Abstract
The typical insect mitochondrial (mt) genome organization, which contains a single chromosome with 37 genes, was found in the infraorder Pentatomomorpha (suborder Heteroptera). The arrangement of mt genes in these true bugs is usually the same as the ancestral mt gene arrangement of insects. Rearrangement of transfer RNA (tRNA) genes, however, has been found in two subfamilies of flat bugs (Mezirinae and Calisiinae, family Aradidae). In this study, we sequenced the complete mt genomes of four species from three other subfamilies (Aradinae, Carventinae and Aneurinae). We found tRNA gene rearrangement in all of these four species. All of the rearranged tRNA genes are located between the mitochondrial control region and cox1, indicating this region as a hotspot for gene rearrangement in flat bugs; the rearrangement is likely caused by events of tandem duplication and random deletion of genes. Furthermore, our phylogenetic and dating analyses indicated that the swap of positions between trnQ and trnI occurred ~162 million years ago (MYA) in the most recent common ancestor of the five subfamilies of flat bugs investigated to date, whereas the swap of positions between trnC and trnW occurred later in the lineage leading to Calisiinae, and the translocation of trnC and trnY occurred later than 134 MYA in the lineage leading to Aradinae.
Mitochondrial (mt) genomes are one of the most explored sources of molecular markers for studying animal phylogeny and phylogeography1,2. In addition to the sequences of mt genes and mt genomes, arrangement of mt genes has also been explored in these studies3,4,5. Like in most other animals, insect mt genomes are circular and contain 37 genes (13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes) and a control region on a single chromosome2,6. The arrangement of genes in mt genomes is usually stable in insects. Indeed, most insects known retained the ancestral mt gene arrangement of Pancrustacea (i.e. hexapods and crustaceans)6,7. However, multipartite and fragmented mt genomes with two or more chromosomes or minichromosomes have been found in sucking lice (order Phthiraptera)8, booklice (Order Psocoptera)9,10 and thrips (order Thysanoptera)11. Extensive rearrangement of mt genes has also been found in these three paraneopteran orders9,12,13,14.
The ancestral mt gene arrangement of insects is retained in most species of the fourth paraneopteran order, Hemiptera15,16,17,18; however, gene rearrangement has been found in the suborder Sternorrhyncha (i.e. whiteflies)19,20. True bugs (suborder Heteroptera), with the largest number of published complete mt genomes in Hemiptera, showed mt gene rearrangement in unique-headed bugs21, pyrrhocoroid bugs22 and flat bugs22,23,24. Flat bugs (family Aradidae) is a relatively large family in the infraorder Pentatomomorpha of Heteroptera, with approximately 1,970 species and 270 genera in eight extant subfamilies and a extinct subfamily Archearadinae25,26,27. Most of the flat bugs are mycophagous (i.e. fungi eater) and are generally found under tree bark28. The sister relationship between Aradidae and Termitaphididae (together comprising the superfamily Aradoidea) is less controversial and the Aradoidea is consistently placed as sister to the rest of Pentatomomorpha29. However, the phylogenetic relationships among the subfamilies of flat bugs are still poorly studied27,30.
Prior to this study, the mt genomes of three flat bugs have been sequenced, representing two subfamilies: Neuroctenus parus and Brachyrhynchus hsiaoi from the subfamily Mezirinae22,24, and Aradacanthia heissi from the subfamily Calisiinae23. All of these three species have tRNA gene rearrangement near the control region in their mt genomes. To understand how tRNA gene rearrangement occurred in the flat bugs (family Aradidae), we sequenced the mt genomes of four more species of flat bugs from three other subfamilies: Aradus compar (Aradinae), Libiocoris heissi (Carventinae), Aneurus similis (Aneurinae) and Aneurus sublobatus (Aneurinae). We compared the mt gene arrangement among the seven species of flat bugs from five subfamilies of the Aradidae. Our results indicated that mt tRNA gene rearrangement in flat bugs was driven by events of tandem duplication and random deletion near the control region. trnQ and trnI swapped their positions in the most recent common ancestor of the five subfamilies of flat bugs ~162 million years ago (MYA); later, trnC and trnW swapped their positions in the lineage leading to the subfamily Calisiinae whereas trnC and trnY were translocated in the lineage leading to the subfamily Aradinae later than 134 MYA.
Results
General features of flat bug mitochondrial genomes
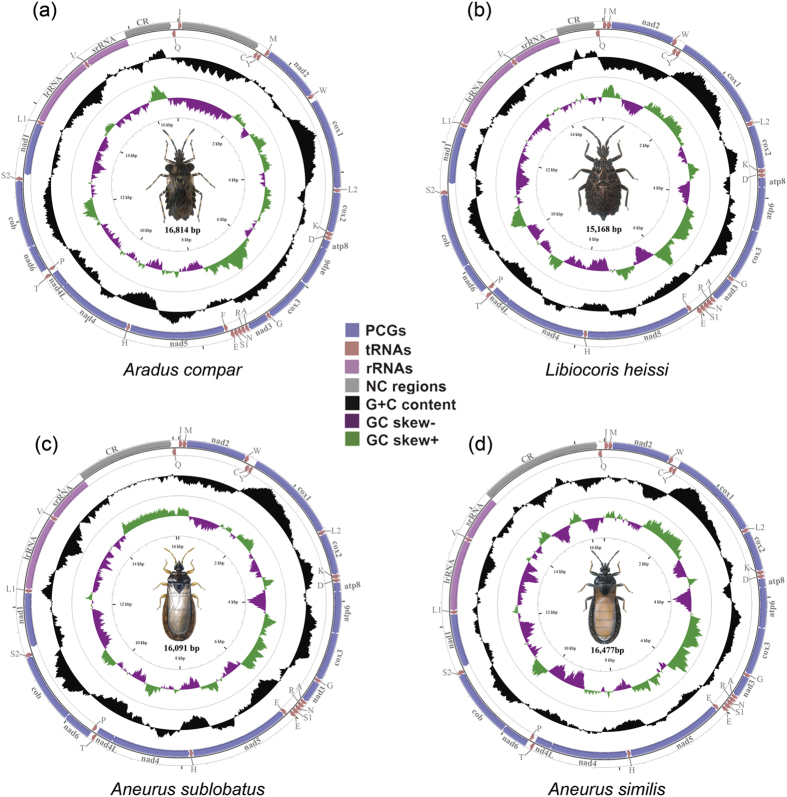
We sequenced the complete mt genomes of four species of flat bugs in the present study (Fig. 1), making mt genome sequences available for seven species of flat bugs representing five subfamilies of the Aradidae (Table 1). The mt genomes of the seven flat bugs range from 15,168 bp (Libiocoris heissi) to 16,814 bp (Aradus compar) in length. Each mt genome of the flat bug contains the 37 genes commonly found in animal mt genomes, as well as a putative control region (CR) which included the presumed origin of replication and promoters for transcription initiation6. All of the protein coding-genes (PCGs) start with ATN codons except for cox1, which start with TTG codon (Supplementary Table S1).
Figure 1. Mitochondrial genomes of four flat bugs sequenced in this study.
Circular maps were drawn with CGView66. Arrows indicate the orientation of gene transcription. Abbreviations of gene names are: atp6 and atp8 for ATP synthase subunits 6 and 8, cox1–3 for cytochrome oxidase subunits 1–3, cob for cytochrome b, nad1–6 and nad4L for NADH dehydrogenase subunits 1–6 and 4L, lrRNA and srRNA for large and small rRNA subunits. tRNA genes are indicated with their one-letter corresponding amino acids. CR for control region. The GC content was plotted using a black sliding window, as the deviation from the average GC content of the entire sequence. GC-skew was plotted as the deviation from the average GC-skew of the entire sequence. The illustrations of four flat bugs were drawn by F.S.
Table 1. Structural features of flat bug mitochondrial genomes.
| Subfamily | Species | Whole genome |
lrRNA |
srRNA |
Control region |
Source | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Size(bp) | A + T(%) | Size(bp) | A + T% | Size(bp) | A + T% | Size(bp) | A + T% | |||
| Mezirinae | Neuroctenus parus | 15354 | 68.9 | 1514 | 71.9 | 785 | 68.8 | 649 | 69.2 | NC_012459 |
| Brachyrhynchus hsiaoi | 15250 | 70.4 | 1245 | 74.2 | 808 | 72.5 | 703 | 69.8 | NC_022670 | |
| Calisiinae | Aradacanthia heissi | 15528 | 74.7 | 1230 | 77.6 | 788 | 74.0 | 1032 | 81.5 | HQ441233 |
| Aradinae | Aradus compar | 16814 | 64.1 | 1267 | 69.8 | 801 | 62.7 | 795 | 64.5 | present study |
| Carventinae | Libiocoris heissi | 15168 | 68.7 | 1242 | 72.0 | 782 | 70.1 | 586 | 66.9 | present study |
| Aneurinae | Aneurus sublobatus | 16091 | 67.7 | 1251 | 70.3 | 790 | 68.9 | 1569 | 68.1 | present study |
| Aneurus similis | 16477 | 68.0 | 1234 | 71.6 | 789 | 68.9 | 2067 | 69.5 | present study | |
The mt genomes of the flat bugs were AT biased and showed the positive AT- and negative GC-skews (Supplementary Table S2), as is usually observed in insect mt genomes31. For PCGs, genes on the J-strand had a positive AT- and negative GC-skews, whereas the opposites were observed on the N-strand. It has been proposed that the strand bias of nucleotide composition might be related to replication and transcription mechanisms32.
Control regions and intergenic spacers
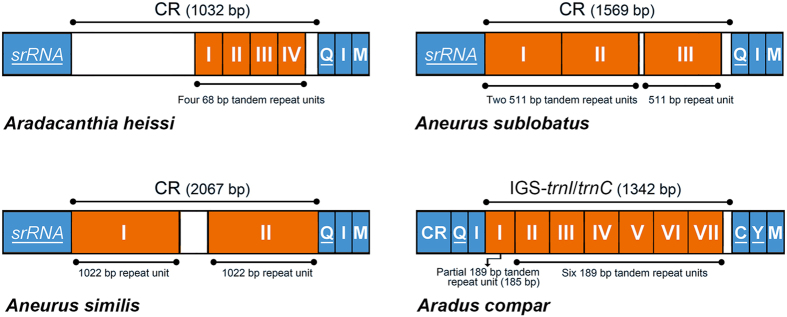
The CRs of the seven flat bugs varied considerably in length, ranging from 586 bp in Libiocoris heissi to 2,067 bp in Aneurus similis. Repeat sequences were found in the CRs of Aradacanthia heissi, Aneurus similis and Aneurus sublobatus: four copies of 68-bp tandem repeat unit in Aradacanthia heissi, two identical 1,022-bp repeats in Aneurus similis, and three identical 511-bp repeat units in Aneurus sublobatus (the first two units are tandem repeats) (Fig. 2).
Figure 2. Repeat sequences in the control region and the intergenic spacer of flat bug mitochondrial genomes.
Abbreviations of gene names follow Fig. 1. Genes are transcribed from left to right except those underlined, which have the opposite transcriptional orientation. The location and copy number of repeat units were shown by colored rectangle with Roman numerals inside. IGS-trnI/trnC for the intergenic spacer between trnI and trnC.
In addition to the CRs, we also identified three intergenic spacers (IGS), between the genes trnQ-trnI, trnS-nad1, and trnP-nad6 that we designate “IGS-trnQ/trnI, IGS-trnS2/nad1 and IGS-trnP/nad6”, respectively, in all of the seven flat bug mt genomes (Table S3). Two long IGSs, between the genes trnI-trnC and trnW-cox1 that we designate “IGS-trnI/trnC and IGS-trnW/cox1”, were found in Aradus compar. Tandem repeat sequences were also found in IGS-trnI/trnC (1,342 bp) in Aradus compar (Fig. 2). Most of the IGSs were located between rearranged genes, e.g. IGS-trnQ/trnI (44–135 bp) in all of the seven species and IGS-trnI/trnC and IGS-trnW/cox1 (56 bp) in Aradus compar (discussed below). In general, the CRs and IGSs varied substantially in both length and sequence among the seven species of flat bugs.
Gene rearrangement
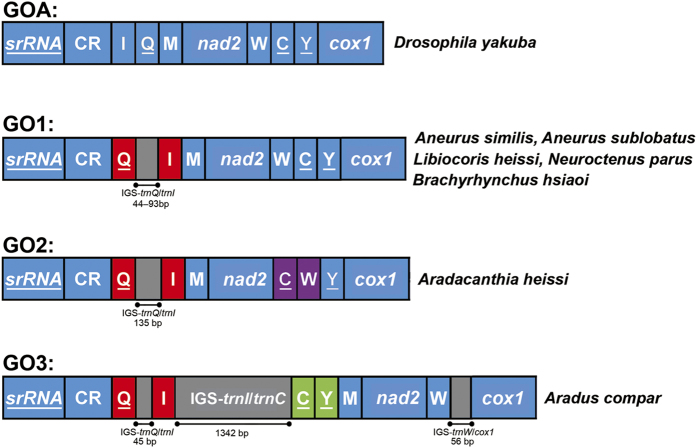
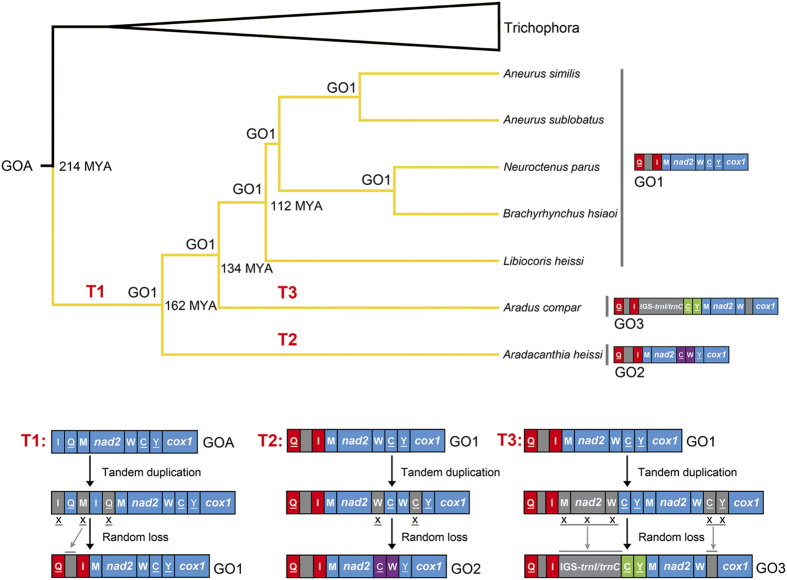
Compared with the ancestral mt gene order of insects (GOA, Fig. 3)6, tRNA gene rearrangement was found in all of the seven flat bugs and resulted in three types of gene orders (GO1–3). The five species from the subfamilies Aneurinae, Carventinae and Mezirinae possessed the same gene order (GO1) with the swap of positions between trnQ and trnI (Fig. 3). GO2 was found in Aradacanthia heissi with the swap of positions between trnQ and trnI, and between trnC and trnW (Fig. 3). The swap of positions between trnQ and trnI and the translocation of trnC and trnY were found in Aradus compar (GO3; Fig. 3).
Figure 3. Mitochondrial gene rearrangements in flat bugs.
Abbreviations of gene names follow Fig. 1. Genes are transcribed from left to right except those underlined, which have the opposite transcriptional orientation. Rearrangements of tRNA genes were highlighted by color. IGS for the intergenic spacer.
Phylogenetic relationship and divergence time reconstruction
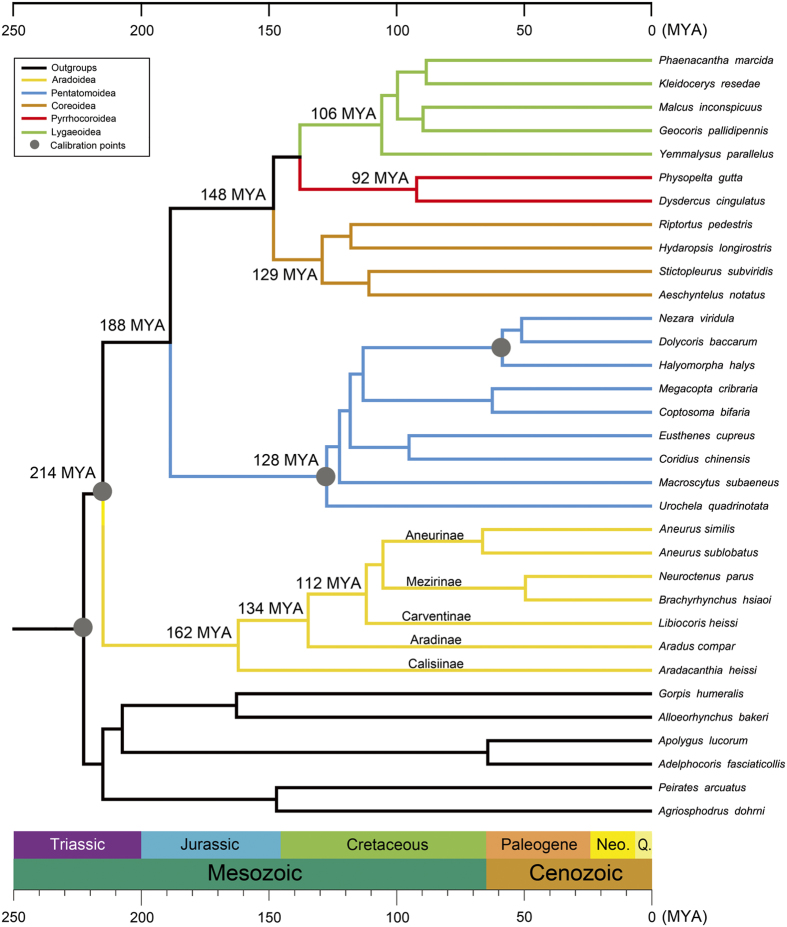
To understand the evolution of tRNA gene rearrangement in flat bugs, we performed phylogenetic analyses and estimated the divergence time for Aradidae and Pentatomomorpha. Twenty-seven Pentatomomorpha species from five superfamilies (Aradoidea, Pentatomoidea, Lygaeoidea, Pyrrhocoroidea and Coreoidea), together with six outgroup species from Cimicomorpha, were included in our phylogenetic analyses. We used maximum likelihood (ML) and Bayesian inference (BI) methods and generated four phylogenetic trees with two data matrices, PCGR and PCG12R (also see Materials and Methods below). The monophyly of each superfamily was well supported in all trees with bootstrap values (BS) 100 and posterior probabilities (PP) 1 (Supplementary Fig. S1). The sister relationship between Aradoidea and other superfamilies was also highly supported (BS = 100 and PP = 1). Pentatomoidea was placed as the sister group to Eutrichophora (Lygaeoidea, Pyrrhocoroidea and Coreoidea). In Eutrichophora, we found conflicting phylogenetic results between different datasets and methods. The sister-group relationship between Pyrrhocoroidea and Lygaeoidea was recovered in two BI trees (PP = 1 and PP = 0.99) and ML analysis of the dataset PCGR (BS = 73), however, ML analysis of the dataset PCG12R supported the sister group of Pyrrhocoroidea and Coreoidea (BS = 49). The relationships among the five subfamilies of flat bugs were recovered as: (Calisiinae + (Aradinae + (Mezirinae + Carventinae + Aneurinae))). Although the relationship among the three subfamilies (Mezirinae, Carventinae and Aneurinae) was not well resolved (BS < 65 and PP < 0.62) based on the present taxon sampling, the close relationship of these three subfamilies to the exclusion of other two subfamilies was strongly supported in both ML and BI analyses (BS = 100 and PP = 1).
Relaxed molecular clock analyses were performed for our two datasets using four fossil calibration points. Results were not significantly different between the two datasets (PCG12R and PCGR; Supplementary Table S4). The most recent common ancestor (MRCA) of Pentatomomorpha was estimated to be at the late Triassic (~214 MYA); the diversification into five superfamilies occurred from the late Jurassic to middle Cretaceous (214 to 92 MYA) (Fig. 4). The MRCA of flat bugs (Aradidae; five of the eight extant subfamilies included in the present study) was dated to be ~162 MYA (confidence interval, 181 to 140 MYA). Aradinae separated from Carventinae, Mezirinae and Aneurinae at around 125 MYA (PCGR) and 134 MYA (PCG12R).
Figure 4. Chronogram showing Pentatomomorpha phylogeny and divergence time.
Consensus tree presenting divergence dates produces by the PhyloBayes analysis of the PCG12R dataset using four fossil calibration points (Supplementary Table S9), the autocorrelated Lognormal relaxed-clock model, the site-heterogeneous mixture CAT + GTR substitution model, and soft bound 10%. A geological time scale is shown at the bottom.
Discussion
Flat bugs (family Aradidae) are a relatively large family in Pentatomomorpha, with eight extant subfamilies and approximately 1,970 species25,26; the monophyly of Aradidae is accepted generally27. The seven species of flat bugs, for which mt genomes have been sequenced, have three types of tRNA gene rearrangement. Our phylogenetic and molecular clock analyses of the mitochondrial genome sequences indicated that the swap of positions between trnQ and trnI is shared by all seven species of flat bugs and thus likely occurred in their MRCA ~162 MYA. trnC and trnW swapped their positions later in the lineage leading to the subfamily Calisiinae, and trnC and trnY were translocated in the lineage leading to the subfamily Aradinae later than 134 MYA. Divergence times of the main lineages of the flat bugs estimated in our analyses are consistent with their fossil records, for example Aradinae separated from Carventinae, Mezirinae and Aneurinae in early Cretaceous, which was in accordance with the oldest Aradinae fossils Aradus nicholasi (125 to 113 MYA)33; and the diversification between the extinct subfamily Archearadinae (represented by two species, Archearadus burmensis34,35 and Microaradus anticus36) and the extant subfamilies also took place in early Cretaceous (~100 MYA)37. Our study provided a preliminary framework for understanding the evolution of the flat bugs and their mitochondrial genomes. Further studies with more complete sampling are needed to reconstruct a comprehensive phylogeny and more accurate molecular dating.
Of the several mechanisms proposed to explain mt gene rearrangement, tandem duplication followed by random gene loss (TDRL) is generally considered the most important in insects38. According to the TDRL, duplication of part of the mt genome was caused by the slipped-strand mispairing or inaccurate termination during replication and novel gene orders generated from random deletion of the supernumerary gene copies3,5. Alternative mechanisms including inversion39, tandem duplication/nonrandom loss (TDNR)40, tRNA duplication/anticodon mutation41,42,43, and recombination44,45 have been proposed to account for mt gene rearrangements that cannot be explained by TDRL model alone.
When mapped on the phylogenetic tree, it was clear that GO1 was an ancestral character for flat bugs and occurred ~162 MYA via a TDRL event (T1: from trnI-trnQ-trnM to trnQ-trnI-trnM) from GOA in the MRCA of Pentatomomorpha. T1 event could be simply a TDRL of trnI and trnQ (from trnI-trnQ-trnI-trnQ to trnQ-trnI). However, the observed IGS-trnQ/trnI in GO1 could not be explained by such process (Fig. 3). The mt genome retained one copy of duplicated genes during subsequent evolution, the other copy was deleted or became pseudogenes or IGSs5,46,47. So, we assumed that the gene cluster of trnI-trnQ-trnM was duplicated in tandem (trnI-trnQ-trnM-trnI-trnQ-trnM) and one copy of each duplicated gene was randomly deleted during T1 event (Fig. 5). The second TDRL event (T2: from trnW-trnC-trnY to trnC-trnW-trnY) changed the GO1 to GO2 in the subfamily Calisiinae. No IGS was found between trnC and trnW in GO2 so T2 event was likely caused by tandem duplication of trnW and trnC followed by random loss of one copy of each duplicated gene. The GO3 of Aradinae was derived from GO1 via another TDRL event (T3: trnM-nad2-trnW-trnC-trnY to trnC-trnY-trnM-nad2-trnW). The IGSs between trnI and trnC, trnW and cox1 in Aradus compar which corresponds to random losses of genes, provided evidence supporting the steps of gene duplication and deletion in the gene rearrangement from GO1 to GO3.
Figure 5. Reconstruction of the mitochondrial rearrangement scenarios in the evolution of flat bugs and proposed mechanism of tRNA gene rearrangement in the model of TDRL.
Phylogenetic tree were simplified from Fig. 4. Genes are transcribed from left to right except those underlined, which have the opposite transcriptional orientation. Rearrangements of tRNA genes were highlighted by color. IGS for the intergenic spacer. Eliminated genes were indicated by crosses and pointed to the relative IGSs by grey arrows.
All of the tRNA gene rearrangement occurred in the region between CR and cox1. Many studies have noted that tandem duplications and gene deletions may be subject to mechanistic constraints such that genes flanking the origins of strand replication (e.g., the CR) are more likely to be duplicated, forming “hotspots” of gene rearrangement that make convergent gene order rearrangement more probable5,48. Various rearrangements of this hotspot region have been found in many insects, e.g., almost all species of Hymenoptera have the rearranged position of trnI and trnQ48; all species of the Lepidoptera suborder Ditrysia have the arrangement trnM-trnI-trnQ49; and most species of Neuroptera have the transposition of trnW and trnC50,51.
Materials and Methods
Samples and DNA extraction
The flat bugs, Aneurus similis, Aneurus sublobatus, Aradus compar and Libiocoris heissi were collected in China, and the sampling information was provided in Supplementary Table S5. Specimens were initially preserved in 100% ethanol in the field and transferred to −20 °C for long-term storage at the Entomological Museum of China Agricultural University (Beijing, China). For each species, the genomic DNA was extracted from one adult’s muscle tissues of the thorax using the DNeasy DNA Extraction kit (QIAGEN).
Genome sequencing, assembly and annotation
The mt genomes were amplified and sequenced as described in Li et al.15,21. Protein-coding genes and two rRNA genes were identified by BLAST searches in NCBI (http://www.ncbi.nlm.nih.gov) and then confirmed by alignment with homologous genes from closely related species. tRNA genes were identified using the tRNAscan-SE52 and ARWEN53 and checked manually. Two tRNAs (trnR and trnS1) not found by the programs were determined based on similarities with sequences from closely related species. The annotated mt genome sequences of four flat bugs have been deposited in GenBank under accession numbers: JQ780816, JQ780817, JQ780818 and JQ780819. The nucleotide composition of each mt genome was calculated using MEGA 6.054. We also measured the AT- and GC-skews for whole mt genome sequences and protein-coding genes.
Sequence alignment and phylogenetic analyses
We retrieved nucleotide sequences of the 13 mt protein-coding genes and two rRNA genes for 29 species of insects from the NCBI. To this initial data set, we added mt genomes of the four flat bugs determined in the current study, thus generating a dataset of 33 taxa (27 Pentatomomorpha species and six Cimicomorpha species as outgroups, see Supplementary Table S6).
Each protein-coding gene was aligned individually based on codon-based multiple alignments using the MAFFT algorithm within the TranslatorX55 online platform. Poorly aligned sites were removed from the protein alignment before back-translate to nucleotides by using GBlocks within the TranslatorX with default settings. Sequences of each rRNA gene were individually aligned using the MAFFT v7.0 online server with G-INS-i strategy56. Ambiguous positions in the alignment of rRNAs were filtered using GBlocks v0.91b57 with default settings. Two datasets were assembled for phylogenetic analyses: 1) the PCGR matrix, including all three codon positions of PCGs and two rRNA genes (total of 12,231 bp); 2) the PCG12R matrix, including the first and the second codon positions of PCGs and two rRNA genes (total of 8,783 bp). The optimal partition strategy and models of each dataset was selected by PartitionFinder v1.1.158. We created an input configuration file that contained 15 pre-define partitions by gene. We used the “greedy” algorithm with branch lengths estimated as “unlinked” and Akaike Information Criterion (AIC) to search for the best-fit scheme (Supplementary Table S7). All these datasets were analyzed under maximum likelihood (ML) framework by using RAxML- HPC2 8.1.1159. Bootstrapping analysis with 1,000 replicates was performed with the fast ML method implemented in RAxML using GTRGAMMA model for nucleotide data.
A 10-fold Bayesian cross-validation was performed to test the fit of the site-heterogeneous mixture models (CAT and CAT + GTR) and “site-homogeneous” models (GTR) to nucleotide dataset using PhyloBayes 3.3 f60 (see PhyloBayes manual). The result showed that the CAT + GTR model was the best fitting model for both datasets (data not shown). We then inferred phylogenies from the PCG12R and PCGR datasets using PhyloBayes MPI 1.4 f61, with the CAT + GTR model. In each individual analysis, two independent chains starting from a random tree were run for 30,000 cycles, with trees being sampled every cycle until 30,000 trees were sampled. The initial 7,500 trees of each MCMC run were discarded as burn-in. A consensus tree was computed from the remaining 45,000 trees combined from two runs.
Divergence time estimation
Divergence time was estimated for each of the two nucleotide datasets using PhyloBayes 3.3 f60, using the best fitting relaxed clock models and the tree generated from the PCG12R dataset and CAT + GTR model. We used Bayes factor (calculated using thermodynamic integration) in PhyloBayes to compare three widely used relaxed models (Lognormal, CIR, and UGAM)62. Bayes factor analysis was conducted by running 10,000 points, sampling every 10 after a burn-in of 1,000. The uncorrelated UGAM model fall into the same category as the models implemented in BEAST63, and this model is shown to fit the data more poorly than two autocorrelated models (CIR and Lognormal). As the Bayes factors for the CIR and Lognormal models were similar, therefore “-auto” analyses (see PhyloBayes manual) were used to compare these two models. Results of Bayes factors are shown in Supplementary Table S8.
For all molecular clock analyses, a birth-death prior on divergence time and fossil calibrations with soft bounds64 were used. Following the recommendations of Benton and Donoghue65, minimum constraints were taken as the upper boundary (youngest) of the time period suggested for the oldest fossil in a crown clade, and maximum as the lower boundary (oldest) of the time period suggested for the nearest well-preserved plesiomorphic relative of the clade. Considering the conflicting relationships among Pyrrhocoroidea, Lygaeoidea and Coreoidea in phylogenetic analyses, we only selected four fossil calibrations for Pentatomidae, Pentatomoidea, Pentatomomorpha and the split of Pentatomomorpha and Cimicomorpha (Supplementary Table S9). The range of fossil age was collected from relevant literature on fossils and a recent version of the Paleobiology Database (https://paleobiodb.org/). We allocated 10% of the probability mass to lie outside each calibration interval. All calculations were performed by running 20,000 generations and sampled every 10 generations (after burn-in of 2,000 generations).
Additional Information
How to cite this article: Song, F. et al. Rearrangement of mitochondrial tRNA genes in flat bugs (Hemiptera: Aradidae). Sci. Rep. 6, 25725; doi: 10.1038/srep25725 (2016).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (No. 2013CB127600), the National Natural Science Foundation of China (Nos 31372229, 31420103902, 31401991), the Beijing Natural Science Foundation (Nos 6144027, 6152016), the National Key Technology R&D Program of the Ministry of Science and Technology (No. 2012BAD19B00), the Chinese Universities Scientific Fund (Nos 2016QC025, 2016QC072, 2016ZB001), and the Special Fund for Scientific Research (No. 2012FY111100). R.S. acknowledges the funding support by the Australian Research Council (DP120100240), and Australia-China Science & Research Fund (ACSRF00980).
Footnotes
Author Contributions F.S., H.L. and W.C. designed and performed the research. F.S., H.L., A.S. and X.Z. performed the molecular work. F.S., H.L., A.S. and R.S. analyzed the data. All authors discussed results and implications. F.S., H.L., R.S., X.Z. and W.C. wrote the manuscript. All authors have read and approved the final manuscript.
References
- Timmermans M. J. T. N. et al. Why barcode? High-throughput multiplex sequencing of mitochondrial genomes for molecular systematics. Nucleic. Acids. Res. 38, e197 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S. L. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu. Rev. Entomol. 59, 95–117 (2014). [DOI] [PubMed] [Google Scholar]
- Moritz C., Dowling T. E. & Brown W. M. Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Annu. Rev. Ecol. Syst. 18, 269–292 (1987). [Google Scholar]
- Inoue J. G., Miya M., Tsukamoto K. & Nishida M. Evolution of the deep-sea gulper eel mitochondrial genomes: large-scale gene rearrangements originated within the eels. Mol. Biol. Evol. 20, 1917–1924 (2003). [DOI] [PubMed] [Google Scholar]
- San Mauro D., Gower D. J., Zardoya R. & Wilkinson M. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol. Biol. Evol. 23, 227–234 (2006). [DOI] [PubMed] [Google Scholar]
- Boore J. L. Animal mitochondrial genomes. Nucleic. Acids. Res. 27, 1767–1780 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanin A. Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol. Phylogenet. Evol. 38, 100–116 (2006). [DOI] [PubMed] [Google Scholar]
- Shao R., Kirkness E. F. & Barker S. C. The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus. Genome Res. 19, 904–912 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D. D. et al. The multipartite mitochondrial genome of Liposcelis bostrychophila: insights into the evolution of mitochondrial genomes in bilateral animals. PLos One 7, e33973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S. J., Hodson C. H., Hamilton P. T., Opit G. P. & Gowen B. E. Maternal transmission, sex ratio distortion, and mitochondria. Proc. Natl. Acad. Sci. USA 18, 10162–10168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey A. M. et al. A novel mitochondrial genome architecture in thrips (Insecta: Thysanoptera): extreme size asymmetry among chromosomes and possible recent control region duplication. BMC Genomics 16, 439 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R., Campbell N. J. & Barker S. C. Numerous gene rearrangements in the mitochondrial genome of the wallaby louse, Heterodoxus macropus (Phthiraptera). Mol. Biol. Evol. 18, 858–865 (2001). [DOI] [PubMed] [Google Scholar]
- Shao R. & Barker S. C. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol. Biol. Evol. 20, 362–370 (2003). [DOI] [PubMed] [Google Scholar]
- Li H. et al. Mitochondrial genomes of two barklice, Psococerastis albimaculata and Longivalvus hyalospilus (Psocoptera: Psocomorpha): contrasting rates in mitochondrial gene rearrangement between major lineages of Psocodea. PLos One 8, e61685 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. Comparative mitogenomic analysis of damsel bugs representing three tribes in the family Nabidae (Insecta: Hemiptera). PLos One 7, e45925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N., Liang A. P. & Bu C. P. A molecular phylogeny of Hemiptera inferred from mitochondrial genome sequences. PLos One 7, e48778 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li H., Wang P., Song F. & Cai W. Z. Comparative mitogenomics of plant bugs (Hemiptera: Miridae): identifying the AGG codon reassignments between serine and lysine. PLos One 9, e101375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Bu C. P., Wipfler B. & Liang A. P. Comparative analysis of the mitochondrial genomes of Callitettixini spittlebugs (Hemiptera: Cercopidae) confirms the overall high evolutionary speed of the AT-rich region but reveals the presence of short conservative elements at the tribal level. PLos One 9, e109140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao M. L., Baumann L. & Baumann P. Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha). BMC Evol. Biol. 4, 25 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. Higher-level phylogeny of paraneopteran insects inferred from mitochondrial genome sequences. Sci. Rep. 5, 8527 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp. (Hemiptera: Enicocephalidae). PLos One 7, e29419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J. M. et al. Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera). BMC Genomics 9, 610 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A. M. et al. The complete mitochondrial genome of the flat bug Aradacanthia heissi (Hemiptera: Aradidae). Zootaxa 3238, 23–38 (2012). [Google Scholar]
- Li H., Shi A. M., Song F. & Cai W. Z. Complete mitochondrial genome of the flat bug Brachyrhynchus hsiaoi (Hemiptera: Aradidae). Mitochondrial DNA, doi: 10.3109/19401736.2013.867437 (2014). [DOI] [PubMed] [Google Scholar]
- Kormilev N. A. & Froeschner R. C. Flat bugs of the world: a synonymic list (Heteroptera, Aradidae). Entomography 5, 1–246 (1987). [Google Scholar]
- Heiss E. Superfamily Aradoidea Brullé 1836 in Catalogue of the Heteroptera of the Palaearctic Region Vol. 4 (eds Aukema B., Rieger C.) 3–34 (The Netherlands Entomological Society, 2001). [Google Scholar]
- Grebennikov V. V. & Heiss E. DNA barcoding of flat bugs (Hemiptera: Aradidae) with phylogenetic implications. Arthropod Syst. Phylo. 72, 213–219 (2014). [Google Scholar]
- Leschen R. A. B. & Taylor S. J. Notes on the biology of Aradus robustus (Hemiptera: Aradidae). Entomol. News 98, 183–185 (1987). [Google Scholar]
- Cassis G. & Schuh R. T. Systematic methods, fossils, and relationships within Heteroptera (Insecta). Cladistics 26, 262–280 (2010). [DOI] [PubMed] [Google Scholar]
- Grozeva S. M. & Kerzhner I. M. On the phylogeny of aradid subfamilies (Heteroptera, Aradidae). Acta Zool. Hung. 38, 199–205 (1992). [Google Scholar]
- Wei S. J. et al. New views on strand asymmetry in insect mitochondrial genomes. PLos One 5, e12708 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanin A., Léger N. & Deutsch J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetics inferences. Syst. Biol. 54, 277–298 (2005). [DOI] [PubMed] [Google Scholar]
- Popov Y. A. New fossil Hemiptera (Heteroptera and Coleorrhyncha) from the Mesozoic of Mongolia. N. Jb. Geol. Paleont. Mh. 3, 166–181 (1989). [Google Scholar]
- Heiss E. & Grimaldi D. Archearadus burmensis gen. n., sp. n., a remarkable Mesozoic Aradidae in Burmese Amber (Heteroptera, Aradidae). Carolinea 59, 99–102 (2001). [Google Scholar]
- Heiss E. & Grimaldi D. The first known female of Archearadus burmensis Heiss & Grimaldi 2001 in Cretaceous Burmese Amber (Heteroptera: Aradidae). Z. Arb. Gem. Öst. Ent. 54, 55–59 (2002). [Google Scholar]
- Heiss E. & Poinar G. O. New Aradidae in Mesozoic Burmese amber (Hemiptera, Heteroptera). Ann. Naturhist. Mus. Wien. 114, 307–316 (2012). [Google Scholar]
- Grimaldi D., Engel M. S. & Nascimbene P. C. Fossiliferous Cretaceous amber from Myanmar (Burma): its discovery, biotic diversity, and paleontological significance. Am. Mus. Novit. 3361, 1–72 (2002). [Google Scholar]
- Boore J. L. The duplication random-loss model for gene rearrangement exemplified by mitochondrial genomes of deuterosome animals in Comparative genomics: empirical and analytical approaches to gene order dynamics, map alignment and the evolution of gene families (eds Sankoff D., Nadeau J. H.) 133–148 (Kluwer Academic Publishers, 2000). [Google Scholar]
- Smith M. J., Banfield D. K., Doteval K., Gorski S. & Kowbel D. J. Gene arrangement in sea star mitochondrial DNA demonstrates a major inversion event during echinoderm evolution. Gene 76, 181–185 (1989). [DOI] [PubMed] [Google Scholar]
- Lavrov D. V., Boore J. L. & Brown W. M. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: duplication and non-random loss. Mol. Biol. Evol. 19, 163–69 (2002). [DOI] [PubMed] [Google Scholar]
- Cantatore P., Gadaleta M. N., Roberti M., Saccone C. & Wilson A. C. Duplication and remoulding of tRNA genes during the evolutionary rearrangement of mitochondrial genomes. Nature 329, 853–855 (1987). [DOI] [PubMed] [Google Scholar]
- Rawlings T. A., Collins T. M. & Bieler R. Changing identities: tRNA duplication and remolding within animal mitochondrial genomes. Proc. Natl. Acad. Sci. USA 100, 15700–15705 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer S. A. & Kumazawa Y. The mitochondrial genome of the lizard Calotes versicolor and a novel gene inversion in South Asian draconine agamids. Mol. Biol. Evol. 24, 1330–1339 (2007). [DOI] [PubMed] [Google Scholar]
- Lunt D. H. & Hyman B. C. Animal mitochondrial DNA recombination. Nature 387, 247 (1997). [DOI] [PubMed] [Google Scholar]
- Dowton M. & Campbell N. J. H. Intramitochondrial recombination — Is it why some mitochondrial genes sleep around? Trends Ecol. Evol. 16, 269–271 (2001). [DOI] [PubMed] [Google Scholar]
- Kurabayashi A. & Sumida M. Afrobatrachian mitochondrial genomes: genome reorganization, gene rearrangement mechanisms, and evolutionary trends of duplicated and rearranged genes. BMC Genomics 14, 633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W. et al. Tandem duplication and random loss for mitogenome rearrangement in Symphurus (Teleost: Pleuronectiformes). BMC Genomics 16, 355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowton M., Cameron S. L., Dowavic J. I., Austin A. D. & Whiting M. F. Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Mol. Biol. Evol. 26, 1607–1617 (2009). [DOI] [PubMed] [Google Scholar]
- Cao Y. Q., Ma C., Chen J. Y. & Yang D. R. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genomics 13, 276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S. L., Sullivan J., Song H., Miller K. B. & Whiting M. F. A mitochondrial genome phylogeny of the Neuropterida (lacewings, alderflies and snakeflies) and their relationship to the other holometabolous insect orders. Zool. Scr. 38, 575–590 (2009). [Google Scholar]
- Zhao J., Li H., Winterton S. L. & Liu Z. Q. Ancestral gene organization in the mitochondrial genome of Thyridosmylus langii (McLachlan, 1870) (Neuroptera: Osmylidae) and implications for lacewing evolution. PLos One 8, e62943 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M. & Eddy S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic. Acids. Res. 25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D. & Canbäck B. ARWEN, a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24, 172–175 (2008). [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abascal F., Zardoya R. & Telford M. J. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic. Acids. Res. 38, W7–W13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K. & Standley D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- Lanfear R., Calcott B., Ho S. Y. W. & Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701 (2012). [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- Lartillot N., Lepage T. & Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288 (2009). [DOI] [PubMed] [Google Scholar]
- Lartillot N., Rodrigue N., Stubbs D. & Richer J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 62, 611–615 (2013). [DOI] [PubMed] [Google Scholar]
- Lepage T., Bryant D., Philippe H. & Lartillot N. A general comparison of relaxed molecular clock models. Mol. Biol. Evol. 24, 2669–2680 (2007). [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Suchard M. A., Xie D. & Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. & Rannala B. Bayesian estimation of species divergence times under a molecular clock using fossil calibrations with soft bounds. Mol. Biol. Evol. 23, 212–226 (2006). [DOI] [PubMed] [Google Scholar]
- Benton M. J. & Donoghue P. C. J. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 24, 26–53 (2007). [DOI] [PubMed] [Google Scholar]
- Stothard P. & Wishart D. S. Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.