Abstract
Internal transcribed spacer 2 (ITS2) sequencing was used to characterize the peanut mycobiome during 90 days storage at five conditions. The fungal diversity in in-shell peanuts was higher with 110 operational taxonomic units (OTUs) and 41 genera than peanut kernels (91 OTUs and 37 genera). This means that the micro-environment in shell is more suitable for maintaining fungal diversity. At 20–30 d, Rhizopus, Eurotium and Wallemia were predominant in in-shell peanuts. In peanut kernels, Rhizopus (>30%) and Eurotium (>20%) were predominant at 10–20 d and 30 d, respectively. The relative abundances of Rhizopus, Eurotium and Wallemia were higher than Aspergillus, because they were xerophilic and grew well on substrates with low water activity (aw). During growth, they released metabolic water, thereby favoring the growth of Aspergillus. Therefore, from 30 to 90 d, the relative abundance of Aspergillus increased while that of Rhizopus, Eurotium and Wallemia decreased. Principal Coordinate Analysis (PCoA) revealed that peanuts stored for 60–90 days and for 10–30 days clustered differently from each other. Due to low aw values (0.34–0.72) and low levels of A. flavus, nine of 51 samples were contaminated with aflatoxins.
Peanuts are important economic crops in the world and are cultivated on a large scale, with Africa, China and India being the greatest producers. In 2013, the annual peanut yield in China was 17.0 million tons (http://data.stats.gov.cn/easyquery.htm?cn=C01), which accounts for more than 45% of the worldwide peanut production (http://faostat.fao.org). Peanut kernels have high nutritional and commercial value due to the presence of calcium, carbohydrates, fatty acids, fibers, phosphorus, proteins and vitamins. Peanuts are mainly used in the manufacture of oil, sweets, candies and pastes. More than 60% of the worldwide peanut yield is destined to oil production, with peanut oil being the fifth most consumed type of oil around the world1.
The contamination of peanuts with Aspergillus flavus and aflatoxins is considered to be one of the most serious safety problems in the world2,3. This fungal pathogen infects peanut kernels before and after harvest4,5. Pre-harvest peanut kernels contain mycelia and spores of aflatoxigenic fungi, which contribute to significant economic losses and aflatoxin accumulation during storage6. Contamination with aflatoxins compromises the quality of the product because aflatoxins are the most toxic and carcinogenic compounds among the toxins. Aflatoxin B1 (AFB1), which is the most toxic, has both teratogenic and mutagenic properties and causes toxic hepatitis, hemorrhage, edema, immunosuppression, and hepatic carcinoma7. AFB1, which poses a health risk in animals and humans, has been classified as a class I human carcinogen by the International Agency for Research on Cancer8,9. More than 100 countries and organizations including the National Health and Family Planning Commission P.R. of China, the European Union and the U.S. Food and Drug Administration, have established limits for total aflatoxins and AFB1 levels in peanuts10,11.
A. flavus growth and subsequently aflatoxin production depend on several factors, including geographical region, season, and the environmental conditions during peanut growth and storage. Tropical and subtropical regions are the most favorable for A. flavus growth and aflatoxin production12. A. flavus growth and aflatoxin production are markedly affected by environmental factors, especially water activity (aw) and temperature13. The Yangtze River zone, especially Hubei province, China, is the major peanut-producing region of the country. However, this region has a subtropical climate with high temperatures and high relative humidity, which is the favorable environmental condition for A. flavus growth and aflatoxins production in stored peanuts. In 2012, the distribution and toxigenicity of A. flavus and A. parasiticus in peanut soils of four agroecological zones in China (Southeast coastal zone, Yangtze River zone, Yellow River valley and Northeast zone) were investigated in our laboratory. Previous findings revealed that Yangtze River zone had the highest concentration of A. flavus (2749.3 CFU/g) and the highest toxigenic potential of aflatoxin production among the four agroecological zones. It is not surprising that peanut contamination with aflatoxins is frequently reported in this region3,14. However, there are other fungal population that can affect A. flavus growth and aflatoxin production. Therefore, it is necessary to characterize the fungal microbiome of peanuts and its variations during storage under the environmental conditions of this region.
High-throughput sequencing technologies have opened new frontiers in microbial community analyses by providing an economic and efficient means of identifying the microbial phylotypes in samples. Furthermore, next-generation sequencing techniques have led to a revolution in microbial ecology by providing opportunities to generate unprecedented numbers of sequences and detect rare or low-abundance organisms15,16. Studies have revolutionized our understanding of the microbial communities present in our bodies17,18,19,20,21, soils22,23 and deep seas24. This revolution in sequencing technology, coupled with the development of advanced computational tools that exploit metadata to relate hundreds of samples to one another in ways that reveal clear biological patterns, has re-invigorated studies focused on the internal transcribed spacer 2 (ITS2) region of rDNA. The ITS2 region, which is an excellent phylogenetic marker suitable for fungal taxon assignment25, has been successfully used in comparative ecology studies where it gives results that are convergent with, if not comparable to, those for other markers25,26. ITS-based surveys are extremely valuable because they allow the assessment of biodiversity and ecological characteristics of whole communities or individual microbial taxa. However, alternative techniques, such as metagenomics can provide insight into all genes and their functions in a given community. ITS2 region phylogenies tend to match trends in overall gene content; the ability to relate at the species level to the host or the environmental parameters has proven immensely powerful.
In the present study, the barcoded Illumina paired-end sequencing (BIPES) technique was used to characterize the mycobiome and its variations in stored in-shell peanuts and peanut kernels under different conditions (i.e. temperature and relative humidity). This study characterizes the mycobiome and its variation in stored peanuts using ITS2 sequencing and provides a direct comparison of the peanut mycobiome diversity during simulated storage. The findings of this study are likely to encourage the implementation and design of mould and aflatoxin management strategies.
Results
Data characteristics
In in-shell peanuts, the average number of raw reads generated per sample was 84,834, of which 56,709 were retained following filtering and denoising steps, and 54,426 reads were subsequently clustered into different operational taxonomic units (OTUs). The average length of each read was 329 bp (range: 307–348 bp) (Table 1). Of 245 OTUs, 6 OTUs cannot be identified with other organisms, 23 OTUs represent peanut and other plants, 14 OTUs represent nematode and other animals, and 3 OTUs represent E. coli. Remaining 199 OTUs represent fungi, and of them 18 OTUs represent uncultured fungi (Table S1).
Table 1. Summary of pyrosequencing analysis.
| Storage |
In-shell peanuts |
Peanut kernels |
|||
|---|---|---|---|---|---|
| Temperature/relative humidity | Time | Number of Reads | Average Read Length | Number of Reads | Average Read Length (bp) |
| 0 | 48352 | 324 | 48352 | 324 | |
| 20 °C/70% | 10 | 36868 | 327 | 10440 | 327 |
| 20 | 44551 | 327 | 49526 | 331 | |
| 30 | 39340 | 332 | 45115 | 334 | |
| 60 | 54865 | 327 | 47447 | 334 | |
| 90 | 8694 | 319 | – | – | |
| 20 °C/75% | 10 | 21355 | 323 | 60505 | 328 |
| 20 | 42635 | 327 | 41625 | 333 | |
| 30 | 23883 | 332 | 22802 | 333 | |
| 60 | 39909 | 325 | 31118 | 323 | |
| 90 | 20617 | 318 | 43916 | 318 | |
| 25 °C/75% | 10 | 8134 | 325 | – | – |
| 20 | 49697 | 323 | 14472 | 327 | |
| 30 | 50917 | 331 | 20643 | 324 | |
| 60 | 22340 | 332 | 70923 | 328 | |
| 90 | 148647 | 333 | 44956 | 324 | |
| 30 °C/75% | 10 | 119292 | 328 | 31395 | 328 |
| 20 | 50890 | 329 | 75468 | 328 | |
| 30 | 63462 | 337 | 13954 | 332 | |
| 60 | 63108 | 331 | 56194 | 346 | |
| 90 | 136917 | 348 | 49436 | 363 | |
| 30 °C/80% | 10 | 19000 | 307 | 17193 | 334 |
| 20 | 90315 | 334 | 41494 | 334 | |
| 30 | 65118 | 334 | 31678 | 334 | |
| 60 | 42468 | 328 | 23112 | 311 | |
| 90 | 163054 | 340 | 29730 | 339 | |
| Average | 56709 | 329 | 38396 | 331 | |
In peanut kernels, the average number of raw reads generated per sample was 61,243, of which 38,396 were retained following filtering and denoising steps, and 36,718 reads were subsequently clustered into 196 OTUs. The average length of each reads was 331 bp (range: 311–363 bp) (Table 1). Of 196 OTUs, 2 OTUs cannot be identified with other organisms, 21 OTUs represent peanut and other plants, 13 OTUs represent nematode and other animals, and 7 OTUs represent bacteria. Remaining 153 OTUs represent fungi, and of them 13 OTUs represent uncultured fungi (Table S2).
Fungal diversity in in-shell peanuts and peanut kernels
The average number of OTUs detected per sample was 110 (range: 81–140). On average, each OTU contained 495 reads. OTUs representing 41 genera of fungi were detected. 61.1% of OTUs and 50.7% of reads belonged to the Ascomycota, 9.4% of OTUs and 13.0% of reads belonged to Basidiomycota, and 7.1% of OTUs and 23.7% of reads belonged to Zygomycota. Dominant orders among Ascomycota included Dothideomycetes, Eurotiomycetes, Saccharomycetes and Sordariomycetes. Dominant Eurotiomycetes groups included the genera Aspergillus (12.8% of OTUs and 12.1% of reads), Eurotium (4.8% of OTUs and 21.6% of reads) and Penicillium (7.7% of OTUs and 10.1% of reads) genera. Dominant genera among Zygomycota and Basidiomycota were Rhizopus (5.2% of OTUs and 23.5% of reads) and Wallemia (5.3% of OTUs and 11.3% of reads), respectively (Table 2). Aspergillus species that found were Aspergillus aculeatus, Aspergillus candidus, Aspergillus flavipes, A. flavus, Aspergillus gracilis, Aspergillus niger, Aspergillus ochraceus, Aspergillus penicillioides (7.19% of reads), Aspergillus restrictus, Aspergillus tamarii, Aspergillus versicolor, Aspergillus vitricola and Aspergillus wentii. The predominant species in Eurotium was Eurotium niveoglaucum (39.78% of reads). The predominant species in Penicillium were Penicillium citrinum, Penicillium pinophilum, and Penicilliium simplicissium. In Wallemia, Wallemia sebi was the only species. The predominant species in Rhizopus was Rhizopus oryzae (46.16% of reads) (Table S1).
Table 2. Distribution of operational taxonomic units (OTUs) among fungal lineages including the 10 most abundant genera detected in in-shell peanuts and peanut kernels.
| In-shell peanuts |
Peanut kernels |
||||
|---|---|---|---|---|---|
| Taxnomic affinity | Percent of OTUs | Percent of reads | Taxnomic affinity | Percent of OTUs | Percent of reads |
| Ascomycota | 61.1 | 50.7 | Ascomycota | 60.5 | 44.2 |
| Eurotiomycetes | 30.1 | 44.1 | Eurotiomycetes | 31.5 | 39.5 |
| Eurotium | 4.8 | 21.6 | Eurotium | 6.1 | 19.6 |
| Aspergillus | 12.8 | 12.1 | Aspergillus | 14.4 | 10.1 |
| Penicillium | 7.7 | 10.1 | Penicillium | 8.4 | 9.7 |
| Dothideomycetes | 8.2 | 0.8 | Dothideomycetes | 7.1 | 1.0 |
| Lasiodiplodia | 1.4 | 0.4 | Lasiodiplodia | 1.1 | 0.8 |
| Cladosporium | 2.0 | 0.2 | Cladosporium | 1.7 | 0.2 |
| Saccharomycetes | 2.1 | 0.9 | Saccharomycetes | 2.1 | 1.3 |
| Candida | 1.8 | 0.9 | Candida | 1.5 | 1.3 |
| Sordariomycetes | 16.9 | 4.7 | Sordariomycetes | 16.5 | 2.2 |
| Fusarium | 3.3 | 2.4 | Fusarium | 3.9 | 0.8 |
| Bionectria | 1.3 | 1.2 | Gibberella | 2.8 | 0.2 |
| Zygomycota | 7.1 | 23.7 | Zygomycota | 7.8 | 27.8 |
| Mucomycotina | 7.1 | 23.7 | Mucomycotina | 7.8 | 27.8 |
| Rhizopus | 5.2 | 23.5 | Rhizopus | 5.9 | 27.8 |
| Basidiomycota | 9.4 | 13.0 | Basidiomycota | 9.7 | 13.5 |
| Wallemiomycetes | 5.3 | 11.3 | Wallemiomycetes | 6.8 | 13.4 |
| Wallemia | 5.3 | 11.3 | Wallemia | 6.8 | 13.4 |
| Tremellomycetes | 2.4 | 0.2 | Tremellomycetes | 1.7 | 0.1 |
| Cryptococcus | 1.1 | 0.2 | Cryptococcus | 0.7 | 0.1 |
The average number of OTUs detected per sample was 91 (range: 69–114). On average, each OTU contained 403 reads. OTUs representing 37 genera of fungi were detected. 60.5% of OTUs and 44.2% of reads belonged to the Ascomycota, 9.7% of OTUs and 13.5% of reads belonged to Basidiomycota, and 7.8% of OTUs and 27.8% of reads belonged to Zygomycota. Dominant orders among Ascomycota were Dothideomycetes, Eurotiomycetes, Saccharomycetes and Sordariomycetes. Dominant Eurotiomycetes groups included the genera Aspergillus (14.4% of OTUs and 10.1% of reads), Eurotium (6.1% of OTUs and 19.6% of reads) and Penicillium (8.4% of OTUs and 9.7% of reads). Dominant genera among Zygomycota and Basidiomycota were Rhizopus (5.9% of OTUs and 27.8% of reads) and Wallemia (6.8% of OTUs and 13.4% of reads) (Table 2). The species in Aspergillus were A. aculeatus, Aspergillus ellipticus, A. flavipes, A. flavus, Aspergillus glaucus, A. niger (10.50% of reads), A. ochraceus, A. penicillioides (2.05% of reads), A. restrictus, A. tamarii, Aspergillus terreus, A. versicolor and A. vitricola. The predominant species in Eurotium was E. niveoglaucum (34.35% of reads). The predominant species in Penicillium were P. citrinum, P. pinophilum, and P. simplicissium. In Wallemia, W. sebi and Walleia sp. F53 were the main species. The predominant species in Rhizopus was R. oryzae (48.79% of reads) (Table S1).
Storage time and fungal diversity
There were significant variation in per-sample OTUs richness based on storage conditions and storage time (Fig. 1). In in-shell peanuts, all denoised reads were clustered into 245 OTUs using a minimum pair-wise identity of 97%. The average number of OTUs detected per sample in in-shell peanuts was 110 (range: 81–140). In general, OTU number decreased at 10 d, reached its highest value at 20 d, and subsequently decreased, except in samples stored at 25 °C with 75% relative humidity. Though there were differences among storage conditions, but no significant difference in the results.
Figure 1.
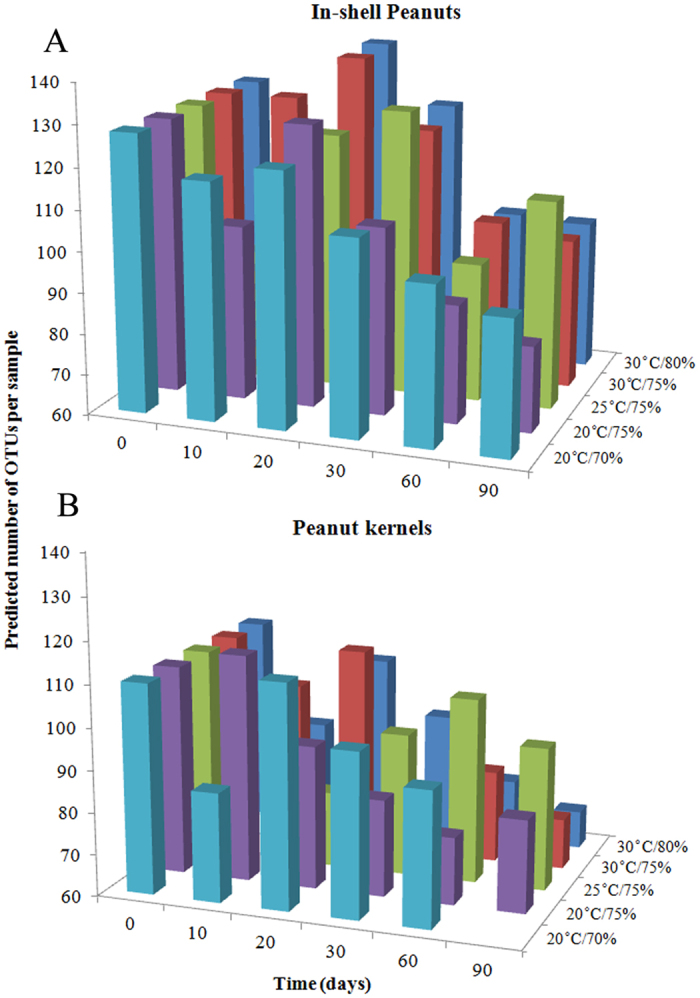
Predicted number of operational taxonomic units (OTUs) per sample in in-shell peanuts (A) and peanut kernels (B) stored for 0–90 d at different conditions.
In peanut kernels, all denoised reads were clustered into 196 OTUs using a minimum pair-wise identity of 97%. The average number of OTUs detected per sample in in-shell peanuts was 91 (range: 69–114). In general, the number of OTUs decreased with storage time, and the differences were seen among the storage conditions but not significant (p > 0.05).
Fungal community variation in phylum across storage time
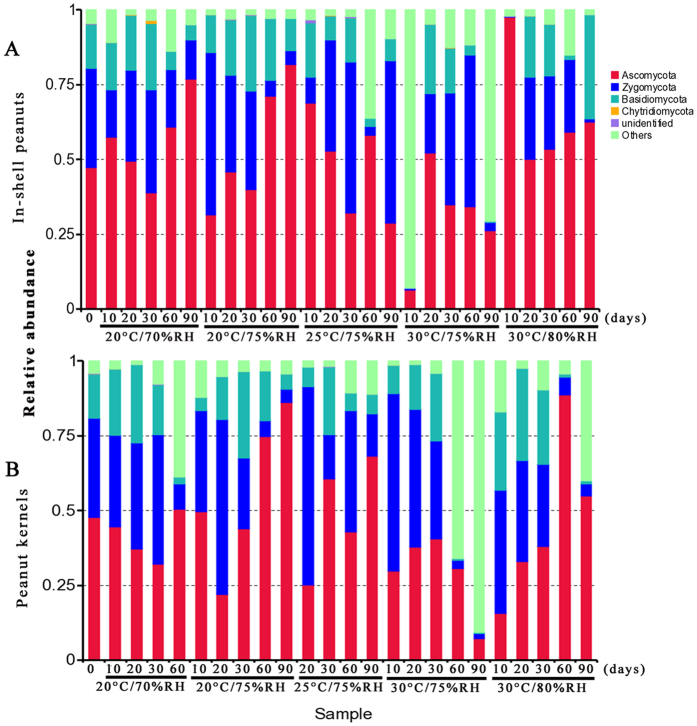
There are obvious variations in the relative abundance of fungal phyla per sample based on storage conditions and storage time (Fig. 2). In in-shell peanuts, four fungal phyla, i.e., Ascomycota, Basidiomycota, Chytridiomycota, and Zygomycota were identified. Of them, Ascomycota, Basidiomycota, and Zygomycota were the predominant phyla, with 50.7%, 13.0% and 23.7% relative abundance, respectively. In peanut kernels, the same four fungal phyla were identified. Of them, Ascomycota, Basidiomycota, and Zygomycota were the main phyla (44.2%, 13.5% and 27.8% relative abundance, respectively).
Figure 2.
Overall distribution of fungi at phylum level in in-shell peanuts (A) and peanut kernels (B) stored for 0–90 d at different conditions. RH: relative humidity.
In in-shell peanuts, the relative abundance of Ascomycota fungi increased from 30 to 90 d at 20 °C with 70% and 75% relative humidity, and from 20 to 90 d at 30 °C with 80% relative humidity. At 25 °C with 75% relative humidity, there were obvious oscillations in the relative abundance of Ascomycota fungi during storage. Similarly, in peanut kernels, there were also obvious oscillations in the relative abundance of Ascomycota fungi, but the regularity was absent.
Fungal community variation in genus level across storage time
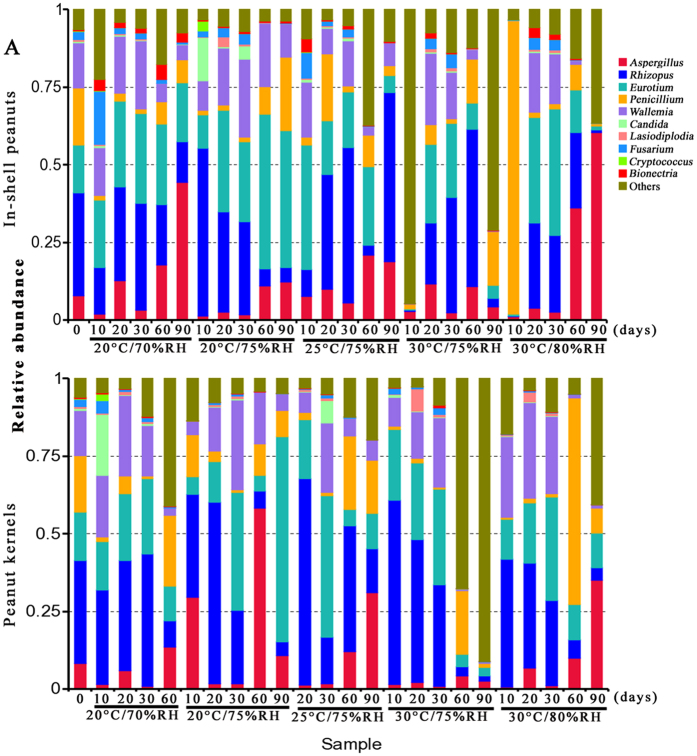
There were obvious variation in the relative abundance of fungal genera per-sample based on storage conditions and storage time (Fig. 3). In in-shell peanuts, the average number of clean reads retained after the filtering and denoising steps was 56,709, of which 54,426 reads were subsequently clustered into 110 OTUs and 41 fungal genera. Of them, Aspergillus, Eurotium, Penicillium, Rhizopus and Wallemia were the main genera, with average relative abundances of 12.1%, 21.6%, 10.1%, 33.1% and 11.3%, respectively. Fusarium had a relative abundance of 2.4%. The relative abundance of Aspergillus fungi at 60 and 90 d was significantly higher than that at 10, 20 or 30 d under all storage conditions (p < 0.05). In general, the relative abundance of Aspergillus fungi increased from 30 to 90 d, except in samples stored at 30 °C with 75% relative humidity. The relative abundance of Aspergillus fungi reached its maximum value (60.3%) at 30 °C with 80% relative humidity. At 20 and 30 d, Eurotium, Rhizopus and Wallemia were the main genera, with relative abundance higher than 15%, 20% and 10%, respectively.
Figure 3.
Overall distribution of fungi at genus level in in-shell peanuts (A) and peanut kernels (B) stored for 0–90 d at different conditions. RH: relative humidity.
In peanut kernels, the average number of clean reads retained after the filtering and denoising steps was 38,936, of which 36,718 were subsequently clustered into 91 OTUs; 37 fungal genera were identified. Of them, Aspergillus, Eurotium, Penicillium, Rhizopus and Wallemia were the most predominant genera (19.6%, 10.1%, 9.7%, 27.8% and 13.4% relative abundance, respectively). The relative abundance of Aspergillus fungi at 60 and 90 d was significantly higher than other days at 10, 20 or 30 d (p < 0.05). The relative abundance of Rhizopus fungi at 10 and 20 d were high (>30%) and decreased from 30 to 90 d. The relative abundance of Wallemia fungi was high at 20 and 30 d but further decreased from 30 to 90 d. The relative abundance of Eurotium fungi reached a maximum value at 30 d (> 20%) and subsequently decreased from 30 to 90 d except in samples stored at 20 °C with 75% relative humidity. The relative abundance of Penicillium reached the highest level at 60 d.
Changes in Mycobiome are associated with storage time
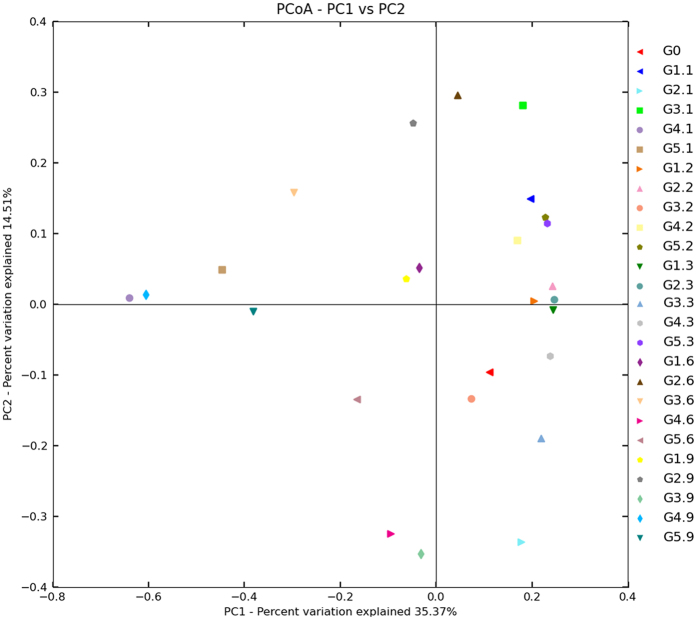
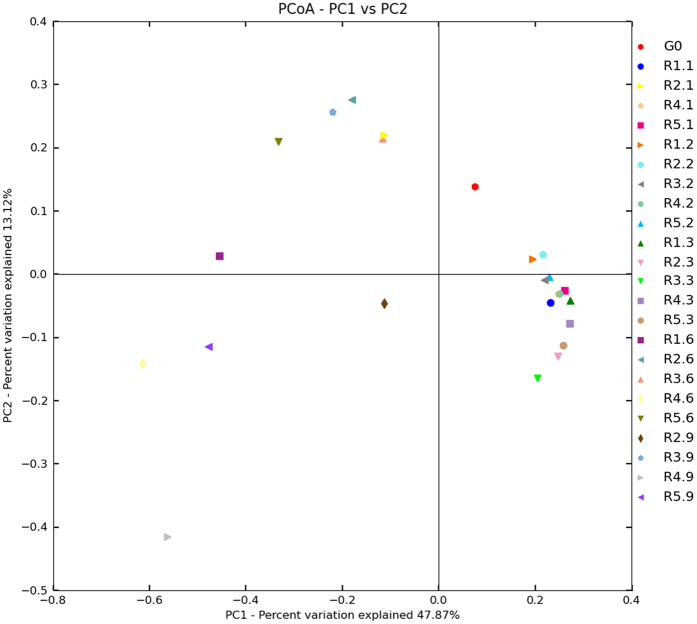
To investigate whether there is an association between any of the subject storage parameters and changes in mycobiome, we performed Principal Coordinate Analysis (PCoA). The results revealed that peanuts stored for 60–90 days and peanuts stored for 10–30 days clustered differently from each other (Figs 4 and 5). In in-shell peanuts, all peanuts at 60 and 90 d clustered in the left and PCoA case scores (Bray Curtis) were less than zero, except for G2.6 (from 60 d at 20 °C with 75% relative humidity); all samples at 10, 20 and 30 d clustered in the right and PCoA case scores were more than zero, except for G4.1 (from 10 d at 30 °C with 75% relative humidity) and G5.1 (from 10 d at 30 °C with 80% relative humidity). In peanuts kernels, all samples at 60 and 90 d clustered in the left and PCoA case scores were less than zero; all samples at 10, 20 and 30 d clustered in the right and PCoA case scores were more than zero, except for R2.1 (from 10 d at 20 °C with 75% relative humidity). These suggest a trend in association between storage time and the peanuts mycobiome.
Figure 4. Principal Coordinate Analysis (PCoA) of distribution of fungi in in-shell peanuts during storage.
G0: raw peanuts; G1.1, G1.2, G1.3, G1.6, G1.9: stored in-shell peanuts at 10, 20, 30, 60, 90 d with 20 °C/70% RH; G2.1, G2.2, G2.3, G2.6, G2.9: stored in-shell peanuts at 10, 20, 30, 60, 90 d with 20 °C/75% RH; G3.1, G3.2, G3.3, G3.6, G3.9: stored in-shell peanuts at 10, 20, 30, 60, 90 d with 25 °C/75% RH; G4.1, G4.2, G4.3, G4.6, G4.9: stored in-shell peanuts at 10, 20, 30, 60, 90 d with 30 °C/75% RH; G5.1, G5.2, G5.3, G5.6, G5.9: stored in-shell peanuts at 10, 20, 30, 60, 90 d with 30 °C/80% RH. RH: relative humidity.
Figure 5. Principal Coordinate Analysis (PCoA) of distribution of fungi in peanut kernels during storage.
G0: raw peanuts; R1.1, R1.2, R1.3, R1.6, R1.9: stored in-shell peanuts at 10, 20, 30, 60, 90 d with 20 °C/70% RH; R2.1, R2.2, R2.3, R2.6, R2.9: stored in-shell peanuts at 10, 20, 30, 60, 90 d with 20 °C/75% RH; R3.1, R3.2, R3.3, R3.6, R3.9: stored in-shell peanuts at 10, 20, 30, 60, 90 d with 25 °C/75% RH; R4.1, R4.2, R4.3, R4.6, R4.9: stored in-shell peanuts at 10, 20, 30, 60, 90 d with 30 °C/75% RH; R5.1, R5.2, R5.3, R5.6, R5.9: stored in-shell peanuts at 10, 20, 30, 60, 90 d with 30 °C/80% RH. RH: relative humidity.
Aflatoxins in stored peanuts
As shown in Table 3, of 25 in-shell peanuts five (20%) were contaminated with AFB1 (ranging from 0.34 to 10.40 μg/kg, four (16%) were contaminated with AFB2 (0.10–1.87 μg/kg), one (4%) were contaminated with AFG1 (0.72 μg/kg), and two (8%) were contaminated with AFG2 (0.15 μg/kg). Of 25 peanut kernels, four (16%) were contaminated with AFB1 (0.34–68.79 μg/kg, three (12%) were contaminated with AFB2 (0.07–6.25 μg/kg), and one (4%) was contaminated with AFG2 (0.24 μg/kg).
Table 3. Mean levels of aflatoxins B1, B2, G1 and G2 in stored peanuts.
| Storage |
In-shell peanuts Aflatoxins (μg/kg) |
Peanut kernels Aflatoxins (μg/kg) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Temperature/relative humidity | Time | AFB1 | AFB2 | AFG1 | AFG2 | AFB1 | AFB2 | AFG1 | AFG2 |
| G0 | 0 | – | – | – | – | ||||
| 20 °C/70% | 10 | 10.40 | 1.87 | – | 0.15 | – | – | – | – |
| 20 | – | – | – | – | – | – | – | – | |
| 30 | – | – | – | – | – | – | – | – | |
| 60 | 0.88 | 0.08 | – | – | – | – | – | – | |
| 90 | 0.35 | 0.07 | – | – | 0.35 | 0.07 | – | – | |
| 20 °C/75% | 10 | – | – | – | – | – | – | – | – |
| 20 | – | – | – | – | – | – | – | – | |
| 30 | – | – | – | – | – | – | – | – | |
| 60 | 0.76 | 0.10 | 0.72 | 0.15 | – | – | – | – | |
| 90 | – | – | – | – | – | – | – | – | |
| 25 °C/75% | 10 | – | – | – | – | – | – | – | – |
| 20 | – | – | – | – | – | – | – | – | |
| 30 | – | – | – | – | 68.79 | 6.25 | – | 0.24 | |
| 60 | – | – | – | – | – | – | – | – | |
| 90 | – | – | – | - | – | – | – | – | |
| 30 °C/75% | 10 | – | – | – | – | 7.65 | 0.54 | – | – |
| 20 | – | – | – | – | – | – | – | – | |
| 30 | – | – | – | – | – | – | – | – | |
| 60 | – | – | – | – | – | – | – | – | |
| 90 | – | – | – | – | – | – | – | – | |
| 30 °C/80% | 10 | – | – | – | – | – | – | – | – |
| 20 | – | – | – | – | – | – | – | – | |
| 30 | – | – | – | – | – | – | – | – | |
| 60 | – | – | – | – | – | – | – | - | |
| 90 | 0.34 | – | – | – | 0.34 | – | – | – | |
Discussion
The average number of raw reads, clean reads, and taxon reads in in-shell peanuts were 84,834, 56,709 and 54,426, respectively, and 61,243, 38,396 and 36,718 in peanut kernels, respectively. This result suggested that the number of fungi in in-shell peanuts was higher than that in peanut kernels. Furthermore, the total number of fungal OTUs in in-shell peanuts was 199, which was higher than in peanut kernels (OTUs: 153). The average number of OTUs detected per sample in in-shell peanuts was 110 (range 81–140), while that in peanut kernels was 91 (range: 69–114). Furthermore, in in-shell peanuts, 41 fungal genera were identified, which was higher than that in peanut kernels. Fungal diversity of in-shell peanuts was significantly higher than in peanut kernels during storage. This indicates that the micro-environment in peanut shell proves favorable for maintaining fungal diversity.
In general, Aspergillus, Eurotium, Penicillium Rhizopus, and Wallemia were predominant genera in both in-shell peanuts and peanut kernels during storage. This result attributes to the greater adaptation of these fungi to the substrate, especially during storage27,28. The occurrence of Aspergillus, Penicillium, and Rhizopus agrees with the findings of other investigators studying peanut kernels from Brazil27,28,29 and India30 using traditional isolation, enumeration and identification methods of the mycoflora on the Dichloran Rose Bengal Chloramphenicol agar (DRBC) or A. flavus and A. parasiticus agar (AFPA) media. Eurotium was not detected in these studies because Eurotium did not grow well on DRBC or AFPA media. Nakai et al.27 found a predominance of Fusarium spp. (67.7% in hulls and 25.8% in kernels) and Aspergillus spp. (10.3% in hulls and 21.8% in kernels). In the previous studies, only eight genera were isolated from peanuts kernels and hulls i.e. Aspergillus, Cladosporium, Drechslera, Fusarium, Penicillium, Phoma, Rhizopus, and Trichoderma. However, in our study, 41 and 37 fungal genera were detected in in-shell peanuts and peanut kernels during storage using high-throughput ITS2 sequencing technologies, respectively. The number of fungal genera reported is about 5-folds increase compared to previous studies. This is because the studies of evaluated the mycoflora in stored peanuts using traditional isolation, enumeration and identification provided only a limited snap shot of the fungal members of the microbiome. While, the barcoded Illumina paired-end sequencing (BIPES) method using in this study31 provided a more in-depth comprehensive profile of the mycobiome.
At 20–30 d, the relative abundance of Eurotium, Rhizopus and Wallemia were higher than that of Aspergillus, because the three genera fungi were xerophilic and grew well on substrates with low aw (Fig. 6). During growth, Eurotium, Rhizopus and Wallemia fungi released metabolic water on substrates with low aw, thereby favoring the growth of Aspergillus, which are less xerophilic fungi. Therefore, from 30 to 90 d, the relative abundance of Aspergillus increased while that of Eurotium, Rhizopus and Wallemia decreased. Similarly, the results from PCoA analysis showed some tendency for stored peanuts at 60–90 d to cluster together, and stored peanuts at 10–30 d to cluster together. These results suggested that storage time plays a vital role in impacting the variation of mycobiome in stored peanuts. However, given the limitation of lesser study samples in the current study, it is difficult to draw definite conclusions regarding association of the mycobiome with storage time and/or conditions. Therefore, further confirmations on researching larger population sizes were needed.
Figure 6. Storage time variation of water activity (aw) in in-shell peanuts (up) and peanut kernels (below) stored for 0–90 d at different conditions.
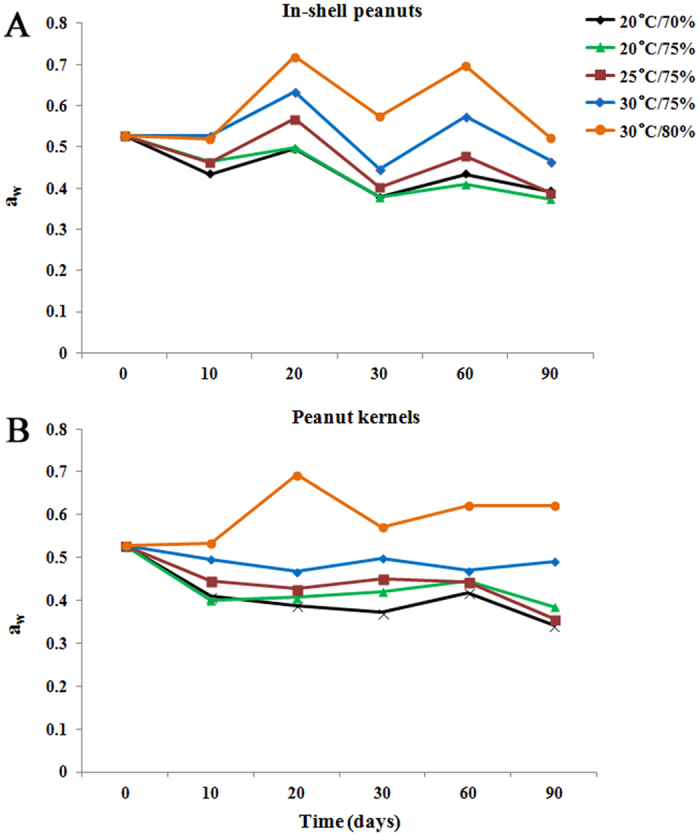
Wallemia is a genus of cosmopolitan xerophilic fungi that are present in several environments characterized by low aw32,33, and is frequently involved in food spoilage. In 2006, Sun et al.34 isolated one W. sebi isolate from the surface of apples. This was the first report of its occurrence in China as a saprophyte on foods. In the present study, Wallemia was identified in peanuts grown in China. The results confirmed that Wallemia grows well on substrates with low aw because aw of peanuts is ≤0.72 (ranging from 0.36 to 0.72) (Fig. 6). In general, the relative abundance of Wallemia in stored in-shell peanuts and peanut kernels were higher at 20–30 d and subsequently decreased from 30 to 90 d with the concomitant increase in Aspergillus.
In both in-shell peanuts and peanut kernels during storage, Aspergillus, Eurotium, Penicillium, Rhizopus and Wallemia were the predominant genera. Of them, Rhizopus was the most abundant genus with a relative abundances >20% as they could grow well on peanuts since it has a low aw of ≤0.72. Rhizopus is common saprobic fungi on plants and specialized parasites on animals. In general, the relative abundance of Rhizopus in stored in-shell peanuts and peanut kernels were high at 20–30 d and decreased from 30 to 90 d with the concomitant increase in Aspergillus. Eurotium is the teleomorph genus associated with Aspergillus, which could grow on substrates with low aw. These organisms are universally distributed in nature and are usually referred to as halophilic or xerophilic. They cause significant damage in stored grains, cereals and food products preserved by drying or salt/sugar addition35,36,37,38. More important, Eurotium release metabolic water on substrates with low aw, thereby creating favorable conditions for less xerophilic fungi (e.g., A. flavus and A. niger) that can produce more hazardous mycotoxins (e.g., aflatoxins and ochratoxins). The results of this study confirmed the above findings. The relative abundance of Aspergillus (with the exception of Eurotium) increased from 30 to 90 d in stored in-shell peanuts and peanut kernels, i.e 2.6% to 60.3%, and 1.1% to 35%, respectively, especially at 30 °C with 80% relative humidity. So we could conclude that rapidly grown Eurotium at initial stage could release metabolic water which in turn increases aw, thereby creating a favorable condition for Aspergillus species growth. Most fungi from grains grow well on DRBC, and Pitt and Hocking37 reported that DRBC is adequate for the numeration of fungi present in food and feed. Eurotium growth was observed on DG18 medium rather than on DRBC. Therefore, Eurotium in peanuts should be determined using DG18, which is the medium for xerophilic fungi32.
The aw in this study (0.37–0.72 in in-shell peanuts and 0.34–0.69 in peanut kernels) (Fig. 6) was below the minimum range of 0.78–0.80 established for the growth of A. flavus39. The low aw in the stored samples is probably due to the previously used process of peanut drying. In the present experiment, the temperature ranged from 20 °C to 30 °C and relative humidity ranged from 70% to 80%. Therefore, the temperature was lower than 32–33 °C, which is the optimum temperature for the growth of A. flavus40. The relative humidity values in the samples were lower than those measured by Christensen et al.41, who reported a relative humidity of approximately 83–85%, which favors the growth of A. flavus. Due to the low aw of stored peanuts, temperature and relative humidity, only nine of the 51 samples were contaminated with aflatoxins at levels ranging from 0.34 to 68.79 μg/kg. And only one sample was contaminated with aflatoxins at levels >20 μg/kg, which is the maximum level allowed by the National Health and Family Planning Commission of the P.R. China for AFB1 (http://www.nhfpc.gov.cn/cmsresources/mohwsjdj/cmsrsdocument/doc11939.pdf). The results of aflatoxins are also in accordance with the results of the mycological analysis, because the percentages of A. flavus, which are well-known aflatoxin-producing species, are lower than 0.01%.
Conclusions
The results of this study revealed that there were more genera, species and number of fungi in in-shell peanuts than in peanut kernels, and suggested that the micro-environment in shell was more suitable for maintaining the fungal biodiversity and resist infection of fungi from outer environment. Aspergillus, Eurotium, Penicillium, Rhizopus and Wallemia were the predominant genera in both in-shell peanuts and peanut kernels during storage. At 20 to 30 d, Eurotium, Rhizopus and Wallemia were the main genera; however, from 30 to 90 d, their relative abundance decreased and that of Aspergillus increased. Due to low aw values (0.34–0.72) of stored peanuts, nine of 51 samples were contaminated with aflatoxins, and only one sample had AFB1 levels >20 μg/kg. This study identified the mycobiome and its variation in stored peanuts during simulated storage using high-throughput ITS2 sequencing, and provided the basis for a detailed characterization and identification of mycobiome in stored peanuts.
Methods
Ethics Statement
Specific permission was not needed for our field studies. The peanuts variety used in our field study was main cultivar name Baisha 1016 in Hubei province. No transgenic or created mutant plant has been used in our study. Also we confirm that the field studies did not involve endangered or protected species.
Sample preparation
Peanuts were obtained from Xiangyang City, Hubei province, which is in the center of Yangtze River valley. After harvest, the peanuts were transported to Beijing in 25-kg bags (a total of five bags) in 3 d. Half of the peanuts were unshelled (peanut kernels). Both peanut kernels and in-shell peanuts were stored at similar conditions. According to climatic conditions (temperature and relative humidity) of Xiangyang City from April to June, both peanut kernels and in-shell peanuts were stored in a ZXMP-A1230 constant temperature and humidity incubator (Zhicheng, Shanghai, China) at five storage conditions: 20 °C with 70% relative humidity, 20 °C with 75% relative humidity, 25 °C with 75% relative humidity, 30 °C with 75% relative humidity, and 30 °C with 80% relative humidity. Samples were collected at 10, 20, 30, 60 and 90 d of storage and analyzed.
Total microbiome genomic DNA extraction
Water was sterilized at 121 °C for 30 min, and then filtered through 0.22 μm filters. The sterile water was used as a negative control for the experiment. In this experiment, peanuts kernels (100 g) were separated from the hulls and washed with 100 ml sterile water. Water samples were collected and vacuum-filtered through 0.22 μm filters within 24 hours. Filters containing sample were placed in 50-ml tubes and stored at −20 °C. Genomic DNA was extracted from the filters using the MoBio PowerWater® DNA Isolation Kit (MoBio Laboratories, Inc., Carlsbad, CA, USA) according to manufacturer’s recommendations. The final DNA elution was performed with sterile deionized water instead of the provided buffer. DNA quality and quantity were measured by spectrophotometric quantification in a Beckman DU800 (Beckman, USA) and NanoDrop 1000 (Thermo Fisher Scientific, USA), and by agarose gel electrophoresis. Extracted DNA was stored at −80 °C prior to amplification and sequencing.
PCR amplification and ITS2 sequencing
PCR was used to amplify the ITS2 region of rDNA. The barcoded primers ITS2F (5′-GCATCGATGAAGAACGCAGC-3′) and ITS2R (5′-TCCTCCGCTTATTGATATGC-3′) were used to amplify fungal ITS2 fragments. PCR reactions were carried out in a total reaction volume of 30 μl consisting 15 μl of Phusion High-Fidelity PCR Master Mix (New England Biolabs Inc. Ipswich, MA, USA), 0.2 μM of forward and reverse primers, and 10 ng of template DNA. The PCR amplification program consisted of a initial heating to 98 °C for 1 min, 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s and extension at 72 °C for 60 s, followed by a 5-min extension at 72 °C prior to storage at 4 °C. Amplified products were cleaned and purified using the GeneJET Gel Extraction Kit (Thermo Scientific, South Logan, UT 84321, USA) according to the manufacturer’s instructions. Of 51 peanut samples, 49 samples were successfully amplified. Amplicons were then quantified and sequencing libraries were generated using NEB Next Ultra DNA library Prep Kit for Illumina (New England Biolabs Inc. USA) according to manufacturer’s recommendations. The amplicon libraries were subsequently sequenced on an Illumina MiSeq platform at Novogene (Novogene, Beijing, P.R. China).
Bioinformatics analyses
Paired-end reads from the original DNA fragments were merged by using FLASH42. Sequences were analyzed with the QIIME43 software package using default parameters for each step. The UCLUST method44,45 was used to cluster the sequences into OTUs at an identity threshold of 97%. Meanwhile, the RDP Classifier46 was used to assign each OTU to a taxonomic level. Other analyses, including rarefaction curves, Shannon index, and Good’s coverage, were performed with QIIME. In addition, the OTU table produced by the QIIME pipeline was imported into MEGAN 4 and mapped on the NCBI taxonomy database47. Abundance-based comparisons were therefore made solely within selected taxonomic groups such as Aspergillus, Eurotium, Penicillium, Rhizopus and Wallemia, using an OTU table that was rarified in QIIME.
PCoA has been recognized as a simple and straight-forward method to group and separate samples in a dataset, and has been used in disease-association, gender-association and ethnicity studies19,48,49. In the current study, PCoA was used to analyze the sequencing results using the Multivariate Statistical Package, MVSP (Kovach, Wales, UK) and SAS (Cary, NC). The PCoA performs an Eigen analysis on the data matrix using a Brays Curtis distance metric.
Determination of aflatoxins
Aflatoxins levels were determined by Chinese standard methods50 and AOAC method 994.0851 with minor modifications. In this experiment, peanut kernels (50 g) were manually de-shelled, ground, mixed to obtain peanut paste, and stored at −20 °C until analysis. Finely ground samples (5.0 g) were extracted with 15 ml of acetonitrile:water (84:16, v/v). Following ultrasonic extraction at 50 °C for 10 min and filtration through double-layer slow quantitative filter paper, 4 ml of the resulting filtrate was mixed with 2 ml petroleum ether. The mixture was mixed on a vortex for 30 s and allowed to stand for 15–20 min. The lower layer (3 ml) was collected, mixed with 8 ml pure water and filtered through a 0.45 μm organic membrane. Extracts (8 ml) were passed through immunoaffinity columns with a flow rate of one droplet per second and eluted with 2 ml of methanol into glass tubes. The eluate was evaporated to dryness under a stream of nitrogen gas at 60 °C. The purified extract was re-dissolved with 1 ml of acetonitrile : water (15 : 85, v/v). The resulting supernatant was collected in glass tubes for high-performance liquid chromatography (HPLC) quantitative analysis.
The determination of aflatoxin levels was performed by HPLC. HPLC analysis was performed with a Waters 2695 (Waters Corporation, Milford, MA, USA) coupled to a Waters 2475 fluorescence detector (λexc 360 nm; λem 440 nm) and a post-column derivation system, and an Agilent TC-C18 column (250 × 4.6 mm, 5 μm particle size). The mobile phase (water : methanol : acetonitrile, 4 : 1 : 1) was pumped at a flow rate of 0.5 mL/min. AFB1, B2, G1 and G2 (Sigma-Aldrich, St. Louis, MO, USA) were used as standards. The mean recovery of the method used was calculated by spiking peanut kernels at different levels ranging from 1 to 100 ng/g of aflatoxins and was estimated at 95.2 ± 8.4%. The lowest detection limit was 1 ng/g.
Statistics
All the experiments results were evaluated using analysis of variance (ANOVA) for multiple comparisons followed by the Turkey test. Differences were considered significant at p < 0.05.
Additional Information
How to cite this article: Xing, F. et al. Variation in fungal microbiome (mycobiome) and aflatoxins during simulated storage of in-shell peanuts and peanut kernels. Sci. Rep. 6, 25930; doi: 10.1038/srep25930 (2016).
Supplementary Material
Acknowledgments
This work was funded by National Basic Research Program of China (973 program) (2013CB127801, 2013CB127805), Special Fund for Agro-scientific Research in the Public Interest (201203037).
Footnotes
Author Contributions The experiments were conceived and designed by Y.L. and F.X. The experiments were performed by N.D. and F.X. The date was analyzed by F.X. The following authors contributed in buying reagents or materials or provided analysis tools: J.N.S., X.L., L.W., L.Z., Y.Z. and Y.W. Finally, F.X. was responsible for writing the paper.
References
- Santos R. C. D. Nova cultivar de amendoim para as condições do nordeste brasileiro. Pesq. Agrop. Bras. 35, 665–670 (2000). [Google Scholar]
- Bankole S. & Adebanjo A. Mycotoxins in food in West Africa: current situation and possibilities of controlling it. Afr. J. Biotechnol. 2, 254–263 (2004). [Google Scholar]
- Williams J. H. et al. Human aflatoxicosis in developing countries: a review of toxicology, exposure, potential health consequences, and interventions. Am. J. Clin. Nutr. 80, 1106–1122 (2004). [DOI] [PubMed] [Google Scholar]
- Barros G., Torres A., Palacio G. & Chulze S. Aspergillus species from section Flavi isolated from soil at planting and harvest time in peanut‐growing regions of Argentina. J. Sci. Food Agric. 83, 1303–1307 (2003). [Google Scholar]
- Passone A., Doprado M. & Etcheverry M. Monitoring of aflatoxin contamination potential risk in sun dried peanuts and stored in big bag. VI Congreso Latinoamericano de Micología, Mar del Plata, Argentina. 204 (2008, Nov. 10th–13th). Available at: http://www.almic.org. (Accessed: 20th February 2014).
- Passone M. A., Rosso L. C., Ciancio A. & Etcheverry M. Detection and quantification of Aspergillus section Flavi spp. in stored peanuts by real-time PCR of nor-1 gene, and effects of storage conditions on aflatoxin production. Int. J. Food Microbiol. 138, 276–281 (2010). [DOI] [PubMed] [Google Scholar]
- Santos C. C. M., Lopes M. R. V. & Kosseki S. Y. Ocorrêcia de aflatoxinas em amendoim e produtos de amendoim comercializados naregiā de Sāo José de Rio Preto/SP. Rev. Inst. Adolfo Lutz 60, 153–157 (2001). [Google Scholar]
- International Agency for Research on Cancer (IARC). Some naturally occurring substances: food items and constituents, heterocyclic aromatic amines and mycotoxins. IARC Monogr. Eval. Carcinog. Risks Hum. 56, 245–395 (1993).8411624 [Google Scholar]
- International Agency for Research on Cancer (IARC). Some traditional herbal medicines, some mycotoxins, naphthalene and styrene. IARC Monogr. Eval. Carcinog. Risks Hum. 82, 1–556 (2002). [PMC free article] [PubMed] [Google Scholar]
- Commission of the European Communities. Commission regulation (EU) No 165/2010 of 26 February 2010 amending Regulation (EC) No 1881/2006 setting maximum levels for certain contaminants in foodstuffs as regards aflatoxins. Off. J. Eur. Union L50, 8–12 (2010). [Google Scholar]
- Food and Drug Administration U.S. Aflatoxin in Peanuts and Peanut Products: 2010; CPG Sec. 570.375. Compliance manuals. (2010) Available at: http://www.fda.gov/ICECI/ComplianceManuals. (Accessed: 12th January 2016).
- Smith J. E. & Ross I. C. The toxigenic Aspergillus. (eds Smith J. E. & Henderson R. S.) 31–61 (CRC press, 1991). [Google Scholar]
- Giorni P., Magan N. & Battilani P. Environmental factors modify carbon nutritional patterns and niche overlap between Aspergillus flavus and Fusarium verticillioides strains from maize. Int. J. Food. Microbiol. 130, 213–218 (2009). [DOI] [PubMed] [Google Scholar]
- Ding X., Li P., Bai Y. & Zhou H. Aflatoxin B1 in post-harvest peanuts and dietary risk in China. Food Control 23, 143–148 (2012). [Google Scholar]
- Begerow D., Nilsson H., Unterseher M. & Maier W. Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl. Microbiol. Biot. 87, 99–108 (2010). [DOI] [PubMed] [Google Scholar]
- Ekblom R. & Galindo J. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 107, 1–15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E. A. et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum M. A. et al. Characterization of the oral fungal microbiome (Mycobiome) in healthy individuals. PLoS Pathog. 6, e1000713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar Rodríguez M. et al. Obesity changes the human gut mycobiome. Sci. Rep. 5, 14600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. et al. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet. 11, e1005614 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch L. F. et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1, 283–290 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C. L., Hamady M., Knight R. & Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microb. 75, 5111–5120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin M. L. et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 103, 12115–12120 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello A. et al. ITS-1 versus ITS-2 pyrosequencing: a comparison of fungal populations in truffle grounds. Mycologia 103, 1184–1193 (2011). [DOI] [PubMed] [Google Scholar]
- Arfi Y., Buée M., Marchand C., Levasseur A. & Record E. Multiple markers pyrosequencing reveals highly diverse and host-specific fungal communities on the mangrove trees Avicennia marina and Rhizophora stylosa. FEMS Microbiol. Ecol. 79, 433–444 (2012). [DOI] [PubMed] [Google Scholar]
- Nakai V. K. et al. Distribution of fungi and aflatoxins in a stored peanut variety. Food Chem. 106, 285–290 (2008). [Google Scholar]
- Rossetto C. A. V., Silva O. F. & Araújo A. E. S. Influência da calagem, da época de colheita e da secagem na incidência de fungos e aflatoxinas em grãos de amendoim armazenados. Cienc. Rural 35, 309–315 (2005). [Google Scholar]
- Fernandez E. M., Rosolem C. A., Maringoni A. C. & Oliveira D. M. T. Fungus incidence on peanut grains as affected by drying method and Ca nutrition. Field Crop Res. 52, 9–15 (1997). [Google Scholar]
- Bhattacharya K. & Raha S. Deteriorative changes of maize, groundnut and soybean seeds by fungi in storage. Mycopathologia 155, 135–141 (2002). [DOI] [PubMed] [Google Scholar]
- Zhou H. W. et al. Bipes, a cost-effective high-throught method for assessing microbial diversity. ISME J. 5, 741–749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R. A., Hoekstra E. S., Lund F., Filtenborg O. & Frisvad J. C. Methods for the detection, isolation and characterization of food-borne fungi in Introduction to food- and airborne fungi (eds Samson R. A. et al.) 283–297 (Centraalbureau voor Schimmelcultures, 2004). [Google Scholar]
- Zalar P., de Hoog G. S., Schroers H. J., Frank J. M. & Gunde-Cimerman N. Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. Et ord. nov.). Antonie van Leeuwenhoek 87, 311–328 (2005). [DOI] [PubMed] [Google Scholar]
- Sun G., Zhang M., Ma H. & Gleason M. L. Wallemia- a genus newly recorded from China. Mycotaxon 95, 277–280 (2006). [Google Scholar]
- Thom C. & Raper K. B. The Aspergillus glaucus group. U.S. Dep. Agric. Misc. Pub. 426, 1–46 (1941). [Google Scholar]
- Raper K. B. & Fennell D. I. The genus Aspergillus (eds Raper K. B. & Fennell D. I.) 686 (Williams & Wilkins, 1965). [Google Scholar]
- Pitt J. I. & Hocking A. D. Fungi and food spoilage 3rd edn (eds Pitt J. I. & Hocking A. D.) 19–51 (Springer, 2009). [Google Scholar]
- Dovičičová M. The study of potentially toxigenic species of the genus Aspergillus and associated perfects isolated from wheat grains of Slovak origin, PhD thesis, Slovak University of Agriculture in Nitra (2010).
- Lancey J., Ramakrishna N., Hamer A., Magan N. & Marfleet C. Grain fungi in Handbook of applied mycology, Vol. 3 (eds Arora D. K. et al.) 12–77 (Marcel Dekker, 1991). [Google Scholar]
- Hocking A. D. Toxigenic Aspergillus species in Food microbiology: Fundamentals and frontiers 2nd edn, (eds Doyle M. P. et al.) 393–405 (ASM Press, 1997). [Google Scholar]
- Christensen C. M., Mirocha C. J. & Meronuck R. A. Molds, mycotoxins and mycotoxicoses (eds Christensen C. M. et al.) 142 (University of Minnesota Press, 1977). [Google Scholar]
- Magoc T. & Salzberg S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillever P., Siskaroodi M., Keshavarzian A. & Mudu E. A. Quantitative assessment of the human gut microbiome using multitag pyrosequencing. Chem. Biodivers. 7, 1065–1075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D., Mitra S., Ruscheweyh H. J., Weber N. & Schuster S. C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21, 1552–1560 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L. et al. Principal component analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- McVean G. A genealogical interpretation of principal components analysis. PLoS Genet. 5, e1000686 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N. et al. Variation in fungal microbiome (mycobiome) and aflatoxin in stored in-shell peanuts at four different areas of China. Front. Microbiol. 6, 1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- AOAC International. Natural contaminants in Official methods of the Association of Official Analytical Chemists 17th edn, (ed Cunnif P.) Ch. 49 (AOAC International, 2000). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.