Introduction
History
The story of microRNAs (miRNA) begins with the decade long investigation by the Ruvkun laboratory into the regulation of lin-14 expression during nematode (C.elegans) development. In a series of studies that spanned the `90s, Ruvkun and colleagues provided evidence demonstrating that lin-14 expression was regulated by a post-transcriptional mechanism requiring the interaction between the lin-14 3′-UTR and the non-coding RNA lin-4 [1–6]. In late 2000, Pasquinelli and colleagues reported that another “small temporal RNA” (~ 21 nucleotides) from C. elegans, let-7, down-regulated lin-41 expression, and unlike lin-4, was detected in a broad range of bilaterian animals including vertebrate, ascidian, hemichordate, mollusc, annelid and arthropod [7]. The finding that let-7 was phylogenetically conserved was a milestone in the history of the field as it strongly suggested the post-transcriptional regulation of gene expression by small RNAs was more wide-spread and not restricted solely to C. elegans. The following year, a series of papers were published describing the identification of 30–50 new miRNAs in the fly, worm and human, providing support for the notion that miRNAs have a prevalent role in the regulation of gene expression in animals [8–11]. As confirmation of this early prediction, the latest release of miRBase (version 19, Aug. 2012; www.mirbase.org) contains 25,141 mature miRNA sequences from 193 species with 2041 and 1281 human and mouse miRNAs, respectively.
Biogenesis
miRNAs are initially transcribed as a primary transcript (pri-miRNA) with the characteristic 5′m7G cap structure and 3′ poly(A) tail of RNA polymerase II transcripts [12, 13]. Genomic mapping has shown that pri-miRNAs are often derived from the intron of either protein-coding or noncoding RNA (ncRNA) transcripts and less frequently from the exon of ncRNAs [14]. Within the nucleus, the pri-miRNA forms a stem-loop structure that is recognized by the microprocessor complex which contains two core components, the RNase III endonuclease Drosha and the double-stranded RNA binding protein DGCR8 (Di George Syndrome critical region gene 8) [15, 16]. DGCR8 binds to the stem-loop structure and then guides Drosha into position, cleaving ~11 base pairs (bp) from the base of the stem-loop to produce a 70–100 bp hairpin RNA molecule designated the precursor miRNA (pre-miRNA) [17]. The pre-miRNA is subsequently transported from the nucleus by Exportin 5, a nuclear export receptor, to the cytoplasm [18, 19]. Once in the cytoplasm, a second RNase III endonuclease, Dicer, cleaves the pre-miRNA to produce ~22 nucleotide double-stranded RNA molecule in which one strand, the mature miRNA, is transferred to the RNA-induced silencing complex (RISC) containing Dicer, a member of the Argonuate protein family and the RNA binding protein Tarbp2 (TAR (HIV) RNA binding protein 2); the other strand is typically targeted for degradation [20]. The mature miRNA directs RISC to 3′-UTR of target mRNA through complementary binding of the miRNA seed sequence which results in inhibition of translation and/or degradation of the target transcript [21].
Tissue-specific expression
With the identification of new miRNAs came the realization that some miRNAs were not ubiquitously expressed as let-7, but instead appeared to have a restricted pattern of expression. For example, the mature form of miRNA-1 (miR-1) was found to be expressed only in the human heart but not in the brain, kidney, liver, lung or HeLa cells [8, 10]. The concept that some miRNAs were expressed in a tissue-specific fashion was confirmed in a study by Lagos-Quintana and coworkers (2002) showing that miR-1, -122a and -124a expression was restricted to striated muscle, liver and brain, respectively [11]. In a screen for new miRNAs, Sempere et al. (2004) identified 30 miRNAs that were enriched or specifically expressed within a particular tissue [22]. This study also provided the first description of striated muscle-specific miR-1, -133a and -206, which were later designated as myomiRs [22, 23]. The myomiR family has expanded to include miR-208a, miR-208b, miR-499 and, most recently, miR-486 (see Table 1) [24–26]. Northern blot analyses showed that these new members of the myomiR family are strictly striated muscle-specific (miR-208a, miR-208b and miR-499), being derived from the intron of different muscle-specific myosin heavy chain genes, or highly enriched in muscle (miR-486) [24, 25]. Most myomiR family members are expressed in both the heart and skeletal muscle except for miR-208a, which is cardiac-specific, and miR-206, which is skeletal muscle-specific and enriched in slow-twitch muscles such as the soleus (see Table 1) [27].
Table 1.
Members of the MyomiR family of muscle-specific microRNAs.
| myomiR(mouse) | Expression pattern | Host gene | Knockout phenotype | Reference |
|---|---|---|---|---|
| miR-1a-1 | heart, skeletal muscle | intergenic | no knockout | ▬ |
| miR-1a-2 | heart, skeletal muscle | intergenic | 50% lethal, cardiac defect | 36 |
| miR-133a-1 | heart, skeletal muscle | intergenic | no overt phenotype | 41 |
| miR-133a-2 | heart, skeletal muscle | intergenic | no overt phenotype | 41 |
| miR-133a dKO | septal defect, myopathy | 41, 42 | ||
| miR-133b | skeletal muscle, nerve | intergenic | no overt phenotype | 37 |
| miR-206 | skeletal muscle (type I) | intergenic | no overt phenotype | 35 |
| miR-208a | heart | Myh6 | blunted stress response | 26 |
| conduction defects | 33 | |||
| miR-208b | heart, skeletal muscle (type I) | Myh7 | no overt phenotype | 25 |
| miR-486 | heart, skeletal muscle | Ank1 | no knockout | ▬ |
| miR-499 | heart, skeletal muscle (type I) | Myh7b/14 | no overt phenotype | 25 |
Note. dKO, double knockout of miR-133a-1 and miR-133a-2.
MyomiRs
From the time of their original description, much effort has gone into determining the function of myomiRs in striated muscle (reviewed in Liu and Olson, 2010) [28]. The finding that over-expression of miR-1 caused a shift toward a myogenic profile in HeLa cells suggested myomiRs may have a fundamental role in promoting a muscle cell identity [29]. The importance of myomiRs in myogenesis was first demonstrated by the finding of Sokol and Ambros (2005) who reported the deletion of miR-1 in the fly resulted in premature death from a failure of skeletal muscle to properly grow during early development [30]. Table 1 lists the known myomiRs, their host gene, expression pattern and whether or not they have been knocked out in the mouse. Interestingly, myomiRs appear to have either uniform expression throughout the muscle (miR-1 and miR-133a), independent of fiber-type, or are enriched in slow-twitch, type I muscles (miR-206, miR-208b and miR-499) [25, 27]; to date, no myomiR has been reported to be enriched specifically in fast-twitch, type II muscle. However, RNA-seq analysis of porcine and bovine skeletal muscle has identified non-myomiRs that are relatively enriched in a muscle of a particular fiber type [31, 32]. For instance, Muroya and colleagues reported that in steers miR-885 and miR-196a were highly enriched in the fast-twitch semitendinosus muscle relative to the slow-twitch masseter muscle [32].
As detailed in Table 1, most of the myomiRs have been deleted in the mouse with surprisingly little impact on skeletal muscle phenotype [25, 26, 33–37]. For example, deletion of skeletal muscle-specific miR-206 resulted in no obvious phenotype as reflected by no change in soleus muscle weight, morphology or fiber-type distribution; however, recovery from denervation was delayed in muscle of the miR-206 knockout [35]. In a similar manner, the Olson laboratory showed that miR-208a was necessary for the stress response involved in cardiac hypertrophy [26]. Collectively, these findings are consistent with the idea that a primary function of miRNAs is to mediate the stress response of the cell by helping to restore homeostasis through regulating gene expression [38]. It is not exactly clear why these knockout mice do not show a more dramatic phenotype as would be predicted from in vitro studies [39, 40]. One possibility is the overlap in gene targets between myomiR family members, such as miR-1 and miR-206, which could rescue any deleterious phenotype resulting from the miRNA knockout. This idea is supported by the double knockout of miR-133a-1 and miR-133a-2 in which mice showed septal defects and skeletal muscle myopathy that was not present in the single miR-133a knockout mice [41, 42].
The purpose of this review is to present what is currently known about the role of myomiRs, as well as other miRNAs, in skeletal muscle in response to both resistance and endurance exercise following either an acute bout or with training. The review will also cover the function of miRNAs in satellite cells, muscle injury and repair, aging and disease.
MicroRNAs in exercise
It is well established that exercise of sufficient intensity will place mechanical and/or metabolic stress on the contracting muscle. Accordingly, it is reasonable to speculate a priori that miRNAs will have a role in the stress response of skeletal muscle to changes in contractile activity. Defining the role of miRNAs in skeletal muscle plasticity is still in its infancy such that many of the exercise studies provide data that is purely correlative in nature. For the field to continue moving forward, future studies will need to provide more mechanistic data through the validation of punitive miRNA target genes and incorporate such genes into a regulatory network.
Exercise has repeatedly been shown to be a potent activator of gene expression, with expression patterns varying considerably depending on the mode of exercise [43]. Furthermore, exercise, particularly resistance exercise, results in the activation of various signaling cascades which in turn promotes an increase in protein synthesis [44–46]. Some of these alterations in gene expression may be attributable to changes in miRNA levels, as recent investigations have demonstrated that exercise induces changes in miRNA levels within multiple cell types [47–49]. These fluctuations in miRNA levels may augment training adaptations by regulating specific genes involved in those adaptations (see Table 2).
Table 2.
The effect of exercise on microRNA expression.
| Mode of Exercise | Species | microRNA expression | Target gene | Outcome | Ref |
|---|---|---|---|---|---|
| Resistance Exercise | |||||
| Functional overload (c) | m | ↓miR-1,-133a | IGF-1, IGF-1R | hypertrophy | 27 |
| Traditional RE (a) | h | ↓miR-1 | IGF-1, IGF-1R | hypertrophy | 50 |
| Traditional RE (c) | h | ↓miR-378 ↑miR-451 | growth pathways | low response to RE | 48 |
| Endurance Exercise | |||||
| Running (a) | m | ↓miR-23 ↑miR-1, -181, -107 | PGC-1α* | mito. biogenesis | 59 |
| Running (c) | m | ↓miR-696 | PGC-1α* | mito. biogenesis | 60 |
| Cycling (a) | h | ↑miR-1, -133 | MAPK & TGF-β pathways | modulate response to training | 62 |
| Cycling (c) | h | ↓miR-1, -133a, -133b, -206 | |||
| Cycling (c) | h | ↓miR-1, -101, -133, -455 | RUNX*, SOX9*, PAX3* | angiogenesis | 63 |
| Swimming (a) | m | ↓miR-494 | mtTFA, FOXJ3 | mito. biogenesis | 64 |
| Swimming (c) | r | ↓miR-1, -133, -133b ↑miR-29c | COL1A1, COL3A1 | incr. compliance | 49 |
| Swimming (c) | r | ↑miR-129 | SPRED-1, PI3KR2 | angiogenesis | 67 |
| Swimming (c) | r | ↓miR-143 ↑miR-27a, -27b | ACE, ACE2 | hypertrophy | 65 |
| Swimming (c) | r | ↓miR-16 | VEGF, VEGFR2 | angiogenesis | 68 |
Note: a, acute; c, chronic; m, mouse; h, human; r, rat; mito, mitochondrial; incr, increase;
predicted target.
Resistance Exercise and Skeletal Muscle miRNA Expression
Resistance exercise places a mechanical strain on the muscle resulting in a number of different adaptations, the most notable being a hypertrophy of the muscle; this hypertrophic growth occurs primarily through the accretion of contractile proteins thus increasing the capacity to generate greater force. A commonly used animal model of muscle hypertrophy is the synergist ablation model. This model involves the surgical removal of two primary synergist muscles thereby placing a mechanical overload on the remaining plantaris muscle and inducing a robust hypertrophy of the muscle. Following seven days of synergist ablation, expression of miR-1 and miR-133a were down-regulated by approximately 50% in the mouse plantaris muscle [27]. In humans, resistance exercise has been shown to modulate the expression of various miRNAs. In young men, an acute bout of resistance exercise coupled with ingestion of amino acids resulted in a significant decrease in miR-1 expression in skeletal muscle, in agreement with the loss of miR-1 expression in response to a hypertrophic stimulus observed in the mouse [27, 50]. This decrease in miR-1 expression following exercise may result in the modulation of protein synthesis, as miR-1 has been shown to directly target and inhibit various factors in the IGF1/AKT signaling pathway [51]. Though speculative at this time, it is thought that the changes in miRNA expression following an acute bout of resistance exercise are an integral aspect of the molecular changes underlying the adaptations observed following chronic resistance exercise training (see Figure 1).
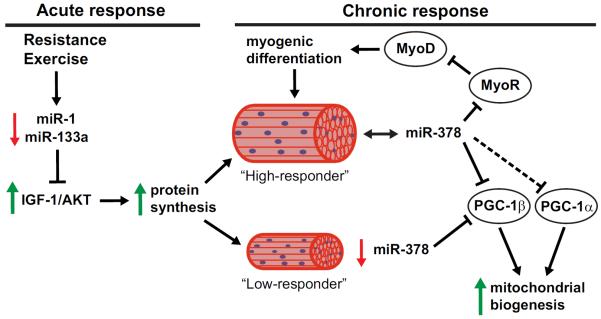
Figure 1. MicroRNA regulation of resistance exercise adaptations.
Acute resistance exercise decreases miR-1 in skeletal muscle, leading to increased IGF1/AKT signaling and therefore, increased protein synthesis. In response to chronic resistance exercise, low-responders showed a decrease in miR-378 expression, a miRNA shown to regulate mitochondrial biogenesis via upregulation of PGC-1β expression. Conversely, the high responders maintained their levels of miR-378, which is proposed to regulate myogenic differentiation by increased MyoD expression via repression of MyoR expression. Red arrow indicates downregulation of miRNA or gene expression. Solid line indicates validated gene target of miRNA, while dashed line indicates predicted gene target of miRNA.
In a study by Davidsen and colleagues (2011), a cohort of young, adult males underwent 12 weeks of resistance training with post-hoc analysis separating the group into “low-responders” and “high-responders” based on each subject's change in lean body mass [48]. The authors then examined whether expression levels of the most abundant miRNAs were different between the groups. Twenty-one miRNAs were profiled, showing that miRNA expression was unaffected in the vastus lateralis muscle in the high-responder group whereas the low-responder group showed a significant change in miR-451 and miR-378, with a downward trend for miR-26a and miR-29a. Interestingly, the change in miR-378 expression showed a significant correlation (R2=0.51) to the change in lean body mass, leading the authors to speculate that maintenance of miR-378 levels were required for changes in lean body mass [48]. This idea is supported by in vitro data that shows that miR-378 promotes myoblast differentiation by targeting MyoR, a negative regulator of the myogenic transcription factor MyoD [52, 53]. In humans, it has been suggested that this myogenic differentiation and addition of nuclei to existing skeletal muscle positively influence muscle hypertrophy [54]. In addition, recently miR-378 has been shown to control mitochondrial metabolism and bioenergetics through the regulation of PGC1-β, and in the context of exercise, would likely be a physiological response observed with endurance exercise [55]. Therefore, the blunted hypertrophic growth in “low responders” to resistance exercise may be that these individuals are predisposed to an aerobic phenotype relative to a hypertrophic phenotype, i.e., loss of miR-378 expression lead to the upregulation of MyoR and PGC-1β (see Figure 1).
Endurance Exercise and Skeletal Muscle miRNA Expression
In contrast to resistance exercise, endurance exercise generally brings about metabolic adaptations with little change in skeletal muscle mass [56–58]. These metabolic adaptations are characterized by an increase in mitochondria, a fiber- type shift to a more oxidative profile and an increase in capillary density. A number of studies have reported changes in miRNA expression following endurance exercise suggesting miRNAs may have a role in the metabolic adaptations that occur with this type of exercise. For example, rectus femoris muscle of mice subjected to a 90 minute bout of forced running showed an upregulation of miR-1, miR-181 and miR-107 and downregulation of miR-23, with the decrease in miR-23 corresponding in an increase in both the mRNA and protein levels of one of its predicted target genes, PGC1-α [59]. Similarly, Aoi et al. (2010) demonstrated that four weeks of running resulted in the downregulation of another miRNA predicted to target PGC1-α, miR-696 [60]. Consistent with regulation by miR-696, PGC1-α protein expression was elevated whereas PGC-1α mRNA levels were unchanged suggesting miR-696 was acting by blocking translation [60]. PGC1-α is thought to have an important role skeletal muscle adaptation to exercise as it has been shown to be involved in mitochondrial biogenesis coupled with the control of energy expenditure, including gluconeogenesis, glycolysis and fatty acid oxidation [61] (see Figure 2). The findings from these studies suggest that the mechanism of miRNA regulation of PGC-1α expression is different in response to an acute bout or chronic training [59, 60]. Given that miR-696 is only expressed in the mouse, it remains to be determined if a similar mechanism via a different miRNA is operative in human skeletal muscle in response to endurance exercise.
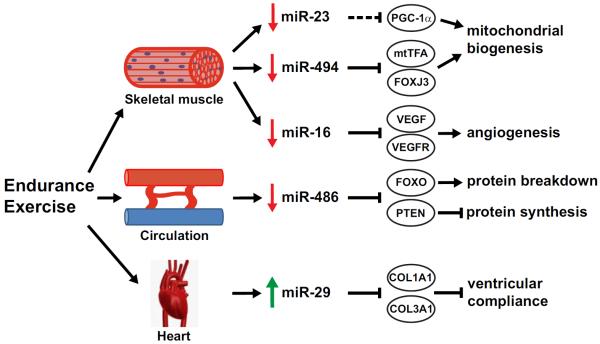
Figure 2. MicroRNA regulation of endurance exercise adaptations.
Endurance exercise results in the downregulation of miR-23 and miR-494 expression in skeletal muscle which is predicted to promote mitochondrial biogenesis via upregulation of PGC-1α, mtTFA and Foxj3. In Addition, miR-19 is downregulated which is predicted in increase angiogenesis by upregulation of Vegf and Vegfr. Endurance exercise decreases circulating levels of miR-486 which could promote a negative protein balance via regulation of Pten and Foxo. Finally, miR-29 levels are increased in the left ventricle following endurance exercise which is proposed to increase ventricular compliance by repressing multiple collagens. Green arrow indicates upregulation of miRNA or gene expression. Red arrow indicates downregulation of miRNA or gene expression. Solid line indicates validated gene target of miRNA, while dashed line indicates predicted gene target of microRNA.
In humans, 60 minutes of endurance exercise (cycling) was reported to increase the expression of miR-1 and miR-133 in the vastus lateralis muscle in untrained individuals, whereas following 12 weeks of training the resting levels of miR-1, miR-133a, miR-133b and miR-206 were lower than pre-training. [62]. The training resulted in a blunted response to the acute bout, which showed no changes in miRNA levels after training. Similarly, Keller et al. (2011) demonstrated that 6 weeks of cycling decreased the expression of miR-1 and miR-133, along with miR-101 and miR-455 in human skeletal muscle [63]. These results suggest that miRNA expression levels may change as training status changes in order to regulate training adaptations; however, to provide a causative role for miRNAs in skeletal muscle adaptation to exercise will require the use of gain- and loss-of-function animal models.
In another mode of endurance exercise, Yamamoyo et al. (2012) recently reported that an acute bout of swimming decreased the level of miR-494 in the gastrocnemius muscle of mice [64]. The authors went on the validate miR-494 regulation of two genes known to be involved in mitochondrial biogenesis, mtTFA and Foxj3. Another group examined miRNA levels in the soleus muscle of rats following swim training and found that miR-16 levels significantly decreased, paralleled by an increase in protein levels for VEGF and its respective type 2 receptor [65], with VEGF subsequently being validated as a miR-16 target by another research group [66]. These alterations within the VEGF pathway were thought to modulate angiogenesis, as there was a concomitant increase in capillarization within the muscle (see Figure 2). Collectively, the results of these studies suggest that miRNAs may have a role in modulating those adaptations that underlie the increase in oxygen delivery and utilization that occur with endurance exercise.
Endurance Exercise and Cardiac Muscle miRNA Expression
Two of the primary determinates of aerobic capacity is oxygen delivery to the working skeletal muscle and the ability of the muscle to take up and utilize that oxygen. Therefore, changes must occur in the cardiovascular system in order to effectively deliver oxygen, some of which may be regulated by local expression of miRNAs.
In rats, 10 weeks of low-intensity swimming produced changes in miRNA expression within the left ventricle. Swimming produced decreased expression of miR-1, miR-133a and miR-133b, accompanied by an increase in the expression of miR-29c. This corresponded with a decrease in collagen type I and collagen type III, both known targets of miR-29c, which may be a possible mechanism relating the benefits of endurance exercise with ventricular compliance [49]. In addition, swim training increased the expression of miR-126 in the left ventricle promoting angiogenesis by regulation of repressive genes (Spred-1, Pi3kr2) within the vascular endothelial growth factor (Vegf) pathway [67]. Similarly, swimming increased miR-27a and miR-27b which targets angiotensin-converting enzyme (Ace), while decreasing miR-143 which targets Ace2 in the heart [68]. Collectively, these adaptations would have a positive influence on cardiovascular function.
Exercise and MicroRNA Levels in the Circulation
In addition to striated muscle (heart and skeletal muscle), endurance exercise has also been shown to alter miRNA expression within the circulation. Plasma samples taken from human subjects showed that an acute bout of endurance exercise elevated circulating levels of miR-146a, miR-222, miR-21 and miR-221 [47]. Following a 90 day training period, the basal level of these miRNAs remained elevated in addition to miR-20a. Interestingly, even after the training period, circulating levels of these miRNAs still showed the transient increase following an acute bout of exercise [47]. Similarly, Aoi et al. (2013) recently showed that both acute and chronic endurance exercise decreased circulating levels of the muscle-enriched miR-486, however all other circulating myomiRs levels were below detectable limits [69]. The decrease of miR-486 levels in circulation may contribute to the negative protein balance typically associated with endurance exercise [70], as miR-486 suppresses factors that promote protein breakdown (Foxo) and inhibit protein synthesis (Pten) [71]. These studies all employed a candidate approach when assessing miRNA levels in circulation such that a more comprehensive screen may lead to the identification of additional miRNAs in circulation that are involved in the response and/or adaptation of muscle, and possibly other tissues, to exercise.
A recent study sought to determine if there was any relationship between circulating miRNAs levels and aerobic capacity [72]. The authors reported that individuals with a low aerobic capacity (VO2max) had elevated levels of miR-21, miR-210 and miR-222; however, the elevated levels of these miRNAs in circulation did not show a correlation with risk factors associated with cardiovascular disease. In contrast, miRNAs do appear to be useful as a biomarker for exercise-induced muscle damage. Uhlemann and colleagues (2012) found that circulating levels of miR-133a were elevated following an exercise bout designed to damage skeletal muscle, while miR-126 was elevated following exercise that would damage the endothelial layer within the vasculature [73]. The findings from these studies indicate that a bout of endurance exercise is capable of eliciting a change in serum miRNA levels, which are specific to the form of exercise, though the physiological implications remain to be understood. One possible mechanism may involve the release of miRNAs containing vesicles (known as exosomes) by the active skeletal muscle thereby acting as a paracrine factor influencing the activity of other tissues [74]. In addition, it is easy to imagine that circulating miRNAs may one day prove to be useful as a biomarker for assessing training status, or to serve as a predictor of disease progression (see Myopathies section).
A better understanding of how exercise, whether acute or chronic, is able to alter miRNA expression, will provide a more complete picture of the molecular mechanisms that underlie exercise-induced adaptations. This knowledge may offer insight into what training modalities are most beneficial, particularly in the context of disease prevention or treatment. Furthermore, miRNAs may themselves represent therapeutic targets that can be manipulated to augment training adaptations.
Muscle Atrophy
Disuse
In contrast to increased mechanical load, prolonged periods of mechanical unloading of skeletal muscle leads to muscle atrophy. This can occur as a result of prolonged bed rest [75], space flight [76] or denervation [75]. Allan and colleagues (2009) demonstrated that ~12 days of space flight produced a significant decrease in miR-206 expression in the mouse gastrocnemius muscle [76]. Using a rodent model of space flight, McCarthy and coworkers (2009) reported that seven days of hind limb unloading decreased miR-107, miR-221 and miR-499 expression; moreover, when unloading was carried out to 28 days, miR-499 was further decreased along with a 60% decrease in miR-208b [77]. Given that miR-499 and miR-208b are components of the myomiR network in skeletal muscle, these findings suggested the network is operative during skeletal muscle atrophy [25]. The authors provided evidence that supported the idea that miR-499 was involved in regulating the fiber-type shift associated with atrophy through the regulation of Sox6, a transcription factor known to repress slow myosin expression [78]. Studies have now shown that a miR-499/Sox6 pathway acts as a master regulator in setting the fiber-type composition of a muscle [79, 80]. Unlike hind limb unloading induced muscle atrophy, following a month of denervation, the expression of miR-1 and miR-133a in the rat soleus was decreased by ~70% compared to sham control; surprisingly, after four month of denervation the expression of miR-1 and miR-133a was actually 2-fold higher than control [81]. This depression of miR-1 and miR-133 during an acute period of disuse was also observed following seven days of bed rest in the vastus lateralis of human subjects [82]. These findings indicate that skeletal muscle atrophy in both rodents and humans is associated with a loss of myomiR expression, though the mechanism of how these miRNAs may regulate atrophy remains to be resolved.
Aging
The change in gene expression in response to exercise has been shown to be significantly different in the elderly [83, 84]. This alteration in gene transcription may in part be modulated by differentially expressed miRNAs which have been shown to occur in aged skeletal muscle [85, 86]. When compared to younger individuals, skeletal muscle from older individuals showed a significant elevation in the miRNAs let-7b and let-7e under resting conditions, which was proposed to be involved in regulating the cell cycle based on let-7 target genes [85]. Older subjects also exhibited increased expression of primary miRNAs-1-1, -1-2, -133a-1 and -133a-2, although these changes did not correspond to an elevations in miRNA levels. Furthermore, following an acute bout of resistance exercise older subjects did not show the same decrease in miR-1 expression as observed in the young subjects [50]. Conversely, when older subjects participated in chronic resistance training, miR-1 levels were significantly decreased following the training period [87]. One explanation for the disparity between the findings of the two previous studies may be the timing of when the muscle was sampled post-exercise. The latest time point examined by Drummond et al. (2008) was 6 hours post-exercise, which could explain why they only saw a decrease in the primary miRNA and not the mature miRNA which could require a longer period or repeated exercise bouts, as used in the Mueller et al. (2011) study, before a significant decrease will be observed.
Muscle Function
One of the primary outcomes associated with chronic exercise is alterations in muscle function, however few investigations have examined what impact miRNA regulation of gene expression has on muscle function. In cell culture, Rhim et al., (2010) inhibited miR-133a activity and then bioengineered artificial skeletal muscle preparations [88]. Artificial muscle devoid of miR-133a showed a 20% increase in peak force that was paralleled by a greater myotube diameter relative to control myotubes [88].
Two of the primary motor proteins in skeletal muscle, α- and β-myosin heavy chain (MHC), have been shown to encode miR-208a and miR-208b, respectively. In addition, the Myh7b gene, a homolog of the β-MHC gene, is expressed in cardiac and slow-twitch muscles and encodes miR-499. Over-expression of miR-499, in conjunction with miR-208b, was shown to increase the number of type-I myofibers within a muscle. Consistent with the increase in type-I fibers, mice over-expressing miR-499 showed greater muscular endurance capacity as assessed by time to exhaustion with treadmill running [25]. Germline knockout of another myomiR, miR-206, however had no impact on downhill running time or distance [34]. Finally, over-expression of miR-1 had a negative effect on contractile function in cardiac muscle by disrupting sarcomere architecture, possible through targeting cardiac myosin light chain kinase and calmodulin expression [89]. These findings highlight the fact that manipulating the levels of a single myomiR may result in altered muscle function. Altering muscle function, particularly in cardiac muscle, can have deleterious outcomes, so any research aimed at potential clinical use of miRNAs needs to include some form of functional outcome measurement.
Satellite Cell Fate and Myogenesis
Satellite cells are mesenchymal stem cells within skeletal muscle that are located between the basal lamina and the sarcolemma. Under resting conditions these cells are normally quiescent, however, following an injury to the muscle they have the ability to re-enter the cell cycle, proliferate and either fused to an existing myofiber or fuse together to form a de novo myofiber; for a more comprehensive review on satellite cell dynamics the reader is referred to two recent reviews [90, 91]. A complex network of transcription factors control satellite cell quiescence, proliferation and differentiation. During quiescence, satellite cells express paired-box 7 (Pax7) transcription factor and following commitment to the myogenic lineage, express myogenic factor 5 (Myf5). Upon activation and proliferation, Myf5 expression is up-regulated, along with another transcription factor, myogenic differentiation 1 (MyoD) [90]. Finally, during terminal differentiation Pax7 is down-regulated and the transcription factor myogenin is up-regulated, along with muscle regulatory factor 4 (MRF4), both factors being required for terminal myogenic differentiation [90]. It should be noted that the most common cell line for studying myogenesis and satellite cells in vitro, is the C2C12 myogenic cell line which has been shown to mimic many of the same processes that occur in vivo [92].
Quiescence
Only recently has the molecular mechanism underlying satellite cell quiescence begun to emerge, revealing that quiescence is an active process that requires the integration of various signaling pathways [93]. Recently, Cheung and colleagues (2012) used a satellite-cell specific Dicer knockout to profile all of the miRNAs expressed in quiescent satellite cells [94]. These authors reported that miR-489 was required for maintaining quiescence by directly repressing the oncogene Dek, a protein required for re-entry into the cell cycle [94]. In addition to miR-489, miR-31 was found to contribute to satellite cell quiescence by suppressing the translation of Myf5, which was shown to be necessary for proliferation and differentiation of satellite cells [95]. In a recent study by Koning et al. (2012) they demonstrated that there is a global downregulation of miRNAs in human satellite cells during quiescence compared to proliferation, in particular miR-106b, miR-25, miR-29c and miR-320c [96].
Proliferation
In addition to quiescence, miRNAs have also been shown to regulate the proliferation of satellite cells. In one of the earliest reports, Chen and coworkers (2006) showed that miR-133a was capable of promoting proliferation by repressing the expression of SRF (serum response factor), a transcription factor known to block cell proliferation [40]. Similar to miR-133a, miR-682 was found to be highly up-regulated during myoblast proliferation, both in vitro and in vivo, with inhibition of miR-682 activity resulting in reduced myoblast proliferation [97]. In vitro, miR-27a has been shown to be involved in promoting myoblast proliferation by targeting myostatin [98].
Differentiation
Numerous groups have investigated the role of miRNAs in myogenic differentiation and, to date, these studies have provided evidence showing that the myomiRs have an important role in this process. Kim et al., (2006) were one of the first groups to show that miR-206 was required for C2C12 myoblast differentiation, primarily through the regulation of DNA polymerase [39]. Similarly, miR-1 was found to promote myogenic differentiation by down-regulating histone deacetylase complex 4 (HDAC4) expression which led to the de-repression of follistatin expression, an antagonist to myostatin activity [40, 99]. In addition to miR-1, miR-206 and miR-29 were also shown to modulate the inhibitory effects of TGFβ signaling on myogenic differentiation through altered HDAC4 expression [100]. Specifically, Winbanks and colleagues (2011) reported that miR-206 and miR-29 were capable of inhibiting TGF-β signaling by repressing Smad3 expression, which in turn resulted in the downregulation of HDAC4 expression [100]. More recently, Dey and coworkers (2012) demonstrated that miR-26a repression of Smad1 and Smad4 expression, confirming the importance of inhibition of TGF-β signaling by miRNAs for myogenic differentiation [101]. MyomiRs miR-1 and miR-206 have also been found to promote myogenic differentiation by regulating the expression of Pax7 [102]. Conversely, the inhibition of miR-1 and miR-206 activity by an antimiR resulted in an increase in Pax7 expression and cell proliferation [102]. Pax7 has also been shown to be a direct target of the newest member of the myomiR family, miR-486, whose expression has been shown to be upregulated during differentiation [103].
Recently, Goljanek-Whysall et al. (2012) used microarray analysis to identify other punitive targets of miR-206 that could be involved in myogenesis. They identified and validated numerous targets of miR-206, including known myogenic factors Meox2, RARB, Fzd7, MAP4K3, CLCN3 and NFAT5, and went on to demonstrate that downregulation of these factors was required for differentiation to proceed in C2C12 cells [104]. MyomiR-133a has been shown to promote differentiation by regulating the IGF-1 pathway through downregulation of the IGF-1 receptor [105]. Other roles for miR-133a have been shown to include promoting sarcomeric actin organization [106] and inhibiting brown-adipocyte formation [107]; although miR-133a was originally shown to have a role in regulating cell proliferation, these more recent studies demonstrate miR-133a controls numerous molecular events which appear to be context dependent. While the previous miRNAs have been shown to promote differentiation, miRNAs can also function to inhibit myogenic differentiation. Recently, Wang et al., (2012) showed that miR-23a inhibited differentiation by directly regulating the expression of myosin heavy chain genes, proteins required for contractile function in mature myofibers [108].
Regeneration
One of the defining characteristics of muscle is its remarkable ability to regenerate following injury. Based on the absolute requirement for satellite cell participation in regeneration many of the miRNAs discussed previously have been implicated in controlling regeneration [109–112]. However, significant injury to muscle has been shown to alter expression of a wide number of miRNAs, many of which were not reported in the aforementioned satellite cell studies [113–115] (see Figure 3).
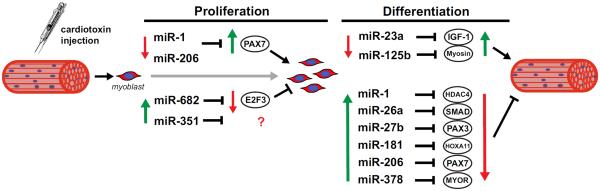
Figure 3. MicroRNAs and target genes involved in muscle regeneration.
In response to cardiotoxin injury, activated satellite cells begin to proliferate and then proceed through myogenic differentiation as part of the regenerative process. Satellite cell proliferation and differentiation are associated with changes in miRNA expression that result in an alteration in target gene expression that have been shown to be important for proper muscle regeneration. Green arrow indicates upregulation of miRNA or gene expression. Red arrow indicates downregulation of miRNA or gene expression.
The majority of the investigations examining miRNAs involvement in satellite cells dynamics utilized in vitro models, whereas studies focused on skeletal muscle regeneration require the use of in vivo model. This requirement is an important distinction because it allows for a comparison to the in vitro findings and whether they are of any biological relevance, since the in vivo model takes into consideration the involvement of other cells type that could provide further regulation of miRNAs expressed in muscle cells. Such comparisons between in vitro and in vivo finding, however, must be tempered by the fact that the most common models of muscle regeneration induce supra-physiological levels of damage. Moreover, it is important to keep in mind that other cell types (macrophages), in addition to satellite cells, are necessary for adequate regeneration to take place and also may express specific miRNAs [116]. One of the first reports to show in vivo changes in miRNAs during regeneration observed an increase in miR-181 [115]. The authors showed that miR-181 was strongly upregulated during the late-phase of muscle regeneration and that it targeted Hox-A11, a negative regulator of MyoD and terminal differentiation. This concomitant increase in MyoD resulted in an increase in miR-1 expression, which in turn suppressed the action of HDAC4 and follistatin [99]. In the early time points following cardiotoxin injury, miR-351 expression increased transiently, allowing for satellite cell proliferation by suppressing of E2f3 expression, an inhibitor of cell cycle progression [113]. Following cardiotoxin injury, miR-206 is highly upregulated, suggesting a role in muscle regeneration, which is consistent with the impaired regeneration, reported in the miR-206 knockout mouse [34]. The increase in miR-206 expression is required to downregulate various genes including Pax7, Notch3, IGFBP5 [34] and Hmgb3 [117], all of which would be inhibitory to the differentiation process. In addition, the increase in miR-1 expression during regeneration further contributes to the downregulation of Pax7 expression [102]. Similarly, miR-26a is upregulated following cardiotoxin-induced muscle injury and local knockdown in the tibialis anterior results in delayed regeneration [101]. In the same model, miR-125b is downregulated following injury resulting in increased expression of one of its direct targets, insulin-like growth factor two (IGF-II)[114]. IGF-II regulates myogenesis and the suppression of IGF-II expression by exogenous administration of miR-125 impairs regeneration [114]. Furthermore, during regeneration, increased expression of miR-133 is required to prevent satellite cells from adopting a brown adipogenic lineage [107].
With literature continuing to emerge defining the role of miRNAs in regeneration, additional studies are examining the effect of exogenous miRNAs administration on the regeneration process. This could have potential clinical implications as a future therapy following muscle injury. Nosaka and colleagues (2010) demonstrated that exogenous administration of miR-1, miR-133 and miR-206 mixture enhanced regeneration following laceration injury, by attenuating fibrosis formation, increasing myogenic factor expression and promoting angiogenesis [118].
Myopathies
Accumulating evidence is showing possible links between miRNA dysregulation and muscular disorders. The most commonly studied group of disorders related to muscle disease is muscular dystrophy. Muscular dystrophies have multiple etiologies, however individuals almost always present with muscle weakness and muscle loss, both resulting in impaired locomotion [119]. Eisenberg and colleagues (2007) were the first to profile the expression pattern of miRNA in a cohort of human subjects that had various types of muscular dystrophy [120]. They found that in the dystrophic group, there was a dysregulation in multiple miRNAs, with the majority of differences being an upregulation in miRNA expression. Interestingly, mir-146b, miR-221m miR-155, miR-214 and miR-222 were consistently dysregulated in almost all of the subjects. In a follow-up study by the same group, miR-486 expression was shown to be decreased in patients with Duchenne muscular dystrophy (DMD) and target members of the Akt/Pten signaling pathways [71]. In particular, miR-486 regulated platelet-derived growth factor receptor β (Pdgfrb) expression which was associated with a compromised regenerative response [71]. In the mdx mouse, an animal model DMD, miR-206 expression was significantly upregulated only in the diaphragm and not limb musculature [121]. In the same mouse model, muscle from mdx mice displayed a significant elevation in miR-31 which was shown to target the dystrophin gene, possible exacerbating the phenotype [122]. The reason for the discrepancy between the human and the mouse findings with respect to miRNA expression remain unknown, though later studies reported upregulation of miR-206 in muscle of both the mdx mouse and DMD patients [123, 124]. Recently, Cacchiarelli et al. [125] utilized a low-density array to profile miRNAs that were dysregulated in skeletal muscle of the mdx mice. They demonstrated that miR-1, miR-29, miR-30c, miR-133a and miR-206 were all differentially expressed in the gastrocnemius muscle, with the downregulation of miR-1 and miR-29 a direct result of reduced dystrophin/nNOS signaling leading to increased activity of HDAC2. The loss of miR-1 expression resulted in an increase in the level of glucose-6-phosphate dehydrogenase, a validated target, which negatively affected the redox state of the cell making it more susceptible to oxidative damage. Furthermore, loss of miR-29 expression was shown to promote fibrosis in mdx muscle [125].
ALS
Amyotrophic lateral sclerosis (ALS) is the most common motor neuron disease that affects skeletal muscle and is characterized by loss of motor neurons, denervation of target muscles, muscle atrophy, and ultimately, paralysis [126]. Williams and colleagues (2008) demonstrated that miR-206 was drastically up-regulated in lower limb muscles of G93A-SOD1 transgenic mice, a model of ALS [35]. Inactivation of miR-206 revealed that miR-206 is involved in a retrograde mechanism that promotes reinnervation through downregulation of HDAC4 [35]. This data is supported by a recent report looking at miRNA levels in muscle samples taken from human patients with ALS [127]. This group reported that in addition to miR-206, the expression of miR-23a, miR-29b, miR-31 and miR-455 was also upregulated in skeletal muscle from ALS patients. Of interest was the finding that miR-23a was shown to directly regulate PGC1-α, one of many genes important for mitochondrial function that was down-regulated in ALS patients. These findings suggest that the mitochondrial dysfunction commonly observed in ALS patients may in part be mediated by mis-regulation of miR-23a, though this mechanisms remains to be tested [127].
Rhabdomyosarcoma
Rhabdomyosarcoma (RMS) is the most common pediatric soft-tissue sarcoma derived from skeletal muscle progenitor cells that continue to proliferate, having reduced capacity to differentiate into mature muscle cells [128, 129]. Given miRNAs clear role in the regulation of myogenesis, specifically differentiation, it seems plausible that miRNAs may play a role in development of specific types of RMS. The muscle-specific nature of the myomiRs made them obvious targets for potential deregulation in RMS. Support for this idea was first reported by Yan and coworkers (2009) who showed that in RMS cell lines and primary tumors miR-1 and miR-206 levels are repressed resulting in increased levels of the oncogene c-Met, a confirmed target gene [130]. Similarly, Roa et al., (2010) showed that miR-1, along with miR-133, were down-regulated in RMS cell lines and that this was paralleled by an upregulation in many of their predicted target genes [131]. These findings were confirmed in a large cohort of tissue from primary RMS tumors, showing that miR-1, miR-133a, miR-133b and miR-206 expression were all reduced when compared to healthy skeletal muscle levels [132]. Interestingly, there was a significant inverse relationship between miR-206 expression and survival along with metastasis at diagnosis. Since miR-206 is involved in repressing proliferation and inducing differentiation of myogenic progenitors, it is conceivable that this loss of miR-206 expression is aiding in the proliferative nature of tumor formation. This was demonstrated by overexpressing miR-206 in multiple RMS cell lines which resulted in reduced proliferation and cell-cycle progression, accompanied by increased apoptosis [132].
Non-myomiRs have also been shown to contribute to tumor formation. Wang and colleagues (2008) showed that epigenetic silencing of miR-29 by the NF-κB pathway was maintained in a RMS cell line thereby impairing differentiation [133]. In addition, downregulation of miR-29 in two other RMS cell lines resulted in an increase in two target genes (cyclin D2 and E2F transcription factor 7) both of which are involved in cell cycle progression [93]. Similarly, miR-26a has been shown to be down-regulated in both RMS cells line and primary tumors [134]. Recently, Sarver and colleagues (2010) reported that miR-183 is over-expressed in RMS and possessed oncogenic potential by targeting two tumor-suppressors, EGR1 and PTEN [135].
MicroRNA Regulation of Oxidative Stress in Skeletal Muscle
The aforementioned studies have clearly demonstrated that exercise is able to induce changes in miRNA expression; however, the underlying mechanisms that control these changes remain to be defined. One plausible scenario is the change in miRNA expression in response to exercise is in part brought about by the change in the redox status of the muscle fiber. Exercise has been shown to dramatically alter the redox environment within muscle [136], therefore it is conceivable that some of the changes in miRNA expression are being driven by altered redox signaling in the cell. This idea is not without precedence, as there are many examples of redox-sensitive miRNAs in other cell types such as endothelial [137], neurons [138] and cardiomyocytes [139]. Therefore, the oxidative stress brought about during exercise may be one mechanism that is able to induce changes in miRNA expression in skeletal muscle.
Similarly, microRNAs themselves may be functioning to alter the redox state of the cell. A recent investigation showed that miR-31 was decreased both after an acute bout of endurance exercise and following a short-term training period [140]. The authors showed that miR-31 targets the transcription factor nuclear respiratory factor 1 (Nrf1), a key transcription factor involved in regulating the expression of mitochondrial-related genes during periods of oxidative stress [141, 142]. Therefore, these results suggest that a decrease in miR-31 levels may act as a protective mechanism within the cell to maintain mitochondrial function during periods of oxidative stress, such as during endurance exercise. Alternatively, miRNA levels may lead to increased levels of oxidative stress with in the muscle. One example of this would be the increased expression of miR-21 that is observed after a bout of endurance exercise [47], as well following a period of functional overload (McCarthy, unpublished data). MiR-21 has been shown to inhibit the conversion of superoxide to hydrogen peroxide by directly targeting SOD3 and TNFα [143]. While increasing reactive oxygen species (ROS) in the cell following exercise has been thought to lead to deleterious effects, recent evident suggests that ROS generation actually promotes some of the desirable physiological adaptations associated with exercise [144, 145]. Overall, these studies support the idea that miRNAs may regulate and be regulated by oxidative stress following exercise in skeletal muscle, though this exciting possibility awaits experimental validation.
Conclusion
MiRNAs have emerged as important players in the regulation of gene expression, being involved in most biological processes examined to date. The concept that miRNAs are primarily involved in the stress response of the cell makes them ideal candidates for mediating the response of skeletal muscle to changes in contractile activity. Although the field is still in its infancy, the studies presented in this review offer great promise that miRNAs will have an important role in mediating the response of skeletal muscle to different forms of exercise. This optimism, however, is tempered by the recognition that the findings to date supporting a role for miRNAs in skeletal muscle plasticity remain for the most part correlative. The challenge for the field moving forward will be to move beyond these descriptive studies by using available genetic mouse models and antimiR technology to perform gain- and loss-of-function studies. Moreover, to obtain a more complete understanding of the molecular mechanism through which miRNAs regulate skeletal muscle gene expression in response to exercise, it will be essential to identify and validate predicted target genes.
MicroRNA expression changes in response to both acute and chronic exercise.
Expression of muscle-specific microRNAs known as myomiRs change in response to exercise.
The microRNAs involved in the adaptation to exercise appear to be specific for the type and intensity of exercise.
MicroRNAs have a role in skeletal muscle development and muscle diseases such as muscular dystrophy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting galley proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arasu P, Wightman B, Ruvkun G. Temporal regulation of lin-14 by the antagonistic action of two other heterochronic genes, lin-4 and lin-28. Genes Dev. 1991;5(10):1825–33. doi: 10.1101/gad.5.10.1825. [DOI] [PubMed] [Google Scholar]
- 2.Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 1996;10(23):3041–50. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Ruvkun G. Isoform-specific mutations in the Caenorhabditis elegans heterochronic gene lin-14 affect stage-specific patterning. Genetics. 2001;157(1):199–209. doi: 10.1093/genetics/157.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruvkun G, Giusto J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 1989;338(6213):313–9. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- 5.Ruvkun G, et al. Dominant gain-of-function mutations that lead to misregulation of the C. elegans heterochronic gene lin-14, and the evolutionary implications of dominant mutations in pattern-formation genes. Dev Suppl. 1991;1:47–54. [PubMed] [Google Scholar]
- 6.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 7.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 8.Lagos-Quintana M, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 9.Lau NC, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 10.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 12.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez A, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 16.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14(23):2162–7. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 18.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi R, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311(5758):195–8. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 21.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–82. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 22.Sempere LF, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy JJ. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta. 2008;1779(11):682–91. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Small EM, et al. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci U S A. 2010;107(9):4218–23. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Rooij E, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17(5):662–73. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–9. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 27.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102(1):306–13. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 28.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18(4):510–25. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 30.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19(19):2343–54. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, et al. Identification of differences in microRNA transcriptomes between porcine oxidative and glycolytic skeletal muscles. BMC Mol Biol. 2013;14:7. doi: 10.1186/1471-2199-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muroya S, et al. Profiling of differentially expressed microRNA and the bioinformatic target gene analyses in bovine fast- and slow-type muscles by massively parallel sequencing. J Anim Sci. 2013;91(1):90–103. doi: 10.2527/jas.2012-5371. [DOI] [PubMed] [Google Scholar]
- 33.Callis TE, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119(9):2772–86. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu N, et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest. 2012;122(6):2054–65. doi: 10.1172/JCI62656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams AH, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326(5959):1549–54. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129(2):303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 37.Heyer MP, et al. Normal midbrain dopaminergic neuron development and function in miR-133b mutant mice. J Neurosci. 2012;32(32):10887–94. doi: 10.1523/JNEUROSCI.1732-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40(2):205–15. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HK, et al. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174(5):677–87. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22(23):3242–54. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu N, et al. Mice lacking microRNA 133a develop dynamin 2-dependent centronuclear myopathy. J Clin Invest. 2011;121(8):3258–68. doi: 10.1172/JCI46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stepto NK, et al. Global gene expression in skeletal muscle from well-trained strength and endurance athletes. Med Sci Sports Exerc. 2009;41(3):546–65. doi: 10.1249/MSS.0b013e31818c6be9. [DOI] [PubMed] [Google Scholar]
- 44.Dreyer HC, et al. Resistance exercise increases leg muscle protein synthesis and mTOR signalling independent of sex. Acta Physiol (Oxf) 2010;199(1):71–81. doi: 10.1111/j.1748-1716.2010.02074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayhew DL, et al. Translational signaling responses preceding resistance training-mediated myofiber hypertrophy in young and old humans. J Appl Physiol. 2009;107(5):1655–62. doi: 10.1152/japplphysiol.91234.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker DK, et al. Exercise, amino acids, and aging in the control of human muscle protein synthesis. Med Sci Sports Exerc. 2011;43(12):2249–58. doi: 10.1249/MSS.0b013e318223b037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baggish AL, et al. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J Physiol. 2011;589(Pt 16):3983–94. doi: 10.1113/jphysiol.2011.213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davidsen PK, et al. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J Appl Physiol. 2011;110(2):309–17. doi: 10.1152/japplphysiol.00901.2010. [DOI] [PubMed] [Google Scholar]
- 49.Soci UP, et al. MicroRNAs 29 are involved in the improvement of ventricular compliance promoted by aerobic exercise training in rats. Physiol Genomics. 2011;43(11):665–73. doi: 10.1152/physiolgenomics.00145.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drummond MJ, et al. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am J Physiol Endocrinol Metab. 2008;295(6):E1333–40. doi: 10.1152/ajpendo.90562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elia L, et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120(23):2377–85. doi: 10.1161/CIRCULATIONAHA.109.879429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gagan J, et al. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J Biol Chem. 2011;286(22):19431–8. doi: 10.1074/jbc.M111.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu J, et al. MyoR: a muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD. Proc Natl Acad Sci U S A. 1999;96(2):552–7. doi: 10.1073/pnas.96.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petrella JK, et al. Potent myofiber hypertrophy during resistance training in humans is associated with satellite cell-mediated myonuclear addition: a cluster analysis. J Appl Physiol. 2008;104(6):1736–42. doi: 10.1152/japplphysiol.01215.2007. [DOI] [PubMed] [Google Scholar]
- 55.Carrer M, et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378. Proc Natl Acad Sci U S A. 2012;109(38):15330–5. doi: 10.1073/pnas.1207605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carter SL, et al. Changes in skeletal muscle in males and females following endurance training. Can J Physiol Pharmacol. 2001;79(5):386–92. [PubMed] [Google Scholar]
- 57.Hawley JA. Adaptations of skeletal muscle to prolonged, intense endurance training. Clin Exp Pharmacol Physiol. 2002;29(3):218–22. doi: 10.1046/j.1440-1681.2002.03623.x. [DOI] [PubMed] [Google Scholar]
- 58.White AT, Schenk S. NAD(+)/NADH and skeletal muscle mitochondrial adaptations to exercise. Am J Physiol Endocrinol Metab. 2012;303(3):E308–21. doi: 10.1152/ajpendo.00054.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Safdar A, et al. miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS One. 2009;4(5):e5610. doi: 10.1371/journal.pone.0005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aoi W, et al. The microRNA miR-696 regulates PGC-1{alpha} in mouse skeletal muscle in response to physical activity. Am J Physiol Endocrinol Metab. 2010;298(4):E799–806. doi: 10.1152/ajpendo.00448.2009. [DOI] [PubMed] [Google Scholar]
- 61.Patti ME, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100(14):8466–71. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nielsen S, et al. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J Physiol. 2010;588(Pt 20):4029–37. doi: 10.1113/jphysiol.2010.189860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keller P, et al. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J Appl Physiol. 2011;110(1):46–59. doi: 10.1152/japplphysiol.00634.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoyo H, et al. MicroRNA (miRNA)-494 regulates mitochondrial biogenesis in skeletal muscle through mitochondrial transcriptional factor A (mtTFA) and forkhead box j3 (Foxj3) Am J Physiol Endocrinol Metab. 2012 doi: 10.1152/ajpendo.00097.2012. [DOI] [PubMed] [Google Scholar]
- 65.Fernandes T, et al. Exercise training prevents the microvascular rarefaction in hypertension balancing angiogenic and apoptotic factors: role of microRNAs-16, -21, and -126. Hypertension. 2012;59(2):513–20. doi: 10.1161/HYPERTENSIONAHA.111.185801. [DOI] [PubMed] [Google Scholar]
- 66.Sun CY, et al. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis. 2013;34(2):426–35. doi: 10.1093/carcin/bgs333. [DOI] [PubMed] [Google Scholar]
- 67.D. A. Silva ND J, et al. Swimming training in rats increases cardiac MicroRNA-126 expression and angiogenesis. Med Sci Sports Exerc. 2012;44(8):1453–62. doi: 10.1249/MSS.0b013e31824e8a36. [DOI] [PubMed] [Google Scholar]
- 68.Fernandes T, et al. Aerobic exercise training-induced left ventricular hypertrophy involves regulatory MicroRNAs, decreased angiotensin-converting enzyme-angiotensin ii, and synergistic regulation of angiotensin-converting enzyme 2-angiotensin (1–7) Hypertension. 2011;58(2):182–9. doi: 10.1161/HYPERTENSIONAHA.110.168252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aoi W, et al. Muscle-enriched microRNA miR-486 decreases in circulation in response to exercise in young men. Front Physiol. 2013;4:80. doi: 10.3389/fphys.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koopman R, et al. Combined ingestion of protein and carbohydrate improves protein balance during ultra-endurance exercise. Am J Physiol Endocrinol Metab. 2004;287(4):E712–20. doi: 10.1152/ajpendo.00543.2003. [DOI] [PubMed] [Google Scholar]
- 71.Alexander MS, et al. Regulation of DMD pathology by an ankyrin-encoded miRNA. Skelet Muscle. 2011;1:27. doi: 10.1186/2044-5040-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bye A, et al. Circulating microRNAs and aerobic fitness--the HUNT-Study. PLoS One. 2013;8(2):e57496. doi: 10.1371/journal.pone.0057496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uhlemann M, et al. Circulating microRNA-126 increases after different forms of endurance exercise in healthy adults. Eur J Prev Cardiol. 2012 doi: 10.1177/2047487312467902. [DOI] [PubMed] [Google Scholar]
- 74.Gallo A, et al. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7(3):e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Belavy DL, et al. Differential atrophy of the lower-limb musculature during prolonged bed-rest. Eur J Appl Physiol. 2009;107(4):489–99. doi: 10.1007/s00421-009-1136-0. [DOI] [PubMed] [Google Scholar]
- 76.Allen DL, et al. Effects of spaceflight on murine skeletal muscle gene expression. J Appl Physiol. 2009;106(2):582–95. doi: 10.1152/japplphysiol.90780.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCarthy JJ, et al. Evidence of MyomiR network regulation of beta-myosin heavy chain gene expression during skeletal muscle atrophy. Physiol Genomics. 2009;39(3):219–26. doi: 10.1152/physiolgenomics.00042.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hagiwara N, Yeh M, Liu A. Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev Dyn. 2007;236(8):2062–76. doi: 10.1002/dvdy.21223. [DOI] [PubMed] [Google Scholar]
- 79.Quiat D, et al. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proc Natl Acad Sci U S A. 2011;108(25):10196–201. doi: 10.1073/pnas.1107413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X, et al. Prdm1a and miR-499 act sequentially to restrict Sox6 activity to the fast-twitch muscle lineage in the zebrafish embryo. Development. 2011;138(20):4399–404. doi: 10.1242/dev.070516. [DOI] [PubMed] [Google Scholar]
- 81.Jeng SF, et al. Profiling muscle-specific microRNA expression after peripheral denervation and reinnervation in a rat model. J Neurotrauma. 2009;26(12):2345–53. doi: 10.1089/neu.2009.0960. [DOI] [PubMed] [Google Scholar]
- 82.Ringholm S, et al. Bed rest reduces metabolic protein content and abolishes exercise-induced mRNA responses in human skeletal muscle. Am J Physiol Endocrinol Metab. 2011;301(4):E649–58. doi: 10.1152/ajpendo.00230.2011. [DOI] [PubMed] [Google Scholar]
- 83.Raue U, et al. Myogenic gene expression at rest and after a bout of resistance exercise in young (18–30 yr) and old (80–89 yr) women. J Appl Physiol. 2006;101(1):53–9. doi: 10.1152/japplphysiol.01616.2005. [DOI] [PubMed] [Google Scholar]
- 84.Raue U, et al. Proteolytic gene expression differs at rest and after resistance exercise between young and old women. J Gerontol A Biol Sci Med Sci. 2007;62(12):1407–12. doi: 10.1093/gerona/62.12.1407. [DOI] [PubMed] [Google Scholar]
- 85.Drummond MJ, et al. Aging and microRNA expression in human skeletal muscle: a microarray and bioinformatics analysis. Physiol Genomics. 2011;43(10):595–603. doi: 10.1152/physiolgenomics.00148.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hamrick MW, et al. The adipokine leptin increases skeletal muscle mass and significantly alters skeletal muscle miRNA expression profile in aged mice. Biochem Biophys Res Commun. 2010;400(3):379–83. doi: 10.1016/j.bbrc.2010.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mueller M, et al. Different molecular and structural adaptations with eccentric and conventional strength training in elderly men and women. Gerontology. 2011;57(6):528–38. doi: 10.1159/000323267. [DOI] [PubMed] [Google Scholar]
- 88.Rhim C, et al. Effect of microRNA modulation on bioartificial muscle function. Tissue Eng Part A. 2010;16(12):3589–97. doi: 10.1089/ten.tea.2009.0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ai J, et al. Overexpression of microRNA-1 impairs cardiac contractile function by damaging sarcomere assembly. Cardiovasc Res. 2012;95(3):385–93. doi: 10.1093/cvr/cvs196. [DOI] [PubMed] [Google Scholar]
- 90.Brack AS, Rando TA. Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell Stem Cell. 2012;10(5):504–14. doi: 10.1016/j.stem.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yablonka-Reuveni Z. The skeletal muscle satellite cell: still young and fascinating at 50. J Histochem Cytochem. 2011;59(12):1041–59. doi: 10.1369/0022155411426780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burattini S, et al. C2C12 murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. Eur J Histochem. 2004;48(3):223–33. [PubMed] [Google Scholar]
- 93.Bjornson CR, et al. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 2012;30(2):232–42. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cheung TH, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482(7386):524–8. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11(1):118–26. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 96.Koning M, et al. A global downregulation of microRNAs occurs in human quiescent satellite cells during myogenesis. Differentiation. 2012;84(4):314–21. doi: 10.1016/j.diff.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 97.Chen Y, et al. Temporal microRNA expression during in vitro myogenic progenitor cell proliferation and differentiation: regulation of proliferation by miR-682. Physiol Genomics. 2011;43(10):621–30. doi: 10.1152/physiolgenomics.00136.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang Z, et al. MicroRNA-27a promotes myoblast proliferation by targeting myostatin. Biochem Biophys Res Commun. 2012;423(2):265–9. doi: 10.1016/j.bbrc.2012.05.106. [DOI] [PubMed] [Google Scholar]
- 99.Sun Y, et al. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J Cell Biol. 2010;189(7):1157–69. doi: 10.1083/jcb.200912093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Winbanks CE, et al. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J Biol Chem. 2011;286(16):13805–14. doi: 10.1074/jbc.M110.192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dey BK, et al. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26(19):2180–91. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen JF, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190(5):867–79. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31(1):203–14. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goljanek-Whysall K, et al. Regulation of multiple target genes by miR-1 and miR-206 is pivotal for C2C12 myoblast differentiation. J Cell Sci. 2012;125(Pt 15):3590–600. doi: 10.1242/jcs.101758. [DOI] [PubMed] [Google Scholar]
- 105.Huang MB, et al. Insulin-like growth factor-1 receptor is regulated by microRNA-133 during skeletal myogenesis. PLoS One. 2011;6(12):e29173. doi: 10.1371/journal.pone.0029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mishima Y, et al. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23(5):619–32. doi: 10.1101/gad.1760209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yin H, et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. 2013;17(2):210–24. doi: 10.1016/j.cmet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang L, et al. MiR-23a inhibits myogenic differentiation through down regulation of fast myosin heavy chain isoforms. Exp Cell Res. 2012;318(18):2324–34. doi: 10.1016/j.yexcr.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 109.Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138(17):3639–46. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McCarthy JJ, et al. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development. 2011;138(17):3657–66. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Murphy MM, et al. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138(17):3625–37. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sambasivan R, et al. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138(17):3647–56. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- 113.Chen Y, et al. MiR-351 transiently increases during muscle regeneration and promotes progenitor cell proliferation and survival upon differentiation. Physiol Genomics. 2012;44(21):1042–51. doi: 10.1152/physiolgenomics.00052.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol. 2011;192(1):69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Naguibneva I, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8(3):278–84. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 116.Perdiguero E, et al. p38/MKP-1-regulated AKT coordinates macrophage transitions and resolution of inflammation during tissue repair. J Cell Biol. 2011;195(2):307–22. doi: 10.1083/jcb.201104053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maciotta S, et al. Hmgb3 is regulated by microrna-206 during muscle regeneration. PLoS One. 2012;7(8):e43464. doi: 10.1371/journal.pone.0043464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nakasa T, et al. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med. 2010;14(10):2495–505. doi: 10.1111/j.1582-4934.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Emery AE. The muscular dystrophies. Lancet. 2002;359(9307):687–95. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- 120.Eisenberg I, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci U S A. 2007;104(43):17016–21. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McCarthy JJ, Esser KA, Andrade FH. MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am J Physiol Cell Physiol. 2007;293(1):C451–7. doi: 10.1152/ajpcell.00077.2007. [DOI] [PubMed] [Google Scholar]
- 122.Cacchiarelli D, et al. miR-31 modulates dystrophin expression: new implications for Duchenne muscular dystrophy therapy. EMBO Rep. 2011;12(2):136–41. doi: 10.1038/embor.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Greco S, et al. Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J. 2009;23(10):3335–46. doi: 10.1096/fj.08-128579. [DOI] [PubMed] [Google Scholar]
- 124.Yuasa K, et al. MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct Funct. 2008;33(2):163–9. doi: 10.1247/csf.08022. [DOI] [PubMed] [Google Scholar]
- 125.Cacchiarelli D, et al. MicroRNAs involved in molecular circuitries relevant for the Duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab. 2010;12(4):341–51. doi: 10.1016/j.cmet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 126.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–49. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 127.Russell AP, et al. Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol Dis. 2012;49C:107–117. doi: 10.1016/j.nbd.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 128.De Giovanni C, et al. Molecular and cellular biology of rhabdomyosarcoma. Future Oncol. 2009;5(9):1449–75. doi: 10.2217/fon.09.97. [DOI] [PubMed] [Google Scholar]
- 129.Tapscott SJ, Thayer MJ, Weintraub H. Deficiency in rhabdomyosarcomas of a factor required for MyoD activity and myogenesis. Science. 1993;259(5100):1450–3. doi: 10.1126/science.8383879. [DOI] [PubMed] [Google Scholar]
- 130.Yan D, et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284(43):29596–604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rao PK, et al. Distinct roles for miR-1 and miR-133a in the proliferation and differentiation of rhabdomyosarcoma cells. FASEB J. 2010;24(9):3427–37. doi: 10.1096/fj.09-150698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Missiaglia E, et al. MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br J Cancer. 2010;102(12):1769–77. doi: 10.1038/sj.bjc.6605684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang H, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14(5):369–81. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ciarapica R, et al. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. 2009;8(1):172–5. doi: 10.4161/cc.8.1.7292. [DOI] [PubMed] [Google Scholar]
- 135.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70(23):9570–80. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 136.Powers SK, Jackson MJ. Exercise-induced oxidative stress: cellular mechanisms and impact on muscle force production. Physiol Rev. 2008;88(4):1243–76. doi: 10.1152/physrev.00031.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Marin T, et al. Mechanosensitive microRNAs-role in endothelial responses to shear stress and redox state. Free Radic Biol Med. 2013 doi: 10.1016/j.freeradbiomed.2013.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xu S, et al. Oxidative Stress Mediated-Alterations of the MicroRNA Expression Profile in Mouse Hippocampal Neurons. Int J Mol Sci. 2012;13(12):16945–60. doi: 10.3390/ijms131216945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li R, et al. MicroRNA-145 protects cardiomyocytes against hydrogen peroxide (H(2)O(2))-induced apoptosis through targeting the mitochondria apoptotic pathway. PLoS One. 2012;7(9):e44907. doi: 10.1371/journal.pone.0044907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Russell AP, et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short term endurance training. J Physiol. 2013 doi: 10.1113/jphysiol.2013.255695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dhar SS, Ongwijitwat S, Wong-Riley MT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283(6):3120–9. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Piantadosi CA, Suliman HB. Mitochondrial transcription factor A induction by redox activation of nuclear respiratory factor 1. J Biol Chem. 2006;281(1):324–33. doi: 10.1074/jbc.M508805200. [DOI] [PubMed] [Google Scholar]
- 143.Zhang X, et al. MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFalpha. Cancer Res. 2012;72(18):4707–13. doi: 10.1158/0008-5472.CAN-12-0639. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 144.Petersen AC, et al. Infusion with the antioxidant N-acetylcysteine attenuates early adaptive responses to exercise in human skeletal muscle. Acta Physiol (Oxf) 2012;204(3):382–92. doi: 10.1111/j.1748-1716.2011.02344.x. [DOI] [PubMed] [Google Scholar]
- 145.Ristow M, et al. Antioxidants prevent health-promoting effects of physical exercise in humans. Proc Natl Acad Sci U S A. 2009;106(21):8665–70. doi: 10.1073/pnas.0903485106. [DOI] [PMC free article] [PubMed] [Google Scholar]