Abstract
This study was conducted to investigate whether polymorphisms of genes involved in immune checkpoints can predict the clinical outcomes of patients with advanced stage non-small cell lung cancer (NSCLC) after 1st line paclitaxel-cisplatin chemotherapy. A total of 379 NSCLC patients were enrolled. Twelve single nucleotide polymorphisms (SNPs) of PD-1, PD-L1, and CTLA-4 genes were selected and genotyped. The associations of SNPs with chemotherapy response and overall survival (OS) were analyzed. Among the 12 SNPs investigated, PD-L1 rs2297136T > C and rs4143815C > G were significantly associated with clinical outcomes after chemotherapy. The rs2297136T > C was significantly associated with both better chemotherapy response and better OS, and the rs4143815C > G had a significantly better response to chemotherapy. Consistent with the individual genotype analyses, rs2297136C-rs4143815G haplotype (ht4) carrying variant alleles at both loci was significantly associated with better chemotherapy response and OS compared with combined other haplotypes. Patients with at least one ht4 had significantly better chemotherapy response and OS compared to those without ht4. PD-L1 rs2297136T > C and rs4143815C > G polymorphisms may be useful for the prediction of clinical outcome of 1st line paclitaxel-cisplatin chemotherapy in NSCLC. Further studies are needed to confirm our findings and to understand the role of PD-L1 in the chemotherapy outcome of NSCLC patients.
Lung cancer is the leading cause of cancer deaths worldwide, with an average 5-year survival rate of 16%1. Non-small cell lung cancer (NSCLC), about two thirds of which present with advanced disease, comprises about 85% of primary lung cancers2. During the last decade, target agents such as EGFR-tyrosine kinase inhibitors or ALK inhibitors have led a major advance in the treatment of NSCLC3,4. However, the benefit has been confined to a subset of NSCLC patients who have an active target for the drugs. Platinum-based chemotherapy still plays an important role in the treatment of the majority of patients with advanced NSCLC, but provides only modest benefits with an objective response rate of 30–40% and median survival time of 8–11 months5,6. Treatment outcomes after chemotherapy vary widely among patients with similar clinical characteristics including stage, the best prognostic index for NSCLC. Therefore, there have been numerous investigations to identify molecular biomarkers that can better predict a patient’s clinical outcome.
Recently, immune checkpoints blockade has emerged as a novel therapy against cancer7. T cells are key players in host antitumor immune function: They can recognize cancer cells, generating cytotoxic T cell populations that can infiltrate the tumor and kill tumor cells. The checkpoints of T cell activation are crucial for maintaining self-tolerance and avoiding immune response-related tissue damage7. Cancer cells can take advantage of this inhibitory mechanism to evade host immune defense by expressing ligands which interact with co-inhibitory receptors on cytotoxic T-cells. Therefore, blocking the specific ligand or receptor may reactivate host immune responses and antitumor functions.
Cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) is an inhibitory receptor which can downregulate T-cell activation at the priming phase. A monoclonal antibody blocking CTLA-4 from its ligands expressed on antigen presenting cells has been shown to improve overall survival (OS) in patients with advanced melanoma8. It has also been investigated in combination with paclitaxel and carboplatin for the treatment of NSCLC in a phase II clinical trial with a promising result9, prompting an ongoing phase III trial. Programmed cell death-1 (PD-1) pathway is a major immune checkpoint by which tumors may inactivate tumor-infiltrating lymphocytes (TILs) in the tumor microenvironment. PD-1 is expressed in various immune cell types including cytotoxic T-cells and its activation inhibits T-cell function, proliferation, and survival10,11. The PD-1 ligand, PD-L1 is expressed on activated T cells, B cells, dendritic cells, and macrophages, and can be induced in a variety of tissue types by inflammatory cytokines, such as IFN-γ12,13. PD-L1 expression has been reported in many types of cancer including lung cancer14,15,16. Antibody blockade against PD-1 and PD-L1 has shown substantial survival benefit in several types of cancer including NSCLC17,18,19,20,21. Notably, tumor PD-L1 expression by immunohistochemistry (IHC) has been found to be predictive of response to anti-PD-1 or anti-PD-L1 treatment17,18,19. In addition, the prognostic role of PD-L1 expression has also been investigated in different types of cancer, although the results are inconsistent22,23.
Because the host immune system plays an important role in killing cancer cells after chemotherapy, altered immune checkpoint function may influence chemotherapy outcomes. In addition, accumulating evidence indicates that cancer chemotherapy may modulate the immune function, which may enhance the effect of chemotherapeutic drugs24,25,26. For example, platinum drugs enhance T-cell activation by dendritic cells, and tumor cell recognition and killing by tumor-specific T cells through downregulation of PD-L1 and PD-L227. In this study, we hypothesized that polymorphisms of genes involved in immune checkpoints may affect the antitumor immune activity, thereby influencing clinical outcomes of chemotherapy in patients with NSCLC. To test this hypothesis, we evaluated the association of polymorphisms in PD-1, PD-L1, and CTLA-4 genes with the chemotherapy response and survival of NSCLC patients undergoing 1st line paclitaxel-cisplatin chemotherapy.
Results
Patient Characteristics and Clinical Predictors
The association between clinicopathological characteristics and the chemotherapy response and OS are shown in Table 1. The overall response rate was 47.5%. We observed events (deaths) in 347 of the 379 patients (91.6%) and median survival time (MST) was 13.2 months (95% CI = 12.5–14.7 months). Only tumor histology was significantly associated with response to chemotherapy. The OS was significantly associated with age, gender, smoking status, tumor histology, weight loss, and second line chemotherapy (Table 1).
Table 1. Univariate analysis for response to chemotherapy and overall survival by clinical variables.
| Variables | No. of | Response to chemotherapy | Overall survival | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| responders | nonresponders | OR (95% CI) | P | MST | 95% CI | Log-Rank | HR (95% CI) | P | ||
| cases | (CR + PR) | (SD + PD) | (months) | P | ||||||
| Overall | 379 | 180 (47.5)a | 199 (52.5) | 13.2 | 12.5–14.7 | |||||
| Age (years) | ||||||||||
| <65 | 179 | 93 (52.0) | 86 (48.0) | 1.00 | 15.7 | 13.7–17.7 | 1.00 | |||
| ≥65 | 200 | 87 (43.5) | 113 (56.5) | 0.71 (0.48–1.07) | 0.10 | 11.9 | 10.8–13.2 | 0.003 | 1.38 (1.11–1.70) | 0.003 |
| Gender | ||||||||||
| Male | 309 | 153 (49.5) | 156 (50.5) | 1.00 | 12.8 | 11.9–14.3 | 1.00 | |||
| Female | 70 | 27 (38.6) | 43 (61.4) | 0.64 (0.38–1.09) | 0.10 | 16.8 | 12.8–22.6 | 0.02 | 0.73 (0.56–0.96) | 0.02 |
| Smoking status | ||||||||||
| Never | 63 | 27 (42.9) | 36 (57.1) | 1.00 | 19.6 | 13.7–30.4 | 1.00 | |||
| Ever | 316 | 153 (48.4) | 163 (51.6) | 1.25 (0.73–2.16) | 0.42 | 12.8 | 11.7–14.2 | 0.001 | 1.59 (1.20–2.12) | 0.002 |
| Histological type | ||||||||||
| Squamous cell ca. | 184 | 98 (53.3) | 86 (46.7) | 1.00 | 13.2 | 11.7–14.4 | 1.00 | |||
| Adenoca. | 172 | 71 (41.3) | 101 (58.7) | 0.62 (0.41–0.94) | 0.02 | 15.1 | 12.1–17.5 | 0.74 (0.59–0.92) | 0.01 | |
| NSCLC-NOS | 23 | 11 (47.8) | 12 (52.2) | 0.80 (0.34–1.92) | 0.62 | 11.4 | 7.4–12.9 | 0.01 | 1.25 (0.80–1.99) | 0.34 |
| Clinical stage | ||||||||||
| III | 159 | 82 (51.6) | 77 (48.4) | 1.00 | 14.7 | 12.8–17.4 | 1.00 | |||
| IV | 220 | 98 (44.6) | 122 (55.5) | 0.75 (0.50–1.14) | 0.18 | 12.7 | 10.8–14.2 | 0.12 | 1.18 (0.96–1.47) | 0.12 |
| PS ECOG | ||||||||||
| 0–1 | 310 | 149 (48.1) | 161 (51.9) | 1.00 | 14.1 | 12.6–15.7 | 1.00 | |||
| 2 | 69 | 31 (44.9) | 38 (55.1) | 0.88 (0.52–1.49) | 0.64 | 12.6 | 9.7–13.2 | 0.42 | 1.12 (0.85–1.48) | 0.42 |
| Weight loss | ||||||||||
| No | 233 | 116 (49.8) | 117 (50.2) | 1.00 | 14.4 | 12.5–16.6 | 1.00 | |||
| Yes | 146 | 64 (43.8) | 82 (56.2) | 0.79 (0.52–1.19) | 0.26 | 12.9 | 11.6–14.0 | 0.001 | 1.47 (1.18–1.83) | 0.001 |
| 2nd line Chemotherapy | ||||||||||
| No | 132 | 11.0 | 8.1–12.8 | 1.00 | ||||||
| Yes | 247 | 15.1 | 13.2–16.6 | 0.02 | 0.76 (0.61–0.95) | 0.02 | ||||
| Radiation to tumor | ||||||||||
| No | 340 | 12.9 | 11.6–14.3 | 1.00 | ||||||
| Yes | 39 | 18.5 | 14.0–23.9 | 0.19 | 0.80 (0.57–1.12) | 0.19 | ||||
Abbreviation: OR, odds ratio; MST, median survival time; CI, confidence interval; HR, hazard ratio; PS, performance status; ECOG, Eastern Cooperative Oncology Group.
aRow percentage.
The genotypes of rs2297136T > C and rs4143815C > G and clinical outcomes
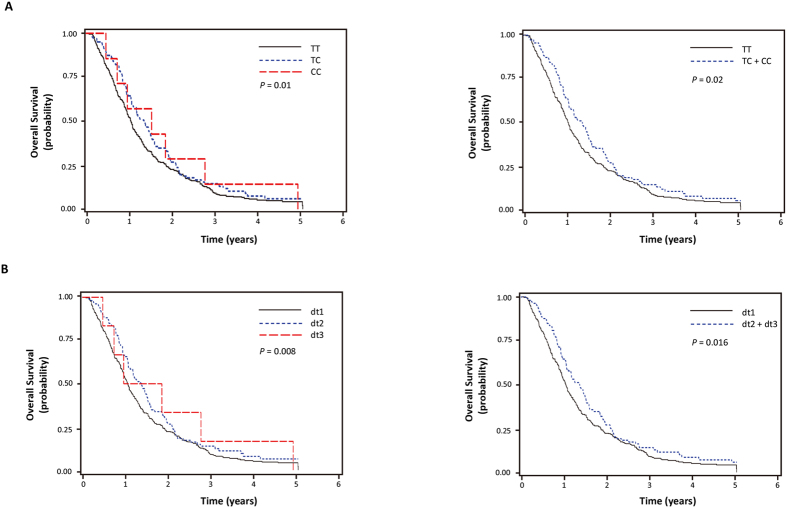
The SNP ID, gene information, and minor allele frequencies are shown in Table 2. Of the 12 SNPs analyzed, the rs2297136T > C and rs4143815C > G SNPs in the 3′UTR of PD-L1 were significantly associated with clinical outcomes after chemotherapy. The rs2297136T > C was significantly associated with both better chemotherapy response and better OS under additive model for the variant C allele (adjusted odds ratio [aOR] = 1.82, 95% CI = 1.19–2.77, P = 0.01; adjusted hazard ratio [aHR] = 0.76, 95% CI = 0.61–0.94, P = 0.01, respectively), and the rs4143815C > G was significantly associated with better response under additive model for the variant G allele (aOR = 1.42, 95% CI = 1.05–1.93, P = 0.02) although not significantly associated with OS (Table 3 and Fig. 1A).
Table 2. Summary of twelve SNPs and response to chemotherapy and overall survival.
| ID No.a | Gene | location | Alleles | CR (%) | MAF | HWP | P for responseb | P for overall survivalc | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dominant | recessive | additive | dominant | recessive | additive | |||||||
| rs2297136 | PD-L1 | 3′UTR | TC | 99.2 | 0.16 | 0.35 | 0.01 | 0.20 | 0.01 | 0.02 | 0.15 | 0.01 |
| rs4143815 | PD-L1 | 3′UTR | CG | 96.6 | 0.40 | 0.67 | 0.21 | 0.01 | 0.02 | 0.47 | 0.18 | 0.23 |
| rs822336 | PD-L1 | promoter | GC | 99.5 | 0.26 | 0.73 | 0.67 | 0.11 | 0.31 | 0.68 | 0.95 | 0.72 |
| rs822337 | PD-L1 | promoter | TA | 96.6 | 0.34 | 0.88 | 0.75 | 0.16 | 0.36 | 0.30 | 0.27 | 0.19 |
| rs822338 | PD-L1 | intron | CT | 98.9 | 0.44 | 0.60 | 0.87 | 0.35 | 0.67 | 0.56 | 0.34 | 0.35 |
| rs36084323 | PD1 | promoter | GA | 97.6 | 0.47 | 0.31 | 0.13 | 0.19 | 0.08 | 0.99 | 0.42 | 0.62 |
| rs2227982 | PD1 | missence | CT | 98.9 | 0.46 | 0.15 | 0.20 | 0.09 | 0.07 | 0.96 | 0.53 | 0.74 |
| rs10204525 | PD1 | 3′UTR | AG | 99.2 | 0.25 | 0.38 | 0.33 | 0.09 | 0.15 | 0.76 | 0.60 | 0.66 |
| rs231775 | CTLA-4 | missence | GA | 99.2 | 0.28 | 0.52 | 0.97 | 0.26 | 0.61 | 0.71 | 0.93 | 0.80 |
| rs5742909 | CTLA-4 | promoter | CT | 97.9 | 0.12 | 0.26 | 0.41 | 0.58 | 0.58 | 0.98 | 0.11 | 0.74 |
| rs733618 | CTLA-4 | promoter | AG | 98.7 | 0.45 | 0.91 | 0.37 | 0.80 | 0.47 | 0.46 | 0.70 | 0.48 |
| rs11571316 | CTLA-4 | promoter | CT | 98.2 | 0.16 | 0.56 | 0.91 | 0.34 | 0.86 | 0.89 | 0.95 | 0.91 |
Abbreviation: CR, call rate; MAF, minor allele frequency; and HWE, Hardy-Weinberg equilibrium.
aInformation about SNPs and SNP ID were obtained from NCBI database (http://ncbi.nih.gov).
bP values were calculated by multivariate regression analysis, adjusted for age, gender, smoking status, tumor histology, stage, ECOG performance status, and weight loss.
cP-values were calculated using multivariate Cox proportional hazard models, adjusted for age, gender, smoking status, tumor histology, stage, ECOG performance status, weight loss, 2nd line chemotherapy and radiation to primary tumor.
Table 3. Genotypes of rs2297136 and rs4143815 polymorphisms and their associations with the response to chemotherapy and overall survival.
| Polymorphism/Genotype | Location | No. of case (%)a | Response to chemotherapy | Overall survival | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Responders (%)b | Non-responders (%)b | OR (95% CI)c | Pc | MST (95% CI)d | L-R P | HR (95% CI)d | Pd | |||
| rs2297136e | 3′UTR | |||||||||
| TT | 264 (70.2) | 115 (43.6) | 149 (56.4) | 1.00 | 12.6 (11.4–13.9) | 0.19 | 1.00 | |||
| TC | 105 (27.9) | 60 (57.1) | 45 (42.9) | 1.79 (1.12–2.87) | 0.01 | 16.3 (12.8–18.5) | 0.77 (0.60–0.98) | 0.04 | ||
| CC | 7 (1.9) | 5 (71.4) | 2 (28.6) | 3.60 (0.66–19.8) | 0.14 | 18.4 (5.7–33.3) | 0.52 (0.24–1.12) | 0.10 | ||
| Dominant | 1.87 (1.18–2.96) | 0.01 | 16.7 (12.8–18.4) | 0.07 | 0.75 (0.59–0.95) | 0.02 | ||||
| Recessive | 3.05 (0.56–16.6) | 0.20 | 13.2 (12.5–14.7) | 0.61 | 0.57 (0.26–1.23) | 0.15 | ||||
| Additive | 1.82 (1.19–2.77) | 0.01 | 0.76 (0.61–0.94) | 0.01 | ||||||
| rs4143815e | 3′UTR | |||||||||
| CC | 133 (36.3) | 58 (43.6) | 75 (56.4) | 1.00 | 13.5 (11.6–16.0) | 0.71 | 1.00 | |||
| CG | 172 (47.0) | 78 (45.4) | 94 (54.7) | 1.10 (0.69–1.77) | 0.69 | 13.2 (11.3–14.8) | 0.97 (0.76–1.24) | 0.81 | ||
| GG | 61 (16.7) | 38 (62.3) | 23 (37.7) | 2.26 (1.19–4.28) | 0.01 | 14.6 (11.9–19.4) | 0.80 (0.57–1.11) | 0.18 | ||
| Dominant | 1.33 (0.85–2.08) | 0.21 | 13.3 (12.1–15.2) | 0.88 | 0.92 (0.73–1.16) | 0.47 | ||||
| Recessive | 2.14 (1.20–3.82) | 0.01 | 13.2 (12.0–14.8) | 0.41 | 0.81 (0.60–1.10) | 0.18 | ||||
| Additive | 1.42 (1.05–1.93) | 0.02 | 0.91 (0.78–1.06) | 0.23 | ||||||
Abbreviation: OR, odds ratio; CI, confidence interval; MST, median survival time (months); L-R P, Log-rank P; HR, hazard ratio.
aColumn percentage.
bRow percentage.
cOR, 95% CI, and their corresponding P values were calculated by multivariate regression analysis, adjusted for age, gender, smoking status, tumor histology, stage, ECOG performance status, and weight loss.
dHRs, 95% CIs and their corresponding P-values were calculated using multivariate Cox proportional hazard models, adjusted for age, gender, smoking status, tumor histology, stage, ECOG performance status, weight loss, 2nd line chemotherapy and radiation to primary tumor.
eGenotype failure in 3 cases for rs2297136T > C and 13 cases for rs4143815C > G.
Figure 1.
Kaplan-Meier plots of overall survival according to PD-L1 rs2297136T > C (A), and diplotypes of rs2297136T > C and rs4143815C > G (B). P values in the multivariate Cox proportional hazard model.
The haplotypes of rs2297136T > C and rs4143815C > G and clinical outcomes
The two SNPs, rs2297136T > C and rs4143815C > G, were in LD (|D’| = 0.9 and r2 = 0.22) with three predominant haplotypes accounting for 99.3% of the haplotypes in the subjects. Consistent with the individual genotype analyses, rs2297136C-rs4143815G haplotype (haplotype 4, ht4) carrying variant alleles at both loci was significantly associated with better chemotherapy response and OS compared with combined other haplotypes, rs2297136T-rs4143815C (ht1), rs2297136T-rs4143815G (ht2), and rs2297136C-rs4143815C (ht3) (aOR = 1.92, 95% CI = 1.27–2.91, P = 0.002; aHR = 0.76, 95% CI = 0.61–0.94, P = 0.01, respectively, Table 4). Therefore, we next examined the survival outcome of the patients carrying ht4. For this analysis, the diplotypes resulting from the four haplotypes were categorized into three groups according to the presence of zero, one, or two ht4 (i.e., diplotype 1 [dt1], ht1-3/ht1-3; dt2, ht1-3/ht4; dt3, ht4/ht4). The dt2 or dt3 with at least one ht4 had significantly better chemotherapy response and survival compared to the dt1 without ht4 (aOR = 2.13, 95% CI = 1.33–3.40, P = 0.002; aHR = 0.74, 95% CI = 0.58–0.95, P = 0.02, respectively, Table 4 and Fig. 1B).
Table 4. Response to chemotherapy and overall survival according to PD-L1 rs2297136T > C and rs4143815C > G haplotypes and diplotypes.
| Response to chemotherapy | Overall survival | |||||||
|---|---|---|---|---|---|---|---|---|
| No.(%)a | Response | Non-response | OR (95% CI)b | Pb | Log-rank p | HR (95% CI)c | Pc | |
| Haplotype | ||||||||
| ht1. TC | 446 (58.8) | 199 (44.6) | 247 (55.4) | 1.00 | 0.32 | 1.00 | ||
| ht2. TG | 193 (25.5) | 91 (47.2) | 102 (52.9) | 1.13 (0.80–1.60) | 0.49 | 1.03 (0.86–1.23) | 0.74 | |
| ht3. CC | 5 (0.7) | 1 (20.0) | 4 (80.0) | 0.31 (0.03–2.84) | 0.30 | 0.97 (0.40–2.38) | 0.95 | |
| ht4. CG | 114 (15.0) | 69 (60.5) | 45 (39.5) | 1.98 (1.29–3.04) | 0.002 | 0.76 (0.61–0.95) | 0.02 | |
| Ptrend | 0.003 | 0.02 | ||||||
| ht1-3 | 644 (85.0) | 291 (45.2) | 353 (54.8) | 1.00 | 0.08 | 1.00 | ||
| ht4 | 114 (15.0) | 69 (60.5) | 45 (39.5) | 1.92 (1.27–2.91) | 0.002 | 0.76 (0.61–0.94) | 0.01 | |
| Diplotype | ||||||||
| dt1(ht1-3/ht1-3) | 271 (71.5) | 115 (42.4) | 156 (57.6) | 1.00 | 0.18 | 1.00 | ||
| dt2(ht1-3/ht4) | 102 (26.9) | 61 (59.8) | 41 (40.2) | 2.08 (1.29–3.35) | 0.003 | 0.77 (0.60–0.98) | 0.04 | |
| dt3(ht4/ht4) | 6 (1.6) | 4 (66.7) | 2 (33.3) | 3.21 (0.55–18.73) | 0.20 | 0.47 (0.20–1.08) | 0.07 | |
| Ptrend | 0.002 | 0.01 | ||||||
| dt1 | 271 (71.5) | 115 (42.4) | 156 (57.6) | 1.00 | 0.07 | 1.00 | ||
| dt2 + dt3 | 108 (28.5) | 65 (60.2) | 43 (39.8) | 2.13 (1.33–3.40) | 0.002 | 0.74 (0.58–0.95) | 0.02 | |
Abbreviation: HR, hazard ratio; CI, confidence interval; ht, haplotype; dt, diplotype.
tumor histology, stage, ECOG, weight loss, 2nd line chemotherapy and radiation to primary tumor.
aColumn percentage.
baORs, 95% CIs and their corresponding P-values described the risk of being a responder and were calculated by multivariate regression analysis, adjusted for age, gender, smoking status, tumor histology, stage, ECOG and weight loss.
caHRs, 95% CIs and their corresponding P-values were calculated using multivariate Cox proportional hazard models, adjusted for age, gender, smoking status.
The predicted effect of rs2297136T > C and rs4143815C > G on the binding of miRNAs and PD-L1 mRNA
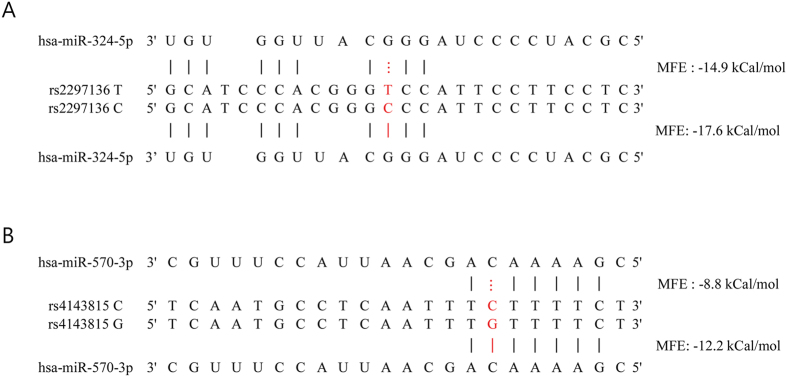
Using RNAhybrid2.2 (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html), we predicted minimum free energy (MFE) for the binding of miRNAs to PD-L1 mRNA for both wild and variant alleles. The predicted effect of allelic variants at PD-L1 rs2297136T > C and rs4143815C > G on miRNA binding was shown in Fig. 2. The MFE were −14.9 kCal/mol and −17.6 kCal/mol for the binding of miR-324-5p to rs2297136 T and C alleles, respectively, and −8.8 kCal/mol and −12.2 kCal/mol for the binding of miR-570-3p to rs4143815 C and G alleles, respectively. This result suggests that rs2297136T > C and rs4143815C > G may affect PD-L1 expression by altered miRNA binding to 3′UTR of PD-L1 mRNA.
Figure 2.
The predicted effect of allelic variants at PD-L1 rs2297136T > C on hsa-miR-324-5p (A) and PD-L1 rs4143815C > G on hsa- miR-570-3p (B) recognition. Minimum free energy (MFE) was predicted using RNAhybrid (http://bibiserv.techfak.uni-biele feld.de/rnahybrid/submission.html).
Discussion
Immune checkpoint blockade, targeting not the cancer cell itself but the host immune system, represents a paradigm shift in cancer therapy. It is evident that the interaction between tumors and their microenvironment including host immune system is critical not only in tumor development and progression, but also in the context of cancer therapy24. The intratumoral immune infiltrate indicative of an ongoing immune response has been correlated with therapeutic outcome28,29. In addition, systemic immune biomarkers such as polymorphisms in genes encoding immune modulators, e.g. interleukin-4 or -6, may reveal the way the host immune system responds to malignancies, thereby determining prognosis30,31. Likewise, genetic variations in immune checkpoint genes may be used as a potential predictor of cancer treatment outcome. This is the first study to investigate the effect of genetic polymorphisms in immune checkpoints on the outcomes of chemotherapy in treatment-naïve patients with advanced stage NSCLC. Two potentially functional polymorphisms, rs2297136T > C and rs4143815C > G, in PD-L1 were significantly associated with 1st line paclitaxel-cisplatin chemotherapy response and/or survival in NSCLC. The SNPs of PD-L1 may be used to predict the clinical outcomes of NSCLC patients receiving 1st line chemotherapy, thereby helping to identify subgroups of patients who would benefit from chemotherapy and to save patients from unnecessary toxicities.
In the present study, PD-L1 rs2297136T > C and rs4143815C > G genotypes and their haplotypes were associated with clinical outcomes of paclitaxel-cisplatin chemotherapy in NSCLC. Because PD-L1 rs2297136T > C and rs4143815C > G are located in 3′-UTR, the SNPs may cause a change in miRNA binding and PD-L1 expression. Functional prediction of the SNPs using RNAhybrid suggested that rs2297136T > C and rs4143815C > G may affect PD-L1 expression by modulating the miRNA-mRNA interaction. In addition, the rs4143815C > G has been reported to alter binding efficiency of miR-570 to PD-L1 mRNA32. Altered PD-L1 expression may affect immune checkpoint function, which consequently may influence chemotherapy outcomes because host immune system plays an important role in killing cancer cells after chemotherapy. Cancer chemotherapy mainly targets tumor cells, but accumulating evidence indicates that it also affects the immune system25,26. Although immunosuppressive properties of chemotherapy are well known, many anticancer agents stimulate tumor-specific immune responses either by inducing immunogenic cancer cell death or by modulating immune effector mechanisms, which may boost host immunity and the effect of chemotherapy24,25. Apoptosis, a non-immunogenic cell death modality, is a frequent mechanism of chemotherapy-induced tumor cell death. However, many chemotherapeutic drugs can elicit specific cellular responses that cause tumor cell death to be immunogenic, leading to T cell mediated tumor-specific immune responses25,33. In addition, some anticancer drugs reportedly have immunomodulatory functions that contribute to the anticancer effect of the drugs. For example, paclitaxel has been reported to specifically impair cytokine production and viability in CD4+ FOXP3+ regulatory T cells but not in CD4+ FOXP3− effector T cells, independent of Toll-like receptor 4 signaling34. In addition, platinum-based chemotherapeutics not only stimulate class I HLA expression but also inhibit signal transducer and activator of transcription 6 (STAT6)-regulated expression of PD-L2, thus limiting immunosuppression by both dendritic cells and tumor cells35. Given the immunological consequence of chemotherapy as a mechanism of killing cancer cells, the host immune system may play important roles in determining chemotherapy outcomes. Recent studies reported that the presence of TILs predicts response to neoadjuvant chemotherapy36. In addition, the presence of TILs has been correlated to PD-L1 expression in the tumor microenvironment as well as in tumor cells37, suggesting an association between PD-L1 expression in response to immune infiltrate and the outcome of chemotherapy. Taken together, immune checkpoint may be a potential determinant of the clinical outcome of chemotherapy. Our findings suggest that the PD-1/PD-L1 pathway plays a critical role in determining the response to chemotherapy and survival in NSCLC, and that PD-L1 polymorphisms may be predictive of the chemotherapy outcome in NSCLC, which is biologically plausible.
In conclusion, polymorphisms in the PD-L1 gene were found to be independent predictive biomarkers for clinical outcomes of NSCLC patients receiving 1st line paclitaxel-cisplatin chemotherapy. The SNPs of PD-L1 may be useful in helping to refine therapeutic decisions in the treatment of NSCLC. Further studies with a larger population are needed to confirm our findings and to understand the role of PD-L1 in determining the clinical outcome of anticancer therapy.
Methods
Study populations
The study population has been described in our previous study38. In brief, 379 patients with stage III or IV NSCLC, who received at least two cycles of paclitaxel-cisplatin chemotherapy as a first-line treatment at Kyungpook National University Hospital (KNUH) in Daegu, Korea between August 2005 and December 2008, were enrolled. Patients who underwent radiotherapy concurrently with chemotherapy as a first treatment modality were excluded to avoid the confounding effect of radiation on the response to chemotherapy. The chemotherapy regimen included paclitaxel 175 mg/m2 administered i.v. over 3 h, and cisplatin 60 mg/m2 infused over 60 min given on day 1, every 3 weeks. Treatment was discontinued in case of disease progression, major toxicities, or according to patient’s or physician’s decision. Assessment of tumor response was carried out by computed tomography scan every two cycles. Responses were assessed using Response Evaluation Criteria in Solid Tumors39. The best overall response for each patient was reported and all responses were reviewed by an independent radiologist. Patients with a complete response (CR) or a partial response (PR) were defined as responders, and patients having stable disease (SD) or progressive disease (PD) were defined as non-responders. For the assessment of survival outcomes, OS, defined as the time between the date of chemotherapy start and the date of death or last follow-up, were recorded. Genomic DNA samples from the patients were provided by the National Biobank of Korea, KNUH, which is supported by the Ministry of Health, Welfare and Family Affairs. Written informed consent was obtained from all patients. This study was approved by the institutional review board of the KNUH and carried out in accordance with the institutional review board-approved guidelines.
Selection of SNPs and genotyping
To collect potentially functional polymorphisms in PD-1, PD-L1, and CTLA-4 genes, we searched the public database (http://www.ncbi.nlm.nih.gov/SNP) and related literature. The SNP selection favored those located in the promoter or untranslated region or coding region of the gene, those previously evaluated in relation to cancer, or those with evidence of functional significance. A total of 12 SNPs with the minor allele frequency (MAF) ≥0.05 in the HapMap JPT data were selected after excluding those in linkage disequilibrium (LD, r2 ≥0.8). Genotyping was performed using SEQUENOM’s MassARRAY® iPLEX assay (SEQUENOM Inc., San Diego, CA) for 11 SNPs other than rs4143815, which was genotyped using the TaqMan® assay (Applied Biosystems, Foster City, CA).
Statistical analysis
Hardy-Weinberg equilibrium was tested using a goodness-of-fit χ2 test with 1 degree of freedom. The linkage disequilibrium among polymorphisms was measured by using HaploView (http://broad.mit.edu/mpg/haploview). The haplotypes and their frequencies were estimated using the Phase program40. The genotypes for each SNP were analyzed as a three-group categorical variable, and also analyzed under dominant, recessive, and additive genetic models. The association between clinical variables or genotypes and chemotherapy response was tested by odds ratio (OR) and 95% confidence intervals (CIs) using unconditional logistic regression analysis. Kaplan-Meier method was used to calculate survival estimates. The difference in OS according to different clinical variables or genotypes was compared using log-rank tests. Cox’s proportional hazard regression model was used for the multivariate survival analyses. The hazard ratio (HR) and 95% confidence interval (CI) were also estimated. A cut-off p-value of 0.05 was adopted for all the statistical analyses. The statistical data were obtained using Statistical Analysis System for Windows, version 9.2 (SAS Institute, Cary, NC, USA).
Additional Information
How to cite this article: Lee, S. Y. et al. PD-L1 polymorphism can predict clinical outcomes of non-small cell lung cancer patients treated with first-line paclitaxel-cisplatin chemotherapy. Sci. Rep. 6, 25952; doi: 10.1038/srep25952 (2016).
Acknowledgments
This study was supported in part by the R&D program of MKE/KEIT (10040393, Development and commercialization of molecular diagnostic technologies for lung cancer through clinical validation) and in part by the National Research Foundation of Korea (NRF) grant funded by the Korea government (2014R1A5A2009242).
Footnotes
Author Contributions Conceived and designed the experiments: S.Y.L., D.K.J. and J.Y.P. Performed the experiments: D.K.J., J.E.C., C.C.J., M.J.H., S.K.D. and H.G.K. Analyzed the data and statistical analyses: S.Y.L., D.K.J., W.K.L. and J.Y.P. Contributed reagents/material/analysis tools: S.Y.L., E.B.L., S.S.Y., K.M.S., Y.S., J.Y.J., J.L., S.I.C., C.H.K. and J.Y.P. Wrote the main manuscript text: S.Y.L., D.K.J. and J.Y.P. All authors reviewed the manuscript.
References
- Siegel R., Naishadham D. & Jemal A. Cancer statistics, 2013. CA Cancer J Clin 63, 11–30 (2013). [DOI] [PubMed] [Google Scholar]
- Molina J. R., Yang P., Cassivi S. D., Schild S. E. & Adjei A. A. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 83, 584–594 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok T. S. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361, 947–957 (2009). [DOI] [PubMed] [Google Scholar]
- Solomon B. J. et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 371, 2167–2177 (2014). [DOI] [PubMed] [Google Scholar]
- Goffin J., Lacchetti C., Ellis P. M., Ung Y. C. & Evans W. K. First-line systemic chemotherapy in the treatment of advanced non-small cell lung cancer: a systematic review. J Thorac Oncol 5, 260–274 (2010). [DOI] [PubMed] [Google Scholar]
- Schiller J. H. et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 346, 92–98 (2002). [DOI] [PubMed] [Google Scholar]
- Pardoll D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 12, 252–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 363, 711–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch T. J. et al. Ipilimumab in combination with paclitaxel and carboplatin as first-line treatment in stage IIIB/IV non-small-cell lung cancer: results from a randomized, double-blind, multicenter phase II study. J Clin Oncol 30, 2046–2054 (2012). [DOI] [PubMed] [Google Scholar]
- Blank C. & Mackensen A. Contribution of the PD-L1/PD-1 pathway to T-cell exhaustion: an update on implications for chronic infections and tumor evasion. Cancer Immunol Immunother 56, 739–745 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir M. E., Butte M. J., Freeman G. J. & Sharpe A. H. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol 26, 677–704 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H. et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 8, 793–800 (2002). [DOI] [PubMed] [Google Scholar]
- Zou W. & Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol 8, 467–477 (2008). [DOI] [PubMed] [Google Scholar]
- Sznol M. & Chen L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clin Cancer Res 19, 1021–1034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomi T. et al. Clinical significance and therapeutic potential of the programmed death-1 ligand/programmed death-1 pathway in human pancreatic cancer. Clin Cancer Res 13, 2151–2157 (2007). [DOI] [PubMed] [Google Scholar]
- Thompson R. H. et al. Tumor B7-H1 is associated with poor prognosis in renal cell carcinoma patients with long-term follow-up. Cancer Res 66, 3381–3385 (2006). [DOI] [PubMed] [Google Scholar]
- Soria J. C., Marabelle A., Brahmer J. R. & Gettinger S. Immune checkpoint modulation for non-small cell lung cancer. Clin Cancer Res 21, 2256–2262 (2015). [DOI] [PubMed] [Google Scholar]
- Topalian S. L. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst R. S. et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garon E. B. et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 372, 2018–2028 (2015). [DOI] [PubMed] [Google Scholar]
- Brahmer J. et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 373, 123–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velcheti V. et al. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest 94, 107–116 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalper K. A. et al. In situ tumor PD-L1 mRNA expression is associated with increased TILs and better outcome in breast carcinomas. Clin Cancer Res 20, 2773–2782 (2014). [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Senovilla L., Zitvogel L. & Kroemer G. The secret ally: immunostimulation by anticancer drugs. Nat Rev Drug Discov 11, 215–233 (2012). [DOI] [PubMed] [Google Scholar]
- Zitvogel L., Apetoh L., Ghiringhelli F. & Kroemer G. Immunological aspects of cancer chemotherapy. Nat Rev Immunol 8, 59–73 (2008). [DOI] [PubMed] [Google Scholar]
- Kepp O. et al. Molecular determinants of immunogenic cell death elicited by anticancer chemotherapy. Cancer Metastasis Rev 30, 61–69 (2011). [DOI] [PubMed] [Google Scholar]
- Hato S. V., Khong A., de Vries I. J. & Lesterhuis W. J. Molecular pathways: the immunogenic effects of platinum-based chemotherapeutics. Clin Cancer Res 20, 2831–2837 (2014). [DOI] [PubMed] [Google Scholar]
- Halama N. et al. Localization and density of immune cells in the invasive margin of human colorectal cancer liver metastases are prognostic for response to chemotherapy. Cancer Res 71, 5670–5677 (2011). [DOI] [PubMed] [Google Scholar]
- Ladoire S. et al. Pathologic complete response to neoadjuvant chemotherapy of breast carcinoma is associated with the disappearance of tumor-infiltrating foxp3 + regulatory T cells. Clin Cancer Res 14, 2413–2420 (2008). [DOI] [PubMed] [Google Scholar]
- Schoof N. et al. Favorable impact of the interleukin-4 receptor allelic variant I75 on the survival of diffuse large B-cell lymphoma patients demonstrated in a large prospective clinical trial. Ann Oncol 20, 1548–1554 (2009). [DOI] [PubMed] [Google Scholar]
- DeMichele A. et al. Host genetic variants in the interleukin-6 promoter predict poor outcome in patients with estrogen receptor-positive, node-positive breast cancer. Cancer Res ; 69, 4184–4191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet 132, 641–648 (2013). [DOI] [PubMed] [Google Scholar]
- Lake R. A. & Robinson B. W. Immunotherapy and chemotherapy–a practical partnership. Nat Rev Cancer 5, 397–405 (2005). [DOI] [PubMed] [Google Scholar]
- Zhu Y. Liu N., Xiong S. D., Zheng Y. J. & Chu Y. W. CD4 + Foxp3 + regulatory T-cell impairment by paclitaxel is independent of toll-like receptor 4. Scand J Immunol 73, 301–308 (2011). [DOI] [PubMed] [Google Scholar]
- Lesterhuis W. J. et al. Platinum-based drugs disrupt STAT6-mediated suppression of immune responses against cancer in humans and mice. J Clin Invest 121, 3100–3108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkert C. et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol 28, 105–113 (2010). [DOI] [PubMed] [Google Scholar]
- Wimberly H. et al. PD-L1 Expression Correlates with Tumor-Infiltrating Lymphocytes and Response to Neoadjuvant Chemotherapy in Breast Cancer. Cancer Immunol Res 3, 326–332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y. et al. Polymorphisms in DNA repair and apoptosis-related genes and clinical outcomes of patients with non-small cell lung cancer treated with first-line paclitaxel-cisplatin chemotherapy. Lung Cancer 82, 330–339 (2013). [DOI] [PubMed] [Google Scholar]
- Eisenhauer E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45, 228–247 (2009). [DOI] [PubMed] [Google Scholar]
- Stephens M., Smith M. J. & Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68, 978–989 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]