Abstract
Breast cancer (BC) is the most common cancer type and the second cause of cancer-related death among women. Therefore, better understanding of breast cancer tumor biology and the identification of novel biomarkers is essential for the early diagnosis and for better disease stratification and management choices. Herein we developed a novel approach which relies on the isolation of circulating microRNAs through an enrichment step using speed-vacuum concentration which resulted in 5-fold increase in microRNA abundance. Global miRNA microarray expression profiling performed on individual samples from 23 BC and 9 normals identified 18 up-regulated miRNAs in BC patients (p(corr) < 0.05). Nine miRNAs (hsa-miR-4270, hsa-miR-1225-5p, hsa-miR-188-5p, hsa-miR-1202, hsa-miR-4281, hsa-miR-1207-5p, hsa-miR-642b-3p, hsa-miR-1290, and hsa-miR-3141) were subsequently validated using qRT-PCR in a cohort of 46 BC and 14 controls. The expression of those microRNAs was overall higher in patients with stage I, II, and III, compared to stage IV, with potential utilization for early detection. The expression of this microRNA panel was slightly higher in the HER2 and TN compared to patients with luminal subtype. Therefore, we developed a novel approach which led to the identification of a novel microRNA panel which was upregulated in BC patients with potential utilization in disease diagnosis and stratification.
Despite recent advances in cancer management and therapy, cancer remains the second leading cause of death worldwide. Breast cancer (BC) is the most common cancer type and the second cause of cancer-related mortality among women. Annually, the number of women who are newly diagnosed with breast cancer is going to exceed 235,000 and there are around 40,000 deaths as a result of breast cancer in the United States1. In the Kingdom of Saudi Arabia, breast cancer is the ninth leading cause of death among women in 20102. Furthermore, breast cancer constitutes around 25% of all new registered cancer incidence among Saudi women whereas 1,308 new breast cancer cases were reported during the year 20092. The incidence rate of breast cancer in Saudi Arabia is also expected to increase over the next decades as a result of the population’s growth and aging.
Better understanding of breast cancer tumor biology and the identification of novel biomarkers is essential for the early diagnosis and for better disease stratification and management choices. During the recent years, microRNA (miRNA) has become increasingly recognized as important regulator of both normal and cancer cell biology3,4,5. Global profiling of cancer tissue versus normal tissue has identified several dysregulated miRNAs in different human cancers6,7,8. However, this approach requires invasive procedures such as biopsy or surgical intervention in order to acquire the representative tissue to be analyzed. Therefore, finding a non-invasive approach for early diagnosis and management of different human disease, such as cancer, has always been challenging task. Several studies have reported the potential utilization of circulating biomarkers in different body fluids, such as serum and plasma, as diagnostic and prognostic tools for different types of cancers, while some studies have reported the use of circulating miRNA for cancer detection and prognostic stratification. However, one limitation of the previous studies is that these studies most often examined the expression of defined set of miRNAs or pooled samples from different patients and then assessed miRNA expression using qRT-PCR or other methods, which limits the novelty of the identified miRNA as well as the inability to perform individual data analysis as related to disease condition9,10,11,12. In current study, we established a novel and unbiased approach where we isolated circulating miRNAs from breast cancer patients, we employed speed-vacuum centrifugation step to increase the abundance of miRNAs and subsequently conducted global miRNA profiling on individual patient specimens. Our approach led to the identification of a novel 18-microRNA panel which was upregulated in breast cancer patients as early as Stage I and Stage II.
Results
Distinct microRNA expression profile in patients with breast cancer compared to healthy controls
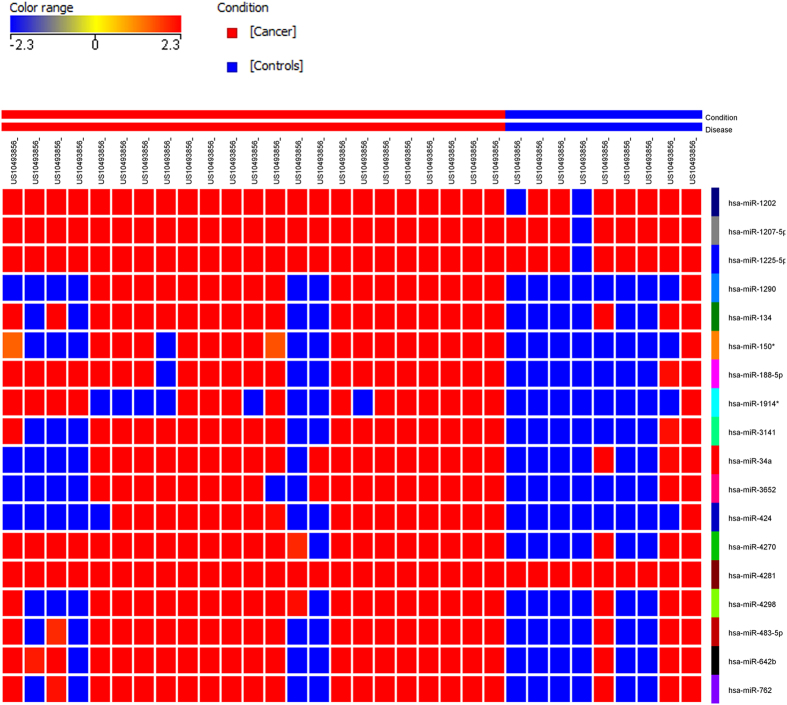
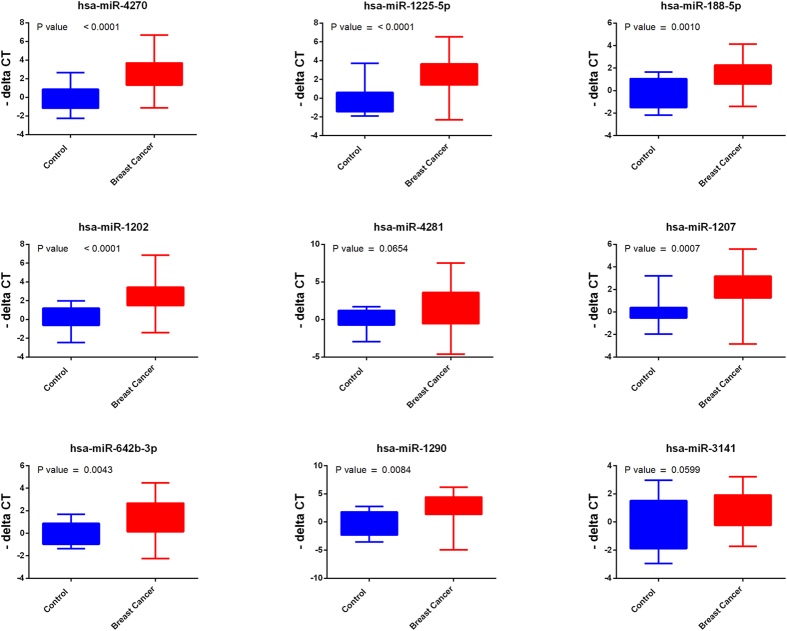
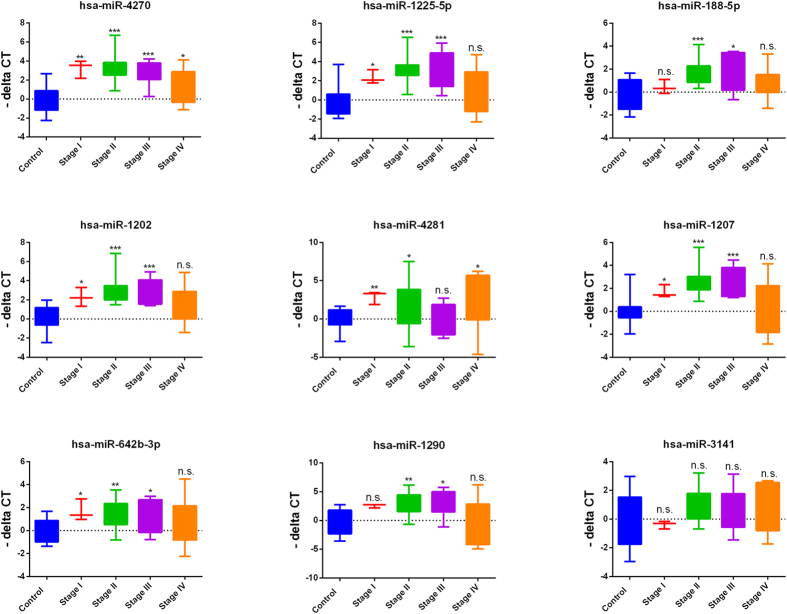
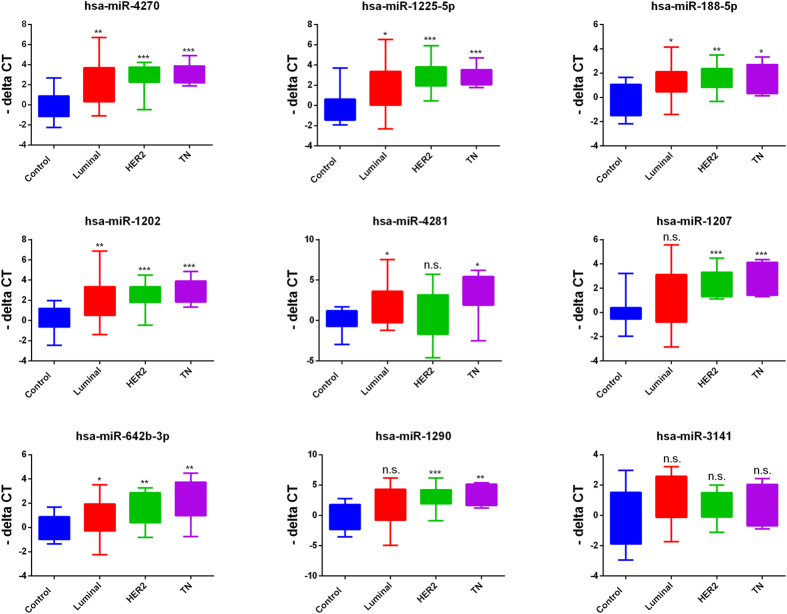
Circulating RNAs were isolated using Norgen’s slurry format kit. To further increase the concentration of isolated RNA, we utilized speed-vacuum concentration approach. As shown in supplementary Fig. 1, this approach increased the detection limit for circulating miRNA in these samples ~4.6 fold. Circulating miRNAs were identified by performing global miRNA microarray expression profiling on 23 breast cancer and 9 normal control samples. Using Benjamini-Hochberg False Discovery Rate (FDR) multiple testing correction method (p(corr) < 0.05) and two-fold change cut-off, we identified 18 up-regulated miRNAs in samples obtained from patients with breast cancer compared to healthy controls. Individual data are presented in the heatmap in Fig. 1. Breast cancer-associated miRNAs identified were: hsa-miR-4270, hsa-miR-1225-5p, hsa-miR-188-5p, hsa-miR-1202, hsa-miR-4281, hsa-miR-1207-5p, hsa-miR-642b-3p, hsa-miR-1290, hsa-miR-3141, hsa-miR-150-3p, hsa-miR-4298, hsa-miR-483-5p, hsa-miR-134, hsa-miR-762, hsa-miR-1914-3p, hsa-miR-34a-5p, hsa-miR-3652, and hsa-miR-424-5p (Table 1). Among the identified miRNAs, hsa-miR-4270 was the most significantly upregulated miRNA in patients’ samples compared to controls (p value = 3.5 × 10−6, p value (Corr) = 0.001). The top nine miRNAs (based on p value, Table 1) : hsa-miR-4270, hsa-miR-1225-5p, hsa-miR-188-5p, hsa-miR-1202, hsa-miR-4281, hsa-miR-1207-5p, hsa-miR-642b-3p, hsa-miR-1290, and hsa-miR-3141 were subsequently chosen for further validation in a cohort of 46 BC and 14 normal controls, including 23 BC and 8 normals which were used for the microarray profiling. Data presented in Fig. 2 collectively validated the microarray data. Interestingly, the expression of the nine miRNAs was slightly higher in Stage I, Stage II, and Stage III compared to Stage IV (Fig. 3). Anova analysis revealed significant difference in the expression of hsa-miR-4270, hsa-miR-1225-5p, hsa-miR-1202, hsa-miR-1207, and hsa-miR-1290 as function of cancer stage. When the expression of those nine miRNAs was plotted as function of breast cancer molecular subtype, similar trends of miRNA expression were overall seen, whereas the expression was slightly higher in the HER2 and TN compared to the luminal cancer subtype. Additionally, patients with the luminal subtype exhibited the largest degree of heterogeneity, possibly due to further luminal A/B sub classification (Fig. 4).
Figure 1. Heatmap depicting the expression of 18 circulating miRNAs in patients with breast cancer (BC) compared to normal healthy controls.
Heatmap of individual plasma samples of 9 control and 23 patients with BC. The miRNA expression levels exhibited ≥2.0 fold changes and p ≤ 0.05 are presented. Each column represents an individual sample and each row represents a single miRNA. Expression level of each miRNA in a single sample is depicted according to the color scale.
Table 1. Differentially expressed circulating plasma miRNAs in 23 breast cancer patients compared to 9 normal controls.
| Updated_systematic_name | mirbase accession No | p (Corr) | p | Regulation | FC |
|---|---|---|---|---|---|
| hsa-miR-4270 | MIMAT0016900 | 0.001193408 | 5.35E-06 | up | 182.9 |
| hsa-miR-1225-5p | MIMAT0005572 | 0.004337585 | 7.78E-05 | up | 22.7 |
| hsa-miR-188-5p | MIMAT0000457 | 0.004337585 | 6.59E-05 | up | 73.0 |
| hsa-miR-1202 | MIMAT0005865 | 0.006776397 | 1.52E-04 | up | 27.3 |
| hsa-miR-4281 | MIMAT0016907 | 0.019025994 | 5.12E-04 | up | 11.6 |
| hsa-miR-1207-5p | MIMAT0005871 | 0.0202241 | 7.26E-04 | up | 14.7 |
| hsa-miR-642b-3p | MIMAT0018444 | 0.0202241 | 6.99E-04 | up | 62.9 |
| hsa-miR-1290 | MIMAT0005880 | 0.022832992 | 9.22E-04 | up | 45.9 |
| hsa-miR-3141 | MIMAT0015010 | 0.029752607 | 0.001334 | up | 50.7 |
| hsa-miR-150-3p | MIMAT0004610 | 0.033017445 | 0.001629 | up | 43.7 |
| hsa-miR-4298 | MIMAT0016852 | 0.03568337 | 0.00192 | up | 40.2 |
| hsa-miR-483-5p | MIMAT0004761 | 0.038129777 | 0.002223 | up | 42.3 |
| hsa-miR-134 | MIMAT0000447 | 0.04201405 | 0.002826 | up | 42.2 |
| hsa-miR-762 | MIMAT0010313 | 0.04201405 | 0.0028 | up | 43.1 |
| hsa-miR-1914-3p | MIMAT0007890 | 0.044369638 | 0.003895 | up | 30.8 |
| hsa-miR-34a-5p | MIMAT0000255 | 0.044369638 | 0.004159 | up | 29.5 |
| hsa-miR-3652 | MIMAT0018072 | 0.044369638 | 0.003801 | up | 44.3 |
| hsa-miR-424-5p | MIMAT0001341 | 0.044369638 | 0.003333 | up | 43.0 |
Figure 2. Validation of the expression of 9 miRNAs identified from microarray data employing plasma or serum from patients with breast cancer (BC, n = 46) and healthy controls (n = 14) using quantitative Real-Time PCR.
P values were calculated using unpaired t-test and are indicated on each plot. Data are presented as “−delta CT” using box and whiskers plots.
Figure 3. Expression of 9 circulating miRNAs according to breast cancer stage.
The expression of a group of 9 circulating miRNAs measured using qRT-PCR in normal (n = 14) or patients with breast cancer (BC, n = 46) are plotted as a function of cancer stage. P values were calculated using unpaired t-test. Data are presented as “−delta CT” using box and whiskers plots. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.00005, n.s., not significant.
Figure 4. Expression of 9 circulating miRNAs according to breast cancer molecular subtype.
The expression of a group of 9 circulating miRNAs measured using qRT-PCR in normal (n = 14) or patients with breast cancer (BC, n = 46) are plotted as a function of BC molecular type. P values were calculated using unpaired t-test. Data are presented as “−delta CT” using box and whiskers plots. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.00005, n.s., not significant, TN: triple negative, HER2: HER2+.
Discussion
Identifying reliable blood biomarkers for early diagnosis or prognostic stratification of various human diseases is an area of intensive investigation. Proteins, DNA, and mRNA could be detected in the circulation of cancer patients and have been suggested in some studies to reflect disease activity13,14,15. miRNAs have recently emerged as providing reliable biomarkers for disease status in a number of cancer patients as well as in other diseases, due to their stability and ease of detection10,16,17. Current protocols employed to detect circulating miRNAs rely mostly on using either a PCR-based method for measuring the expression of selected panel of miRNAs or the utilization of pooled serum/plasma samples and running a more robust assay such as miRNA microarrays. However, these approaches in general do not lead to the identification of novel miRNA or studying their relevance at the individual patient’s level. In our studies we have employed individual patients’ and control’s samples and utilized an unbiased discovery approach using miRNA microarray profiling.
Several of the established protocols for miRNA isolation from serum and plasma require a large sample volume to obtain sufficient RNA for down-stream analysis. Thus, clinical serum and plasma samples from several patients are frequently pooled10,18, which may compromise statistical analyses by preventing using the individual patient as the statistical unit, that may lead to erroneous conclusions. In our study, we have avoided this limitation as we developed a novel approach which relies on the isolation of circulating microRNAs through an enrichment step using speed-vacuum concentration that resulted in ~5-fold enrichment. In addition, we employed an unbiased discovery approach through using microarray chips which can detect more than 1700 human-specific miRNAs.
We identified 18 novel miRNAs, which were upregulated in BC patients compared to matched healthy controls. The expression of nine selected miRNAs was subsequently validated in a cohort of 46 breast cancer and 14 normal subjects, which collectively corroborated the microarray data.
Some of the identified miRNAs have previously been reported to be associated with a number of human diseases, hsa-miR-134 is upregulated sera of patients with acute myocardial infarction19 while hsa-miR-483-5p levels were increased in sera of patients with adrenocortical tumors and pulmonary tuberculosis20,21. Elevated expression of hsa-miR-1290 was reported in prostate cancer and in fatty liver disease22,23. To our knowledge, none of the other miRNAs identified in our study has been reported to be enriched in the circulation of other human diseases. Interestingly, those miRNAs were also detected in the circulation of BC patients with early stage (I and II), which suggest potential utilization of this miRNA panel for early detection.
Some miRNAs which have been reported to be enriched in breast cancer tissues, such as hsa-miR-21 and hsa-miR-155, were not significantly upregulated in the circulation of breast cancer patients in our study. Therefore our data suggest that circulating microRNAs are unlikely to be derived from the tumor itself, but rather reflect the generalized homeostatic responses during health and disease. In support of this hypothesis, pathway analysis on the predicted gene targets for the identified miRNAs revealed potential role in important cellular processes (supplementary Fig. 2). Also, hsa-miR-1202 has recently been reported to be down-regulated in patients with depression24 and thus mental state may contribute to some changes in circulating miRNAs profile. In addition, we observed upregulated expression of two HSV2 miRNAs in the circulation of breast cancer patients (supplementary Fig. 3).
In conclusion, we have identified a novel panel consisting of 18 circulating miRNAs that is enriched in patients with breast cancer and their presence was higher in patients with stage I, II, and III suggesting it possible utilization for early diagnosis and disease stratification. Similar strategy and methodology can also be utilized for detecting circulating miRNAs in other human diseases.
Materials and Methods
Ethics statement
The clinical study and blood collection were approved by Institutional Research Ethics Board at the King Saud University College of Medicine (Riyadh, Riyadh, Saudi Arabia). The methods were carried out in accordance with the approved guidelines. Informed consent was obtained from all subjects.
Patient and blood collection
Blood samples from 46 breast cancer patients were obtained from patients undergoing standard treatment at the King Khaled University Hospital (Riyadh, Saudi Arabia). Controls samples were obtained from 14 healthy women. The clinical information of subjects included in current study and their tumor characteristics are listed in Table 2. Samples with signs of hemolysis were excluded from the study.
Table 2. clinical information of patients included in current study and their tumor characteristics and normal control.
| Cancer | N = 46 | % |
|---|---|---|
| Age, y | ||
| Median age | 50 y | |
| Range | 27–71 y | |
| Gender | ||
| Female | 46 | 100% |
| Stage | ||
| I | 3 | 6.5% |
| II | 17 | 37.0% |
| III | 10 | 21.7% |
| IV | 13 | 28.3% |
| NA | 3 | 6.5% |
| Molecular Subtype | ||
| Luminal | 22 | 48% |
| HER2 | 17 | 37% |
| TN | 7 | 15% |
| Recurrence | ||
| Recurrent | 4 | 8.6% |
| Non Recurrent | 42 | 91.4% |
| Normal | N = 14 | |
| Age, y | ||
| Median age | 36.5 y | |
| Range | 26–60 y | |
HER2: HER2+, TN: Triple Negative.
Plasma and serum preparation and total RNA isolation
Plasma and serum were prepared from freshly collected whole blood samples (EDTA-coated tubes for plasma collection or regular tubes for serum collection) by centrifugation at 2,000 rpm for 15 minutes at room temperature. Collected plasma and serum were frozen at −80 °C. Circulating RNAs were extracted from ~3.5 mL of the collected plasma or serum using Norgen’s Plasma/Serum Circulating and Exosomal RNA Purification Kit Slurry Format (Norgen Biotek, Thorold, Ontario, Canada) according to the manufacturer’s protocol, followed by RNA elution in 100 μl of elution buffer. Twenty five μl of the purified total RNA were dried by speed-vacuum centrifugation (60 minutes at RT), and were then reconstituted in five μl of DNAse and RNAse-free water (5-fold enrichment). Normalization across samples was done by using equal volume of input plasma or serum (~3.5 ml) for miRNA isolation from each sample. Isolated miRNA fraction was subsequently used for global miRNA profiling by microarray (after drying and reconstitution) or by qRT-PCR (without drying and reconstitution).
miRNA expression profiling
miRNA expression profiling was conducted on the isolated miRNA fraction from 23 patients with breast cancer and nine normals. Two microliter of the extracted RNA (after 5-fold concentration) was labeled and subsequently hybridized to the Agilent Human SurePrint G3 8 × 60 k v21 miRNA microarray chip as described before6. Subsequently, data were imported into GeneSpring 13.0 software for analysis (Agilent Technologies). Differentially expressed miRNAs in cancer patients versus healthy control samples were determined using a corrected p-value (≤0.05, Benjamini-Hochberg multiple testing correction method). MicroRNAs showing ≥2.0 fold change were considered significant.
miRNA validation by qRT-PCR
Validation of selected number of miRNA was carried out on circulating miRNA fraction isolated from either plasma or serum samples from 46 BC and 14 controls. Validation was conducted using the miProfile™ Custom miRNA qPCR Array (GeneCopoeia, Rockville, MD, United States). For miRNA expression detection, five μl of the purified total RNA were used as input according to the manufacturer’s protocol. All primers used in the current study were provided by GeneCopoeia, except for hsa-miR-21 primers which were obtained from Applied Biosystems. The relative expression levels were expressed as “−delta CT” of breast cancer patients compared to normal controls. Quantification of hsa-miR-21 in plasma samples before and after speed vacuum concentration was conducted using Taqman microRNA assay kit (Applied Biosystems, USA) as described before5.
Molecular profiling of breast cancer patients
Molecular profiling of BC patients was conducted in accordance with previously published criteria and based on the pathological characteristics of patients utilized in current study25,26.
Statistical analysis
Statistical analyses and graphing were conducted using GraphPad Prism 6.0 software (GraphPad, San Diego, CA, USA). P-values were calculated using the two-tailed t-test or one way anova analysis.
Additional Information
How to cite this article: Hamam, R. et al. microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci. Rep. 6, 25997; doi: 10.1038/srep25997 (2016).
Supplementary Material
Acknowledgments
We would like to thank the college of medicine research center (CMRC) deanship for scientific research, King Saud University for supporting this work. We would like to thank Ms Dana Hamam for her assistance with RNA isolation.
Footnotes
The authors declare no competing financial interests.
Author Contributions R.H.: Collection and/or assembly of data, M.A.: Conception and design, A.M.A.: Collection and/or assembly of data, K.A.A.: Collection and/or assembly of data; M.K.: Conception and design; A.A.: Conception and design; N.M.A.: Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing, Final approval of manuscript, Obtained funding.
References
- Siegel R., Ma J., Zou Z. & Jemal A. Cancer statistics. CA: a cancer journal for clinicians 64, 9–29, doi: 10.3322/caac.21208 (2014). [DOI] [PubMed] [Google Scholar]
- El Bcheraoui C. et al. Breast cancer screening in saudi arabia: free but almost no takers. Plos one 10, e0119051, doi: 10.1371/journal.pone.0119051 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L. & Hannon G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews. Genetics 5, 522–531, doi: 10.1038/nrg1379 (2004). [DOI] [PubMed] [Google Scholar]
- Alajez N. M. et al. Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell death & disease 1, e85, doi: 10.1038/cddis.2010.64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamam D. et al. microRNA-320/RUNX2 axis regulates adipocytic differentiation of human mesenchymal (skeletal) stem cells. Cell death & disease 5, e1499, doi: 10.1038/cddis.2014.462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnubalaji R. et al. Genome-wide mRNA and miRNA expression profiling reveal multiple regulatory networks in colorectal cancer. Cell death & disease 6, e1614, doi: 10.1038/cddis.2014.556 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M. V. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65, 7065–7070, doi: 10.1158/0008-5472.can-05-1783 (2005). [DOI] [PubMed] [Google Scholar]
- Mattie M. D. et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer 5, 24, doi: 10.1186/1476-4598-5-24 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan H. M., Miller N., Kelly R., Newell J. & Kerin M. J. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist 15, 673–682, doi: 10.1634/theoncologist.2010-0103 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. S. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America 105, 10513–10518, doi: 10.1073/pnas.0804549105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin V. Y., Siu J. M., Cheuk I., Ng E. K. & Kwong A. Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. British journal of cancer 112, 1751–1759, doi: 10.1038/bjc.2015.143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckrath I. et al. Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget 6, 13387–13401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anker P., Mulcahy H., Chen X. Q. & Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer metastasis reviews 18, 65–73 (1999). [DOI] [PubMed] [Google Scholar]
- Petricoin E. F. 3rd et al. Serum proteomic patterns for detection of prostate cancer. Journal of the National Cancer Institute 94, 1576–1578 (2002). [DOI] [PubMed] [Google Scholar]
- Wang G. K. et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. European heart journal 31, 659–666, doi: 10.1093/eurheartj/ehq013 (2010). [DOI] [PubMed] [Google Scholar]
- Chen X. et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell research 18, 997–1006, doi: 10.1038/cr.2008.282 (2008). [DOI] [PubMed] [Google Scholar]
- Creemers E. E., Tijsen A. J. & Pinto Y. M. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circulation research 110, 483–495, doi: 10.1161/CIRCRESAHA.111.247452 (2012). [DOI] [PubMed] [Google Scholar]
- Moussay E. et al. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America 108, 6573–6578, doi: 10.1073/pnas.1019557108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F. et al. Predictive value of circulating miR-328 and miR-134 for acute myocardial infarction. Molecular and cellular biochemistry 394, 137–144, doi: 10.1007/s11010-014-2089-0 (2014). [DOI] [PubMed] [Google Scholar]
- Szabo D. R. et al. Analysis of circulating microRNAs in adrenocortical tumors. Laboratory investigation; a journal of technical methods and pathology 94, 331–339, doi: 10.1038/labinvest.2013.148 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. Plos one 8, e81076, doi: 10.1371/journal.pone.0081076 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. European urology 67, 33–41, doi: 10.1016/j.eururo.2014.07.035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Ge G., Pan T., Wen D. & Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. Plos one 9, e105192, doi: 10.1371/journal.pone.0105192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J. P. et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nature medicine 20, 764–768, doi: 10.1038/nm.3582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzier R. et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clinical cancer research: an official journal of the American Association for Cancer Research 11, 5678–5685, doi: 10.1158/1078-0432.CCR-04-2421 (2005). [DOI] [PubMed] [Google Scholar]
- Perou C. M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752, doi: 10.1038/35021093 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.