Abstract
Although 22q11.2 deletion syndrome (22q11.2DS) is the most recurrent human microdeletion syndrome associated with a highly variable phenotype, little is known about the condition's true incidence and the phenotype at diagnosis. We performed a multicenter, retrospective analysis of postnatally diagnosed patients recruited by members of the Association des Cytogénéticiens de Langue Française (the French-Speaking Cytogeneticists Association). Clinical and cytogenetic data on 749 cases diagnosed between 1995 and 2013 were collected by 31 French cytogenetics laboratories. The most frequent reasons for referral of postnatally diagnosed cases were a congenital heart defect (CHD, 48.6%), facial dysmorphism (49.7%) and developmental delay (40.7%). Since 2007 (the year in which array comparative genomic hybridization (aCGH) was introduced for the routine screening of patients with intellectual disability), almost all cases have been diagnosed using FISH (96.1%). Only 15 cases (all with an atypical phenotype) were diagnosed with aCGH; the deletion size ranged from 745 to 2904 kb. The deletion was inherited in 15.0% of cases and was of maternal origin in 85.5% of the latter. This is the largest yet documented cohort of patients with 22q11.2DS (the most commonly diagnosed microdeletion) from the same population. French cytogenetics laboratories diagnosed at least 108 affected patients (including fetuses) per year from among a national population of ∼66 million. As observed for prenatal diagnoses, CHDs were the most frequently detected malformation in postnatal diagnoses. The most common CHD in postnatal diagnoses was an isolated septal defect.
Introduction
The incidence of chromosome 22q11.2 deletion syndrome (22q11.2DS, also known as DiGeorge syndrome and velocardiofacial syndrome; OMIM 188400 and 192430, respectively) is estimated at between 5.6 and 14.1 per 100 000 live births.1, 2 The phenotype is highly variable – over 180 associated clinical manifestations already described – and ranges from severe, with life-threatening malformations, to nearly asymptomatic cases. The major clinical features include congenital heart defects (CHDs), palate defects (velopharyngeal insufficiency, cleft palate, etc), mild-to-moderate immunodeficiency (because of thymic aplasia or hypoplasia), hypocalcemia caused by hypoparathyroidism, a distinct gestalt, developmental delay (DD), learning disabilities, intellectual disability (ID) and behavioral disturbances.3, 4, 5 Renal, ocular and skeletal anomalies have also been observed.4, 5
According to the literature, the 22q11.2 deletion occurs de novo in 90% of cases and therefore is inherited in the remaining 10%.3, 5, 6, 7, 8 In our prenatal series of 272 fetuses with 22q11.2DS, 27% of the deletions were inherited.9 Of the inherited deletions, 72 to 77% are of maternal origin.3, 8, 10
The 22q11.2 region contains eight low copy repeats (LCRs, referred to as LCR22-A to LCR22-H). The deletion results from nonallelic LCR-mediated, homologous recombination during meiosis.9 Approximately 90% of patients have a 3-Mb deletion spanning LCR22-A to LCR22-D, is referred to as the ‘typically deleted region' (TDR).11, 12, 13 A smaller (1.5 Mb) deletion spanning LCR22-A to LCR22-B is found in 4 to 7% of patients.11, 12, 13 Atypical deletions with at least one breakpoint not mediated by an LCR have also been reported.14, 15, 16 The relationship between the size of the 22q11.2 deletion and the corresponding clinical features is still subject to debate.11 Since 1999, <60 cases with recurrent 22q11.2 distal deletions (between LCR22-D to LCR22-H) of variable size and position have been described.17
The TDR contains over 40 genes that primarily code for transcription factors (TBX1, TUPLE1, etc), cell cycle components (UFD1L and PNUTL1), cell adhesion molecules (DGCR2, DGCR6, etc) and metabolic and enzymatic factors (COMT and PRODH). The molecular substrates underlying the various phenotypes have not been clearly identified. Several genes have been implicated in heart malformations, including the major candidate gene TBX1 but also UFD1L, HIRA, CRKL and, most recently, HIC2.18, 19, 20, 21, 22, 23, 24
In recent decades, the diagnostic procedure most frequently employed to detect the 22q11.2 deletion has been fluorescent in situ hybridization (FISH) using commercial probes for regions between LCR22-A and LCR22-B (ie, in the proximal part of the TDR). In the past 10 years, a number of molecular biology techniques have been developed. In particular, array comparative genomic hybridization (aCGH) has been widely used since 2007 and is now considered to be the first-line technique for diagnosis of genomic rearrangements in patients with DD and in fetuses or patients with congenital malformations of unknown etiology. Other techniques (such as multiplex ligation-dependent probe amplification and BACs-on-Beads) have also been developed.25, 26 These techniques enable the diagnosis of 22q11.2DS in patients with atypical phenotypes and/or who have been missed by conventional cytogenetic techniques.
The primary objective of the present study was to estimate the annual incidence of new cases of 22q11.2DS diagnosed in French cytogenetics laboratories. We also sought to determine (1) the clinical signs and/or malformations that had prompted genetic screening, (2) the inheritance status and (3) the technique used for diagnosis. Next, we focused on the phenotype and the size of the 22q11.2 deletion in cases diagnosed using aCGH. Most of the latter cases had an atypical presentation and smaller-than-usual deletions.
Patients and methods
A multicenter, retrospective study was set up to collect data on patients with 22q11.2DS diagnosed postnatally by members of the Association des Cytogénéticiens de Langue Française (the French-Speaking Cytogeneticists Association) between 1995 and 2013. Thirty-one French laboratories with postnatal diagnosis activity were involved in the collaboration and recorded data on a total of 747 patients. The following data were requested: family history, the indication for genetic testing (clinical, biological and/or imaging data), the technique used for diagnosis, inheritance status, parental phenotype and age at diagnosis. Previously published data on fetuses with 22q11.2DS10 were used to estimate the number of new 22q11.2DS cases per year between 2010 and 2012. Postnatally diagnosed cases were compared with prenatally diagnosed cases in terms of the prenatal ultrasound findings. All samples were referred to laboratories that had obtained informed consent from the patient or the parents, in accordance with French legislation.
For statistical analysis, the χ2 test or Mann–Whitney test was used to compare patients diagnosed using FISH with those diagnosed using aCGH in terms of clinical signs and fetal ultrasound features. The threshold for statistical significance was set to P<0.05.
Data on patients diagnosed with aCGH have been submitted to the Decipher database (https://decipher.sanger.ac.uk/).
Results
The study population
Of the 747 postnatally diagnosed patients, the gender was recorded in 555 cases, and the male/female gender ratio was 0.87 (260 males and 299 females). The patient's age at diagnosis was recorded in 746 cases. The study population comprised 143 newborns (aged below 1 month), 166 infants (aged between 1 month and 2 years), 119 young children (aged between 3 and 6 years), 100 older children (aged between 7 and 12 years), 64 adolescents (aged between 13 and 19 years) and 154 adults (aged ≥20 years). The mean age at diagnosis was 9.8 years (range: birth to 65 years) for the study population as a whole and 4.3 years when excluding adult cases (Table 1).
Table 1. Numbers of patients according to the age at diagnosis.
| Age at diagnosis | Total number of patients | Males | Females | Number with known phenotype | Percentage with a CHD | Percentage with an isolated feature |
|---|---|---|---|---|---|---|
| Newborns (aged <1 month) | 143 | 56 | 56 | 140 | 78.60% | 16.40% |
| Infants (aged 1 month to 2 years) | 166 | 66 | 67 | 161 | 63.10% | 15.50% |
| Young children (aged 3–6 years) | 119 | 46 | 42 | 115 | 29.60% | 13.00% |
| Older children (aged 7–12 years) | 100 | 32 | 36 | 96 | 43.80% | 11.50% |
| Adolescents (aged 13–19 years) | 64 | 25 | 21 | 59 | 32.20% | 15.30% |
| Adults (aged ≥20 years) | 154 | 36 | 77 | 118 | 34.70% | 23.70% |
| TOTAL | 746 | 260 | 299 | 688 | 48.50% | 25.00% |
The indication for genetic screening
Data on the phenotype were available for 688 of the 747 postnatally diagnosed patients. Data on clinical signs are summarized in Table 2.
Table 2. Clinical features observed at diagnosis.
| Whole series (n=688) | Diagnosis using FISH (n=673) | Diagnosis using aCGH (n=15) | ||||
|---|---|---|---|---|---|---|
| % | N | % | N | % | N | |
| Facial dysmorphism | 49.7% | 342 | 49.4% | 334 | 53.3% | 8 |
| Congenital heart defects | 48.5% | 334 | 49.0% | 331 | 20.0%a | 3 |
| Developmental delay/intellectual disability | 40.7% | 280 | 39.9% | 270 | 66.7% | 10 |
| Velopharyngeal insufficiency | 18.3% | 126 | 18.5% | 125 | 6.7% | 1 |
| Hypocalcemia | 14.5% | 100 | 14.8% | 100 | 0.0% | 0 |
| Frequent infections | 7.8% | 54 | 8.0% | 54 | 0.0% | 0 |
| Oral cleft | 7.8% | 54 | 8.0% | 54 | 0.0% | 0 |
| Psychiatric and behavioral disorders | 7.4% | 51 | 7.2% | 49 | 13.3% | 2 |
| Thymus agenesia | 7.1% | 49 | 7.2% | 49 | 0.0% | 0 |
| Kidney abnormalities | 6.0% | 41 | 5.6% | 38 | 20.0% | 3 |
| Feeding difficulties | 5.5% | 38 | 5.6% | 38 | 0.0% | 0 |
| Epilepsy | 5.5% | 38 | 5.6% | 38 | 0.0% | 0 |
| Respiratory disorders | 5.1% | 35 | 5.2% | 35 | 0.0% | 0 |
| Growth retardation | 4.4% | 30 | 3.8% | 26 | 26.7%a | 4 |
| Clubfeet | 3.3% | 23 | 3.3% | 22 | 6.7% | 1 |
| Scoliosis | 2.5% | 17 | 2.5% | 17 | 0.0% | 0 |
| Abnormal male genitalia | 2.3% | 16 | 2.4% | 16 | 0.0% | 0 |
| Deafness | 2.3% | 16 | 2.4% | 16 | 0.0% | 0 |
| Microcephaly | 1.6% | 11 | 1.2% | 8 | 20.0%a | 3 |
| Polydactyly | 1.0% | 7 | 1.0% | 7 | 0.0% | 0 |
| Neural tube defect | 0.7% | 5 | 0.6% | 4 | 6.7% | 1 |
| Central nervous system malformation | 0.7% | 5 | 0.4% | 3 | 13.3%a | 2 |
| Spine bone malformation | 0.6% | 4 | 0.4% | 3 | 6.7% | 1 |
Significantly greater or lower frequency than for cases diagnosed with FISH.
The most common reasons for referral for genetic testing were facial dysmorphism, CHDs and DD, as follows:
Facial dysmorphism was observed in 49.7% of cases (n=342). Features were described in 154 cases. The most common features were abnormal ears (n=84), a small mouth (n=67) and a tubular nose (n=32).
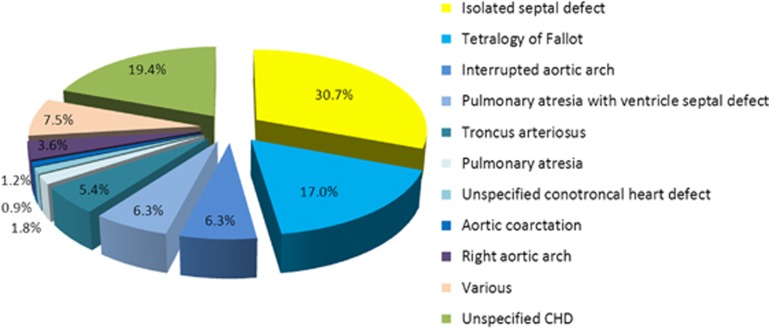
CHDs prompted referral in 48.5% of cases (n=334). Of the CHDs, 39.5% were conotruncal, and the most frequent CHD was an isolated septal defect (30.7%), followed by tetralogy of Fallot (17.0%) (Figure 1).
DD or ID prompted referral in 40.7% of cases (n=280). Some patients merely had difficulties at school (4.2%).
Psychiatric disorders were present in 7.4% of cases (n=51). In all, 32 patients had behavioral problems, 9 had autism, 3 adults had schizophrenia and 4 adults had depressive or manic-depressive syndrome.
Lip and/or palate defects were present in 7.8% of cases (n=54). All types of clefts were observed, with the most frequent being cleft palate (n=24). A lip defect was observed in three cases, including one in the absence of a palate defect.
Urinary abnormalities were present in 6.0% of cases (n=41). The most common reason was renal hypoplasia or agenesis (n=18), followed by cystic kidneys (n=10) and urinary tract anomalies (n=9).
Abnormal male genitalia were present in 2.3% of cases (n=16); these included testis agenesis or microorchidism (n=5), undescended testis (n=5), hypospadias (n=5) and small penis (n=3). In some individual cases, several defects were present.
Respiratory tract defects were present in 6.3% of cases (n=36) and included laryngomalacia or tracheaomalacia (n=12), subglottic stenosis (n=8) and laryngeal webs (n=8).
A few patients had gastrointestinal malformations (n=7), including imperforate anus or anal stenosis (n=4) and esophageal or intestinal stenosis (n=3).
Eight patients (1.4%) were referred by an ophthalmologist, mainly because of myopia (n=4).
Various other abnormalities were observed but in <1% of patients.
Abnormal ultrasound findings were detected during pregnancy for 8.0% of the patients (n=46); these were mainly related to intrauterine growth retardation (n=26) and polyhydramnios (n=12).
Figure 1.
Types of CHD. Type of defect identified in the 562 postnatally diagnosed patients with CHDs. The ‘various' subgroup includes all CHDs found in <1.5% of the affected patients: patent ductus arteriosus (n=5), patent foramen ovale (n=3) and double aortic arch (n=3).
Interestingly, 111 (25.0%) of the patients were referred because of a single feature (Table 1). Of these, 49.5% had a CHD only.
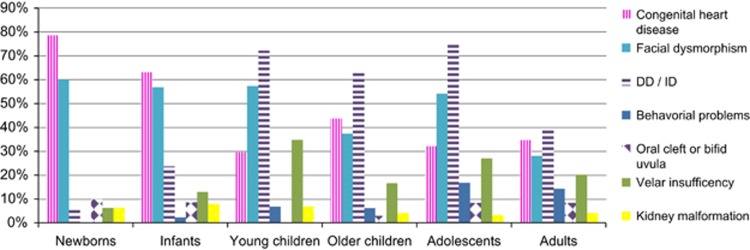
The features observed in patients at diagnosis are summarized as a function of age in Figure 2:
Figure 2.
Features observed at 22q11DS diagnosis according to the age at diagnosis.
For newborns diagnosed in the first month of life, 78.6% of the cases had a CHD, 60.0% had facial dysmorphism, 27.1% had hypocalcemia, 20.7% had thymic agenesis or hypoplasia and 10.0% had a lip and/or palate defect. In addition, 61 newborns (42.7%) had a CHD and facial dysmorphism.
For infants diagnosed at between 1 month and 2 years of age, 62.1% had a CHD, 57.1% had facial dysmorphism, 23.6% had DD (36.8% of whom had language delay alone) and 17.4% had hypocalcemia.
For children diagnosed at between 3 and 6 years of age, 72.2% had DD (48.2% of whom had language delay alone), 57.4% had facial dysmorphism, 34.8% had velar insufficiency and 29.6% had a CHD.
For children diagnosed at between 7 and 12 years of age, 63.5% had abnormal development (including 2 cases with severe ID), 43.8% had a CHD and 37.5% had facial dysmorphism.
For teenagers (aged 13 to 19 years), 74.6% were referred for ID or learning difficulties and 16.9% had behavioral or psychiatric problems.
For adults, 39.0% had ID or DD, 34.7% had a CHD, 28.0% had facial dysmorphism, 20.3% had hypernasal speech and 14.4% had hypocalcemia. The incidence of ID or DD was greater (P=0.0202) in men (44.4%) than in women (23.4%).
There were no significant differences between males and females in terms of the frequency of the various features (Supplementary Data 1), except for DD in adults and velar insufficiency in young children.
Patients diagnosed using aCGH
The vast majority of cases of 22q11.2DS recorded since 2007 (the year in which routine clinical use of this technique began in France) had been diagnosed using FISH (96.1%, n=386); aCGH had been used to diagnose only 15 patients (3.9%) (Supplementary Data 2).
The clinical features of the latter 15 patients are summarized in Table 2. Most often, postnatal genetic testing had been prompted by observation of DD or ID (10 out of 15; 66.67%). A CHD constituted a warning sign in 20.0% of these cases (compared with 48.5% for the study population as a whole; P=0.0355). When facial dysmorphism was present, it was atypical.
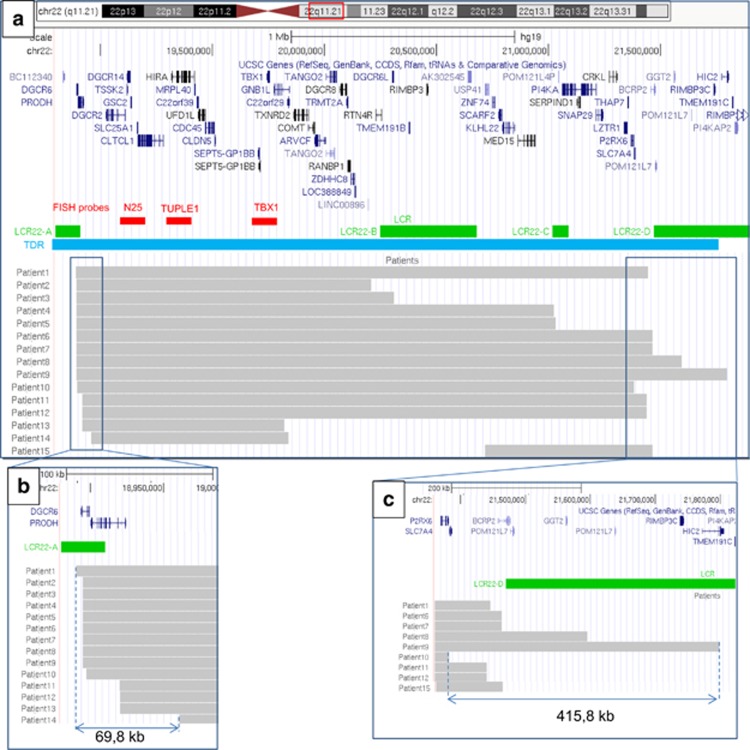
According to the aCGH results, the size of the deletion ranged from 745 to 2904 kb (mean: 2020 kb). In all but one of the patients, the proximal breakpoint was located near the distal part of LCR22-A in a 69.8-kb region (18 890 211 and 18 960 000 hg19). In the remaining patient, the proximal breakpoint was located near the distal part of LCR22-B (Figure 3). In 9 patients, the distal breakpoint was close to the LCR22-D (in a 415.8 kb region located between 21 382 953 and 21 798 755 bases from telomere (hg19)). The distal breakpoint was located near the proximal part of LCR-B in two cases and involved the LCR22-C in two cases. Finally, the distal breakpoint was located away from the LCRs in two cases.
Figure 3.
A map of the 22q11.21 deletions diagnosed using aCGH. The map was built using UCSC Genome Browser Build 37/ hg 19. (a) 22q11.21 deletions diagnosed using aCGH in our 15 patients. Gray bars represent the deleted regions. The TDR is situated between LCR22-A and LCR 22-D. (b) Focus on proximal breakpoints near the distal part of LCR-A. In all, 14 patients had a proximal breakpoint near LCR-A in a small (69.8 kb) region. (c) Focus on distal breakpoints near the proximal part of LCR-D. The distal breakpoint is more variable; 9 patients had a distal breakpoint close to the LCR22-D in a 415.8-kb region. The full colour version of this figure is available at European Journal of Human Genetics online.
Overall, only 8 of the 15 patients had a TDR deletion between the distal LCR22-A and the proximal LCR22-D. The mean deletion size was 2597 kb (Figure 3). When considering these eight patients, one had a CHD, five had DD, none had behavioral problems and three had growth retardation (including one case with no other associated signs). The deletion had occurred between LCR22-A and LCR22-B in two cases, between LCR22-A and LCR22-C in two cases and between LCR22-A and TBX1 (not mediated by LCRs) in two cases.
One patient (presenting with a urinary tract anomaly and a CHD) had an atypical 22q11.2 deletion between LCR22-B and LCR22-D.
Two patients presented with other copy number variations (CNVs) (Supplementary Data 3).
Parental inheritance
Of the 341 postnatally diagnosed patients with a parental analysis, the 22q11.2 deletion was found to be inherited in 70 cases (15.0%). The parental origin was known for 62 patients and was maternal in 53 cases (85.5%).
The number of new cases diagnosed annually
In contrast to Down's syndrome, the total number of diagnosed cases of 22q11.2DS is not specifically recorded by the French health authorities. We therefore sought to estimate the annual incidence of new cases of 22q11.2DS diagnosed by French cytogeneticists between 2010 and 2012 (Table 3). By adding together the previously reported prenatal diagnoses of 22q11.2DS10 and the postnatally diagnoses described here, we arrived at a total of 105 22q11.2DS diagnoses for 2010 (31 prenatally and 74 postnatally), 102 for 2011 (37 prenatally and 65 postnatally) and 119 for 2012 (41 prenatally and 78 postnatally). Hence, an average of 108 new cases of 22q11.2DS was recorded each year (of which 66.2% were diagnosed postnatally) (Table 3).
Table 3. Number of cases of 22q11.2DS diagnosed per year in the collaborating laboratories, compared with the number of cases of 21 trisomy diagnosed in France over the same period.
| 2010 | 2011 | 2012 | Average per year | Average per year (prenatal and PoD) | |
|---|---|---|---|---|---|
| 22q11DS | |||||
| Prenatal diagnosis | 31 | 37 | 41 | 36 | 108 |
| Postnatal diagnosis | 74 | 65 | 78 | 72 | |
| Trisomy 21a | |||||
| Prenatal diagnosis | 2369 | 2477 | 1971 | 2272 | 2758 |
| Postnatal diagnosis | 435 | 535 | 488 | 486 | |
In comparison, 2804 cases of trisomy 21 were recorded in 2010 in France (2369 prenatally and 435 postnatally), with 3012 in 2011 (2477 prenatally and 535 postnatally) and 2459 in 2012 (1971 prenatally and 488 postnatally).27, 28, 29, 30 Hence, an average of 2758 new cases of trisomy 21 were recorded each year (of which 82.4% were diagnosed prenatally) (Table 3).
Considering the total number of karyotypes provided by the participating laboratories (data were available for 32 of the 37 laboratories participating in the prenatal study10 and 27 of the 31 laboratories participating in the present postnatal study; Supplementary Data 4) and the overall number of cytogenetic diagnoses recorded by the French health authorities (both prenatal and postnatal),30 the cytogenetics laboratories participating in the present study accounted for 60.5% of all prenatal diagnoses and 46.3% of postnatally diagnoses. The total annual incidence of new cases of 22q11.2DS is thus estimated to be at least 36 for prenatal diagnoses and 72 for postnatally diagnoses; 22q11.2DS is thus 20 times less frequent than trisomy 21.
Discussion
To the best of our knowledge, our cohort of 22q11.2DS cases (including the prenatal diagnoses described previously10) is the largest yet studied. The male/female gender ratio was lower for postnatal diagnoses (0.87) than for prenatal diagnoses (1.23).10 This difference may tentatively suggest that males have a more severe phenotype than females, and this in turn may be associated with earlier diagnosis in males. Nevertheless, we did not observe any statistically significant differences between males and females in terms of the frequency of congenital malformations (especially for CHD), as also reported by Bassett et al.31
In the present series (postnatally diagnosed patients, together with the fetuses previously described10), the most frequent warning sign for 22q11.2DS was a CHD (in 58.5% of cases). With respect to our previously reported data on prenatal diagnoses,10 CHDs are more frequent in fetuses than in postnatally diagnosed patients (83.8 vs 48.5%, respectively; P<0.0001; Table 4). The frequencies of CHDs in fetuses and newborns are similar (83.8 and 78.6%, respectively10). Even though the CHD was generally a conotruncal disorder (including tetralogy of Fallot, interrupted aortic arch, etc), the most common CHD in our postnatally diagnosed patients was an isolated septal defect. The literature data on cases of 22q11.2DS (most of which are diagnosed postnatally) suggest that the frequency of a CHD is between 31.1 and 80.0%,1, 3, 5, 7, 8, 32, 33 with tetralogy of Fallot and a ventricular septal defect being the most frequent.3, 7, 8, 33, 34 Of the 15 patients diagnosed using aCGH, 3 had a CHD (including 2 with a deletion encompassing the TBX1 gene that is strongly suspected to be involved in CHDs18) and the third patient (patient 15) had a distal deletion encompassing CRKL (also suspected to be involved in CHDs23) (Figure 3). Nevertheless, the presence of a CHD cannot be explained solely by the loss of these genes.
Table 4. Malformations in prenatally and postnatally diagnosed patients detectable in prenatal ultrasound assessment.
| Postnatal diagnoses, n=688 | Prenatal diagnosis (9) (ultrasound findings), n=272 | Total, n=960 | ||||
|---|---|---|---|---|---|---|
| % | N | % | N | % | N | |
| Congenital heart defects | 48.5%a | 334 | 83.8% | 228 | 58.5% | 562 |
| Thymus agenesia | 7.1%a | 49 | 3.7% | 10 | 6.1% | 59 |
| Kidney abnormalities | 6.0% | 41 | 9.2% | 25 | 6.9% | 66 |
| Oral cleft | 7.8% | 54 | 3.3% | 9 | 6.6% | 63 |
| Growth retardation | 4.4% | 30 | 2.9% | 8 | 3.1% | 30 |
| Clubfeet | 3.3% | 23 | 3.7% | 10 | 3.4% | 33 |
| Abnormal male genitalia | 2.3%a | 16 | 0.4% | 1 | 1.8% | 17 |
| Microcephaly | 1.6% | 11 | 0.0% | 0 | 1.1% | 11 |
| Polydactyly | 1.0% | 7 | 0.7% | 2 | 0.9% | 9 |
| Neural tube defect | 0.7%a | 5 | 2.6% | 7 | 1.3% | 12 |
| Spine bone malformation | 0.6% | 4 | 0.0% | 0 | 0.4% | 4 |
| Central nervous system malformation | 0.7% | 5 | 1.8% | 5 | 1.0% | 10 |
Significantly greater frequency than for prenatal diagnoses.
The second most frequent feature observed at referral was facial dysmorphism. According to the literature data, the typical facies in 22q11.2DS is characterized by a long face, a flattened malar region, narrow palpebral fissures, hooded eyelids, a tubular nose and alar hypoplasia.5, 35 In our series, many postnatally diagnosed patients had facial dysmorphism (48.7%), although the latter was not always precisely described by clinicians. The facial dysmorphism was sometimes atypical in patients diagnosed using aCGH. The recognition of fetal facial dysmorphism is problematic and facial dysmorphism was noted in only 2.6% of the fetuses reported on previously.10
In terms of neuropsychiatric disorders, 48.9% of our patients were referred for DD or ID. However, data on intellectual capacities were not available for some patients. Only 3 adults (1.9% of the diagnosed adults) had schizophrenia at the time of diagnosis. The incidence of psychiatric disorders is higher in the literature, with a value of ∼20% for schizophrenia.31, 36, 37
The mean patient age at the time of postnatal diagnosis was quite old (9.8 years, and just 4.3 years when adult patients were excluded). This was probably because of the highly variable phenotype in the postnatally diagnosed cases recorded here. The present retrospective study enabled us to collect information on the most frequent clinical signs or malformations that had prompted genetic testing (usually using FISH). Interestingly, our series showed that the reason for referral varied according to the patient's age (Figure 2). Even though CHDs were the most common feature in fetuses with 22q11.2DS10 and were associated with pregnancy termination rate of 68.9%, the postnatal phenotype might not be the same as the classical phenotype described by DiGeorge and Shprintzen. In our series, CHDs, facial dysmorphism, hypocalcemia and thymus agenesis were the most frequent reasons for referral from birth to 2 years. However, learning difficulties and DD became more frequent in patients aged from 3 to 19 years. Literature data on the adult phenotype are scarce; the most commonly reported signs are minor facial dysmorphism, hypernasal speech and hypocalcemia.31, 38 In our series, the adult phenotype was highly variable. Fifty-four adult cases were diagnosed after one of their children had been diagnosed with 22q11.2DS, and 8 of the parents presented with ID.
In the present study, 15 patients with atypical phenotype had been unexpectedly diagnosed using aCGH. This technique is now replacing FISH in laboratories and is particularly useful in cases with an atypical phenotype. Of the 15 patients diagnosed using aCGH, 14 had a deletion that overlapped with the region targeted by commercially available FISH probes (between LCR22-A and LCR22-B). The remaining patient could not have been diagnosed using FISH (Figure 3). Hence, as previously suggested, FISH should not be used as the only diagnostic method for 22q11.2DS.39
The phenotypic heterogeneity in 22q11.2DS might be because of (1) the various sizes of the deletions and/or (2) rearrangements elsewhere on the genome – especially CNVs, mutations in candidate genes and changes in the three-dimensional structure of the genome that can lead to dysregulation of gene expression. Zeitz et al.40 found that long-range interactions in the COMT gene had an impact on phenotypic variability. The use of aCGH only covers the CNVs and maps deletion breakpoints. In our series, only 8 of the 15 patients diagnosed using aCGH had a common deletion that fully encompassed the TDR (mean deletion size: 2597 kb) (Figure 3). Even though the deleted genes within the 22q11.2 region (such as TBX1, CRKL and PRODH) have been extensively discussed in the literature, we decided to look at whether phenotype variability might be related to the deletion breakpoints. In all cases other than patient 15, the deletion involved the LCR22-A. All of these deletions overlapped with the proximal variable region described by Bittel et al.41 (chr22: 18 656 078–19 018 178). In contrast, the distal breakpoint was more variable; in view of our small series, no conclusions could be reached. Differences between patients could be because of the aCGH design or breakpoints in the distal borderline region.
Only two patients had other CNVs that may have contributed to the final phenotype, including a 16q24.3 deletion encompassing ANKRD1 (which has been linked to autism spectrum disorder, ID and short stature42). Our patient (aged 1 year at the time of diagnosis) had DD but normal growth. Hence, CNVs and proximal breakpoint sites do not appear to account for the clinical variability in 22q11.2DS in the present small series.
In the present series, the 22q11.2 deletion was inherited in 15.0% of cases. By considering the parental analysis for fetuses described previously (189 parental analysis, for which 27.0% of the deletion were inherited),10 we observed that the 22q11.2 deletion was inherited in 22.8% of cases. This value is similar to that reported in the Europe-wide study by Ryan et al. (28%) but is higher than in other recent reports (10–15%).3, 5, 6 Inherited 22q11.2 deletions were mainly of maternal origin in our series, as is also found in the literature.3 Nevertheless, 18.5% of the inherited deletions were of paternal origin in the series as a whole (the fetuses reported in Besseau-Ayasse et al.10 and the cases in the present report); this justifies systematic screening for 22q11.2DS in both parents. Furthermore, a previous study43 of reproductive fitness found that adults with 22q11.2DS had significantly fewer children than healthy controls: respectively 22.5 and 5.7% of the affected women and men had children. Even though the proportion of affected women with children did not appear to depend on the presence of mental disabilities and schizophrenia, the situation was quite different for affected men: 14.8% of the men with mental disabilities and schizophrenia had children, compared with 41.1% of the men without these conditions.43 As also suggested by our present data, it seems that (1) mental retardation and cognitive problems in adulthood are more severe in males than in females44 and (2) poor social integration and/or reduced spermatogenesis might occur in males with 22q11.2DS. Even though our data suggest that the frequency of ID or DD is more frequent in male adults than in female adults (44.4 vs 23.4%, P=0.02), this result may have resulted from bias in our study and therefore needs to be studied with greater accuracy. Studies of the intellectual profile by gender have yielded contrasting results; however, some recent literature data show that the mean full-scale IQ was higher in females than in males.45, 46 This discrepancy could be because of (1) a higher incidence of psychiatric disorders for males (although the relationship between psychiatric disorders and IQ is still subject to debate), (2) a negative age-IQ effect in boys and (3) more normal frontal lobe volumes (which might be related to performance in psychological tests) in girls, as previously suggested.46
The exact incidence of 22q11.2DS has not been assessed but is thought to be between 5.6 and 14.1 per 100 000 live births.1, 2 In a previous study of the same French population, prenatal testing enabled the diagnosis of an average of 36 fetuses per year between 2010 and 2012; however, the parents' right to request an abortion will have decreased the number of neonates actually born with 22q11.2DS.10 The present data were not exhaustive because some French cytogenetic laboratories did not participate in the study. Nevertheless, our results show that on average, 72 postnatal diagnoses were made each year. Given the number of live births in France (an average of 795 000 per year between 2010 and 2012)47 we estimate that there are at least 9 cases of 22q11.2DS per 100 000 live births and thus at least 100 new cases per year in France Even though the frequency of 22q11.2DS is 20 times lower than that of Down's syndrome, this deletion is nevertheless the second most frequently diagnosed chromosomal aberration responsible for ID/malformations (mainly diagnosed during the postnatal period). We suggest that data on the incidence of 22q11.2DS should be collected systematically (as is already the case in France for trisomy 21).
Conclusion
This study highlights the phenotypic variability in 22q11.2DS; the phenotype ranged from multiple congenital anomalies with a CHD (enabling early diagnosis) to a near lack of symptoms (in adults). The most frequent reason for referral for genetic testing for 22q11.2DS in postnatally diagnosed patients was a CHD (mainly septal defects), although this parameter varied strongly with age at diagnosis. Some cases of 22q11.2DS are now fortuitously diagnosed using aCGH. The atypical phenotypes diagnosed using aCGH or as part of family screening (prompted by diagnosis in a child) showed that the incidence of 22q11.2DS is probably underestimated; the clinical criteria may thus need to be reevaluated. Some of the phenotypic variability may be explained by aCGH, analysis of mutations in the contralateral allele and whole-exome sequencing. Understanding this variability should improve genetic counseling. We estimate that 22.8% of cases are inherited (mainly from the mother); this requires systematic screening of family members. In adulthood, the phenotype differs when comparing females and males (with an increased incidence of ID/DD in the latter).
Acknowledgments
We thank the members of the Association des Cytogénéticiens de Langue Française for their collaboration. The present work was funded by the French Ministry of Health (reference: DHOS 2007).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Oskarsdottir S, Vujic M, Fasth A: Incidence and prevalence of the 22q11 deletion syndrome: a population-based study in Western Sweden. Arch Dis Child 2004; 89: 148–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanet Prévalence des maladies rares: données bibliographiques Juin 2013 n°1. Data available from URL http://www.orpha.net/consor/cgi-bin/Education_Home.php?lng=FR.
- Ryan AK, Goodship JA, Wilson DI et al: Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet 1997; 34: 798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BN: Deletion 22q11: spectrum of associated disorders. Semin Pediatr Neurol 2007; 14: 136–139. [DOI] [PubMed] [Google Scholar]
- Monteiro FP, Vieira TP, Sgardioli IC et al: Defining new guidelines for screening the 22q11.2 deletion based on a clinical and dysmorphologic evaluation of 194 individuals and review of the literature. Eur J Pediat 2013; 172: 927–945. [DOI] [PubMed] [Google Scholar]
- Bassett AS, McDonald-McGinn DM, Devriendt K et al: Practical guidelines for managing patients with 22q11.2 deletion syndrome. J Pediatr 2011; 159: 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Emanuel BS, Zackai EH: 22q11.2 Deletion syndrome. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K (eds): Gene Reviews, 1993.
- Cancrini C, Puliafito P, Digilio MC et al: Clinical features and follow-up in patients with 22q11.2 deletion syndrome. J Pediatr 2014; 164: 1475–1480, e2. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Morrow BE: Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am J Hum Genet 1999; 64: 1076–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau-Ayasse J, Violle-Poirsier C, Bazin A et al: A French collaborative survey of 272 fetuses with 22q11.2 deletion: ultrasound findings, fetal autopsies and pregnancy outcomes. Prenatal Diagn 2014; 34: 424–430. [DOI] [PubMed] [Google Scholar]
- Michaelovsky E, Frisch A, Carmel M et al: Genotype-phenotype correlation in 22q11.2 deletion syndrome. BMC Med Genet 2012; 13: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C, Papolos D, Pandita RK et al: Molecular analysis of velo-cardio-facial syndrome patients with psychiatric disorders. Am J Hum Genet 1997; 60: 851–859. [PMC free article] [PubMed] [Google Scholar]
- Saitta SC, Harris SE, Gaeth AP et al: Aberrant interchromosomal exchanges are the predominant cause of the 22q11.2 deletion. Hum Mol Genet 2004; 13: 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira SI, Hacker AM, Bellucco FT et al: Atypical 22q11.2 deletion in a patient with DGS/VCFS spectrum. Eur J Hum Genet 2008; 51: 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen JM, Diderich KE, Oudesluijs G et al: Phenotypic variability of atypical 22q11.2 deletions not including TBX1. Am J Med Genet A 2012; 158A: 2412–2420. [DOI] [PubMed] [Google Scholar]
- Rauch A, Zink S, Zweier C et al: Systematic assessment of atypical deletions reveals genotype-phenotype correlation in 22q11.2. J Med Genet 2005; 42: 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar S, Ou Z, Shaw CA et al: 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet 2008; 82: 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE: DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet 2001; 27: 286–291. [DOI] [PubMed] [Google Scholar]
- Mercher T, Coniat MB, Monni R et al: Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc Natl Acad Sci USA 2001; 98: 5776–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paylor R, Lindsay E: Mouse models of 22q11 deletion syndrome. Biol Psychiatry 2006; 59: 1172–1179. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Garg V, Matsuoka R, Thomas T, Srivastava D: A molecular pathway revealing a genetic basis for human cardiac and craniofacial defects. Science 1999; 283: 1158–1161. [DOI] [PubMed] [Google Scholar]
- Olaopa M, Zhou HM, Snider P et al: Pax3 is essential for normal cardiac neural crest morphogenesis but is not required during migration nor outflow tract septation. Dev Biol 2011; 356: 308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A: Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet 2001; 27: 293–298. [DOI] [PubMed] [Google Scholar]
- Dykes IM, van Bueren KL, Ashmore RJ et al: HIC2 is a novel dosage-dependent regulator of cardiac development located within the distal 22q11 deletion syndrome region. Circ Res 2014; 115: 23–31. [DOI] [PubMed] [Google Scholar]
- Fernandez L, Lapunzina P, Arjona D et al: Comparative study of three diagnostic approaches (FISH, STRs and MLPA) in 30 patients with 22q11.2 deletion syndrome. Clin Genet 2005; 68: 373–378. [DOI] [PubMed] [Google Scholar]
- Vialard F, Simoni G, Aboura A et al: Prenatal BACs-on-Beads: a new technology for rapid detection of aneuploidies and microdeletions in prenatal diagnosis. Prenatal Diagn 2011; 31: 500–508. [DOI] [PubMed] [Google Scholar]
- Le rapport médical et scientifique de l'Agence de la biomédecine 2010. Génétique postnale 2010. URL http://www.agence-biomedecine.fr/annexes/bilan2010/accueil.htm.
- Le rapport médical et scientifique de l'Agence de la biomédecine 2011. Génétique postnatale 2011 et Génétique prénatale 2010. Data available from URL http://www.agence-biomedecine.fr/annexes/bilan2011/accueil.htm.
- Le rapport médical et scientifique de l'Agence de la biomédecine 2012. Génétique postnatale 2012 et Génétique prénatale 2011. Data available from URL http://www.agence-biomedecine.fr/annexes/bilan2012/accueil.htm.
- Le rapport médical et scientifique de l'Agence de la biomédecine 2013. Génétique prénatale 2012. URL http://www.agence-biomedecine.fr/annexes/bilan2013/donnees/diag-prenat/01-diag_prenat/synthese.htm.
- Bassett AS, Chow EW, Husted J et al: Clinical features of 78 adults with 22q11 Deletion Syndrome. Am J Med Genet A 2005; 138: 307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretelle F, Beyer L, Pellissier MC et al: Prenatal and postnatal diagnosis of 22q11.2 Deletion Syndrome. Eur J Med Genet 2010; 53: 367–370. [DOI] [PubMed] [Google Scholar]
- Repetto GM, Guzman ML, Puga A et al: Clinical features of chromosome 22q11.2 microdeletion syndrome in 208 Chilean patients. Clin Genet 2009; 76: 465–470. [DOI] [PubMed] [Google Scholar]
- Momma K: Cardiovascular anomalies associated with chromosome 22q11.2 deletion syndrome. Am J Cardiol 2010; 105: 1617–1624, Review. [DOI] [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg RB, Lewin ML et al: A new syndrome involving cleft palate, cardiac anomalies, typical facies, and learning disabilities: velo-cardio-facial syndrome. Cleft Palate J 1978; 15: 56–62. [PubMed] [Google Scholar]
- Schneider M, Debbané M, Bassett AS et alInternational Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome: Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am J Psychiatry 2014; 171: 627–639, Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks S, Niarchou M, Davies AR et al: Further evidence for high rates of schizophrenia in 22q11.2 deletion syndrome. Schizophr Res 2014; 153: 231–236. [DOI] [PubMed] [Google Scholar]
- Cohen E, Chow EW, Weksberg R, Bassett AS: Phenotype of adults with the 22q11 deletion syndrome: a review. Am J Med Genet 1999; 86: 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmaze C, Scambler P, Prieur M et al: Routine diagnosis of DiGeorge syndrome by fluorescent in situ hybridization. Hum Genet 1993; 90: 663–665. [DOI] [PubMed] [Google Scholar]
- Zeitz MJ, Lerner PP, Ay F et al: Implications of COMT long-range interactions on the phenotypic variability of 22q11.2 deletion syndrome. Nucleus 2013; 4: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel DC, Yu S, Newkirk H et al: Refining the 22q11.2 deletion breakpoints in DiGeorge syndrome by aCGH. Cytogenet Genome Res 2009; 124: 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacharow S, Li D, Fan YS, Tekin M: Familial 16q24.3 microdeletion involving ANKRD11 causes a KBG-like syndrome. Am J Med Genet A 2012; 158A: 547–552. [DOI] [PubMed] [Google Scholar]
- Costain G, Chow EW, Silversides CK, Bassett AS: Sex differences in reproductive fitness contribute to preferential maternal transmission of 22q11.2 deletions. J Med Genet 2011; 48: 819–824. [DOI] [PubMed] [Google Scholar]
- Sobin C, Kiley-Brabeck K, Monk SH, Khuri J, Karayiorgou M: Sex differences in the behavior of children with the 22q11 deletion syndrome. Psychiat Res 2009; 166: 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklasson L, Gillberg C: The neuropsychology of 22q11 deletion syndrome. A neuropsychiatric study of 100 individuals. Res Dev Disabil 2010; 31: 185–194. [DOI] [PubMed] [Google Scholar]
- Antshel KM, AbdulSabur N, Roizen N, Fremont W, Kates WR: Sex differences in cognitive functioning in velocardiofacial syndrome (VCFS). Dev Neuropsychol 2005; 28: 849–869. [DOI] [PubMed] [Google Scholar]
- Démographie – Nombre de naissances vivantes – France métropolitaine. Data available from URL http://www.insee.fr/fr/bases-de-donnees/bsweb/serie.asp?idbank=000436391.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.