Abstract
Learning disabilities (LDs) are a clinically and genetically heterogeneous group of diseases. Array-CGH and high-throughput sequencing have dramatically expanded the number of genes implicated in isolated intellectual disabilities and LDs, highlighting the implication of neuron-specific post-mitotic transcription factors and synaptic proteins as candidate genes. We report a unique family diagnosed with autosomal dominant learning disability and a 6p21 microdeletion segregating in three patients. The 870 kb microdeletion encompassed the brain-expressed gene LRFN2, which encodes for a synaptic cell adhesion molecule. Neuropsychological assessment identified selective working memory deficits, with borderline intellectual functioning. Further investigations identified a defect in executive function, and auditory-verbal processes. These data were consistent with brain MRI and FDG-PET functional brain imaging, which, when compared with controls, revealed abnormal brain volume and hypometabolism of gray matter structures implicated in working memory. We performed electron microscopy immunogold labeling demonstrating the localization of LRFN2 at synapses of cerebellar and hippocampal rat neurons, often associated with the NR1 subunit of N-methyl-D-aspartate receptors (NMDARs). Altogether, the combined approaches imply a role for LRFN2 in LD, specifically for working memory processes and executive function. In conclusion, the identification of familial cases of clinically homogeneous endophenotypes of LD might help in both the management of patients and genetic counseling for families.
Introduction
The genetic bases of learning disabilities (LDs) remain mostly unknown, and knowledge about fully penetrant causal genes responsible for isolated LDs in patients remains limited and needs fine phenotyping. Notably, intra-familial variable expressivity, the contribution of socio-environmental factors and the lack of large family series, together with non-routine cognitive investigations, limits accurate diagnosis. As previously suggested, chromosomal imbalances with a purely cognitive phenotype may provide clues towards candidate genes that can have a direct impact on cognition.1, 2 However, most of the genetic determinants of higher cognitive functions including learning, attention, memory and executive function remain unknown.3, 4
Working memory (WM) is a neural system that provides temporary active maintenance of necessary information while performing complex cognitive tasks such as reasoning, learning, understanding, thinking, decision making, and planning.5, 6, 7 Although selective WM deficit is a rare condition, many studies have shown that poor WM performance can have an impact on academic performance.8, 9, 10 Recent technological advances in genetics and WM sub-tests might help to identify and diagnose such disorders, allowing early identification of patients with selective WM, better-targeted interventions, and improved long-term outcomes.11 For example, a recent study demonstrated that training could enhance WM in children with Down syndrome.12 These findings thus support the importance of early recognition of WM difficulties.
Here, for the first time, we present a unique family with a non-syndromic specific cognitive phenotype mainly characterized by short-term memory and/or WM deficits, without intellectual disability (ID), carrying a familial deletion encompassing LRFN2 (Leucine-rich repeat and fibronectin III domain-containing 2).
Patients and methods
Patients
Patient 1 was the first child of young, non-consanguineous parents (Figure 1a). She was right-handed and 7-years-old at the time of examination. The pregnancy was normal, with a natural delivery at 38 weeks of gestation. Birth measurements were weight, 2680 g (25th percentile); length, 48.5 cm (25–50th percentile) and occipitofrontal circumference, 31 cm (<3rd percentile). Psychomotor acquisitions were within the normal range, with sitting occurring at the age of 9 months and walking at the age of 12 months. Patient 1 was referred because of speech delay, which interfered with her education. Speech was absent at 36 months, but subsequently improved with schooling. No behavioral problems were associated with the LDs. A speech evaluation was performed in her fifth year because of incomprehensible speech. No specific diagnosis could be made. A neuropediatric evaluation identified asymmetric spasticity with brisk patellar reflexes and a positive Babinsky sign. To explore this symptom, a brain MRI was performed and mild atrophy of the superior vermis was diagnosed. Other investigations included a normal standard chromosomal analysis, the absence of fragile-X, as well as normal creatine and guanidoacetate urinary assays, normal thyroid hormone and creatine kinase levels, and normal urinary organic acid and plasma amino-acid chromatography. A neuropsychological evaluation (WISC-IV) showed an IQ of 72 and identified a comprehension score in the low range, with a vocabulary level that was clearly below the normal level for a child of her age (verbal comprehension=79). Her perceptual reasoning was also impaired, with deficits in planning complex spatial actions (visual puzzles=4), deficits in spatial reasoning (matrix=6) and, finally, problems when asked to perceive visual details quickly (picture completion=6). On the other hand, her performance on processing speed tasks (digit symbol-coding=8, symbol search=9) was normal. Finally, the results on the WM tasks indicated clear deficits with performance in the very low range on both tasks (digit span=2, arithmetic=1). Speech evaluation pointed to very low, short-term memory skills, including low repetition scores in both logatomes (−3.09 SD) and sentences (−3.64 SD), as well as low recognition of her own errors (−4.73 SD). These results contrasted with appropriate lexical, comprehensive and syntaxic skills for the patient's age and normal attention levels. The clinical evaluation suggested atypical learning disability with non-specific speech delay.
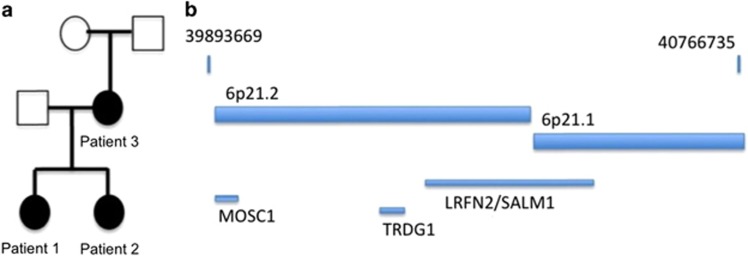
Figure 1.
(a) Family tree of the reported patients. Black boxes represent carriers of the learning disability and the genetic anomaly. (b) Representation of the 6p21 microdeletion with coordinates of the deletion. Three genes were included in the deletion.
Patient 2 (Patient 1's sister) was 5 years of age and right-handed at time of examination. She was born prematurely at 33 weeks of gestation because of a retro-placental hematoma. The immediate APGAR was zero, and was 10 at 5 and 10 min after initial reanimation. Birth weight was 2080 g. She spent 15 days in hospital with no further complications. Psychomotor acquisitions were at the limit of normal range, with sitting acquired at 9 months, crawling at 10 months, and walking at 18 months. Speech was delayed and was still absent when she started school. Mild behavioral troubles, including aggressiveness with frustration, were noted. Speech evaluation was performed and dysphasia was diagnosed. Speech therapy was started, and Patient 2 started standard schooling with a personal assistant. The clinical examination was normal and no facial dysmorphism was noticed. No etiological exam was performed. A neuropsychological evaluation was done when 5-years-old using the WPPSI-III. On the verbal tests, performance was very poor (information=1, vocabulary=1), contrasting with a better achievement in non-verbal tests (block design=12, matrix=12, picture completion=9). She could, however, name pictures, suggesting some intact linguistic abilities.
Patient 3, the mother of the children, was interviewed as a complementary examination following diagnosis. She was 29 years of age and right-handed. She was born at term. She did not mention delayed psychomotor acquisitions, although speech delay was a major concern during her childhood. She was oriented towards a school for special needs before the age of 10. She followed a vocational course to become a horticulturist, but never worked. The clinical examination was normal. Mild, non-specific facial dysmorphism was noticed with hypotelorism and deep-set eyes. A brain MRI was performed and showed superior vermis atrophy, associated with mild bilateral atrophy of cerebellar lobes 6 and 7, and bilateral parietal atrophy (Figure 2a). No hippocampal atrophy was found. The neuropsychological evaluation (WAIS-IV) revealed an IQ of 78, with heterogenous results in the sub-tests (Table 1). While perceptual organization was found to be normal, verbal comprehension was within the inferior normal range, mainly due to low performance on the similarities test. Processing speed and WM were deficient. Scores on all of the tests were in the extremely low range, showing significant impairment (Table 1). Long-term memory was explored using the RLRI test, a task similar in its construction to the California Verbal Learning Test. Overall, recall (as measured by the number of words correctly recalled) and recognition memory (as measured by the number of items correctly recognized) were above average (Table 1), thus showing intact episodic memory function, and therefore a preserved capacity to learn new information.
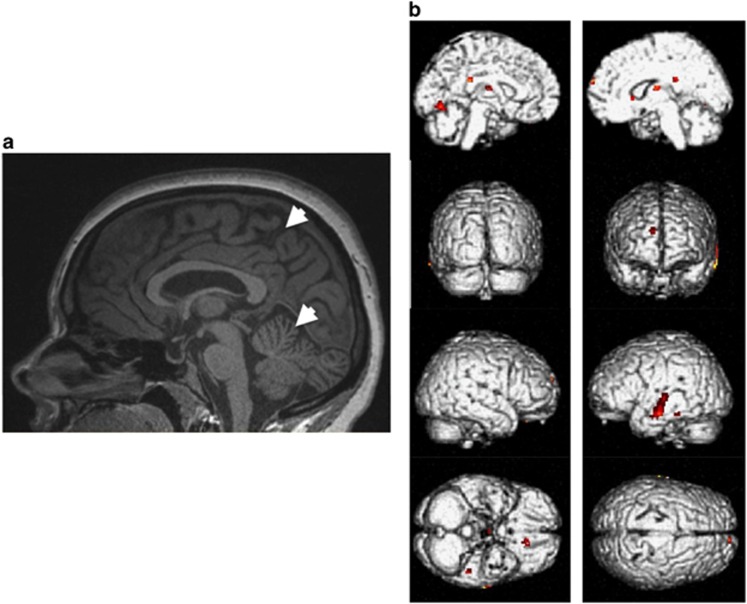
Figure 2.
(a) Brain anatomical imaging of Patient 3 at 29 years of age, showing mild atrophy of the superior vermis and enlargement of parietal sulcus. (b) Functional brain of Patient 3 imaging showing hypometabolism in six clusters: left temporal cortex, left orbito-frontal cortex, left lingual gyrus, right thalamus, right superior and medial frontal gyrus, and left and right posterior cingulate. Hypometabolism is represented by the yellow – red gradient (red for strongly hypometabolic). A full color version of this figure is available at European Journal of Human Genetics online.
Table 1. Neuropsychological Assessment of Patient 3.
| Score | Rank | |
|---|---|---|
| WAIS-IV | ||
| Verbal comprehension | 84 | 14 |
| Information | 8 | Average |
| Similarities | 6 | Low average |
| Vocabulary | 8 | Average |
| Working memory | 68 | 2 |
| Arithmetic | 5 | Low average |
| Digit span | 4 | Low average |
| Perceptual organization | 98 | Average |
| Block design | 10 | Average |
| Matrix | 11 | Average |
| Picture completion | 8 | Average |
| Processing speed | 72 | 3 |
| Digit symbol-coding | 4 | Low average |
| Symbol search | 6 | Low average |
| Long-term memory | ||
| Free recall trial 1 | 8 | Above average |
| Free recall trial 2 | 9 | Low average |
| Free recall trial 3 | 12 | Above average |
| Delayed recall | 12 | Above average |
| Cued-recall rial 1 | 16 | Above average |
| Cued-recall trial 2 | 16 | Above average |
| Cued-recall trial 3 | 16 | Above average |
| Recognition task | 16 | Above average |
| Short-term – working memory | ||
| WMS III-spatial span | 8/19 | Percentile 25 |
| WMS III-spatial span forward | 6/19 | Percentile 9 |
| WMS III-spatial span backward | 10/19 | Percentile 50 |
| TAP-working memory | 716 (324) | Percentile 54 |
| Attention | ||
| D2 test | <Percentile 5 | |
| Total letters identified | 263 | Percentile 25–50 |
| Omissions+errors | 14 (5.32%) | Percentile 5–10 |
| Total identified – errors | 103 | Percentile 34 |
| TAP-alertness | 0.011 | Percentile 24 |
| Intrinsic alertness | 266 (45) | Percentile 18 |
| Attentional focus | 271 (46) | Percentile 54 |
| TAP incompatibility | 0.475 | |
| Executive function | ||
| Trail-making test | ||
| Trail making-part A | 51s | <10th Percentile |
| Trail making-part B | 167s | <10th Percentile |
| Wisconsin card sorting test | ||
| Categories | 5 | >Percentile 16 |
| Total errors | 42 (33%) | Percentile 42 |
| Perseverative errors | 7 (5%) | Percentile 82 |
| Non perseverative errors | 29 (23%) | Percentile 14 |
| Failure to maintain set | 5 | <Percentile 1 |
Abbreviations: D2 test: concentration endurance test; TAP: test of attentional performance; WMS III: Wechsler memory scale-III.
Array-CGH
A diagnostic array comparative genomic hybridization (array-CGH) experiment was conducted to identify a chromosomal rearrangement in Patient 1. The platform used the Human Genome CGH Microarray 180 K from Agilent according to the manufacturer's protocol (Agilent Technologies, Santa Clara, CA, USA). Data were processed with feature extraction (v. 9.1) software and the results were analyzed with CGH analytics (v. 4.0) software (Agilent Technologies) in the Hg19 genome assembly. When a chromosomal rearrangement was identified, the anomaly was confirmed by an independent method (diagnosis was made using quantitative PCR). Array-CGH results were shared on the Decipher database (ID 314446).
WM assessment
Further testing of Patient 3's WM impairment involved standard neuropsychological tests and experimental tasks. WM was first assessed with neuropsychological tests including the Spatial and Digit Span of the WMS-III13 and the WM test of attentional performance (TAP).14 To explore the awareness of her own memory deficits, we asked Patient 3 to predict her memory performance on the standard digit span task of the WAIS-IV. For this purpose, after completing each trial at each sequence length, Patient 3 was asked if she could recall in order the number of items presented.15 Classic span experimental tasks were used. For example, the phonological span and the counting span task were conducted and compared with the literature norms.16, 17
To determine whether or not Patient 3 also presented attention impairment, the alertness and incompatibility subcomponents of the TAP were administered, as was the D2 attention test.18 Executive functioning was assessed using the trail-making test19 and the Wisconsin Card sorting test.20
Functional brain imaging
Functional imaging with (18F) fluoro-2-deoxy-D-glucose positron emission tomography (18FDG-PET/CT) was performed in Patient 3 only. Before undergoing the 18FDG-PET/CT, the patient had fasted for 6 h. After 10 min of rest and wearing an eye mask, she received an injection of 189.5 MBq of F18-FDG. Thirty minutes later, the scan was performed using a dedicated PET/CT system (GEMINI TOF, Philips Medical Systems, Eindhoven, The Netherlands) in the 3D mode. Emission data were corrected for dead time, random and scatter coincidences, and attenuation correction was performed using a low-dose CT scan. The images were first inspected visually and then, after intensity normalization, they were compared with those of 20 controls by a voxel-based method using Statistical Parametric Mapping (SPM8, http://www.fil.ion.ucl.ac.uk/spm).
Cellular experiments
For the electron microscope (EM) studies, DAB (3′,3-diaminobenzidine tetrahydrochloride)/immunoperoxidase-labeled parasagittal brain sections were prepared in several experiments from three animals, as described previously.21, 22, 23 Adult male Sprague-Dawley rats were anesthetized and perfused with paraformaldehyde in phosphate buffer. All animal experiments described in this paper were done according to the National Institutes of Health (NIH) guidelines for animal use (NIH Animal Protocol #1167-07). Fifty-micrometer sections were cryoprotected in 30% sucrose, frozen and stored at −80 °C. For each experiment, a set of sections was thawed, washed and incubated in 10% normal goat serum (NGS), then in primary antibody (in phosphate-buffered saline (PBS)) overnight, and processed with a Vectastain kit and 3′,3-diaminobenzidine tetrachloride (DAB; Vector Laboratories, Burlingame, CA, USA). The sections were washed, fixed in 2% glutaraldehyde, washed, fixed in 1% osmium tetroxide and washed again. Further processing, including alcohol dehydration and embedding in epon was done as described previously. Ultrathin sections were examined with EM. Phosphate-buffered saline control sections that lacked the affinity-purified primary antibodies were always unlabeled.
Electron microscopy of immunogold-labeled sections of the hippocampus and cerebellum from two animals was conducted using previous protocols21, 22, 23 Adult rats were anesthetized, as described above, then perfused and postfixed with 4% paraformaldehyde plus 0.5% glutaraldehyde. Parasagittal sections (300 μm) were cryoprotected and frozen in a Leica EM CPC (cryopreparation chamber; Vienna, Austria). The frozen sections were then immersed in uranyl acetate in methanol at −90 °C in a Leica AFS (automatic freeze-substitution instrument), infiltrated with Lowicryl HM-20 resin, and polymerized with ultraviolet light. The ultrathin sections were incubated in sodium borohydride plus glycine in Tris-buffered saline plus 0.1% Triton X-100 (TBST), followed by 10% NGS in TBST, primary antibody in 1% NGS in TBST overnight at 4 °C, and immunogold (10 nm F(ab')2 gold; Ted Pella, Redding, CA, USA) in 1% NGS in TBST, plus 0.5% polyethylene glycol (20 000 MW). Finally, the sections were stained with uranyl acetate and lead citrate. All labeling used LRFN2 antibodies24 or NR1 antibodies.23, 25 Images were processed in Adobe Photoshop with minimal use of levels, brightness, and contrast (applied evenly throughout the section).
Results
Array-CGH analysis and interpretation
Array-CGH was performed on Patient 1, and found an 876-kb chromosomal microdeletion at the 6p21 locus, mapped to Hg19 chr6:g.(39889216_39893669)_(40766735_40780054)del. No chromosomal rearrangement with a similar length was identified in public databases such as database of genomic variants (DGV) or Decipher (see URLs). Familial segregation showed that both sisters carried the 6p21 microdeletion, inherited from the mother. Familial segregation could not be studied further since the mother had no contact with her parents and other family members. Three genes were included in this deletion, namely TRDG1, LRFN2, and MOCS1 (Figure 1b). The TRDG1 gene is a testicular factor, with no identified brain expression (see URLs). The MOCS1 gene (OMIM *603707) is a gene implicated in severe autosomal recessive neonatal epileptic encephalopathy.
WM assessment (Patient 3)
Tasks assessing short-term and WM were mostly deficient (Table 1). On the WAIS-IV, Patient 3 achieved a digit span forward of 3 and backward of 4, suggesting deficiencies in storing and manipulating verbal information. On the WMS-III, she achieved spatial spans of 4 (forward) and 6 (backward). These results suggest that her spatial WM was less impaired than her verbal WM. To further explore Patient 3's short-term memory and WM, two experimental tasks (phonological and complex span tasks) were administrated and the results were compared with findings in the scientific literature on healthy populations. On the phonological span task, Patient 3 successfully recalled, at best, a sequence of two consecutive pseudo-words (score was less than 1.5 SD below the mean). Patient 3's WM was also measured using a counting span task. During the complex span task, she recalled two series of length two, and one series of length three, giving her a span score of 7 (12.8 SDs lower than the mean (25.7)).17 These results thus confirmed Patient 3's low span performance. Tests on the awareness of memory showed accordance between predictions and performance (predicted 4 and recalled 4), thus showing that Patient 3 was aware of her low span. Performance on the D2 attention test revealed mainly low concentration performance (Table 1). Sub-tests of the TAP showed a relatively intact attentional focus for simple tasks, while the results of the TAP incompatibility and the WCST did not suggest severe inhibition deficits. The performance on the trail-making test showed slow processing speed, probably associated with shifting difficulties.
Functional brain imaging
Visual analysis of the 18FDG-PET/CT images revealed hypometabolism of the left temporal region (Figure 2b). Images from Patient 3 were compared with those of controls using the SPM statistical model with a threshold masking value of P=0.001, intensity peak=3.57 and cluster size=19, using the t-test in the SPM package. As a result, we confirmed the existence of hypometabolism in (i) the left temporal cortex (peak intensity=6.38 and number of voxels=124 for the middle temporal gyrus, and peak intensity=4.11 and number of voxels=20 for the inferior temporal gyrus), (ii) the left orbito-frontal cortex (peak intensity=7.78; number of voxels=31), (iii) the left lingual gyrus (peak intensity=3.93 and number of voxels=61), (iv) the right thalamus (peak intensity=3.93 and number of voxels=45), (v) the right superior and medial frontal gyrus (peak intensity=4.95 and number of voxels=19), and (vi) the left and right posterior cingulate (peak intensity=4.04 and number of voxels=35).
Synaptic localization of LRFN2
To determine the localization of LRFN2 (also known as synaptic adhesion-like molecule 1) in the brain, immunoperoxidase and immunogold labeling were used. Light microscope examination of hippocampus, and both the cerebral and cerebellar cortices, using EM/DAB immunoperoxidase labeling of sections revealed LRFN2 in various neuron populations in these regions (data not shown). We then examined two of these regions, hippocampus and cerebellum, in greater detail using EM; both regions have key roles in different aspects of learning and memory, particularly in relation to the function of N-methyl-D-aspartate (NMDA)-type glutamate receptors.22, 26 Labeling of synapses in the hippocampus CA1 stratum radiatum with EM/DAB immunoperoxidase and immunogold was localized to both the postsynaptic and presynaptic sides (Figure 3). Immunogold distribution was prominent in the postsynaptic membrane/density and the presynaptic membrane, as well as in perisynaptic and extrasynaptic locations on the postsynaptic and presynaptic sides. In addition, labeling for LRFN2 co-localized at the synapse with labeling for NR1. Distinct labeling of the large mossy terminal synapses in the cerebellar granular layer was not common with EM/DAB immunoperoxidase, but was prominent with immunogold, and the distribution (Figure 4) was similar to that seen in the hippocampus. In addition, the granule cell dendrites at mossy terminal synapses have attachment plaques (puncta adherentia) that contain NMDARs and PSD-95 scaffolding proteins,25 and LRFN2 was also localized to these (Figure 4).
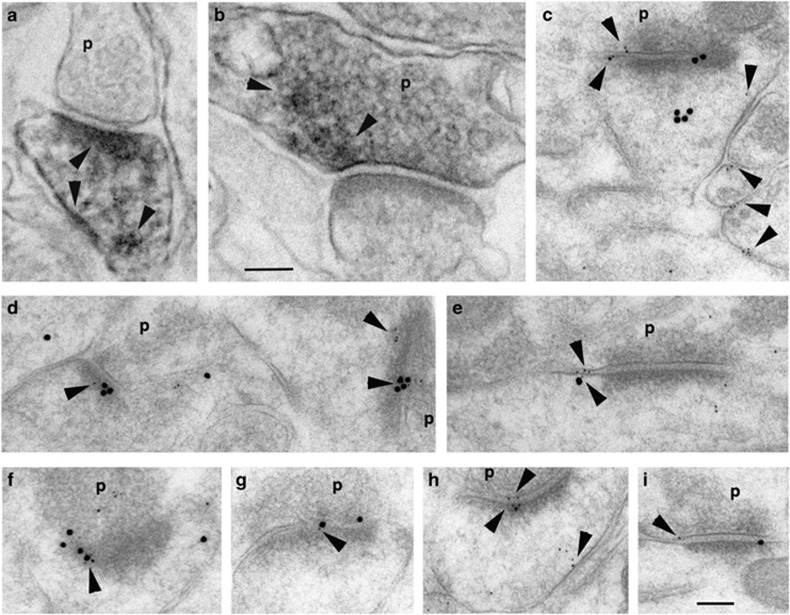
Figure 3.
Localization of LRFN2 at synapses in the hippocampus CA1 stratum radiatum with EM/DAB immunoperoxidase (a, b) or immunogold (c–i). DAB labeling is found in patches throughout the postsynaptic spine (a) and in the presynaptic terminals also (b; arrowheads). Immunogold labeling for LRFN2 (5 nm gold; arrowheads) co-localizes with labeling for the NMDA receptor subunit, NR1 (15 nm gold) in the postsynaptic membrane/density (d, f, g; f is very oblique). Extrasynaptic labeling for LRFN2 also is found (c, e, h, i; co-localized with NR1 in e). p, presynaptic terminal. Scale bar is 100 nm each for EM/DAB and immunogold.
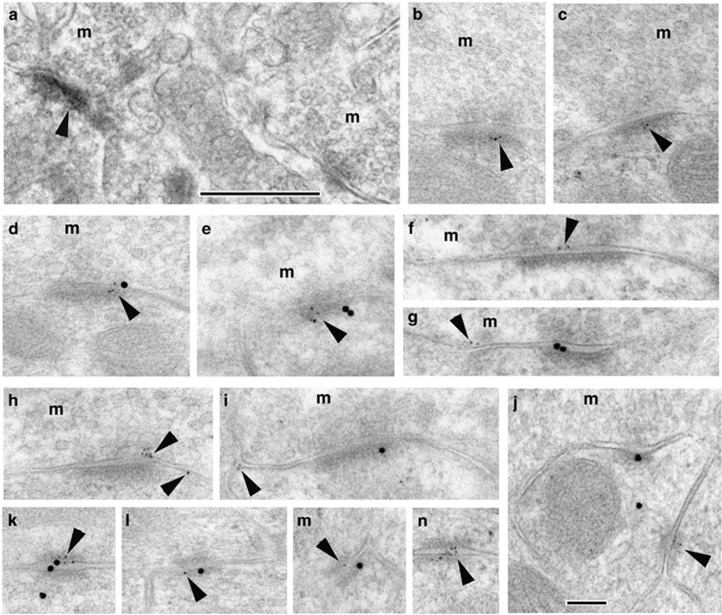
Figure 4.
Localization of LRFN2 at synapses in the cerebellar granular region with EM/DAB immunoperoxidase (a) or immunogold (b–n). DAB labeling is generally weak, but can be found in the postsynaptic density (arrowhead; compare with an unlabeled synapse in the lower right corner). Immunogold labeling for LRFN2 (5 nm gold; arrowheads) is found in the postsynaptic membrane/density (b, c) where it co-localizes with labeling for the NMDA receptor subunit, NR1 (15 nm gold; d, e). Labeling for LRFN2 also is seen on the presynaptic membrane or in extrasynaptic membrane locations (f–i). In addition, labeling for LRFN2 is found in attachment plaques (j–n) where it can be seen to co-localize with NR1 in some examples (k–m). m, mossy fiber terminal. Scale bar is 500 nm for the EM/DAB and 100 nm for immunogold.
Discussion
We report the first family with a chromosomal microrearrangement encompassing LRFN2. We argue that heterozygous LRFN2 deletion likely contributes to the learning disability seen in the family members because: (i) LRFN2 co-localizes with NR1 in the postsynaptic region; and (ii) dysfunction of NMDARs have been shown to alter excitatory synapse functioning and WM processes.27, 28, 29
To date, two cases with deletions encompassing LRFN2 are available in public databases (ClinGen or Decipher (295383)). Their deletions were larger than that reported here (respectively, 7.87 and 3.19 Mb) and included numerous genes in the vicinity of LRFN2. Although they occurred de novo and were considered as causing disease, these variations may not be comparable to the one reported here. Two small deletions were identified in healthy individuals from large CNV-discovery studies referenced in the DGV (essv7032076 and nssv8240). Similarly, 6 loss-of-function alleles (namely 3 nonsense and 3 frameshift) are reported over 121 368 alleles sequenced in ExAC. Overall, the rareness of LRFN2 loss-of-function events suggests dosage sensitivity.
The patients reported here presented an endophenotype of learning disability, predominantly characterized by deficits on a series of tasks, which seem to cluster around WM, attention and executive function, especially in language. While these patients, on the basis of their IQ score, should not be described as presenting with ID, it is worth noting that they are all in the range considered ‘borderline intellectual functioning'. Brain imaging reinforced the neuropsychological findings. Functional imaging in Patient 3 revealed specific hypometabolism of the left pre-frontal and temporal regions, which are known to be involved in WM processes.30 This result, together with the co-localization of LFRN2 with the NMDAR, suggests that LFRN2 can be implicated in learning disability with selective WM deficit.
The delineation of cognitive disorders has been improved through the generalization of neuropsychological assessment of patients. Specific sub-tests investigating WM processes were introduced in 2006.9 To the best of our knowledge, there have been no previous reports of congenital and isolated WM deficit without more severe ID or psychiatric traits. In the literature, WM performance is described as a highly heritable trait, although the underlying molecular factors remain to be determined.31, 32 More generally, the molecular basis of non-syndromic learning disability is still largely unknown. The clinical heterogeneity of such conditions has limited the application of classical genetic studies. The identification of de novo or familial CNV segregating with the phenotype, combined with fine phenotyping and the emergence of clinical learning disorder endophenotypes, will provide a better understanding of LDs. This strategy has facilitated the identification of genes or chromosomal loci implicated in other learning disorders, such as developmental verbal dyspraxia.33, 34 Executive function or WM deficits have been reported in genetic neurodevelopmental disorders associated with ID and physical abnormalities, like Down's syndrome or Williams syndrome.35, 36 Various approaches, including linkage analysis and association studies, have indicated various candidate genes, such as ROBO1 or CHD13, associated with low WM performances as a specific endophenotype of a cognitive dysfunction.37, 38 Remediation of these types of LDs can often be more effectively achieved when the precise cognitive dysfunction has been identified.35
Currently, the functional roles of genes that have been identified in cognitive disorders can be separated into two main functional groups39: (i) neuronal differentiation and specification; and (ii) synaptic scaffolding and functioning. The latter group is involved in a broad spectrum of neurodevelopmental disorders. Hypotheses favor the implication of the scaffolding proteins of the postsynaptic region in higher cognitive processes.40 The leucine-rich repeat (LRR) superfamily of proteins is an example of accelerated evolution involving genomic duplication, with 139 LRR proteins in humans compared with 66 in flies. This observation suggests that these proteins have a critical role in human-specific, higher order cognitive processes.41 More precisely, several LRR genes were diagnosed as deleted or mutated in patients with WM and executive function disorders associated with neurological disorders such as Parkinson's disease (LRRK2),42 and with neuropsychiatric phenotypes (LRFN5, LRRTMs, and LRRNs).43, 44, 45, 46 Moreover, null mice for different LRR proteins interacting with the PSD complex showed LDs associated with behavioral abnormalities.47, 48, 49, 50 Among LRR proteins, LRFNs form a family of transmembrane cell adhesion molecules that promote changes in neuronal morphology, dendritic outgrowth and synapse formation.51, 52, 53 It was further demonstrated that LRFNs form complexes with NMDARs, and promote the specialization of excitatory synapses.24, 52, 53 In particular, the N-terminal domains of LFRN2 and NR1 interact directly, and this interaction regulates the surface expression of NMDARs in hippocampal neurons.24 LRFN2 also associates with scaffolding proteins, such as the PSD-95 family members, through an intracellular PDZ-binding motif.24, 54 Together, these interactions may regulate NMDAR trafficking and surface expression, thereby influencing WM and cognitive functions. In the pre-frontal cortex, which is highly implicated in WM processes, NMDA glutamate receptors are key actors for cellular memory mechanisms such as long-term potentiation. Expression levels of the NR1 subunit, as well as the presence of functional NMDARs, have been shown to be critical for glutamate signaling and WM.27, 28, 29 Dystrobrevin binding protein-1 (dysbindin or DTNBP1) null mice display deficits in WM performance, secondary to NR1 dysfunction.27 Knockdown of NR1 expression in mice28 and blockade of NMDARs in patients29 also cause WM deficits. Furthermore, the cerebellum is involved in WM and is implicated in language functions.55 Therefore, in addition to the hippocampus, the glutamatergic connections in both the pre-frontal cortex and cerebellum are important for different aspects of learning and memory, and dysfunction may contribute to LDs, as well as previously reported cognitive deficits such as ID and autism.55, 56
To summarize, we present a family that harbors a unique microdeletion, notably encompassing LRFN2, which is mainly characterized by LDs and a WM deficit. Thus, neuropsychological evaluation that included experimental WM tasks, as well as brain anatomical and functional imaging together with cellular experiments, has emphasized the role of LRFN2 in learning disability. This study highlights the power of multidisciplinary, fine phenotyping in describing new genetic disorders.
Acknowledgments
We thank the family for their participation in this study. We also thank the regional council of Burgundy for the financial support of the project. GKS, YW, and RSP were supported by the NIDCD/NIH Intramural Research Program.
The authors declare no conflict of interest.
References
- Flint J: The genetic basis of cognition. Brain J Neurol 1999; 122: 2015–2032. [DOI] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP: A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 2001; 413: 519–523. [DOI] [PubMed] [Google Scholar]
- Graham SA, Fisher SE: Decoding the genetics of speech and language. Curr Opin Neurobiol 2013; 23: 43–51. [DOI] [PubMed] [Google Scholar]
- Newbury DF, Fisher SE, Monaco AP: Recent advances in the genetics of language impairment. Genome Med 2010; 2: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddeley AD, Hitch GJ: Development of working memory: should the Pascual-Leone and the Baddeley and Hitch models be merged? J Exp Child Psychol 2000; 77: 128–137. [DOI] [PubMed] [Google Scholar]
- Baddeley A: The concept of working memory: a view of its current state and probable future development. Cognition 1981; 10: 17–23. [DOI] [PubMed] [Google Scholar]
- Khan ZU, Muly EC: Molecular mechanisms of working memory. Behav Brain Res 2011; 219: 329–341. [DOI] [PubMed] [Google Scholar]
- Holmes J, Gathercole SE, Dunning DL: Poor working memory: impact and interventions. Adv Child Dev Behav 2010; 39: 1–43. [DOI] [PubMed] [Google Scholar]
- Gathercole SE, Alloway TP: Practitioner review: short-term and working memory impairments in neurodevelopmental disorders: diagnosis and remedial support. J Child Psychol Psychiatry 2006; 47: 4–15. [DOI] [PubMed] [Google Scholar]
- Gathercole SE, Alloway TP, Willis C, Adams A-M: Working memory in children with reading disabilities. J Exp Child Psychol 2006; 93: 265–281. [DOI] [PubMed] [Google Scholar]
- Kaufman AS, Lichtenberger EO: Assessing Adolescent and Adult Intelligence. John Wiley & Sons, 2005. [Google Scholar]
- Bennett SJ, Holmes J, Buckley S: Computerized memory training leads to sustained improvement in visuospatial short-term memory skills in children with Down syndrome. Am J Intellect Dev Disabil 2013; 118: 179–192. [DOI] [PubMed] [Google Scholar]
- Wechsler D: Wechsler memory scale (WMS-III). Psychological Corporation, 1997. [Google Scholar]
- Zimmermann P, Fimm B: Test for attentional performance (TAP). PsyTest Herzogenrath, 1995. [Google Scholar]
- Murphy MD, Schmitt FA, Caruso MJ, Sanders RE: Metamemory in older adults: the role of monitoring in serial recall. Psychol Aging 1987; 2: 331–339. [DOI] [PubMed] [Google Scholar]
- Case R, Kurland DM, Goldberg J: Operational efficiency and the growth of short-term memory span. J Exp Child Psychol 1982; 33: 386–404. [Google Scholar]
- Conway AR, Cowan N, Bunting MF, Therriault DJ, Minkoff SR: A latent variable analysis of working memory capacity, short-term memory capacity, processing speed, and general fluid intelligence. Intelligence 2002; 30: 163–183. [Google Scholar]
- Brickenkamp R: Test d2. Concentration-Endurence Test: Manual, 5th edn. Gottingen, Germany: Verlag für Psychologie, 1981. [Google Scholar]
- Reitan RM: Trail Making Test: Manual for administration and scoring. Reitan Neuropsychology Laboratory, 1986. [Google Scholar]
- Heaton RK: A manual for the Wisconsin card sorting test. Western Psycological Services, 1981. [Google Scholar]
- Petralia RS, Wenthold RJ: Immunocytochemistry of NMDA receptors. Methods Mol Biol 1999; 128: 73–92. [DOI] [PubMed] [Google Scholar]
- Petralia RS: Distribution of extrasynaptic NMDA receptors on neurons. ScientificWorldJournal 2012; 2012: 267120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Hua F et al: Organization of NMDA receptors at extrasynaptic locations. Neuroscience 2010; 167: 68–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Chang K, Petralia RS, Wang YX, Seabold GK, Wenthold RJ: A novel family of adhesion-like molecules that interacts with the NMDA receptor. J Neurosci 2006; 26: 2174–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wang Y-X, Wenthold RJ: NMDA receptors and PSD-95 are found in attachment plaques in cerebellar granular layer glomeruli. Eur J Neurosci 2002; 15: 583–587. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ et al: Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 2010; 62: 405–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsgodt KH, Robleto K, Trantham-Davidson H et al: Reduced dysbindin expression mediates N-methyl-D-aspartate receptor hypofunction and impaired working memory performance. Biol Psychiatry 2011; 69: 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic M, Mielnik CA, Ramsey AJ: NMDA receptor-deficient mice display sexual dimorphism in the onset and severity of behavioural abnormalities. Genes Brain Behav 2014; 13: 850–862. [DOI] [PubMed] [Google Scholar]
- Driesen NR, McCarthy G, Bhagwagar Z et al: The impact of NMDA receptor blockade on human working memory-related prefrontal function and connectivity. Neuropsychopharmacology 2013; 38: 2613–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collette F, Hogge M, Salmon E, Van der Linden M: Exploration of the neural substrates of executive functioning by functional neuroimaging. Neuroscience 2006; 139: 209–221. [DOI] [PubMed] [Google Scholar]
- Ando J, Ono Y, Wright MJ: Genetic structure of spatial and verbal working memory. Behav Genet 2001; 31: 615–624. [DOI] [PubMed] [Google Scholar]
- Chen L-S, Rice TK, Thompson PA, Barch DM, Csernansky JG: Familial aggregation of clinical and neurocognitive features in sibling pairs with and without schizophrenia. Schizophr Res 2009; 111: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Drayna D: Genetics of speech and language disorders. Annu Rev Genom Hum Genet 2011; 12: 145–164. [DOI] [PubMed] [Google Scholar]
- Thevenon J, Callier P, Andrieux J et al: 12p13.33 microdeletion including ELKS/ERC1, a new locus associated with childhood apraxia of speech. Eur J Hum Genet 2013; 21: 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddeley A, Jarrold C: Working memory and Down syndrome. J Intellect Disabil Res 2007; 51: 925–931. [DOI] [PubMed] [Google Scholar]
- Carney DPJ, Brown JH, Henry LA: Executive function in Williams and Down syndromes. Res Dev Disabil 2013; 34: 46–55. [DOI] [PubMed] [Google Scholar]
- Arias-Vásquez A, Altink ME, Rommelse NNJ et al: CDH13 is associated with working memory performance in attention deficit/hyperactivity disorder. Genes Brain Behav 2011; 10: 844–851. [DOI] [PubMed] [Google Scholar]
- Bates TC, Luciano M, Medland SE, Montgomery GW, Wright MJ, Martin NG: Genetic variance in a component of the language acquisition device: ROBO1 polymorphisms associated with phonological buffer deficits. Behav Genet 2011; 41: 50–57. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Ayub M, Vincent JB: The genetic basis of non-syndromic intellectual disability: a review. J Neurodev Disord 2010; 2: 182–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithianantharajah J, Komiyama NH, McKechanie A et al: Synaptic scaffold evolution generated components of vertebrate cognitive complexity. Nat Neurosci 2013; 16: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan J, Walshe K, Alsbury S et al: The extracellular leucine-rich repeat superfamily; a comparative survey and analysis of evolutionary relationships and expression patterns. BMC Genom 2007; 8: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler A, Mirelman A, Gurevich T et al: LRRK2 Ashkenazi Jewish Consortium. Lower cognitive performance in healthy G2019S LRRK2 mutation carriers. Neurology 2012; 79: 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruijn DRH, van Dijk AHA, Pfundt R et al: Severe progressive autism associated with two de novo changes: a 2.6-Mb 2q31.1 deletion and a balanced t(14;21)(q21.1;p11.2) translocation with long-range epigenetic silencing of LRFN5 expression. Mol Syndr 2010; 1: 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhail FM, Lose EJ, Robin NH et al: Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet A 2011; 155A: 2386–2396. [DOI] [PubMed] [Google Scholar]
- Kleffmann W, Zink AM, Lee JA et al: 5q31 Microdeletions: definition of a critical region and analysis of LRRTM2, a candidate gene for intellectual disability. Mol Syndr 2012; 3: 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa I, Clark TG, Holt R et al: International Molecular Genetic Study of Autism Consortium (IMGSAC). Polymorphisms in leucine-rich repeat genes are associated with autism spectrum disorder susceptibility in populations of European ancestry. Mol Autism 2010; 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle HJ, Luong TN, Medina-Marino A et al: Deletion of densin-180 results in abnormal behaviors associated with mental illness and reduces mGluR5 and DISC1 in the postsynaptic density fraction. J Neurosci Off J. Soc Neurosci 2011; 31: 16194–16207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama K, Yamada K, Ornthanalai VG et al: Slitrk1-deficient mice display elevated anxiety-like behavior and noradrenergic abnormalities. Mol Psychiatry 2010; 15: 177–184. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Katayama K-I, Sohya K et al: Selective control of inhibitory synapse development by Slitrk3-PTPδ trans-synaptic interaction. Nat Neurosci 2012; 15: 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima N, Odaka YS, Sakoori K et al: Impaired cognitive function and altered hippocampal synapse morphology in mice lacking Lrrtm1, a gene associated with schizophrenia. PLoS One 2011; 6: e22716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabold GK, Wang PY, Petralia RS et al: Dileucine and PDZ-binding motifs mediate synaptic adhesion-like molecule 1 (SALM1) trafficking in hippocampal neurons. J Biol Chem 2012; 287: 4470–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Kim S, Chung HS et al: SALM synaptic cell adhesion-like molecules regulate the differentiation of excitatory synapses. Neuron 2006; 50: 233–245. [DOI] [PubMed] [Google Scholar]
- Mah W, Ko J, Nam J, Han K, Chung WS, Kim E: Selected SALM (synaptic adhesion-like molecule) family proteins regulate synapse formation. J Neurosci 2010; 30: 5559–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimura N, Inoue T, Katayama K, Aruga J: Comparative analysis of structure, expression and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Gene 2006; 380: 72–83. [DOI] [PubMed] [Google Scholar]
- Wang SS, Kloth AD, Badura A: The cerebellum, sensitive periods, and autism. Neuron 2014; 83: 518–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P et al: Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 2011; 474: 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]