Abstract
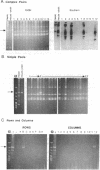
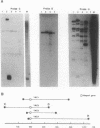
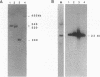
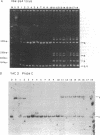
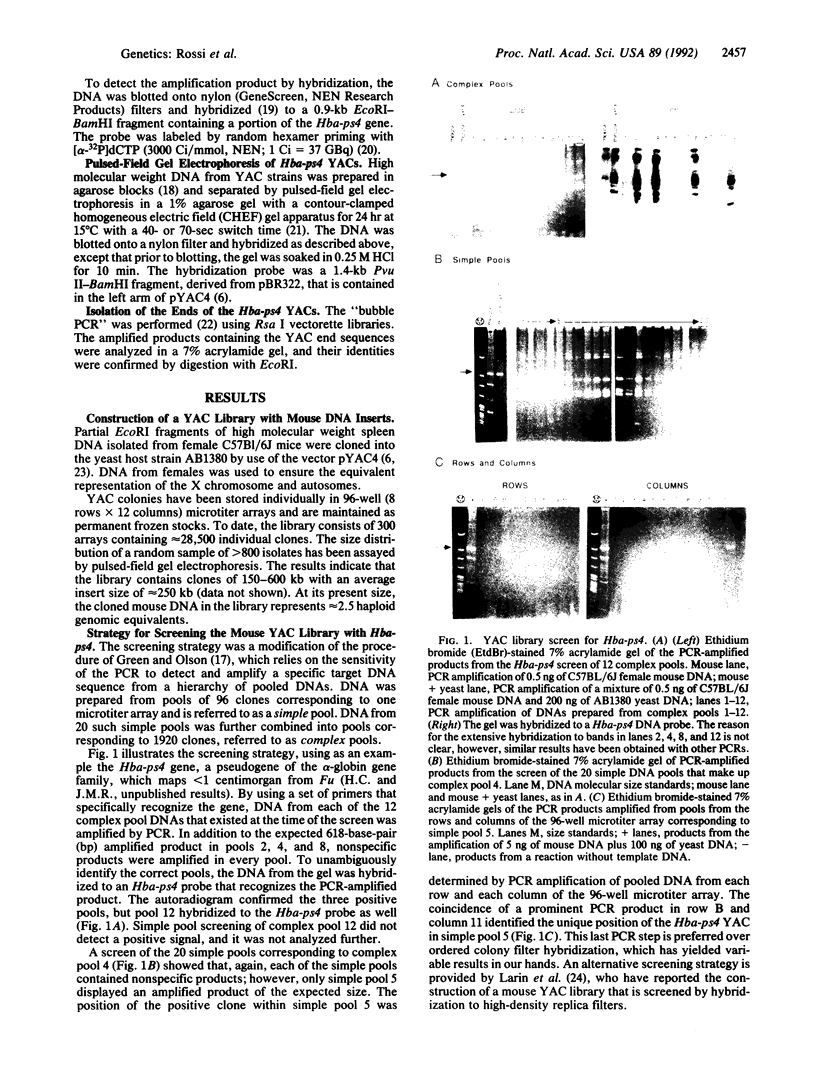
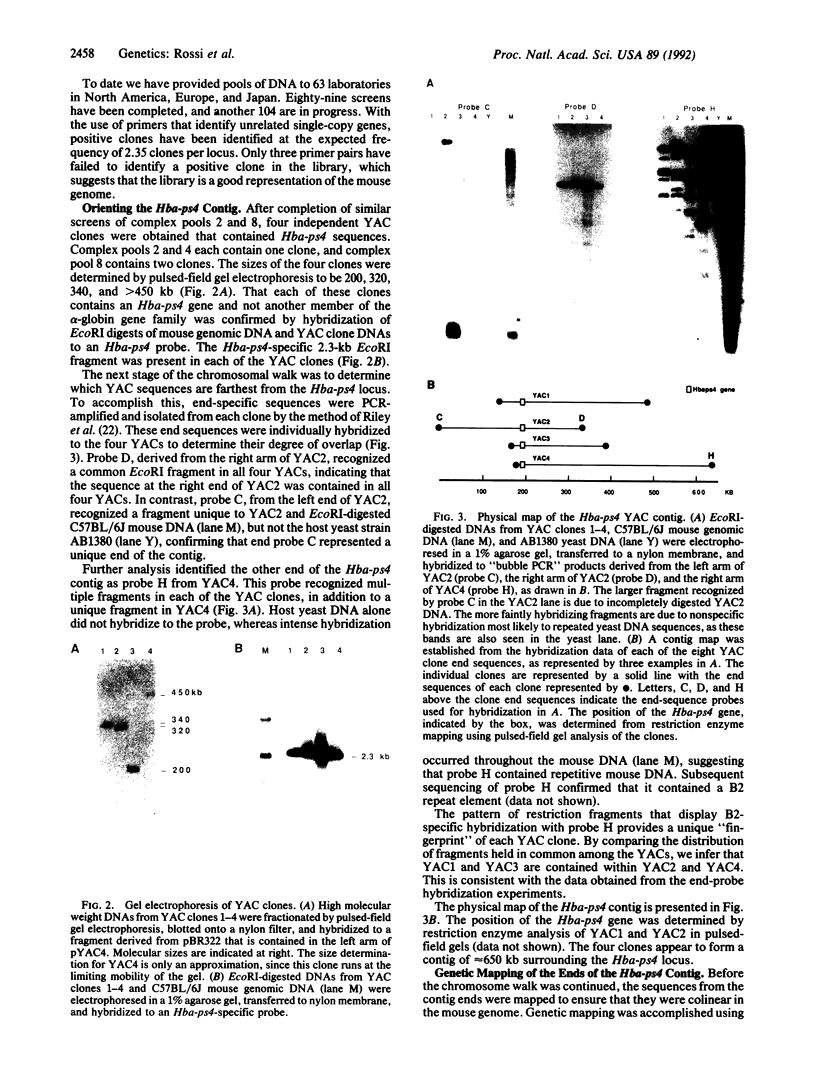
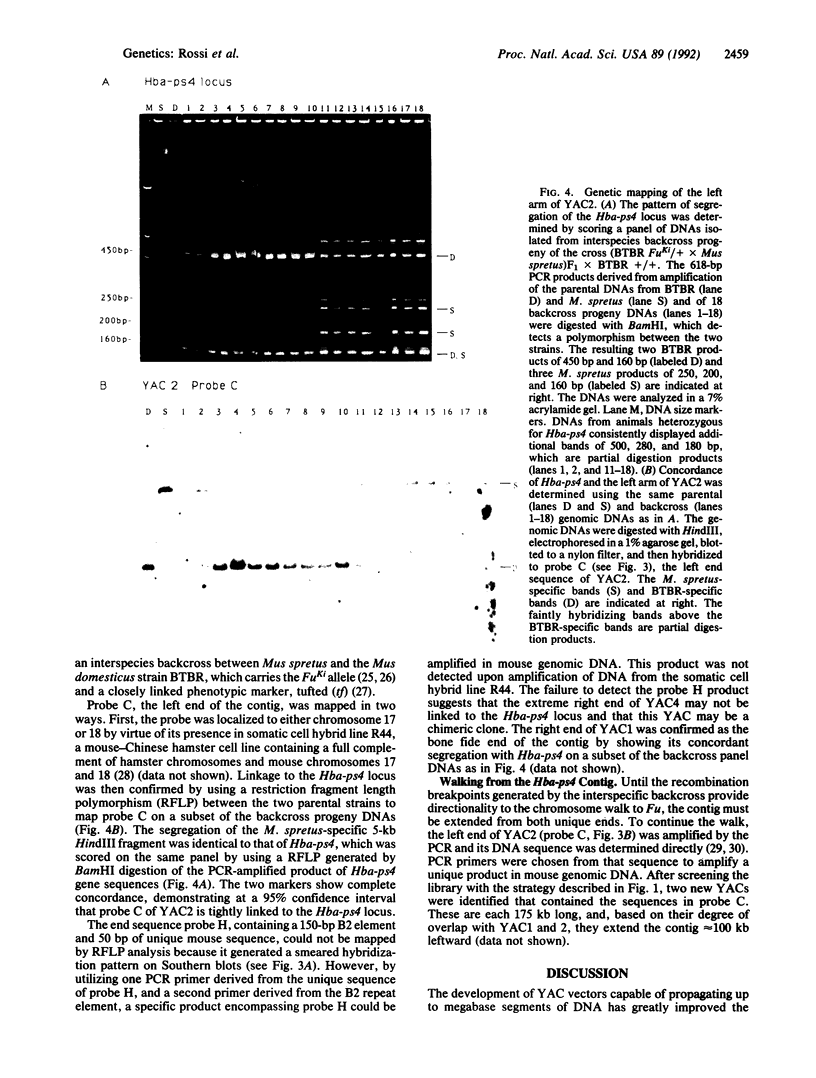
A yeast artificial chromosome library with mouse genomic DNA inserts has been constructed. The library encompasses a 2.5-fold coverage of the mouse genome, with an average insert size of 250 kilobases. The screening strategy uses the polymerase chain reaction on pooled DNAs prepared from individually stored clones. The usefulness of the library for chromosome walking was illustrated by constructing a 600-kilobase-long contig of DNA surrounding Hba-ps4, a DNA marker that is tightly linked to the fused (Fu) locus on chromosome 17.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avner P., Amar L., Dandolo L., Guénet J. L. Genetic analysis of the mouse using interspecific crosses. Trends Genet. 1988 Jan;4(1):18–23. doi: 10.1016/0168-9525(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Brownstein B. H., Silverman G. A., Little R. D., Burke D. T., Korsmeyer S. J., Schlessinger D., Olson M. V. Isolation of single-copy human genes from a library of yeast artificial chromosome clones. Science. 1989 Jun 16;244(4910):1348–1351. doi: 10.1126/science.2544027. [DOI] [PubMed] [Google Scholar]
- Burgers P. M., Percival K. J. Transformation of yeast spheroplasts without cell fusion. Anal Biochem. 1987 Jun;163(2):391–397. doi: 10.1016/0003-2697(87)90240-5. [DOI] [PubMed] [Google Scholar]
- Burke D. T., Carle G. F., Olson M. V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science. 1987 May 15;236(4803):806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- Burke D. T., Olson M. V. Preparation of clone libraries in yeast artificial-chromosome vectors. Methods Enzymol. 1991;194:251–270. doi: 10.1016/0076-6879(91)94020-d. [DOI] [PubMed] [Google Scholar]
- Burke D. T., Rossi J. M., Leung J., Koos D. S., Tilghman S. M. A mouse genomic library of yeast artificial chromosome clones. Mamm Genome. 1991;1(1):65–65. doi: 10.1007/BF00350849. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Fox H. S., Silver L. M., Martin G. R. An alpha globin pseudogene is located within the mouse t complex. Immunogenetics. 1984;19(2):125–130. doi: 10.1007/BF00387855. [DOI] [PubMed] [Google Scholar]
- Green E. D., Olson M. V. Systematic screening of yeast artificial-chromosome libraries by use of the polymerase chain reaction. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1213–1217. doi: 10.1073/pnas.87.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan R. J., O'Brien M. C. Genetic analysis of mutations at the fused locus in the mouse. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4413–4417. doi: 10.1073/pnas.83.12.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénet J. L., Simon-Chazottes D., Avner P. R. The use of interspecific mouse crosses for gene localization: present status and future perspectives. Curr Top Microbiol Immunol. 1988;137:13–17. doi: 10.1007/978-3-642-50059-6_2. [DOI] [PubMed] [Google Scholar]
- Hermanson G. G., Hoekstra M. F., McElligott D. L., Evans G. A. Rescue of end fragments of yeast artificial chromosomes by homologous recombination in yeast. Nucleic Acids Res. 1991 Sep 25;19(18):4943–4948. doi: 10.1093/nar/19.18.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R. G., Ochman H. Production of single-stranded DNA templates by exonuclease digestion following the polymerase chain reaction. Nucleic Acids Res. 1989 Jul 25;17(14):5865–5865. doi: 10.1093/nar/17.14.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs-Cohen R. J., Spiegelman M., Cookingham J. C., Bennett D. Knobbly, a new dominant mutation in the mouse that affects embryonic ectoderm organization. Genet Res. 1984 Feb;43(1):43–50. doi: 10.1017/s0016672300025702. [DOI] [PubMed] [Google Scholar]
- Larin Z., Monaco A. P., Lehrach H. Yeast artificial chromosome libraries containing large inserts from mouse and human DNA. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4123–4127. doi: 10.1073/pnas.88.10.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder A., Swan D., Ruddle F., D'Eustachio P., Leder P. Dispersion of alpha-like globin genes of the mouse to three different chromosomes. Nature. 1981 Sep 17;293(5829):196–200. doi: 10.1038/293196a0. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D., Brinster R. L. Germ-line transformation of mice. Annu Rev Genet. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S C. The Inheritance and Expression of Fused, a New Mutation in the House Mouse. Genetics. 1937 Jan;22(1):1–13. doi: 10.1093/genetics/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J., Butler R., Ogilvie D., Finniear R., Jenner D., Powell S., Anand R., Smith J. C., Markham A. F. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990 May 25;18(10):2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley J. R., Steege D. A., Juricek D. K., Summers W. P., Ruddle F. H. A herpes simplex virus 1 integration site in the mouse genome defined by somatic cell genetic analysis. Cell. 1978 Oct;15(2):455–468. doi: 10.1016/0092-8674(78)90015-6. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R. Introduction of homologous DNA sequences into mammalian cells induces mutations in the cognate gene. Nature. 1986 Nov 6;324(6092):34–38. doi: 10.1038/324034a0. [DOI] [PubMed] [Google Scholar]