ABSTRACT
A novel method for the determination of benzoic acid has been employed to identify carboxypeptidase activities in clinically relevant pathogens. Benzoic acid was determined after chemical derivatization by gas chromatography–mass spectrometry (GC–MS). N-Benzoyl amino acid substrates were evaluated for the detection of carboxypeptidase activities in a number of clinical pathogens. Upon enzymatic hydrolysis of these substrates, benzoic acid was produced which was detected by extraction from the liquid culture supernatant, derivatization as the trimethylsilyl ester, with subsequent analysis by GC–MS. Enzymatic hydrolysis of N-benzoyl glycine was observed for S. agalactiae, M. morganii, and A. baumannii. In addition, P. fluorescens was found to hydrolyze N-benzoyl-L-glutamic acid. Although the method provides an alternative approach for determining carboxypeptidase activity, ultimately it would not be a suitable method in a clinical setting. However, the method is well-suited for identifying carboxypeptidase activities that have not been previously described or to corroborate a carboxypeptidase assay with the ninhydrin reagent.
KEYWORDS: Carboxypeptidase activity, chemical derivatization, enzymatic hydrolysis, gas chromatography–mass spectrometry: N-benzoyl amino acid substrates
Introduction
There are many different classes of enzymes, and their use as taxonomic markers for microorganisms is well documented for esterases, glycosidases, arylamidases, nitroreductases, and aminopeptidases (Orenga et al. 2009), which are all utilized in the detection and enumeration of bacteria. Carboxypeptidases are a class of enzyme that hydrolyzes peptides, dipeptides, or longer homologs from the C-terminal; the human carboxypeptidases are well characterized, but there is a minimal amount of literature precedence on the prevalence of bacterial carboxypeptidases. As such, their potential use as taxonomic markers for clinical pathogens has not been fully explored.
The only enzymatic assay that utilizes a carboxypeptidase substrate currently in widespread use for taxonomic identification of bacteria is the hippuricase assay. Detection of the hippuric acid (N-benzoyl glycine) metabolites benzoic acid and glycine allows species level differentiation of the genera Campylobacter (Morris et al. 1985), Streptococcus (Edberg and Samuels 1976), and Listeria (Feresu and Jones 1988). Several methods of determining hippuricase activity have been described (Ferrieri, Wannamaker, and Nelson 1973). However, these methods generally rely on unspecific colorimetric reactions to determine activity. The prevailing methodology for determining enzymatic activity is by the addition of ninhydrin to incubated samples, which develop a deep purple color upon reaction of the reagent with the free amine moiety of glycine (produced upon enzymatic hydrolysis of hippuric acid). This is a somewhat unspecific means of determining carboxypeptidase activity due to the potential for false positives by reaction of ninhydrin with biogenic amines (Dealler 1993). Previously, Perry et al. (1998) determined carboxypeptidase activity in clinically significant species of Gram-negative bacteria by screening seven N-benzoyl-L-amino acids for activity with a ninhydrin based assay.
This study reports a new method to assess carboxypeptidase activity with greater specificity. The method was developed to analyze the benzoic acid produced upon enzymatic hydrolysis of N-benzoyl amino acid carboxypeptidase substrates by GC–MS. The substrates N-benzoyl glycine and N-benzoyl-L-glutamic acid were assessed against bacterial species that were previously shown by Perry et al. (1998) to exhibit carboxypeptidase activity in 100% of strains tested by the ninhydrin reaction. M. morganii and A. baumannii were assessed for N-benzoyl glycine activity, whereas P. fluorescens was assayed for activity with N-benzoyl-L-glutamic acid. S. agalactiae was assessed as a positive control species for N-benzoyl glycine activity, and E. coli as a negative control. P. aeruginosa was the negative control for N-benzoyl-L-glutamic acid activity. Bacteria were assessed for activity following the methodology described by Perry et al. (1998); in this case, however, the determination of benzoic acid concentration was done by derivatization as the trimethylsilyl ester. Derivatization was necessary to determine trimethylsilyl benzoate by GC–MS. Previous work from this group has identified the range of naturally occurring volatile organic compounds that are produced by a range of Gram-positive bacteria (Staphylococcus aureus) and Gram-negative bacteria (Escherichia coli and Klebsiella pneumonia) resulting from different culture media and different analytical methods using multivariate analysis techniques (Tait et al. 2014b). This area of research was then extended to add specific substrates to samples that would cleave a characteristic volatile organic compound in the presence of the bacteria-containing enzyme. This approach has been successfully applied for the detection of Clostridium difficile in stool samples in a blind study (Tait et al. 2013). The approach was based on the activation of the enzyme p-hydroxyphenylacetate decarboxylase by the addition of the substrate 3-fluoro-4-hydroxyphenyl acetic acid and subsequent detection of the volatile 2-fluoro-4-methylphenol by GC–MS. A similar approach was employed for Listeria monocytogenes in milk (Tait et al. 2014a). In this case, two enzyme substrates were added to utilize the enzyme activity of β-glucosidase and hippuricase enzymes to determine 2-nitrophenol and 3-fluoroaniline by GC–MS. An extension to this research has been to determine exogenous volatile organic compound metabolites by colorimetric analysis (Tait et al. 2015). Two model systems were used to investigate the release of a characteristic volatile organic compound and its subsequent trapping in modified agarose gel. In one model system, Enterococcus faecium and Klebsiella pneumoniae, both with known β-glucosidase activity, generated a yellow color in modified agarose gel as a result of the evolution of volatile 2-nitrophenol. The other model system investigated β-alanyl aminopeptidase activity using Pseudomonas aeruginosa as the test bacteria. In this case, the modified agarose gel turned orange as a result of the hydrolysis of the trifluoroacetic acid salt of 3-amino-N-phenylpropanamide. This study represents a new approach for the characterization of carboxypeptidase enzyme activity.
Materials and methods
Chemicals and reagents
The N-benzoyl glycine, N,O-bis(trimethylsilyl)acetamide, and biphenyl were obtained from Sigma-Aldrich (Poole, UK). Diethyl ether was purchased from Fischer Scientific (Loughborough, UK) and N-benzoyl-L-glutamic acid from TCI UK (Oxford, UK).
Microbiology
Test strains Escherichia coli NCTC 10418, Acinetobacter baumannii ATCC 19606, Morganella morganii WILD, Pseudomonas fluorescens NCTC 10688, Pseudomonas aeruginosa NCTC 10662, and Streptococcus agalactiae NCTC 8181 were obtained from the Freeman Hospital (Newcastle upon Tyne, UK), and maintained by sub-culture on Columbia blood agar supplemented with 5% defibrinated horse blood on a fortnightly basis.
Instrumentation
A Thermo-Finnigan trace GC Ultra coupled with a Polaris Q mass spectrometer was used for all analyses. It was fitted with an Agilent Technologies DB-5 30 m × 25 mm i.d. × 0.25 µm capillary column. The split/splitless injector was set at 230°C with a split ratio of 10, and a helium gas flow rate of 10 mL/min. The oven was programmed with an initial temperature of 50°C and a hold time of 3 min. A temperature gradient of 10°C/min was used with a final temperature of 250°C and a 2 min hold time. Helium was the carrier gas at a flow rate of 1.0 mL/min. The mass spectrometer was operated in electron ionization mode (70 eV), with a mass transfer line temperature of 250°C, and ion source temperature of 260°C. Single ion monitoring of masses at m/z of 135, 154, 155, and 179 was employed for all measurements.
Growth of bacteria and sample preparation
Carboxypeptidase activity was determined by sub-culturing bacteria on Columbia blood agar supplemented with 5% defibrinated horse blood 24 h prior to sample preparation. A 20,000 µg/mL stock solution of the carboxypeptidase substrate was prepared by dissolving 2 g (accurately weighed) of substrate in 100 mL of deionized water. Dissolution was aided by addition of 2 M sodium hydroxide dropwise. The final solution was adjusted to pH 7.0–7.4. The substrate stock solution was sterilized by autoclaving at 121°C for 15 min and 2 mL of this solution was added to a sterile 50 mL PTFE screw cap centrifuge tube. Separately sterilized deionized water was inoculated with the 24 h subculture of the test strain to a turbidity equivalent to a 7.5 McFarland standard; for the positive control S. agalactiae a 0.5 McFarland suspension was prepared. Finally, 2 mL of the bacterial suspension was added to the substrate solution; samples were incubated for 24 h (and 48 h as necessary). Blanks containing only bacteria and only substrate were also prepared and analyzed. Samples were centrifuged after incubation at 5000 g for 10 min, and 2 mL of the supernatant was transferred to a sterile 50 mL centrifuge tube. The supernatant was then acidified with dilute HCl, and extracted with 2 mL of diethyl ether, and inverted approximately eighty times. Samples were centrifuged again at 5,000 g for 10 min. Finally, 500 µL of the organic phase was transferred to a 1.0-mL Thermo Scientific reacti-vial, and evaporated to dryness under a stream of dry nitrogen. An amount of 100 µL of N, O-bis(trimethylsilyl)acetamide was added to the reacti-vial, vortexed for 60 sec, and heated at 70°C overnight. A biphenyl internal standard stock solution was prepared by dissolving 0.1 g (accurately weighed) of biphenyl in acetonitrile and diluting to 10 mL (10,000 µg/mL) to make the stock solution; this was further diluted to an internal standard working solution by taking 100 µL in 10 mL (100 µg/mL). The 50-µL working internal standards were added to the reacti-vials, which were diluted to 0.5 mL by addition of 350 µL of acetonitrile. Samples were vortexed and transferred to GC autosampler vials for analysis.
Quantification of trimethylsilyl benzoate
Derivatization of benzoic acid, as the trimethylsilyl ester, was initially confirmed by scanning in TIC mode in the mass range from 50 to 650 amu, and library matching the mass spectrum with the National Institute of Standards and Technology (NIST) mass spectral library, version 2.0a. The molecular ion m/z 194, as well as fragment ions 179, 135, 105, and 77 were consistent with the library entry. When analyzing routine samples, m/z 179 and 135 were scanned in SIM mode. Trimethylsilyl benzoate was quantified by external calibration with the internal standard. A range of benzoic acid standards in acetonitrile were prepared and 50 µL was transferred to reacti-vials, following the same derivatization procedure as routine samples.
Results and discussion
Various protocols were attempted for the derivatization of benzoic acid, originally with a focus on achieving full conversion of benzoic acid to the trimethysilyl ester rapidly. However, with reaction times of 1 h and 1.25 h, it was found that reproducibility was poor. By performing the derivatization step overnight, acceptable repeatability was achieved (typical relative standard deviation < 14%, n = 5). Various solvents were used to enhance the recovery of benzoic acid after the drying step; however, dimethylformamide (DMF) and pyridine provided no significant advantages over N,O-bis(trimethylsilyl)acetamide (BSA) alone. Therefore, no additional solvent was used in the derivatization process. Temperature was not investigated, but it may be possible to improve the efficiency of derivatization by optimizing the temperature. Following this protocol, a calibration plot (linear equation y = 0.0565x −0.1678; R 2 = 0.9937) was obtained to determine the concentration of the derivatized compounds.
The results based on N-benzoyl glycine activity are shown in Table 1, whereas those for N-benzoyl-L-glutamic acid are in Table 2. Although some benzoic acid was observed in all blanks, a significant increase in trimethylsilyl benzoate concentration was observed in all strains that display carboxypeptidase activity; this was in agreement with results published by Perry et al. (1998). Table 1 shows that the concentration of trimethylsilyl benzoate was greater than the reagent blank for M. morganii and considerably greater for A. baumannii; the reagent blank subtracted concentrations of 7.08 µg/mL and 110.2 µg/mL, respectively. These positive results indicate the presence of carboxypeptidase based on N-benzoyl glycine activity. The considerable difference in reagent blank subtracted concentration of trimethylsilyl benzoate obtained for A. baumannii, with the substrate N-benzoyl glycine, suggests that the substrate may have relevance as a taxonomic marker for this species. The positive control species S. agalactiae (in the case of N-benzoyl glycine) also produced a positive reaction as expected. However, given that S. agalactiae was the only strain tested in which hippuricase activity is used as a taxonomic marker, it did not exhibit stronger activity than the other positive strains (and in the case of A. baumannii much weaker activity). In the case of E. coli, no discernible reagent blank subtracted concentration was obtained for trimethylsilyl benzoate. This was as expected and was in agreement with the result for E. coli obtained by Perry et al. (1998), who tested 42 strains, using the nonspecific ninhydrin test. Figure 1 shows an example chromatograph for the determination of trimethylsilyl benzoate (retention time 13.10 min) from A. baumannii using the substrate N-benzoyl glycine. Figure 1 includes the presence of the internal standard (biphenyl, retention time 15.55 min) and derivatized substrate (21.82 min).
Table 1. Trimethylsilyl benzoate concentrations (µg/mL) after 24 h incubation with N-benzoyl glycine (n = 2).
| Test strain | Bacteria blank | N-benzoyl glycine blank | N-benzoyl glycine + bacteria |
|---|---|---|---|
| S. agalactiae NCTC 8181 | 3.11 (3.10, 3.12) | 4.14 (4.71, 3.59) | 10.08 (10.01, 10.15) |
| A. baumannii ATCC 19606 | 3.01 (3.00, 3.03) | 3.11 (3.14, 3.08) | 113.28 (106.00, 120.56) |
| M. morganii WILD | 3.36 (3.37, 3.36) | 3.34 (3.55, 3.14) | 10.42 (10.97, 9.87) |
| E. coli NCTC 10418 | 3.32 (3.57, 3.07) | 3.37 (3.41, 3.33) | 3.19 (3.21, 3.17) |
Table 2. Trimethylsilyl benzoate concentrations (µg/mL) after 24–48 h incubation with N-benzoyl-L-glutamic acid (n = 2).
| Test strain | Bacteria blank | N-benzoyl-L- glutamate blank | N-benzoyl-L-glutamic acid + bacteria |
|---|---|---|---|
| P. aeruginosa NCTC 10662 (24 h) | 3.37 (3.48, 3.25) | 6.89 (7.11, 6.68) | 6.06 (5.87, 6.24) |
| P. aeruginosa NCTC 10662 (48 h) | – | – | 3.78 (3.77, 3.80) |
| P. fluorescens NCTC 10688 (24 h) | 2.99 (2.99, 2.99) | 5.11 (5.25, 4.98) | 12.14 (11.49, 12.79) |
| P. fluorescens NCTC 10688 (48 h) | – | – | 25.11 (25.66, 24.57) |
Figure 1.
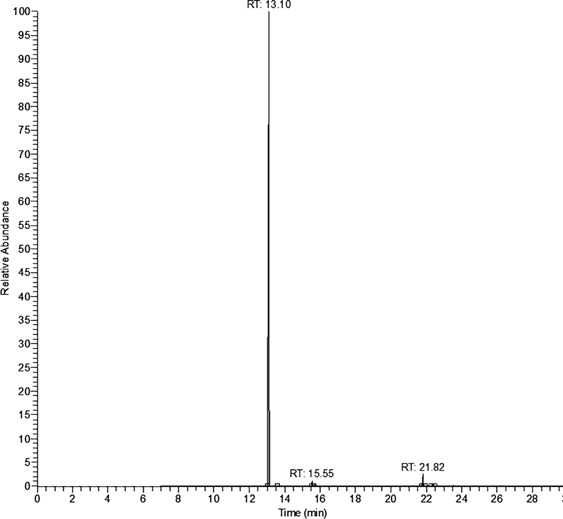
Trimethylsilyl benzoate (retention time 13.10 min) derived from benzoic acid extracted from Acinetobacter baumannii ATCC 19606 samples. Other peaks: biphenyl (retention time 15.55 min) and derivatized substrate (retention time 21.82 min).
The P. fluorescens may be distinguished from P. aeruginosa on the basis of N-benzoyl-L-glutamic acid activity (Table 2). The reagent blank subtracted concentration for trimethylsilyl benzoate was 7.03 µg/mL at 24 h and 20.0 µg/mL at 48 h. No discernible reagent blank subtracted concentration of trimethylsilyl benzoate was obtained for P. aeruginosa. Although carboxypeptidase G2 activity has been widely reported for P. fluorescens (Rowsell et al. 1997), it has not been sought as a mean of distinguishing species of Pseudomonas until now. We have shown that carboxypeptidase may be used as a taxonomic marker.
References
- Dealler S. F. Chromogenic and fluorogenic indicators and substrates in diagnostic microbiology. Reviews in Medical Microbiology. 1993;4:198–206. doi: 10.1097/00013542-199310000-00003. [DOI] [Google Scholar]
- Edberg S. C., Samuels S. Rapid, colorimetric test for the determination of hippurate hydrolysis by group B streptococcus. Journal of Clinical Microbiology. 1976;3:49–50. doi: 10.1128/jcm.3.1.49-50.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feresu S. B., Jones D. Taxonomic studies on brochothrix, erysipelothrix, listeria and atypical lactobacilli. Microbiology. 1988;134:1165–83. doi: 10.1099/00221287-134-5-1165. [DOI] [PubMed] [Google Scholar]
- Ferrieri P., Wannamaker L. W., Nelson J. Localization and characterization of the hippuricase activity of group B streptococci. Infection and Immunity. 1973;7:747–52. doi: 10.1128/iai.7.5.747-752.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. K., Sherbeeny M. R., Patton C. M., Kodaka H., Lombard G. L., Edmonds P., Hollis D. G., Brenner D. J. Comparison of four hippurate hydrolysis methods for identification of thermophilic Campylobacter spp. Journal of Clinical Microbiology. 1985;22:714–18. doi: 10.1128/jcm.22.5.714-718.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenga S., James A. L., Manafi M., Perry J. D., Pincus D. H. Enzymatic substrates in microbiology. Journal of Microbiological Methods. 2009;79:139–55. doi: 10.1016/j.mimet.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Perry J. D., Archer L., Jones A. L., James A. L., Ford M., Gould F. K. Detection of carboxypeptidases as taxonomic markers for gram-negative bacteria. Letters in Applied Microbiology. 1998;26:359–62. doi: 10.1046/j.1472-765x.1998.00350.x. [DOI] [PubMed] [Google Scholar]
- Rowsell S., Pauptit R. A., Tucker A. D., Melton R. G., Blow D. M., Brick P. Crystal structure of carboxypeptidase G2, a bacterial enzyme with applications in cancer therapy. Structure. 1997;5:337–47. doi: 10.1016/s0969-2126(97)00191-3. [DOI] [PubMed] [Google Scholar]
- Tait E., Hill K. A., Perry J. D., Stanforth S. P., Dean J. R. Development of a novel method for detection of clostridium difficile using HS-SPME-GC-MS. Journal of Applied Microbiology. 2013;116:1010–19. doi: 10.1111/jam.12418. [DOI] [PubMed] [Google Scholar]
- Tait E., Perry J. D., Stanforth S. P., Dean J. R. Bacteria detection based on the evolution of enzyme-generated volatile organic compounds: Determination of Listeria monocytogenes in milk samples. Analytica Chimica Acta. 2014a;848:80–87. doi: 10.1016/j.aca.2014.07.029. [DOI] [PubMed] [Google Scholar]
- Tait E., Perry J. D., Stanforth S. P., Dean J. R. Identification of volatile organic compounds produced by bacteria using HS-SPME-GC-MS. Journal of Chromatographic Science. 2014b;52:363–73. doi: 10.1093/chromsci/bmt042. [DOI] [PubMed] [Google Scholar]
- Tait E., Stanforth S. P., Reed S., Perry J. D., Dean J. R. Analysis of pathogenic bacteria using exogenous volatile organic compound metabolites and optical sensor detection. RSC Advances. 2015;5:15494–99. doi: 10.1039/c4ra13914c. [DOI] [Google Scholar]


