Abstract
Sudden infant death syndrome (SIDS) is the most frequent manner of post-perinatal death among infants. One of the suggested causes of the syndrome is inherited cardiac diseases, mainly channelopathies, that can trigger arrhythmias and sudden death. The purpose of this study was to investigate cases of sudden unexpected death in infancy (SUDI) for potential causative variants in 100 cardiac-associated genes. We investigated 47 SUDI cases of which 38 had previously been screened for variants in RYR2, KCNQ1, KCNH2 and SCN5A. Using the Haloplex Target Enrichment System (Agilent) and next-generation sequencing (NGS), the coding regions of 100 genes associated with inherited channelopathies and cardiomyopathies were captured and sequenced on the Illumina MiSeq platform. Sixteen (34%) of the SUDI cases had variants with likely functional effects, based on conservation, computational prediction and allele frequency, in one or more of the genes screened. The possible effects of the variants were not verified with family or functional studies. Eight (17%) of the SUDI cases had variants in genes affecting ion channel functions. The remaining eight cases had variants in genes associated with cardiomyopathies. In total, one third of the SUDI victims in a forensic setting had variants with likely functional effect that presumably contributed to the cause of death. The results support the assumption that channelopathies are important causes of SUDI. Thus, analysis of genes associated with cardiac diseases in SUDI victims is important in the forensic setting and a valuable supplement to the clinical investigation in all cases of sudden death.
Introduction
Sudden unexpected death in infancy (SUDI) is defined as the unexpected and initially unexplained death of infants <1 year. SUDI comprises the sudden infant death syndrome (SIDS), one of the most investigated, unidentified causes of death in the young population. SIDS is a diagnosis of exclusion and is defined as unexpected death of an infant <1 of age that remains unexplained after thorough investigations including performance of complete autopsy, examination of the death scene and review of the clinical history.1 The number of SIDS events has decreased drastically in the past decades because of the recommendations of infants sleeping in back position.2 Hereafter, the prevalence of SIDS has remained unchanged and has been reported to be in the range of 0.1–0.8 per 1000 live births.3, 4 SIDS has most likely a multifactorial aetiology and several risk factors have been suggested in this regard. Extrinsic factors such as smoke exposure, certain sleep practices and intrinsic factors such as metabolic as well as genetic disorders have been suspected to be of importance.3 More recently, special attention has been directed towards the genetic aetiology of inherited cardiac diseases as a cause of SIDS, especially channelopathies including long QT syndrome (LQTS), Brugada syndrome (BrS) and catecholaminergic polymorphic ventricular tachycardia (CPVT).5, 6, 7 In these diseases, the structures and the functions of the ion channels are affected in the cardiac cells leading to perturbed ion channel functions, disrupted action potential propagation and development of arrhythmias.8 Given that changes in the functions of the ion channels are not macro- or microscopically visible in the heart with standard techniques, the cause of death in cases with perturbed ion channel functions is not observed at conventional medicolegal investigations, and genetic investigations will be needed to detect the cause of death.
The frequency of disease-causing variants in genes associated with LQTS and CPVT have previously been investigated in cases of SUDI. Rare, possibly functional variants were found in nearly 10% of the investigated cases.5, 6, 7 Recently, de novo calmodulin variants in infants with severe arrhythmogenic symptoms were discovered and assumed to be the cause of death of some SUDI victims.9 Besides the screening of channelopathic-associated genes, 4% of the cases in a large SIDS cohort (n=286) had variants in one of the sarcomeric genes associated with hypertrophic cardiomyopathy, indicating that structural defects may also cause SIDS.10 With next-generation sequencing (NGS), large-scale gene sequencing can efficiently and rapidly be applied. This has increased the use of genetics as a supplementary, diagnostic tool.
Based on the available literature, it is very likely that inherited cardiac diseases are contributing factors to the cause of death in infancy. The aim of this study was to investigate a cohort of SUDI victims using NGS for possible disease-causing variations in the exons and the promoter regions of 100 genes associated with inherited cardiomyopathies and channelopathies.
Materials and methods
Materials
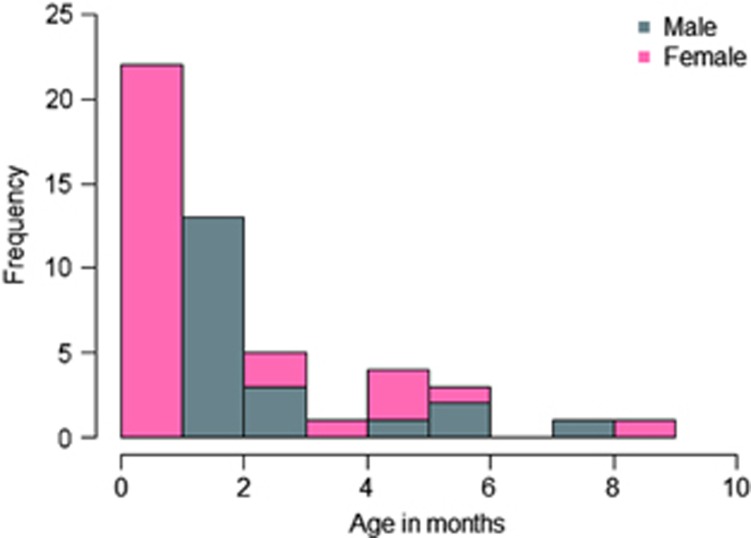
In total, we investigated 47 SUDI victims deceased in Denmark between 1998 and 2011. We included 38 SUDI victims from the Section of Forensic Pathology, Department of Forensic Medicine, Faculty of Health Sciences, Aarhus University, Denmark. The infants were previously investigated for variants of RYR2, KCNQ1, KCNH2 and SCN5A.7, 11 Two deceased individuals had missense variants with possible functional effect in RYR2.7 Nine SUDI victims from the Section of Forensic Pathology, Department of Forensic Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Denmark, were included. The inclusion of deceased infants was based on autopsy reports, police reports and hospital records, if relevant. All autopsies were performed by forensic pathologists according to a specific protocol including radiological imaging, toxicology, microbiological investigation as well as general autopsy with thorough macro- and microscopic investigations of the heart. The cases were included if no other cause of death could be established at the medicolegal investigation. The median age of all the included infants was 2 months (range 0–9 months). Twenty-seven (57%) were females (Figure 1). In total, 40 cases fulfilled the San Diego Criteria of SIDS1 and 42 fulfilled the Nordic Criteria12 (Table 1).
Figure 1.
Age and gender distribution of SUDI cases.
Table 1. Classification of SUDI cases.
|
Nordic criteria |
|||
|---|---|---|---|
| SIDS | Unknown | Other | |
| San Diego criteria | |||
| IA | 0 | ||
| IB | 7 | ||
| II | 33 | ||
| USID | 2 | 4 | 1 |
Genetic investigation
DNA was extracted from blood using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) for 42 of the samples. For one and four samples, DNA was extracted from fresh frozen muscle and spleen, respectively, using the Biorobot EZ1 and the EZ1 DNA Investigator Kit (Qiagen). The extracted DNA was quantified with the Quantifiler Human DNA Quantification Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's protocol.
The exons and promoter regions of 100 genes (Table 2) were investigated using NGS. The genes were selected according to the available literature and OMIM (Online Mendelian Inheritance in Man)13 and were all reported to be associated with cardiomyopathies or channelopathies (Table 2).
Table 2. Investigated genes and associated cardiac diseases.
| Disease | Genes | Other associated diseases |
|---|---|---|
| ARVC | DSC2 (NM_024422.3), DSG2a (NM_001943.3), DSPa (NM_004415.2), JUP (NM_021991.2), PKP2b (NM_004572.3), RPSA (NM_001012321.1), TGFB3 (NM_003239.2), TMEM43 (NM_024334.2) | aDCM, bBrS |
| BrS | CACNA1C (NM_199460.3), CACNB2 (NM_201596.2), GPD1Lc (NM_015141.3), HCN4d (NM_005477.2), KCND3 (NM_004980.4), KCNE3 (NM_005472.4), KCNJ8e (NM_004982.3), MOG1 (NM_016492.4), SCN1Bf,g (NM_199037.3), SCN3Bf (NM_018400.3), SCN4Bf,h (NM_174934.3), SCN5Aa,d,e,f,g,h,i (NM_001099404.1), TRPM4g (NM_017636.3) | aDCM, cRBBB, dSSS, eSIDS, fFAF, gCCD, hLQTS, iPFVF |
| CPVT | CASQ2 (NM_001232.3), RYR2j (NM_001035.2) | jARVC |
| CTD | SLC22A5 (NM_003060.3) | |
| DCM | ABCC9f (NM_005691.3), ACTN2k (NM_001103.3), BAG3 (NM_004281.3), CTF1 (NM_001330.3), CRYAB (NM_001289807.1), DES (NM_001927.3), DMD (NM_004006.2), EYA4 (NM_172105.3), FHL2 (NM_001450.3), FKTN (NM_006731.2), GAA (NM_000152.3), ILK (NM_001014794.2), LAMA4 (NM_001105206.2), LDB3k,l (NM_001171610.1), LMNAj (NM_170707.2), MYPNk (NM_001256267.1), NEBL (NM_006393.2), NEXNk (NM_144573.3), PLNk (NM_002667.3), PRDM16l (NM_022114.3), PSEN1 (NM_000021.3), PSEN2 (NM_000447.2), RBM20 (NM_001134363.2), RPS7 (NM_001011.3), SDHA (NM_004168.2), SGCD (NM_000337.5), STARD3 (NM_006804.3), TAZl (NM_000116.3), TCAP (NM_003673.3), TMPO (NM_003276.2), TNNC1k (NM_003280.2), TNNI3k (NM_000363.4) | fFAF, jARVC, kHCM, lLVNC |
| FAF | CACNA1D (NM_000720.3), GJA5 (NM_005266.6), HCN1d (NM_021072.3), KCNA5 (NM_002234.3), NPPA (NM_006172.3), RANGRF (NM_016492.4), SCN2B (NM_004588.4) | dSSS |
| FD | GLA (NM_000169.2) | |
| HCM | ACTC1a,l (NM_005159.4), ANKRD1 (NM_014391.2), CALR3 (NM_145046.4), CAV3m (NM_001234.4), CSRP3a (NM_003476.4), JPH2 (NM_020433.4), LAMP2 (NM_001122606.1), MYBPC3a,l (NM_000256.3), MYH6a,d (NM_002471.3), MYH7a,l (NM_000257.2), MYL2 (NM_000432.3), MYL3 (NM_000258.2), MYLK2 (NM_033118.3), MYOZ2 (NM_016599.4), PRKAG2n (NM_016203.3), SLC25A4 (NM_001151.3), TNNT2a,l (NM_001276345.1), TPM1a,l (NM_000366.5), TTNa (NM_001267550.1), VCLa (NM_014000.2) | aDCM, dSSS, lLVNC, mLQTS, nWPW |
| LQTS | AKAP9 (NM_005751.4), ANK2 (NM_001148.4), CALM1e,p (NM_006888.4), CALM2e,p (NM_001743.4), CALM3e (NM_005184.2), KCNE1 (NM_000219.5), KCNE1Lf (NM_012282.2), KCNE2f (NM_172201.1), KCNE4 (NM_080671.3), KCNH2o (NM_000238.3), KCNJ2f,o (NM_000891.2), KCNJ5 (NM_000890.3), KCNQ1f,o (NM_000218.2), SNTA1 (NM_003098.2) | eSIDS, fFAF, oSQTS, pCPVT |
| LVNC | DTNA (NM_001390.4) | |
| PFVF | DPP6 (NM_130797.3) |
Abbreviations: ARVC: arrhythmogenic right ventricular cardiomyopathy, BrS: Brugada syndrome, CCD: cardiac conduction disease, CPVT: catecholaminergic polymorphic ventricular tachycardia, CTD: carnitine transporter deficiency, DCM: dilated cardiomyopathy, FAF: familial atrial fibrillation, HCM: hypertrophic cardiomyopathy, LQTS: long QT syndrome, LVNC: left ventricular non-compaction, PFVF: paroxysmal familial ventricular fibrillation, SIDS: sudden infant death syndrome, SSS: sick sinus syndrome, SQTS: short QT syndrome, WPW: Wolff–Parkinson–White syndrome.
Genes analysed and the associated cardiac diseases.
The relevant DNA regions were isolated by a capture method using the custom design of the Haloplex Target Enrichment system (Agilent Technologies, Santa Clara, CA, USA) with 150 bp read lengths. The design included all coding exons, 25 bp of adjacent introns and 5′- and 3′-UTR regions of the 100 investigated genes, to a total size of 787 943 bp. The Haloplex Target Enrichment protocol was used according to the manufacturer's version D.5 of the protocol. In brief, the following was performed. (1) Digestion of 200 ng of genomic DNA with different restriction enzymes in eight tubes and analysis of the fragments using the 2100 Bioloanalyzer (Agilent Technologies). (2) Hybridization of digested DNA to Haloplex probes. The ends of the probes were complementary to the fragments of the target regions. In the hybridization, the fragments were circularized. Indices with barcodes, sequencing motifs and biotin were incorporated. (3) The target DNA was captured with Haloplex magnetic beads. (4) The nicks in the circularized DNA fragments were repaired by ligation. (5) The captured target DNA was eluted, PCR amplified and purified. After library build, the amount of DNA was measured using a Qubit Fluorometer 2.0 with the dsDNA HS assay (Invitrogen, Waltham, MA, USA). The size distribution of the DNA was analysed using a 2100 Bioanalyzer and the High Sensitivity DNA kit (Agilent Technologies). The DNA was sequenced on a MiSeq (Illumina, San Diego, CA, USA) according to the manufacturer's instructions with 150 bp paired-end sequencing using the MiSeq Reagent Kit V2 (300 cycles).
Custom panels designed with AmpliSeq were used to verify variants that were likely to have functional effects or of unknown significance. Libraries were prepared using the Ion AmpliSeq Library Kit with Ion AmpliSeq Custom Primer Pool protocol (Thermo Fisher Scientific). An Ion Chef (Life Technologies, Carlsbad, CA, USA) was used to prepare libraries that were then sequenced (NGS) using a Personal Genome Machine (PGM) (Life Technologies).
Data analysis and bioinformatics
Fastq files generated by MiSeq were used to analyse each sample. SureCall (Agilent Technologies) was used for post-base-calling analysis using default settings with an algorithm that was not described in detail. In brief, SureCall trimmed the reads to remove adapter sequences and low-quality reads and aligned the reads to the human genome reference (GRCh37/hg19)14 using BWA MEM.15 Variant calling was done by BAQ SNP Caller using SAMtools16 for identifying single-nucleotide polymorphisms (SNPs), multiple-nucleotide polymorphisms (MNPs) and indels (<100 bp). SureCall filtered out false positive SNPs, MNPs and indels based on read depth and variant quality. A complete list of identified variants was created in Variant Caller Format (VCF). The target size of the designed gene panel included 788 kbp. Overall, 99.6% of the bases were covered >50 ×. Two exons were not covered sufficiently (SCN5A exon 1b NG_008934.1 and JPH2 exon 3 NG_031867.1) and seven exons were not sequenced because of missing probe designs (TTN exon 174, 180,183, 189, 193 NG_011618.3, RPSA exon 1 NG_033234.1 and MYL2 exon 6 NG_007554.1). Details of the genomic regions not covered sufficiently are available online in (Supplementary Table S1). Median coverage for all samples and regions was 327 (range: 253–567). Variants with a coverage <50 were excluded in the further evaluation.
Alamut Batch v. 1.2.0 and Alamut Visual v.2.5 (Interactive Biosoftware, Rouen, France) were used for the annotation and evaluation of missense, nonsense, splice site variants and small indels. Variants were selected by (1) an in-house in silico analysis tool developed for determining the likelihood of variant pathogenicity, (2) frequency <1% in the relevant reference population17, 18 and associations to cardiac diseases reported in HGMD.19 The in silico analysis was based on parameters of conservation (Grantham distance, AlignGVGD class, BLOSUM62, orthologues), computational prediction (MAPP, SIFT), frequency in the relevant reference population (dbSNP build 137,18 ESP17), the location in the genome (distance to nearest splice site) and the coding effect. Details of the scoring of variants in the in silico analysis is described by Hertz et al.20 The variants were independently evaluated and classified as likely, unknown or unlikely to have functional effects by two medical doctors (CLH and MD). The available databases and literature were reviewed for each variant in order to determine the possible effects. Variants were categorized as having likely functional effects based on the genomic alterations and the literature.21 Evidence for pathogenicity included null variants (nonsense, frameshift, near splice sites, initiation codon), known disease-causing amino acid changes or residues, functional studies, prevalence of the variants in affected individuals with the associated diseases, location in exon and/or functional domains of known disease-causing variants (assumed de novo), co-segregation studies and minor allele frequencies (MAFs) below the disease prevalence.21, 22, 23 Given that rare variants are also subject to population stratification,24 we identified a local Danish reference population (n=2000) that was whole-exome sequenced, of which one half comprised metabolically healthy individuals and the other half had patients with type II diabetes.25 In addition, allele frequencies from ESP (European-American population)17 and dbSNP18 were used.
Sequencing results from the verification of variants from the PGM were analysed using the Torrents Suite v4.0.2 with the variantCaller v4.2.1.0 plugin (Life Technologies).
The software, R v. 2.11.0,26 was used to perform calculations of median and mean values and for the scoring of variants in the in silico analysis using a custom script. The coverage information was calculated using a custom script in python with the docopt packages and R v.3.1,26 with dplyr, magrittr, gplots and tools packages and the Bioconductor v.2.14,27, 28 with packages biomaRt and rtracklayer.
DNA variants were numbered according to reference sequence applied in Table 2 using HGVS nomenclature (www.HGVS.org). Variants have been submitted to the Leiden Open Variation Database (http://databases.lovd.nl, Individual IDs: 00043775–00043790).
Ethical standards
The study was approved by the National Committee on Health Research Ethics (1402655) and the Danish Data Protection Agency (2011-54-1262).
Results
Of the 47 investigated SUDI victims, 16 (43%) had one or several variants likely to have functional effects. In total, 19 variants were observed. Seven of these were assumed to be de novo variants as they have not been reported previously (Table 3). All variants were suspected to be pathogenic in the in silico evaluation, had evidence supporting pathogenicity21 and had frequencies below the disease prevalence of the diseases in the Danish control population25 and/or the European-American population provided by ESP.17 Details of the variants are available online in (Supplementary Table S2). In 8 of the 16 positive cases (50%), the observed variants were in genes that regulate ion channel functions and that are associated with channelopathies. Two cases had variants in PRDM16 and six cases had variants in MYH6, MYPN, DSG2, LAMA4, LDB3 and TAZ respectively. Of the 16 cases with variants with likely functional effects, 10 (63%) were females, and the median age of death was 1 month (range: 0–6 months). Following the San Diego criteria, 13 of the cases with variants with likely functional effects were classified as SIDS II, one case as IB and two cases as USID because of lacking information or investigations. With the Nordic classification, 14 of the cases with variants with likely functional affects were categorized as SIDS and two as unknown because of missing data. Eight cases had only variants of unknown functional significance (Table 3). The variants were classified as unknown either because of high prevalence in the control population, nonconserved amino acid substitution or described as variants of unknown significance in the available literature. No coding variant was found in the exons of CALM1, 2 or 3 although one case had a synonymous variant (c.183C>T, p.(N61N)) in CALM3 unlikely to be of significance.
Table 3. Variants with likely functional effects in the SUDI cases.
| Case | Sex | Age (months) | San Diego criteria1 | Nordic criteria12 | Risk factors | Gene | Function | Nucleotide | Amino acid | MAF in Danish population | MAFa | Evidence of pathogenicity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 0 | II | 1 | 2, 3 | PRDM16 (NM_022114.3) | Proliferation of cardiomyocytes | c.2468G>C | p.(R823P) | − | 0.2% | Known disease-associated variants in exon |
| 2 | M | 2 | II | 1 | 3 | TRPM4 (NM_017636.3) | Calcium channel | c.1575G>A | p.(W525*) | 0.2% | 0.1% | Null variant |
| 3 | M | 2 | IB | 1 | 4 | ANK2 (NM_001148.4) | Channel-interacting | c.4373A>G | p.(E1458G) | 0.05% | 0.02% | Seen in LQTS family, functional evidence43 |
| 4 | F | 1 | II | 1 | 1 | KCNJ5 (NM_000890.3) | Potassium channel | c.148C>Tb | p.(R50C) | − | − | Known disease-associated variants in exon, assumed de novo |
| 5 | F | 1 | II | 1 | 7,9c | RYR2 (NM_001035.2) | Intracellular Ca2+ homeostasis | c.7099G>Ad | p.(G2367R) | − | 0.01% | Seen in ARVC patient44 |
| 6 | F | 2 | II | 1 | 3 | MYH6 (NM_002471.3) | Sarcomeric | c.756C>G | p.(H252Q) | − | − | Functional evidence45 |
| 7 | M | 1 | II | 1 | 2,3 | SLC22A5 (NM_003060.3) | Carnitine transporter | c.34G>A | p.(G12S) | − | 0.2% | Seen in CTD patient46 |
| RYR2 (NM_001035.2) | Intracellular calcium homeostasis | c.10742G>Ab | p.(R3581K) | − | − | Assumed de novo | ||||||
| 8 | M | 1 | II | 1 | 3 | LAMA4 (NM_001105206.2) | Cytoskeletal | c.1901T>Ab | p.(V634D) | − | − | Assumed de novo |
| 10 | M | 6 | II | 1 | 1 | DSG2 (NM_001943.3) | Desmosomal | c.1370C>Gb | p.(S457C) | − | − | Functional protein domain, assumed de novo |
| 11 | F | 0 | USID | 18 | 7 | TRPM4 (NM_017636.3) | Calcium channel | c.1294G>A | p.(A432T) | 0.05% | 0.1% | Seen in CCD family, functional evidence47 |
| TRPM4 (NM_017636.3) | Calcium channel | c.1744G>A | p.(G582S) | 0.05% | 0.1% | Seen in CCD patient48 | ||||||
| 13 | F | 1 | II | 1 | 2,3 | KCNJ5 (NM_000890.3) | Potassium channel | c.631C>T | p.(R211W) | − | 0.02% | Functional protein domain |
| RYR2 (NM_001035.2) | Intracellular calcium homeostasis | c.5614G>Ad | p.(D1872N) | 0.05% | − | Functional protein domain | ||||||
| 14 | M | 2 | II | 1 | 3 | PRDM16 (NM_022114.3) | Proliferation of cardiomyocytes | c.2576C>T | p.(S859L) | − | 0% | Functional protein domain |
| 15 | F | 1 | II | 1 | 2,3,8 | KCNE4 (NM_080671.3) | Potassium channel | c.380A>Gb | p.(E127D) | − | − | Assumed de novo |
| 16 | F | 1 | II | 1 | 3,9e | MYPN (NM_001256267.1) | Z-disc | c.662A>T | p.(D221V) | 0.05% | 0.2% | Known disease-associated variants in exon |
| 18 | F | 0 | USID | 18 | 9f | LDB3 (NM_001171610.1) | Z-disc | c.1691G>Ab | p.(R564Q) | − | − | Functional protein domain, assumed de novo |
| 24 | F | 0 | II | 1 | 3 | TAZ (NM_000116.3) | Mitochondrial | c.535C>Gb | p.(P179A) | − | − | Functional protein domain, assumed de novo |
Abbreviations: MAF: minor allele frequency in the Danish population,25 LQTS: long QT syndrome, ARVC: arrhythmogenic right ventricular cardiomyopathy, CTD: carnitine transporter deficiency, CCD: cardiac conduction disease.
Clinical data and classification of the SUDI cases with variants with likely functional effects. Included are details of the functions of the gene, nucleotide and amino acid changes, frequencies in healthy populations and evidence of pathogenicity.
The San Diego criteria: IA: SIDS (>21 days, <9 months, normal clinical history, normal growth/development, no similar deaths in siblings/close relatives, no suspicion regarding death scene investigation, absence of potentially pathological abnormalities at autopsy including toxicology, microbiology, radiologic, vitreous chemistry and metabolic screening); IB: death scene not investigated and/or ≥1 of following not performed: microbiology, radiologic, vitreous chemistry and metabolic screening; II: ≥1 of following not fulfilled: age range, death among siblings/close relatives, neonatal/perinatal conditions, mechanical asphyxia not excluded with certainty, abnormal growth/development, inflammatory changes/abnormalities not sufficient to be the cause of death. USID: Not fulfilling criteria for SIDS I or II.
The Nordic criteria: 1: unknown cause of death, infant. 2: Brain/neurological disorders. 3: Pulmonary/respiratory disorders. 4: Heart/vessel disorders. 5: Gastrointestinal disorders. 6: Sepsis. 7: Intoxication. 8: Unspecified systemic disorders. 9: Asphyxia, traumatic. 10: Asphyxia, drowning. 11: Asphyxia, other. 12: Severe trauma. 13–16: Trauma (13: neurological system, 14: respiratory system, 15: cardiovascular system, 16: gastrointestinal system). 17: Exsanguination. 18: Unknown cause of death. 19: Other systematic disorders. 20: Combustion. 21: Blood disorders. 22–26: Unspecified disorders (22: Neurological system, 23: Respiratory system, 24: Cardiovascular system, 25: Gastrointestinal system, 26: Urogenital system). 27: Urogenital disorders.
Risk factors/other symptoms: 1: Missing information of birth. 2: Preterm/prenatal problems solved. 3: Mechanical asphyxia cannot be excluded. 4: One or several investigations missing. 5: Age 0–21 days or 270–265 days. 6: Unexplained trauma/abuse/neglect/unintended trauma. 7: Similar cases in siblings, not suspected to be homicide or of genetic aetiology. 8: Pronounced inflammatory changes or abnormalities not sufficient to be cause of death. 9: Other. 10: Death scene investigation not performed.
Minor allele frequency in European-American population in NHLBI GO Exome Sequencing Project ESP17 or dbSNP18 was used.
Novel variant.
Not normal growth/development.
Previously identified by Larsen et al.7
Jaundice.
Home delivery, infant buried in backyard.
Discussion
We investigated 47 SUDI cases for putative, pathogenic variants in the exons and promoter regions of 100 genes associated with inherited cardiac channelopathies and cardiomyopathies. In 34% of the cases, one or more variants with likely functional effects were identified. Half of the variants were in genes coding for ion channels in the cardiomyocytes and, therefore, possibly leading to perturbation of cardiac conduction, in accordance with the fact that no macro- or microscopic alteration of the heart was observed with conventional forensic investigations.
In the present cohort, we found a significantly higher frequency of variants in genes with likely functional effects that previously have been suspected or are known to be associated with cardiac diseases compared with the frequencies previously described in SUDI or SIDS cohorts.5, 6, 7 This is probably mainly because of the fact that the new NGS-based method makes it possible to analyse a larger number of genes than previously possible.
In contrast to Crotti et al,9 no variant suspected of having functional effect was found in CALM1-3. Crotti et al9 investigated two infants with recurrent cardiac arrest and 82 patients with congenital LQTS for variants in the three calmodulin-coding genes. Three likely pathogenic variants were found in four cases, including the two initially investigated infants. We only identified a synonymous variant in CALM3 not likely to be of significance for the cause of death. The investigated cohort in this study is not large enough to exclude that variants in the calmodulin genes can have an effect on the risk of SUDI. We identified five variants in TRPM4. Four of the variants are thought to have functional effects. TRPM4 encodes for a protein in calcium-activated nonselective ion channels that mediates membrane depolarization and plays a central role in the pacing of the cardiomyocytes.29 The function of the gene is associated with calmodulin as the deletion of one of the three C-terminal Ca2+-calmodulin binding sites strongly reduces the Ca2+ sensitivity in the ion channel.30 This suggests that calmodulin may contribute to the pathogenesis of SUDI. Recently, Le Scouarnec et al31 reported the occurrence of rare, coding variants in 21 arrhythmia-susceptibility genes in a large cohort of BrS patients and in controls. They observed a significantly increased frequency of variants in SCN5A in the patients compared with the controls. No difference in the frequencies of the other investigated genes previously suspected of being associated with BrS, including TRPM4, was found. Thus, the impact of genetic variants in these genes should be cautiously interpreted. The study only included individuals with BrS-specific ECG patterns. It is plausible that the genes have functional effects in individuals with different phenotypic expressions.
Two variants were found in PRDM16. The gene was previously associated with left ventricular non-compaction (LVNC) and dilated cardiomyopathy (DCM).32 The gene is expressed in heart tissue in fetal life. Animal studies with gene knockout showed development of bradycardia, reduced cardiac output and decrease in cardiomyocytes and cell proliferation.32 A variant with likely functional effects in the same gene was found in an autopsied 13-month-old deceased boy with unknown cause of death (unpublished data). The boy was not categorized as SIDS or SUDI because of his age at the time of death (>12 months). To our knowledge, no data indicating association between SUDI and PRDM16 have been published. The function of PRDM16 and the frequency of variants in the gene in this cohort support the hypothesis of a possible aetiology of PRDM16 in sudden death in infants. In addition, an infant with a variant in LDB3 with likely functional effects, which also has been associated with LVNC,33 was observed.
In six of the SUDI cases, we found variants with likely functional effects located in genes (MYH6, MYPN, DSG2, LAMA4, LDB3 and TAZ) associated with the structure of the heart and with diseases with distinct phenotypic traits such as arrhythmogenic right ventricular cardiomyopathy (ARVC), DCM and congenital heart disease. Brion et al10 studied HCM-associated variants in a large SIDS cohort comprising 286 cases and found that 4% had a possibly damaging variant. The gene with the highest proportion of variants was MYBPC3. Brion et al10 proposed that the observed genotypes were the causes of death. However, they also emphasized that the variants could simply be rare variants and incidental findings. We did not observe any variants in MYBPC3. In a single SIDS case, Campuzano et al34 found variants in PKP2, TTN and VCL that are associated with cardiomyopathies. It has been shown that variants in the desmosomal genes PKP2, DSG2 and DSP may cause sodium current dysfunction, arrhythmias and possibly sudden death.35, 36, 37 PKP2 variants have been identified in 2.5% of clinically diagnosed BrS patients who did not have any pathogenic variants in SCN5A, CACNA1C, GDP1L and MOG1.38 Four of the investigated SUDI cases in this study had variants with likely functional effects or variants of unknown significance in one of the desmosomal genes. Although the impact of the sodium current is unknown for all of the genes in the desmosome complex, the close interaction could implicate that there is a similar effect among the encoded proteins. Thus, it cannot be excluded that the variants associated with cardiomyopathies are of importance in SUDI.
This study enables us to explore the genetic background of inherited cardiac diseases in a cohort of SUDI victims. However, there are several limitations that need to be acknowledged. There is a limitation in this study in terms of lack of co-segregation evidence. It was not possible to genetically investigate the families of the deceased. Hence, it is unknown whether the observed variants are inherited from the parents or occurred de novo in the infants. Family follow-up, including genetic and clinical investigations would potentially have validated or rejected some of the variants with likely functional effects. SIDS does not segregate in families,1 contradicting a genetic disposition. However, SIDS is multifactorial and occurs in infants at a vulnerable stage.39 Therefore, a variant affecting the pacing of the heart could contribute to the cause of sudden death in these cases. The confirmation of functional effects provides strong evidence of pathogenicity.21 Many of the variants observed in this study have not been functionally verified, and hence the possible effects of the variants are solely supported by the location in the genome, the conservation sites and other variants nearby with similar impact.
The infants in our study were slightly younger and the percentage of females was higher compared with those generally seen in SIDS cohorts.39 However, during the past decades, there has been a tendency towards a younger age distribution in SIDS victims.40, 41, 42 Our cohort is relatively small and we included both SIDS and SUDI victims that may have influenced the observed differences.
To our knowledge, this is the first NGS-based genetic study of a large cohort of SUDI cases investigating a large number of genes associated with inherited cardiac diseases. Rare genes that have not previously been investigated in SUDI cases have now proven to be of importance for the aetiology of sudden unexpected death in infants. There is a strong indication that channelopathies explain the death in some of the SUDI cases. Therefore, we feel that cases of sudden and unexpected deaths of infants with no objective findings at conventional forensic autopsy should be investigated for possible disease-causing variants in the cardiac-associated genes. Genetic investigation in forensic medicine has the potential of supporting the clinical investigation and increase the molecular diagnostic rate. This will be of benefit for the families at risk as focussed cardiac evaluations and prophylactic treatments can be offered.
Acknowledgments
We thank Francisc-Raul Kantor for bioinformatics support. For the screening of variants among Danish controls, we thank LuCamp, The Lundbeck Foundation Centre for Applied Medical Genomics in Personalized Disease Prediction, Prevention, and Care (www.lucamp.org) and the Novo Nordisk Foundation Center for Basic Metabolic Research, an independent Research Centre at the University of Copenhagen partially supported by an unrestricted donation from the Novo Nordisk Foundation (www.metabol.ku.dk). This work was supported by Ellen and Aage Andersen's Foundation and Arvid Nilssons's Foundation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Krous HF, Beckwith JB, Byard RW et al: Sudden infant death syndrome and unclassified sudden infant deaths: a definitional and diagnostic approach. Pediatrics 2004; 114: 234–238. [DOI] [PubMed] [Google Scholar]
- Changing concepts of sudden infant death syndrome: implications for infant sleeping environment and sleep position. American Academy of Pediatrics. Task Force on Infant Sleep Position and Sudden Infant Death Syndrome. Pediatrics 2000; 105 (3 Pt 1): 650–656. [DOI] [PubMed] [Google Scholar]
- Moon RY, Horne RS, Hauck FR: Sudden infant death syndrome. Lancet 2007; 370: 1578–1587. [DOI] [PubMed] [Google Scholar]
- Winkel BG, Holst AG, Theilade J et al: Sudden unexpected death in infancy in Denmark. Scand Cardiovasc J 2011; 45: 14–20. [DOI] [PubMed] [Google Scholar]
- Millat G, Kugener B, Chevalier P et al: Contribution of long-QT syndrome genetic variants in sudden infant death syndrome. Pediatr Cardiol 2009; 30: 502–509. [DOI] [PubMed] [Google Scholar]
- Arnestad M, Crotti L, Rognum TO et al: Prevalence of long-QT syndrome gene variants in sudden infant death syndrome. Circulation 2007; 115: 361–367. [DOI] [PubMed] [Google Scholar]
- Larsen MK, Berge KE, Leren TP et al: Postmortem genetic testing of the ryanodine receptor 2 (RYR2) gene in a cohort of sudden unexplained death cases. Int J Legal Med 2012; 127: 139–144. [DOI] [PubMed] [Google Scholar]
- Schwartz PJ, Crotti L, Insolia R: Long-QT syndrome: from genetics to management. Circ Arrhythm Electrophysiol 2012; 5: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti L, Johnson CN, Graf E et al: Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation 2013; 127: 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion M, Allegue C, Santori M et al: Sarcomeric gene mutations in sudden infant death syndrome (SIDS). Forensic Sci Int 2012; 219: 278–281. [DOI] [PubMed] [Google Scholar]
- Larsen MK.: Sudden unexpected death and genetic heart disease: a molecular autopsy study. PhD dissertation, Aarhus University: Denmark, 2012. [Google Scholar]
- Vege Å, Rognum T, Løberg E et alDiagnosis of sudden infant death in the Nordic countries since 1970, revised (1995); in: Rognum TO (ed): Sudden Infant Death Syndrome: New Trends in the Nineties. Scandinavian University Press, 1995, pp 67–69. [Google Scholar]
- OMIMOnline Mendelian Inheritance in Man. Baltimore, MD: McKusick-Nathans Institute of Genetic Medicine, John Hopkins University. [Google Scholar]
- Kent W, Sugnet C, Furey T et al: The human genome browser at USCS. Genome Res. 2002; 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A et al1000 Genome Project Data Processing Subgroup: The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESP. Exome Variant Server [updated 2014/01/02/10:04:37]. Available from http://evs.gs.washington.edu/EVS/.
- NCBI. National Center for Biotechnology Information [updated 2014/01/02/10:23:41]. Available from http://www.ncbi.nlm.nih.gov/.
- Stenson P, Ball E, Mort M et al: The Human Gene Mutation Database (HGMD). Hum Mutat 2003; 21: 577–581. [DOI] [PubMed] [Google Scholar]
- Hertz CL, Christiansen SL, Ferrero-Miliani L et al: Next-generation sequencing of 34 genes in sudden unexplained death victims in forensics and in patients with channelopathic cardiac diseases. Int J Legal Med 2014; 129: 793–800. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S et al: Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D, Johnston JJ, Teer JK et alNIH Intramural Sequencing Center (NISC) Comparative Sequencing Program: Interpreting secondary cardiac disease variants in an exome cohort. Circ Cardiovasc Genet 2013; 6: 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner MO, Amendola LM, Turner EH et alNational Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project: Actionable, pathogenic incidental findings in 1,000 participants' exomes. Am J Hum Genet 2013; 93: 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I, McVean G: Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012; 44: 243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Sparso T, Li Q et al: Whole-exome sequencing of 2,000 Danish individuals and the role of rare coding variants in type 2 diabetes. Am J Hum Genet 2013; 93: 1072–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing 2014, Available from http://www.R-project.org.
- Huber W, Carey VJ, Gentleman R et al: Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 2015; 12: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM et al: Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launay P, Fleig A, Perraud AL, Scharenberg AM, Penner R, Kinet JP: TRPM4 is a Ca2+-activated nonselective cation channel mediating cell membrane depolarization. Cell 2002; 109: 397–407. [DOI] [PubMed] [Google Scholar]
- Nilius B, Prenen J, Tang J et al: Regulation of the Ca2+ sensitivity of the nonselective cation channel TRPM4. J Biol Chem 2005; 280: 6423–6433. [DOI] [PubMed] [Google Scholar]
- Le Scouarnec S, Karakachoff M, Gourraud JB et al: Testing the burden of rare variation in arrhythmia-susceptibility genes provides new insights into molecular diagnosis for Brugada syndrome. Hum Mol Genet 2015; 24: 2757–2763. [DOI] [PubMed] [Google Scholar]
- Arndt AK, Schafer S, Drenckhahn JD et al: Fine mapping of the 1p36 deletion syndrome identifies mutation of PRDM16 as a cause of cardiomyopathy. Am J Hum Genet 2013; 93: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Ichida F, Matsuoka T et al: Genetic analysis in patients with left ventricular noncompaction and evidence for genetic heterogeneity. Mol Genet Metab 2006; 88: 71–77. [DOI] [PubMed] [Google Scholar]
- Campuzano O, Allegue C, Sarquella-Brugada G et al: The role of clinical, genetic and segregation evaluation in sudden infant death. Forensic Sci Int 2014; 242: 9–15. [DOI] [PubMed] [Google Scholar]
- Cerrone M, Noorman M, Lin X et al: Sodium current deficit and arrhythmogenesis in a murine model of plakophilin-2 haploinsufficiency. Cardiovasc Res 2012; 95: 460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo S, Lodder EM, Verkerk AO et al: Intercalated disc abnormalities, reduced Na(+) current density, and conduction slowing in desmoglein-2 mutant mice prior to cardiomyopathic changes. Cardiovasc Res 2012; 95: 409–418. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Deng C, Rao F et al: Silencing of desmoplakin decreases connexin43/Nav1.5 expression and sodium current in HL1 cardiomyocytes. Mole Med Rep 2013; 8: 780–786. [DOI] [PubMed] [Google Scholar]
- Cerrone M, Lin X, Zhang M et al: Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation 2014; 129: 1092–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming PJ, Blair PS, Pease A: Sudden unexpected death in infancy: aetiology, pathophysiology, epidemiology and prevention in 2015. Arch Dis Childh 2015, e-pub ahead of print 19 February 2015 doi:10.1136/archdischild-2014-306424. [DOI] [PubMed]
- Blair PS, Sidebotham P, Berry PJ, Evans M, Fleming PJ: Major epidemiological changes in sudden infant death syndrome: a 20-year population-based study in the UK. Lancet 2006; 367: 314–319. [DOI] [PubMed] [Google Scholar]
- Blair PS, Sidebotham P, Evason-Coombe C, Edmonds M, Heckstall-Smith EM, Fleming P: Hazardous cosleeping environments and risk factors amenable to change: case-control study of SIDS in south west England. BMJ 2009; 339: b3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollborg P, Wennergren G, Almqvist P, Alm B: Bed sharing is more common in sudden infant death syndrome than in explained sudden unexpected deaths in infancy. Acta Paediatr 2015; 104: 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler PJ, Schott J-J, Gramolini AO et al: Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature 2003; 421: 634–639. [DOI] [PubMed] [Google Scholar]
- Roux-Buisson N, Gandjbakhch E, Donal E et al: Prevalence and significance of rare RYR2 variants in arrhythmogenic right ventricular cardiomyopathy/dysplasia: results of a systematic screening. Heart Rhythm 2014; 11: 1999–2009. [DOI] [PubMed] [Google Scholar]
- Granados-Riveron JT, Ghosh TK, Pope M et al: Alpha-cardiac myosin heavy chain (MYH6) mutations affecting myofibril formation are associated with congenital heart defects. Hum Mol Genet 2010; 19: 4007–4016. [DOI] [PubMed] [Google Scholar]
- Li FY, El-Hattab AW, Bawle EV et al: Molecular spectrum of SLC22A5 (OCTN2) gene mutations detected in 143 subjects evaluated for systemic carnitine deficiency. Hum Mutat 2010; 31: E1632–E1651. [DOI] [PubMed] [Google Scholar]
- Liu H, El Zein L, Kruse M et al: Gain-of-function mutations in TRPM4 cause autosomal dominant isolated cardiac conduction disease. Circ Cardiovasc Genet 2010; 3: 374–385. [DOI] [PubMed] [Google Scholar]
- Stallmeyer B, Zumhagen S, Denjoy I et al: Mutational spectrum in the Ca(2+)—activated cation channel gene TRPM4 in patients with cardiac conductance disturbances. Hum Mutat 2012; 33: 109–117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.