proteins), which might need to be optimized for expression in Chlamydia.
Despite the technical challenges of routine genetic manipulation in Chlamydia, several milestones have been achieved. Foremost among these was the development of a plasmid transformation system. This transformation system has enabled the use of mobile retrohoming introns (TargeTron technology) for gene inactivation by targeted gene knockouts. The low cost of wholesale genome sequencing now permits the use of chemical mutagenesis for genome-wide genetic analyses. Whole-genome sequencing of large collections of mutants coupled with the use of temperature-sensitive and conditional mutant alleles will also enable the identification of genes essential for intracellular growth and host colonization. As the chlamydial toolbox continues to expand, new considerations will also arise, such as selecting appropriate animal models for testing the virulence of genetically modified strains, the mode of inoculation, and the use of appropriate chlamydial strains.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health awards AI085238, AI100759, and AI081694.
ADDENDUM IN PROOF
A recent study reported the development of a novel system that allows targeting of chlamydial genes for deletion or allelic exchange as well as curing of plasmids from C. trachomatis serovars. The system (fluorescence-reported allelic exchange mutagenesis [FRAEM]) (K. E. Mueller, K. Wolf, and K. A. Fields, mBio 7:e01817-15, 2016, http://dx.doi.org/10.1128/mBio.01817-15) is based on a novel C. trachomatis L2 programmable suicide vector that allows for allelic exchange mutagenesis and selection of exchange events through monitoring of fluorescent markers. This new tool now fully transforms C. trachomatis from a genetically recalcitrant pathogen to a fully genetically tractable model organism.
REFERENCES
- 1.Horn M. 2008. Chlamydiae as symbionts in eukaryotes. Annu Rev Microbiol 62:113–131. doi: 10.1146/annurev.micro.62.081307.162818. [DOI] [PubMed] [Google Scholar]
- 2.Greub G. 2010. International Committee on Systematics of Prokaryotes. Subcommittee on the taxonomy of the Chlamydiae: minutes of the inaugural closed meeting, 21 March 2009, Little Rock, AR, USA. Int J Syst Evol Microbiol 60:2691–2693. doi: 10.1099/ijs.0.028225-0. [DOI] [PubMed] [Google Scholar]
- 3.Stephens RS, Myers G, Eppinger M, Bavoil PM. 2009. Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved. FEMS Immunol Med Microbiol 55:115–119. doi: 10.1111/j.1574-695X.2008.00516.x. [DOI] [PubMed] [Google Scholar]
- 4.Harris SR, Clarke IN, Seth-Smith HM, Solomon AW, Cutcliffe LT, Marsh P, Skilton RJ, Holland MJ, Mabey D, Peeling RW, Lewis DA, Spratt BG, Unemo M, Persson K, Bjartling C, Brunham R, de Vries HJ, Morre SA, Speksnijder A, Bebear CM, Clerc M, de Barbeyrac B, Parkhill J, Thomson NR. 2012. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet 44:413–419. doi: 10.1038/ng.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peipert JF. 2003. Clinical practice. Genital chlamydial infections. N Engl J Med 349:2424–2430. [DOI] [PubMed] [Google Scholar]
- 6.Burton MJ. 2007. Trachoma: an overview. Br Med Bull 84:99–116. doi: 10.1093/bmb/ldm034. [DOI] [PubMed] [Google Scholar]
- 7.Mabey D, Peeling RW. 2002. Lymphogranuloma venereum. Sex Transm Infect 78:90–92. doi: 10.1136/sti.78.2.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falkow S. 1988. Molecular Koch's postulates applied to microbial pathogenicity. Rev Infect Dis 10(Suppl 2):S274–S276. doi: 10.1093/cid/10.Supplement_2.S274. [DOI] [PubMed] [Google Scholar]
- 9.Tang FF, Chang HL, Huang YT, Wang KC. 1957. Studies on the etiology of trachoma with special reference to isolation of the virus in chick embryo. Chin Med J 75:429–447. [PubMed] [Google Scholar]
- 10.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 11.DeMars R, Weinfurter J. 2008. Interstrain gene transfer in Chlamydia trachomatis in vitro: mechanism and significance. J Bacteriol 190:1605–1614. doi: 10.1128/JB.01592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demars R, Weinfurter J, Guex E, Lin J, Potucek Y. 2007. Lateral gene transfer in vitro in the intracellular pathogen Chlamydia trachomatis. J Bacteriol 189:991–1003. doi: 10.1128/JB.00845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffrey BM, Suchland RJ, Quinn KL, Davidson JR, Stamm WE, Rockey DD. 2010. Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect Immun 78:2544–2553. doi: 10.1128/IAI.01324-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binet R, Maurelli AT. 2009. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc Natl Acad Sci U S A 106:292–297. doi: 10.1073/pnas.0806768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerard HC, Mishra MK, Mao G, Wang S, Hali M, Whittum-Hudson JA, Kannan RM, Hudson AP. 2013. Dendrimer-enabled DNA delivery and transformation of Chlamydia pneumoniae. Nanomedicine 9:996–1008. doi: 10.1016/j.nano.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Kannan RM, Gerard HC, Mishra MK, Mao G, Wang S, Hali M, Whittum-Hudson JA, Hudson AP. 2013. Dendrimer-enabled transformation of Chlamydia trachomatis. Microb Pathog 65:29–35. doi: 10.1016/j.micpath.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Mishra MK, Gerard HC, Whittum-Hudson JA, Hudson AP, Kannan RM. 2012. Dendrimer-enabled modulation of gene expression in Chlamydia trachomatis. Mol Pharm 9:413–421. doi: 10.1021/mp200512f. [DOI] [PubMed] [Google Scholar]
- 18.Tam JE, Davis CH, Wyrick PB. 1994. Expression of recombinant DNA introduced into Chlamydia trachomatis by electroporation. Can J Microbiol 40:583–591. doi: 10.1139/m94-093. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agaisse H, Derre I. 2013. A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS One 8:e57090. doi: 10.1371/journal.pone.0057090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong S, Yang Z, Lei L, Shen L, Zhong G. 2013. Characterization of Chlamydia trachomatis plasmid-encoded open reading frames. J Bacteriol 195:3819–3826. doi: 10.1128/JB.00511-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, Watkins H, Zhou B, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD. 2013. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect Immun 81:636–644. doi: 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Cutcliffe LT, Skilton RJ, Persson K, Bjartling C, Clarke IN. 2013. Transformation of a plasmid-free, genital tract isolate of Chlamydia trachomatis with a plasmid vector carrying a deletion in CDS6 revealed that this gene regulates inclusion phenotype. Pathog Dis 67:100–103. doi: 10.1111/2049-632X.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Persson K, Bjartling C, Clarke IN. 2013. Genetic transformation of a clinical (genital tract), plasmid-free isolate of Chlamydia trachomatis: engineering the plasmid as a cloning vector. PLoS One 8:e59195. doi: 10.1371/journal.pone.0059195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickstrum J, Sammons LR, Restivo KN, Hefty PS. 2013. Conditional gene expression in Chlamydia trachomatis using the tet system. PLoS One 8:e76743. doi: 10.1371/journal.pone.0076743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding H, Gong S, Tian Y, Yang Z, Brunham R, Zhong G. 2013. Transformation of sexually transmitted infection-causing serovars of Chlamydia trachomatis using blasticidin for selection. PLoS One 8:e80534. doi: 10.1371/journal.pone.0080534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu S, Battaglia L, Bao X, Fan H. 2013. Chloramphenicol acetyltransferase as a selection marker for chlamydial transformation. BMC Res Notes 6:377. doi: 10.1186/1756-0500-6-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agaisse H, Derre I. 2014. Expression of the effector protein IncD in Chlamydia trachomatis mediates recruitment of the lipid transfer protein CERT and the endoplasmic reticulum-resident protein VAPB to the inclusion membrane. Infect Immun 82:2037–2047. doi: 10.1128/IAI.01530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kokes M, Dunn JD, Granek JA, Nguyen BD, Barker JR, Valdivia RH, Bastidas RJ. 2015. Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia. Cell Host Microbe 17:716–725. doi: 10.1016/j.chom.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen BD, Valdivia RH. 2012. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc Natl Acad Sci U S A 109:1263–1268. doi: 10.1073/pnas.1117884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Virok D, Rajaram K, Endresz V, McClarty G, Nelson DE, Caldwell HD. 2011. Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci U S A 108:7189–7193. doi: 10.1073/pnas.1102229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson CM, Fisher DJ. 2013. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS One 8:e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barry CE III, Brickman TJ, Hackstadt T. 1993. Hc1-mediated effects on DNA structure: a potential regulator of chlamydial development. Mol Microbiol 9:273–283. doi: 10.1111/j.1365-2958.1993.tb01689.x. [DOI] [PubMed] [Google Scholar]
- 35.Barry CE III, Hayes SF, Hackstadt T. 1992. Nucleoid condensation in Escherichia coli that express a chlamydial histone homolog. Science 256:377–379. doi: 10.1126/science.256.5055.377. [DOI] [PubMed] [Google Scholar]
- 36.Brickman TJ, Barry CE III, Hackstadt T. 1993. Molecular cloning and expression of hctB encoding a strain-variant chlamydial histone-like protein with DNA-binding activity. J Bacteriol 175:4274–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hackstadt T, Baehr W, Ying Y. 1991. Chlamydia trachomatis developmentally regulated protein is homologous to eukaryotic histone H1. Proc Natl Acad Sci U S A 88:3937–3941. doi: 10.1073/pnas.88.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pedersen LB, Birkelund S, Christiansen G. 1994. Interaction of the Chlamydia trachomatis histone H1-like protein (Hc1) with DNA and RNA causes repression of transcription and translation in vitro. Mol Microbiol 11:1085–1098. doi: 10.1111/j.1365-2958.1994.tb00385.x. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen LB, Birkelund S, Christiansen G. 1996. Purification of recombinant Chlamydia trachomatis histone H1-like protein Hc2, and comparative functional analysis of Hc2 and Hc1. Mol Microbiol 20:295–311. doi: 10.1111/j.1365-2958.1996.tb02618.x. [DOI] [PubMed] [Google Scholar]
- 40.Perara E, Ganem D, Engel JN. 1992. A developmentally regulated chlamydial gene with apparent homology to eukaryotic histone H1. Proc Natl Acad Sci U S A 89:2125–2129. doi: 10.1073/pnas.89.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao S, Kaul R, Wenman WM. 1991. Identification and nucleotide sequence of a developmentally regulated gene encoding a eukaryotic histone H1-like protein from Chlamydia trachomatis. J Bacteriol 173:2818–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moulder JW. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol Rev 55:143–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hybiske K, Stephens RS. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci U S A 104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatch TP. 1996. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J Bacteriol 178:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatch TP, Miceli M, Sublett JE. 1986. Synthesis of disulfide-bonded outer membrane proteins during the developmental cycle of Chlamydia psittaci and Chlamydia trachomatis. J Bacteriol 165:379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raulston JE, Davis CH, Paul TR, Hobbs JD, Wyrick PB. 2002. Surface accessibility of the 70-kilodalton Chlamydia trachomatis heat shock protein following reduction of outer membrane protein disulfide bonds. Infect Immun 70:535–543. doi: 10.1128/IAI.70.2.535-543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, Maurelli AT. 2014. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506:507–510. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. 2012. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc Natl Acad Sci U S A 109:19781–19785. doi: 10.1073/pnas.1212831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Workowski KA, Berman S. 2010. Sexually transmitted diseases treatment guidelines, 2010. MMWR Recomm Rep 59(RR-12): 1–110. [PubMed] [Google Scholar]
- 50.Suchland RJ, Sandoz KM, Jeffrey BM, Stamm WE, Rockey DD. 2009. Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob Agents Chemother 53:4604–4611. doi: 10.1128/AAC.00477-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lenart J, Andersen AA, Rockey DD. 2001. Growth and development of tetracycline-resistant Chlamydia suis. Antimicrob Agents Chemother 45:2198–2203. doi: 10.1128/AAC.45.8.2198-2203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson CC, Griffiths C, Nicod SS, Lowden NM, Wigneshweraraj S, Fisher DJ, McClure MO. 2015. The Rsb phosphoregulatory network controls availability of the primary sigma factor in Chlamydia trachomatis and influences the kinetics of growth and development. PLoS Pathog 11:e1005125. doi: 10.1371/journal.ppat.1005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lowden NM, Yeruva L, Johnson CM, Bowlin AK, Fisher DJ. 2015. Use of aminoglycoside 3′ adenyltransferase as a selection marker for Chlamydia trachomatis intron-mutagenesis and in vivo intron stability. BMC Res Notes 8:570. doi: 10.1186/s13104-015-1542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li CH, Cheng YW, Liao PL, Yang YT, Kang JJ. 2010. Chloramphenicol causes mitochondrial stress, decreases ATP biosynthesis, induces matrix metalloproteinase-13 expression, and solid-tumor cell invasion. Toxicol Sci 116:140–150. doi: 10.1093/toxsci/kfq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maurin M, Bryskier A, Raoult D. 2002. Antibiotic susceptibilities of Parachlamydia acanthamoeba in amoebae. Antimicrob Agents Chemother 46:3065–3067. doi: 10.1128/AAC.46.9.3065-3067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gieffers J, Belland RJ, Whitmire W, Ouellette S, Crane D, Maass M, Byrne GI, Caldwell HD. 2002. Isolation of Chlamydia pneumoniae clonal variants by a focus-forming assay. Infect Immun 70:5827–5834. doi: 10.1128/IAI.70.10.5827-5834.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsumoto A, Izutsu H, Miyashita N, Ohuchi M. 1998. Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma. J Clin Microbiol 36:3013–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Somboonna N, Mead S, Liu J, Dean D. 2008. Discovering and differentiating new and emerging clonal populations of Chlamydia trachomatis with a novel shotgun cell culture harvest assay. Emerg Infect Dis 14:445–453. doi: 10.3201/eid1403.071071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alzhanov DT, Suchland RJ, Bakke AC, Stamm WE, Rockey DD. 2007. Clonal isolation of Chlamydia-infected cells using flow cytometry. J Microbiol Methods 68:201–208. doi: 10.1016/j.mimet.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 60.Suchland RJ, Stamm WE. 1991. Simplified microtiter cell culture method for rapid immunotyping of Chlamydia trachomatis. J Clin Microbiol 29:1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beare PA, Sandoz KM, Omsland A, Rockey DD, Heinzen RA. 2011. Advances in genetic manipulation of obligate intracellular bacterial pathogens. Front Microbiol 2:97. doi: 10.3389/fmicb.2011.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Podgorny OV, Polina NF, Babenko VV, Karpova IY, Kostryukova ES, Govorun VM, Lazarev VN. 2015. Isolation of single Chlamydia-infected cells using laser microdissection. J Microbiol Methods 109:123–128. doi: 10.1016/j.mimet.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 63.Ferreira R, Borges V, Nunes A, Borrego MJ, Gomes JP. 2013. Assessment of the load and transcriptional dynamics of Chlamydia trachomatis plasmid according to strains' tissue tropism. Microbiol Res 168:333–339. doi: 10.1016/j.micres.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 64.Pickett MA, Everson JS, Pead PJ, Clarke IN. 2005. The plasmids of Chlamydia trachomatis and Chlamydophila pneumoniae (N16): accurate determination of copy number and the paradoxical effect of plasmid-curing agents. Microbiology 151:893–903. doi: 10.1099/mic.0.27625-0. [DOI] [PubMed] [Google Scholar]
- 65.O'Connell CM, Nicks KM. 2006. A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology 152:1601–1607. doi: 10.1099/mic.0.28658-0. [DOI] [PubMed] [Google Scholar]
- 66.Carlson JH, Whitmire WM, Crane DD, Wicke L, Virtaneva K, Sturdevant DE, Kupko JJ III, Porcella SF, Martinez-Orengo N, Heinzen RA, Kari L, Caldwell HD. 2008. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect Immun 76:2273–2283. doi: 10.1128/IAI.00102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frazer LC, Darville T, Chandra-Kuntal K, Andrews CW Jr, Zurenski M, Mintus M, AbdelRahman YM, Belland RJ, Ingalls RR, O'Connell CM. 2012. Plasmid-cured Chlamydia caviae activates TLR2-dependent signaling and retains virulence in the guinea pig model of genital tract infection. PLoS One 7:e30747. doi: 10.1371/journal.pone.0030747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Connell CM, AbdelRahman YM, Green E, Darville HK, Saira K, Smith B, Darville T, Scurlock AM, Meyer CR, Belland RJ. 2011. Toll-like receptor 2 activation by Chlamydia trachomatis is plasmid dependent, and plasmid-responsive chromosomal loci are coordinately regulated in response to glucose limitation by C. trachomatis but not by C. muridarum. Infect Immun 79:1044–1056. doi: 10.1128/IAI.01118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kari L, Whitmire WM, Olivares-Zavaleta N, Goheen MM, Taylor LD, Carlson JH, Sturdevant GL, Lu C, Bakios LE, Randall LB, Parnell MJ, Zhong G, Caldwell HD. 2011. A live-attenuated chlamydial vaccine protects against trachoma in nonhuman primates. J Exp Med 208:2217–2223. doi: 10.1084/jem.20111266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Connell CM, Ingalls RR, Andrews CW Jr, Scurlock AM, Darville T. 2007. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J Immunol 179:4027–4034. doi: 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- 71.Olivares-Zavaleta N, Whitmire W, Gardner D, Caldwell HD. 2010. Immunization with the attenuated plasmidless Chlamydia trachomatis L2(25667R) strain provides partial protection in a murine model of female genitourinary tract infection. Vaccine 28:1454–1462. doi: 10.1016/j.vaccine.2009.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nordstrom K, Austin SJ. 1989. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet 23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 73.Farencena A, Comanducci M, Donati M, Ratti G, Cevenini R. 1997. Characterization of a new isolate of Chlamydia trachomatis which lacks the common plasmid and has properties of biovar trachoma. Infect Immun 65:2965–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peterson EM, Markoff BA, Schachter J, de la Maza LM. 1990. The 7.5-kb plasmid present in Chlamydia trachomatis is not essential for the growth of this microorganism. Plasmid 23:144–148. doi: 10.1016/0147-619X(90)90033-9. [DOI] [PubMed] [Google Scholar]
- 75.Stothard DR, Williams JA, Van Der Pol B, Jones RB. 1998. Identification of a Chlamydia trachomatis serovar E urogenital isolate which lacks the cryptic plasmid. Infect Immun 66:6010–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rothstein R, Michel B, Gangloff S. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev 14:1–10. [PubMed] [Google Scholar]
- 77.Camerini-Otero RD, Hsieh P. 1995. Homologous recombination proteins in prokaryotes and eukaryotes. Annu Rev Genet 29:509–552. doi: 10.1146/annurev.ge.29.120195.002453. [DOI] [PubMed] [Google Scholar]
- 78.Hintz NJ, Ennis DG, Liu WF, Larsen SH. 1995. The recA gene of Chlamydia trachomatis: cloning, sequence, and characterization in Escherichia coli. FEMS Microbiol Lett 127:175–180. doi: 10.1111/j.1574-6968.1995.tb07470.x. [DOI] [PubMed] [Google Scholar]
- 79.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Read TD, Joseph SJ, Didelot X, Liang B, Patel L, Dean D. 2013. Comparative analysis of Chlamydia psittaci genomes reveals the recent emergence of a pathogenic lineage with a broad host range. mBio 4:e00604-12. doi: 10.1128/mBio.00604-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Read TD, Myers GS, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, Holtzapple E, Khouri H, Federova NB, Carty HA, Umayam LA, Haft DH, Peterson J, Beanan MJ, White O, Salzberg SL, Hsia RC, McClarty G, Rank RG, Bavoil PM, Fraser CM. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res 31:2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang DJ, Fan H, McClarty G, Brunham RC. 1995. Identification of the Chlamydia trachomatis RecA-encoding gene. Infect Immun 63:676–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pennini ME, Perrinet S, Dautry-Varsat A, Subtil A. 2010. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog 6:e1000995. doi: 10.1371/journal.ppat.1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brinkman FS, Blanchard JL, Cherkasov A, Av-Gay Y, Brunham RC, Fernandez RC, Finlay BB, Otto SP, Ouellette BF, Keeling PJ, Rose AM, Hancock RE, Jones SJ, Greberg H. 2002. Evidence that plant-like genes in Chlamydia species reflect an ancestral relationship between Chlamydiaceae, cyanobacteria, and the chloroplast. Genome Res 12:1159–1167. doi: 10.1101/gr.341802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivares CP. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 86.Ball SG, Subtil A, Bhattacharya D, Moustafa A, Weber AP, Gehre L, Colleoni C, Arias MC, Cenci U, Dauvillee D. 2013. Metabolic effectors secreted by bacterial pathogens: essential facilitators of plastid endosymbiosis? Plant Cell 25:7–21. doi: 10.1105/tpc.112.101329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Becker B, Hoef-Emden K, Melkonian M. 2008. Chlamydial genes shed light on the evolution of photoautotrophic eukaryotes. BMC Evol Biol 8:203. doi: 10.1186/1471-2148-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang J, Gogarten JP. 2007. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol 8:R99. doi: 10.1186/gb-2007-8-6-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moustafa A, Reyes-Prieto A, Bhattacharya D. 2008. Chlamydiae has contributed at least 55 genes to Plantae with predominantly plastid functions. PLoS One 3:e2205. doi: 10.1371/journal.pone.0002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Subtil A, Collingro A, Horn M. 2014. Tracing the primordial Chlamydiae: extinct parasites of plants? Trends Plant Sci 19:36–43. doi: 10.1016/j.tplants.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 91.Collingro A, Tischler P, Weinmaier T, Penz T, Heinz E, Brunham RC, Read TD, Bavoil PM, Sachse K, Kahane S, Friedman MG, Rattei T, Myers GS, Horn M. 2011. Unity in variety—the pan-genome of the Chlamydiae. Mol Biol Evol 28:3253–3270. doi: 10.1093/molbev/msr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hefty PS, Stephens RS. 2007. Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J Bacteriol 189:198–206. doi: 10.1128/JB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Richmond SJ, Stirling P, Ashley CR. 1982. Virus infecting the reticulate bodies of an avian strain of Chlamydia psittaci. FEMS Microbiol Lett 14:31–36. doi: 10.1111/j.1574-6968.1982.tb08629.x. [DOI] [Google Scholar]
- 94.Hsia R, Ohayon H, Gounon P, Dautry-Varsat A, Bavoil PM. 2000. Phage infection of the obligate intracellular bacterium, Chlamydia psittaci strain guinea pig inclusion conjunctivitis. Microbes Infect 2:761–772. doi: 10.1016/S1286-4579(00)90356-3. [DOI] [PubMed] [Google Scholar]
- 95.Hsia RC, Ting LM, Bavoil PM. 2000. Microvirus of Chlamydia psittaci strain guinea pig inclusion conjunctivitis: isolation and molecular characterization. Microbiology 146:1651–1660. doi: 10.1099/00221287-146-7-1651. [DOI] [PubMed] [Google Scholar]
- 96.Everson JS, Garner SA, Lambden PR, Fane BA, Clarke IN. 2003. Host range of chlamydiaphages phiCPAR39 and Chp3. J Bacteriol 185:6490–6492. doi: 10.1128/JB.185.21.6490-6492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu BL, Everson JS, Fane B, Giannikopoulou P, Vretou E, Lambden PR, Clarke IN. 2000. Molecular characterization of a bacteriophage (Chp2) from Chlamydia psittaci. J Virol 74:3464–3469. doi: 10.1128/JVI.74.8.3464-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sait M, Livingstone M, Graham R, Inglis NF, Wheelhouse N, Longbottom D. 2011. Identification, sequencing and molecular analysis of Chp4, a novel chlamydiaphage of Chlamydophila abortus belonging to the family Microviridae. J Gen Virol 92:1733–1737. doi: 10.1099/vir.0.031583-0. [DOI] [PubMed] [Google Scholar]
- 99.Garner SA, Everson JS, Lambden PR, Fane BA, Clarke IN. 2004. Isolation, molecular characterisation and genome sequence of a bacteriophage (Chp3) from Chlamydophila pecorum. Virus Genes 28:207–214. doi: 10.1023/B:VIRU.0000016860.53035.f3. [DOI] [PubMed] [Google Scholar]
- 100.Cherwa JE, Fane BA. 16 May 2011. Microviridae: microviruses and gokushoviruses. eLS doi: 10.1002/9780470015902.a0000781.pub2. [DOI] [Google Scholar]
- 101.Rank RG, Bowlin AK, Cane S, Shou H, Liu Z, Nagarajan UM, Bavoil PM. 2009. Effect of chlamydiaphage phiCPG1 on the course of conjunctival infection with “Chlamydia caviae” in guinea pigs. Infect Immun 77:1216–1221. doi: 10.1128/IAI.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Salim O, Skilton RJ, Lambden PR, Fane BA, Clarke IN. 2008. Behind the chlamydial cloak: the replication cycle of chlamydiaphage Chp2, revealed. Virology 377:440–445. doi: 10.1016/j.virol.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 103.Andersen AA, Rogers DG. 1998. Resistance to tetracycline and sulfadiazine in swine C. trachomatis isolates, p 313 In Stephens RS. (ed), Chlamydia infections: proceedings of the 9th International Symposium on Human Chlamydial Infection. International Chlamydia Symposium, San Francisco, CA. [Google Scholar]
- 104.Di Francesco A, Donati M, Rossi M, Pignanelli S, Shurdhi A, Baldelli R, Cevenini R. 2008. Tetracycline-resistant Chlamydia suis isolates in Italy. Vet Rec 163:251–252. doi: 10.1136/vr.163.8.251. [DOI] [PubMed] [Google Scholar]
- 105.Dugan J, Andersen AA, Rockey DD. 2007. Functional characterization of IScs605, an insertion element carried by tetracycline-resistant Chlamydia suis. Microbiology 153:71–79. doi: 10.1099/mic.0.29253-0. [DOI] [PubMed] [Google Scholar]
- 106.Dugan J, Rockey DD, Jones L, Andersen AA. 2004. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob Agents Chemother 48:3989–3995. doi: 10.1128/AAC.48.10.3989-3995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dean D, Patton M, Stephens RS. 1991. Direct sequence evaluation of the major outer membrane protein gene variant regions of Chlamydia trachomatis subtypes D′, I′, and L2′. Infect Immun 59:1579–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dean D, Schachter J, Dawson CR, Stephens RS. 1992. Comparison of the major outer membrane protein variant sequence regions of B/Ba isolates: a molecular epidemiologic approach to Chlamydia trachomatis infections. J Infect Dis 166:383–392. doi: 10.1093/infdis/166.2.383. [DOI] [PubMed] [Google Scholar]
- 109.Hayes LJ, Bailey RL, Mabey DC, Clarke IN, Pickett MA, Watt PJ, Ward ME. 1992. Genotyping of Chlamydia trachomatis from a trachoma-endemic village in the Gambia by a nested polymerase chain reaction: identification of strain variants. J Infect Dis 166:1173–1177. doi: 10.1093/infdis/166.5.1173. [DOI] [PubMed] [Google Scholar]
- 110.Lampe MF, Suchland RJ, Stamm WE. 1993. Nucleotide sequence of the variable domains within the major outer membrane protein gene from serovariants of Chlamydia trachomatis. Infect Immun 61:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brunham R, Yang C, Maclean I, Kimani J, Maitha G, Plummer F. 1994. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J Clin Invest 94:458–463. doi: 10.1172/JCI117347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fitch WM, Peterson EM, de la Maza LM. 1993. Phylogenetic analysis of the outer-membrane-protein genes of Chlamydiae, and its implication for vaccine development. Mol Biol Evol 10:892–913. [DOI] [PubMed] [Google Scholar]
- 113.Hayes LJ, Yearsley P, Treharne JD, Ballard RA, Fehler GH, Ward ME. 1994. Evidence for naturally occurring recombination in the gene encoding the major outer membrane protein of lymphogranuloma venereum isolates of Chlamydia trachomatis. Infect Immun 62:5659–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Millman K, Black CM, Johnson RE, Stamm WE, Jones RB, Hook EW, Martin DH, Bolan G, Tavare S, Dean D. 2004. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J Bacteriol 186:2457–2465. doi: 10.1128/JB.186.8.2457-2465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Millman KL, Tavare S, Dean D. 2001. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J Bacteriol 183:5997–6008. doi: 10.1128/JB.183.20.5997-6008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Batteiger BE, Wan R, Williams JA, He L, Ma A, Fortenberry JD, Dean D. 2014. Novel Chlamydia trachomatis strains in heterosexual sex partners, Indianapolis, Indiana, USA. Emerg Infect Dis 20:1841–1847. doi: 10.3201/2011.140604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dean D, Bruno WJ, Wan R, Gomes JP, Devignot S, Mehari T, de Vries HJ, Morre SA, Myers G, Read TD, Spratt BG. 2009. Predicting phenotype and emerging strains among Chlamydia trachomatis infections. Emerg Infect Dis 15:1385–1394. doi: 10.3201/eid1509.090272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gomes JP, Bruno WJ, Borrego MJ, Dean D. 2004. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J Bacteriol 186:4295–4306. doi: 10.1128/JB.186.13.4295-4306.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gomes JP, Bruno WJ, Nunes A, Santos N, Florindo C, Borrego MJ, Dean D. 2007. Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hotspots. Genome Res 17:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gomes JP, Nunes A, Bruno WJ, Borrego MJ, Florindo C, Dean D. 2006. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J Bacteriol 188:275–286. doi: 10.1128/JB.188.1.275-286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pannekoek Y, Morelli G, Kusecek B, Morre SA, Ossewaarde JM, Langerak AA, van der Ende A. 2008. Multi locus sequence typing of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis. BMC Microbiol 8:42. doi: 10.1186/1471-2180-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Somboonna N, Wan R, Ojcius DM, Pettengill MA, Joseph SJ, Chang A, Hsu R, Read TD, Dean D. 2011. Hypervirulent Chlamydia trachomatis clinical strain is a recombinant between lymphogranuloma venereum (L(2)) and D lineages. mBio 2:e00045-11. doi: 10.1128/mBio.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Spaargaren J, Schachter J, Moncada J, de Vries HJ, Fennema HS, Pena AS, Coutinho RA, Morre SA. 2005. Slow epidemic of lymphogranuloma venereum L2b strain. Emerg Infect Dis 11:1787–1788. doi: 10.3201/eid1111.050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Van der Bij AK, Spaargaren J, Morre SA, Fennema HS, Mindel A, Coutinho RA, de Vries HJ. 2006. Diagnostic and clinical implications of anorectal lymphogranuloma venereum in men who have sex with men: a retrospective case-control study. Clin Infect Dis 42:186–194. doi: 10.1086/498904. [DOI] [PubMed] [Google Scholar]
- 125.Weir E. 2005. Lymphogranuloma venereum in the differential diagnosis of proctitis. CMAJ 172:185. doi: 10.1503/cmaj.045191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Joseph SJ, Didelot X, Gandhi K, Dean D, Read TD. 2011. Interplay of recombination and selection in the genomes of Chlamydia trachomatis. Biol Direct 6:28. doi: 10.1186/1745-6150-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Joseph SJ, Didelot X, Rothschild J, de Vries HJ, Morre SA, Read TD, Dean D. 2012. Population genomics of Chlamydia trachomatis: insights on drift, selection, recombination, and population structure. Mol Biol Evol 29:3933–3946. doi: 10.1093/molbev/mss198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Joseph SJ, Li B, Ghonasgi T, Haase CP, Qin ZS, Dean D, Read TD. 2014. Direct amplification, sequencing and profiling of Chlamydia trachomatis strains in single and mixed infection clinical samples. PLoS One 9:e99290. doi: 10.1371/journal.pone.0099290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dean D, Kandel RP, Adhikari HK, Hessel T. 2008. Multiple Chlamydiaceae species in trachoma: implications for disease pathogenesis and control. PLoS Med 5:e14. doi: 10.1371/journal.pmed.0050014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brunham RC, Kimani J, Bwayo J, Maitha G, Maclean I, Yang C, Shen C, Roman S, Nagelkerke NJ, Cheang M, Plummer FA. 1996. The epidemiology of Chlamydia trachomatis within a sexually transmitted diseases core group. J Infect Dis 173:950–956. doi: 10.1093/infdis/173.4.950. [DOI] [PubMed] [Google Scholar]
- 131.Dean D, Oudens E, Bolan G, Padian N, Schachter J. 1995. Major outer membrane protein variants of Chlamydia trachomatis are associated with severe upper genital tract infections and histopathology in San Francisco. J Infect Dis 172:1013–1022. doi: 10.1093/infdis/172.4.1013. [DOI] [PubMed] [Google Scholar]
- 132.Dean D, Suchland RJ, Stamm WE. 2000. Evidence for long-term cervical persistence of Chlamydia trachomatis by omp1 genotyping. J Infect Dis 182:909–916. doi: 10.1086/315778. [DOI] [PubMed] [Google Scholar]
- 133.Molano M, Meijer CJ, Weiderpass E, Arslan A, Posso H, Franceschi S, Ronderos M, Munoz N, van den Brule AJ. 2005. The natural course of Chlamydia trachomatis infection in asymptomatic Colombian women: a 5-year follow-up study. J Infect Dis 191:907–916. doi: 10.1086/428287. [DOI] [PubMed] [Google Scholar]
- 134.Barnes RC, Rompalo AM, Stamm WE. 1987. Comparison of Chlamydia trachomatis serovars causing rectal and cervical infections. J Infect Dis 156:953–958. doi: 10.1093/infdis/156.6.953. [DOI] [PubMed] [Google Scholar]
- 135.Bax CJ, Quint KD, Peters RP, Ouburg S, Oostvogel PM, Mutsaers JA, Dorr PJ, Schmidt S, Jansen C, van Leeuwen AP, Quint WG, Trimbos JB, Meijer CJ, Morre SA. 2011. Analyses of multiple-site and concurrent Chlamydia trachomatis serovar infections, and serovar tissue tropism for urogenital versus rectal specimens in male and female patients. Sex Transm Infect 87:503–507. doi: 10.1136/sti.2010.048173. [DOI] [PubMed] [Google Scholar]
- 136.Boisvert JF, Koutsky LA, Suchland RJ, Stamm WE. 1999. Clinical features of Chlamydia trachomatis rectal infection by serovar among homosexually active men. Sex Transm Dis 26:392–398. doi: 10.1097/00007435-199908000-00006. [DOI] [PubMed] [Google Scholar]
- 137.Dang T, Jaton-Ogay K, Flepp M, Kovari H, Evison JM, Fehr J, Schmid P, Boffi El Amari E, Cavassini M, Odorico M, Tarr PE, Greub G. 2009. High prevalence of anorectal chlamydial infection in HIV-infected men who have sex with men in Switzerland. Clin Infect Dis 49:1532–1535. doi: 10.1086/644740. [DOI] [PubMed] [Google Scholar]
- 138.Klint M, Lofdahl M, Ek C, Airell A, Berglund T, Herrmann B. 2006. Lymphogranuloma venereum prevalence in Sweden among men who have sex with men and characterization of Chlamydia trachomatis ompA genotypes. J Clin Microbiol 44:4066–4071. doi: 10.1128/JCM.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Quint KD, Bom RJ, Quint WG, Bruisten SM, van der Loeff MF, Morre SA, de Vries HJ. 2011. Anal infections with concomitant Chlamydia trachomatis genotypes among men who have sex with men in Amsterdam, the Netherlands. BMC Infect Dis 11:63. doi: 10.1186/1471-2334-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rank RG, Yeruva L. 2014. Hidden in plain sight: chlamydial gastrointestinal infection and its relevance to persistence in human genital infection. Infect Immun 82:1362–1371. doi: 10.1128/IAI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ridderhof JC, Barnes RC. 1989. Fusion of inclusions following superinfection of HeLa cells by two serovars of Chlamydia trachomatis. Infect Immun 57:3189–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rockey DD, Viratyosin W, Bannantine JP, Suchland RJ, Stamm WE. 2002. Diversity within inc genes of clinical Chlamydia trachomatis variant isolates that occupy non-fusogenic inclusions. Microbiology 148:2497–2505. doi: 10.1099/00221287-148-8-2497. [DOI] [PubMed] [Google Scholar]
- 143.Jeffrey BM, Suchland RJ, Eriksen SG, Sandoz KM, Rockey DD. 2013. Genomic and phenotypic characterization of in vitro-generated Chlamydia trachomatis recombinants. BMC Microbiol 13:142. doi: 10.1186/1471-2180-13-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Suchland RJ, Rockey DD, Weeks SK, Alzhanov DT, Stamm WE. 2005. Development of secondary inclusions in cells infected by Chlamydia trachomatis. Infect Immun 73:3954–3962. doi: 10.1128/IAI.73.7.3954-3962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Draskovic I, Dubnau D. 2005. Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulphide bonds. Mol Microbiol 55:881–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hahn J, Albano M, Dubnau D. 1987. Isolation and characterization of Tn917lac-generated competence mutants of Bacillus subtilis. J Bacteriol 169:3104–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci U S A 100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Seth-Smith HM, Harris SR, Persson K, Marsh P, Barron A, Bignell A, Bjartling C, Clark L, Cutcliffe LT, Lambden PR, Lennard N, Lockey SJ, Quail MA, Salim O, Skilton RJ, Wang Y, Holland MJ, Parkhill J, Thomson NR, Clarke IN. 2009. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics 10:239. doi: 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mueller KE, Fields KA. 2015. Application of β-lactamase reporter fusions as an indicator of effector protein secretion during infections with the obligate intracellular pathogen Chlamydia trachomatis. PLoS One 10:e0135295. doi: 10.1371/journal.pone.0135295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kemege KE, Hickey JM, Barta ML, Wickstrum J, Balwalli N, Lovell S, Battaile KP, Hefty PS. 2015. Chlamydia trachomatis protein CT009 is a structural and functional homolog to the key morphogenesis component RodZ and interacts with division septal plane localized MreB. Mol Microbiol 95:365–382. doi: 10.1111/mmi.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mirrashidi KM, Elwell CA, Verschueren E, Johnson JR, Frando A, Von Dollen J, Rosenberg O, Gulbahce N, Jang G, Johnson T, Jäger S, Gopalakrishnan AM, Sherry J, Dunn JD, Olive A, Penn B, Shales M, Cox JS, Starnbach MN, Derre I, Valdivia R, Krogan NJ, Engel J. 2015. Global mapping of the Inc-human interactome reveals that retromer restricts Chlamydia infection. Cell Host Microbe 18:109–121. doi: 10.1016/j.chom.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Weber MM, Bauler LD, Lam J, Hackstadt T. 2015. Expression and localization of predicted inclusion membrane proteins in Chlamydia trachomatis. Infect Immun 83:4710–4718. doi: 10.1128/IAI.01075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller JE, Yang J, Mills D, Manias D, Dunny G, Lambowitz AM, Belfort M. 1998. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell 94:451–462. doi: 10.1016/S0092-8674(00)81586-X. [DOI] [PubMed] [Google Scholar]
- 154.Lambowitz AM, Zimmerly S. 2011. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol 3:a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Perutka J, Wang W, Goerlitz D, Lambowitz AM. 2004. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J Mol Biol 336:421–439. doi: 10.1016/j.jmb.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 156.Fields KA, Fischer E, Hackstadt T. 2002. Inhibition of fusion of Chlamydia trachomatis inclusions at 32°C correlates with restricted export of IncA. Infect Immun 70:3816–3823. doi: 10.1128/IAI.70.7.3816-3823.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Suchland RJ, Rockey DD, Bannantine JP, Stamm WE. 2000. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect Immun 68:360–367. doi: 10.1128/IAI.68.1.360-367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Enyeart PJ, Chirieleison SM, Dao MN, Perutka J, Quandt EM, Yao J, Whitt JT, Keatinge-Clay AT, Lambowitz AM, Ellington AD. 2013. Generalized bacterial genome editing using mobile group II introns and Cre-lox. Mol Syst Biol 9:685. doi: 10.1038/msb.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Colbert T, Till BJ, Tompa R, Reynolds S, Steine MN, Yeung AT, McCallum CM, Comai L, Henikoff S. 2001. High-throughput screening for induced point mutations. Plant Physiol 126:480–484. doi: 10.1104/pp.126.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Brown HM, Knowlton AE, Snavely E, Nguyen BD, Richards TS, Grieshaber SS. 2014. Multinucleation during C. trachomatis infections is caused by the contribution of two effector pathways. PLoS One 9:e100763. doi: 10.1371/journal.pone.0100763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen YS, Bastidas RJ, Saka HA, Carpenter VK, Richards KL, Plano GV, Valdivia RH. 2014. The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Pathog 10:e1003954. doi: 10.1371/journal.ppat.1003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Snavely EA, Kokes M, Dunn JD, Saka HA, Nguyen BD, Bastidas RJ, McCafferty DG, Valdivia RH. 2014. Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog Dis 71:336–351. doi: 10.1111/2049-632X.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gopalakrishnan AM, Kundu AK, Mandal TK, Kumar N. 2013. Novel nanosomes for gene delivery to Plasmodium falciparum-infected red blood cells. Sci Rep 3:1534. doi: 10.1038/srep01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Thomason LC, Costantino N, Shaw DV, Court DL. 2007. Multicopy plasmid modification with phage λ Red recombineering. Plasmid 58:148–158. doi: 10.1016/j.plasmid.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bhaya D, Davison M, Barrangou R. 2011. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 167.Joung JK, Sander JD. 2013. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kim H, Kim JS. 2014. A guide to genome engineering with programmable nucleases. Nat Rev Genet 15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]