Abstract
Inherited cardiomyopathy is the major cause of sudden cardiac death (SCD) and heart failure (HF). The disease is associated with extensive genetic heterogeneity; pathogenic mutations in cardiac sarcomere protein genes, cytoskeletal protein genes and nuclear envelope protein genes have been linked to its etiology. Early diagnosis is conducive to clinical monitoring and allows for presymptomatic interventions as needed. In the present study, the entire coding sequences and flanking regions of 12 major disease (cardiomyopathy)-related genes [namely myosin, heavy chain 7, cardiac muscle, β (MYH7); myosin binding protein C, cardiac (MYBPC3); lamin A/C (LMNA); troponin I type 3 (cardiac) (TNNI3); troponin T type 2 (cardiac) (TNNT2); actin, α, cardiac muscle 1 (ACTC1); tropomyosin 1 (α) (TPM1); sodium channel, voltage gated, type V alpha subunit (SCN5A); myosin, light chain 2, regulatory, cardiac, slow (MYL2); myosin, heavy chain 6, cardiac muscle, α (MYH6); myosin, light chain 3, alkali, ventricular, skeletal, slow (MYL3); and protein kinase, AMP-activated, gamma 2 non-catalytic subunit (PRKAG2)] in 8 patients with dilated cardiomyopathy (DCM) and in 8 patients with hypertrophic cardiomyopathy (HCM) were amplified and then sequenced using the Ion Torrent Personal Genome Machine (PGM) system. As a result, a novel heterozygous mutation (MYH7, p.Asn885Thr) and a variant of uncertain significance (TNNT2, p.Arg296His) were identified in 2 patients with HCM. These 2 missense mutations, which were absent in the samples obtained from the 200 healthy control subjects, altered the amino acid that was evolutionarily conserved among a number of vertebrate species; this illustrates that these 2 non-synonymous mutations play a role in the pathogenesis of HCM. Moreover, a double heterozygous mutation (PRKAG2, p.Gly100Ser plus MYH7, p.Arg719Trp) was identified in a patient with severe familial HCM, for the first time to the best of our knowledge. This patient provided us with more information regarding the genotype-phenotype correlation between mutations of MYH7 and PRKAG2. Taken together, these findings provide insight into the molecular mechanisms underlying inherited cardiomyopathy. The mutations identified in this study may be further investigated in the future in order to improve the diagnosis and treatment of patients with inherited cardiomyopathy. Furthermore, our findings indicated that sequencing using the Ion Torrent PGM system is a useful approach for the identification of pathogenic mutations associated with inherited cardiomyopathy, and it may be used for the risk evaluation of individuals with a possible susceptibility to inherited cardiomyopathy.
Keywords: hypertrophic cardiomyopathy, dilated cardiomyopathy, gene mutation, genetic testing, Ion Torrent PGM system
Introduction
Inherited cardiomyopathies are divided into the following 4 categories according to alterations in ventricular morphology and function: hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), arrhythmogenic right ventricular cardiomyopathy (ARVC) and restrictive cardiomyopathy (RCM) (1,2). It is associated with extensive genetic heterogeneity; inherited cardiomyopathy-linked mutations have been found in over 100 disease-causing genes, including mutations in the genes encoding the following: cardiac sarcomere proteins, cytoskeletal proteins and nuclear envelope proteins (3–5), cardiac development and structural remodeling proteins (6–8), cardiac transcription factors (9–14), Tax-1-binding protein (15) and the RAS-mitogen-activated protein kinase pathway (16). As the leading cause of sudden cardiac death (SCD) in adolescents and young athletes, HCM is the most common type of inherited cardiomyopathy with a morbidity rate of approximately 1 in 500 individuals worldwide, which is characterized by unexplained left ventricular hypertrophy (17,18). DCM is characterized by left ventricular dilatation and systolic dysfunction [with a left ventricular ejection fraction (LVEF) of <50%], and affects at least 1/2,500 individuals in the general population (3); it is primarily caused by pathogenic gene mutations inherited in a Mendelian autosomal dominant pattern (19). Molecular genetic testing is crucial for selecting the correct therapy and management strategies for the disease, as well as to evaluate the prognosis of patients with inherited cardiomyopathy and of their family members.
Currently, conventional capillary-based sequencing is the gold standard approach for detecting mutations associated with this disease. However, this method has drawbacks, as not all types of genetic variation are detectable and it is a time-consuming and costly technique. In a striking technological development, the Ion Torrent Personal Genome Machine (PGM) system was launched in 2011 by Life Technologies, and it has been demonstrated to be a more rapid, more sensitive and less costly system, and it also allows the scalable sequencing of samples (20).
In the present study, we enrolled 16 Chinese patients diagnosed with inherited cardiomyopathy (either HCM or DCM) and performed bioinformatics and molecular genetic analyses of the entire coding sequence and flanking regions of 12 major disease (cardiomyopathy)-related genes using the Ion Torrent PGM system. As a result, we identified a novel [myosin, heavy chain 7, cardiac muscle, β (MYH7), p.Asn885Thr] mutation, a variant of uncertain significance [troponin T type 2 (cardiac) (TNNT2), p.Arg296His] and a double heterozygous [protein kinase, AMP-activated, gamma 2 non-catalytic subunit (PRKAG2), p.Gly100Ser plus MYH7, p.Arg719Trp] mutation. The findings of our study expand the mutational spectrum of MYH7 and TNNT2 which are associated with HCM, and enhance our understanding of the molecular mechanisms underlying inherited cardiomyopathy. Furthermore, we present a useful approach for the genetic testing of patients with inherited cardiomyopathy.
Subjects and methods
Patients and healthy controls
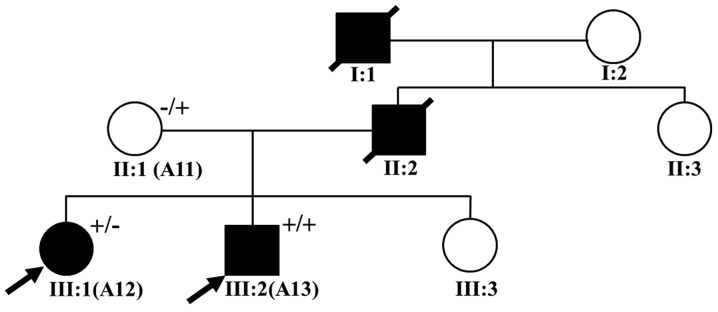
A total of 16 patients with inherited cardiomyopathy were recruited, namely 8 patients with DCM and 8 patients with HCM, who were traditionally diagnosed according to the criteria specified in the American College of Cardiology Foundation/American Heart Association (ACCF/AHA) guideline for the diagnosis and treatment of hypertrophic cardiomyopathy (21) and the European guidelines for the study of familial dilated cardiomyopathies (22). Of the 16 patients, 2 (patients A12 and A13) were considered to have familial HCM (Fig. 1). The demographic and clinical characteristcs of the patients, including family history, clinical symptoms, echocardiography results, and 12-lead electrocardiography (ECG) records, were collected. In addition, 100 healthy individuals without any symptoms of cardiovascular disease were enrolled into this study as healthy control subjects. All of the subjects provided written informed consent prior to participating voluntarily in this study. The study protocol was approved by the Ethics Committee of The Affiliated Hospital of Kunming University of Science and Technology (Kunming, China) and complied with the principles of the Declaration of Helsinki.
Figure 1.
Pedigree of the family with hypertrophic cardiomyopathy (HCM). Male family members are indicated by squares; female family members are indicated by circles, deceased individuals are indicated by symbols with a strikethrough, the unaffected individuals are represented by open symbols, and the solid symbols represent affected individuals. In addition, the probands are marked with a black arrow. The presence of a mutation is indicated by a (+) sign and the absence of mutations is indicated by a (−) sign. III:1 and III:2 are the probands, I:1 and II:2 died of sudden cardiac death, and clinical data were unavailable for the other members; II:1 possesses the mutation but the individual is clinically unaffected, and clinical data were unavailable for the other members.
DNA extraction, genomic library construction and template preparation/amplification
Peripheral whole blood samples (~2 ml) from each subject were collected in Vacutainer tubes coated with EDTA (BD Biosciences, Franklin Lakes, NJ, USA) and stored at 4°C until DNA extraction. Genomic DNA was extracted from anticoagulated whole blood of each sample using a commercial Blood Genomic DNA Miniprep kit (Axygen, Union City, CA, USA). The majority of the primers were designed using the freely available Lasergene PrimerSelect software (DNASTAR, Madison, WI, USA) and Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA, USA), and then 116 pairs of primer sets were synthesized and we amplified 226 coding exons of the following genes: MYH7; myosin binding protein C, cardiac (MYBPC3); TNNT2; troponin I type 3 (cardiac) (TNNI3); myosin, light chain 2, regulatory, cardiac, slow (MYL2); lamin A/C (LMNA); myosin, light chain 3, alkali, ventricular, skeletal, slow (MYL3); PRKAG2; sodium channel, voltage gated, type V alpha subunit (SCN5A); myosin, heavy chain 6, cardiac muscle, α (MYH6); actin, α, cardiac muscle 1 (ACTC1); and tropomyosin 1 (α) (TPM1); (data available upon request). From this panel, approximately 15 pairs of primer sets having similar annealing temperatures and of similar amplicon size were combined in one reaction pool in order to reduce the reaction times and reagent costs. Polymerase chain reaction (PCR) amplification was performed using PrimeSTAR GXL DNA polymerase (Takara, Otsu, Japan) under the following conditions: DNA denaturation at 98°C for 3 min, followed by 30 cycles of denaturing at 98°C for 10 sec, annealing at 54 or 57°C for 15 sec and extension at 68°C for 1–3 min, and finalized with one extension cycle of 68°C for 5 min (data available upon request). Finally, the multiplex PCR products from each sample were mixed in microtubes at equal concentrations which were determined using the SequalPrep Normalization kit (Invitrogen, Carlsbad, CA, USA). Genomic library construction was performed using the manual (Publi cation no. 4471989, Revision N) and DNA template preparation was conducted using an Ion OneTouch 2 instrument and an Ion OneTouch enrichment system (ES) (both from Life Technologies, Carlsbad, CA, USA).
Ion Torrent PGM sequencing and bioinformatics analysis
DNA high-throughput sequencing was performed using reagents from the Ion PGM Sequencing 400 kit (obtained from Life Technologies). The prepared samples of Ion Sphere Particles (ISP) were loaded onto an Ion 314 sequencing chip (Life Technologies), and DNA sequencing was performed in the Ion PGM instrument using the Ion PGM 400 sequencing kit set at 640 flows for 160 runs. Raw data from the PGM runs were processed using the Ion Torrent platform-specific pipeline software Torrent Suite v4.0.2 (Life Technologies) to generate sequence reads. The FastQC (v3.4.1.1) plug-in software was used in order to perform the analysis of the mean read depth and alignment quality. The short reads alignment was rapidly and accurately achieved using the Burrows Wheeler Aligner (BWA) Multi-Vision software package. Coverage Analysis (v4.0-r77897) plug-in software was used to assess the number of mapped bases, the percentage of coverage on the target gene.
The sequence variants in the 12 genes in each sample were identified using the Torrent Suite Variant Caller (TSVC; v4.0-r76860) plug-in and browser extensible data (BED) files (chromosome coordinates) that specify the coding regions of the target genes within the human reference genome (hg19) retrieved from the NCBI database (build 37) as a reference. The TSVC plug-in generated files in variant caller format (VCF) were further annotated using online Ion Reporter software (https://ionreporter.lifetechnologies.com/ir/secure/home.html), and Integrated Genomic Viewer (IGV) software (23) was then used to complete the visualization and to eliminate false-positive variants.
Molecular genetic analysis
We analyzed mutations that have been reported to be associated with the disease phenotype in the PubMed Database (http://www.ncbi.nlm.nih.gov/pubmed/) or by the Human Gene Mutation Database (HGMD; http://www.hgmd.cf.ac.uk). In addition to the above, the mutations meeting the following criteria were putatively considered pathogenic (24): i) if the mutation had a minor allele frequency (MAF) of <10% in the NCBI dbSNP137 (http://www.ncbi.nlm.nih.gov/projects/SNP/), the 1000 Genomes Project (http://www.1000genomes.org/) and NHLBI Grand Opportunity Exome Sequencing Project (ESP; https://esp.gs.washington.edu/drupal/) databases; ii) pathogenicity prediction programs that assess the functional significance of amino acid substitutions were used, including PolyPhen-2 (25), SIFT (26) or MutationTaster algorithms (27), which labeled the mutation as pathogenic; iii) the novel mutations were absent from the 100 unrelated healthy control subjects in order to rule out the possibility of the polymorphisms existing in the normal population; iv) evolutionary conservation analysis of the novel mutation was performed in a number of vertebrate species (namely, Macaca mulatta, Felis catus, Mus musculus, Gallus gallus, Danio rerio and Xenopus tropicalis). The Clustal W alignment program (http://www.genome.jp/tools/clustalw/) was used to align orthologs from eukaryotes, and the weblogo (http://weblogo.berkeley.edu/logo.cgi) format was used for aligning eukaryotic species. All of the putative pathogenic mutations were verified by PCR and conventional capillary-based sequencing analysis using an ABI 3730 automatic sequencer (Applied Biosystems, Foster City, CA, USA).
Results
Demographic and clinical characteristics of the patients with DCM and HCM
The recorded demographic and clinical characteristics of all 16 patients (8 patients diagnosed with DCM and 8 patients diagnosed with HCM) are presented in Table I. The patients with DCM had a median age at diagnosis of 44 years (ranging from 16 to 70 years and SD of 17.2 years) and those with HCM had a median age at diagnosis of 37.1 years (ranging from 10 to 57 years and SD of 19.2 years). Echocardiography revealed a mildly dilated left ventricular end-systolic diameter (LVESD, an average of 57.8 mm) and left ventricular end-diastolic diameter (LVED, an average of 68.0 mm) and LVEF (an average of 32.6%) of <50% in the patients with DCM. Echocardiography also revealed that the patients with HCM experienced significant concentric left ventricular hypertrophy (LVH) with severe alterations in interventricular septum thickness (IVST, average of 19.2 mm) and left ventricular posterior wall thickness (LVPWT; average of 11.5 mm).
Table I.
Available demographic and clinical characteristics of patients with inherited cardiomyopathy.
| Subject no. | Gender | Age (years) | Disease | Family History | Echocardiogram
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| IVST (mm) | LVED (mm) | LVESD (mm) | LVPWT (mm) | LA (mm) | LVEF (%) | |||||
| 19 | Male | 70 | DCM | Negative | N/A | N/A | N/A | N/A | N/A | N/A |
| A7 | Male | 16 | DCM | Negative | 8 | 61 | N/A | 8.8 | 35 | 45.0 |
| A63 | Male | 41 | DCM | Positive | 9.3 | 60.6 | 48 | 8.6 | 40 | 38.0 |
| A64 | Female | 56 | DCM | Positive | 9.9 | 67.4 | 59.6 | 8.5 | 58.2 | 24.0 |
| 38 | Male | 40 | DCM | Negative | 9.2 | 90.1 | 81.6 | 8.5 | 61.8 | 19.0 |
| A51 | Male | 59 | DCM | Negative | N/A | N/A | N/A | N/A | N/A | N/A |
| A88 | Male | 30 | DCM | Negative | 7.0 | N/A | N/A | 8.0 | 40 | 22.0 |
| A37 | Female | 40 | DCM | Positive | 7.9 | 61.3 | 42 | 6.8 | 32.6 | 48.0 |
| Mean ± SD | N/A | 44.0±17.2 | N/A | N/A | 8.5±1.0 | 68.0±12.6 | 57.8±17.5 | 8.2±0.7 | 44.6±12.3 | 32.6±12.5 |
| A36 | Female | 49 | HCM | Positive | 22 | 44.2 | 31.7 | 10.9 | 32 | 54.0 |
| A15 | Male | 14 | HCM | Positive | 18.8 | 37.2 | 24.0 | 16.0 | 46.0 | 65.0 |
| 51 | Female | 57 | HCM | Negative | 11.7 | 37.4 | 26.1 | 11.5 | 29.1 | 58.0 |
| A94 | Male | 57 | HCM | Positive | 16.3 | 49.6 | 26.2 | 13.5 | 52.5 | 78.0 |
| A12 | Female | 21 | HCM | Positive | 25.9 | 32.0 | 18.4 | 9.4 | 26.9 | 75.0 |
| A13 | Male | 10 | HCM | Positive | 21.0 | N/A | N/A | 12.0 | 40.0 | 48.0 |
| 64 | Male | 42 | HCM | Negative | 18.1 | 51.7 | 34.4 | 9.8 | 44.0 | 61.0 |
| A86 | Male | 47 | HCM | Positive | 20.0 | 49.2 | 32.3 | 8.8 | 32.0 | 63.0 |
| Mean ± SD | N/A | 37.1±19.2 | N/A | N/A | 19.2±4.2 | 43.0±7.6 | 27.6±5.6 | 11.5±2.4 | 37.8±9.2 | 62.6±10.1 |
HCM, hypertrophic cardiomyopathy; DCM, dilated cardiomyopathy; IVST, interventricular septum thickness; LVED, left ventricular end-diastolic diameter; LVESD, left ventricular end-systolic diameter; LVPWT, left ventricular posterior wall thickness; LA, left atrium; LVEF, left ventricular ejection fraction; N/A, not applicable.
Among the patients with familial HCM (Fig. 1), the proband of this family (A13, III:2) was a young child, who at the age of 10 years was diagnosed with a malignant HCM phenotype that manifested as a greater LVPWT with evident septal asymmetry, an enlarged left atrium (LA), decreased left ventricular systolic and diastolic function (IVST, 21 mm; LVPWT, 12 mm; LA, 40 mm; and LVEF, 48%); at the time of the initial diagnosis the child was experiencing chest pain and syncope. The family history revealed that the grandfather of the proband had died from heart disease at age 42 and that his father had succumbed to sudden cardiac death at age 40. In the absence of clinical data, the child's mother (A11, II:1) was assumed to be free of clinical symptoms. Patient A12 (III:1), who is the proband's elder sister, was diagnosed at the age of 21 with a severe HCM phenotype (IVST, 25.9 mm; LVPWT, 9.4 mm; and LA, 26.9 mm).
Analysis of raw data collected by performing multiplex PCR amplification followed by Ion Torrent PGM sequencing
To rapidly detect HCM- and DCM-related mutations of the 12 target genes, 116 primer pairs were designed for multiplex PCR in order to amplify a total of 154,537 base pairs (bp): the PCR fragment size ranged from 245 to 2906 bp (data available upon request). To distinguish the sequence data of each individual sample, we linked an Ion Xpress Barcode Adapter sequence to each fragment in the library. Following the optimization of PCR amplification conditions, we used only 6 microtubes containing multiplexed PCR pools in order to perform 116 PCR reactions on each sample (data available upon request).
In general, an Ion 314 semiconductor chip can generate an amount of 300 kb data on the microporous surface, and as it only utilizes 50% of the microporous. we assumed that the average reads length was 200 bp, and each of the nucleotide sequencing has a depth of 30x (Q30=99.9% certainty that the correct base was called). Therefore, a pool of library DNA from 5 to 6 patient samples was amplified using an Ion OneTouch 2 system and then loaded onto an Ion 314 semiconductor chip. Finally, runs of all 16 samples were performed in 3 independent replicates. The average 314 semiconductor chip loading obtained was 77.6% (ranging from 75 to 81%), and an average PGM run generated raw data of approximately 49.9 Mb on the 314 semiconductor chip (ranging from 49.1 to 50.5 Mb) (data available upon request) and a mean read length of 160 bp (ranging from 147 to 181 bp) (data available upon request). The sequencing data quality was assessed using the FastQC plug-in software, which revealed that the average Q value was approximately 30 (Q30=99.9% certainty that the correct base was called) (data available upon request).
Finally, upon completion of the analysis, we obtained a total of 783,648 raw reads, including 112,378,736 raw bases, the average for 7,023,671 bases/sample (Table II). This resulted in an average of 89% of bases per sample sequenced, indicative of a quality of >Q20 (where Q20=99% certainty that the correct base was called). Total reads were mapped uniquely to the human reference genome (hg19) retrieved from the NCBI database (build 37) using the Burrows Wheeler Aligner (BWA) Multi-Vision software package. Approximately 96.6% of the bases were matched to hg19 (Table II), and approximately 90% of the bases were matched to coding regions of the target gene(s) using the Coverage Analysis plug-in. It was found that the average depth of coverage of all the exons was at least 28-fold across all 16 samples (Table II). According to previous research, the majority of the raw sequencing data were considered as qualified (28,29).
Table II.
Ion Torrent PGM run statistics and potential disease mutations in patients with inherited cardiomyopathy.
| Subject no. | Total bases | ≥Q20 bases | Reads | Mapped reads | Mean depth | Variants | Synonymous | Insertions and deletions | Non-synonymous |
|---|---|---|---|---|---|---|---|---|---|
| 19 | 14,418,988 | 13,103,929 | 107,304 | 103,943 | 50.06 | 90 | 5 | 1 | 1 |
| A15 | 7,791,816 | 7,066,013 | 58,703 | 56,546 | 29.74 | 36 | 7 | 0 | 3 |
| A36 | 5,324,605 | 4,823,023 | 39,257 | 37,978 | 14.74 | 16 | 5 | 0 | 1 |
| A37 | 5,831,466 | 5,307,572 | 40,876 | 39,480 | 21.54 | 37 | 8 | 0 | 2 |
| A7 | 4,812,050 | 4,088,627 | 43,384 | 41,417 | 21.88 | 98 | 4 | 0 | 4 |
| A13 | 6,089,346 | 5,237,958 | 46,431 | 44,261 | 23.60 | 89 | 5 | 0 | 2 |
| A63 | 7,376,806 | 6,320,407 | 53,364 | 51,338 | 32.39 | 84 | 5 | 1 | 0 |
| A64 | 8,285,404 | 7,132,742 | 61,327 | 58,623 | 37.20 | 116 | 6 | 1 | 1 |
| A94 | 4,111,436 | 3,522,751 | 32,388 | 30,602 | 15.55 | 89 | 7 | 1 | 1 |
| A12 | 5,626,106 | 5,101,170 | 39,302 | 38,165 | 26.36 | 91 | 6 | 0 | 3 |
| 38 | 7,807,093 | 7,039,285 | 48,538 | 47,368 | 33.25 | 128 | 7 | 0 | 1 |
| 51 | 7,785,380 | 7,012,358 | 46,493 | 45,430 | 27.94 | 124 | 5 | 0 | 3 |
| 64 | 7,546,099 | 6,803,657 | 46,726 | 45,536 | 27.51 | 127 | 8 | 0 | 2 |
| A51 | 6,792,213 | 6,086,301 | 40,887 | 39,770 | 31.94 | 93 | 5 | 0 | 1 |
| A86 | 5,261,059 | 4,714,393 | 32,785 | 31,808 | 22.73 | 76 | 6 | 0 | 1 |
| A88 | 7,518,870 | 6,751,598 | 45,884 | 44,622 | 31.58 | 105 | 8 | 0 | 1 |
| Mean | 7,023,671 | 6,256,987 | 48,978 | 47,305 | 28 | 87 | – | – | – |
Identification of pathogenic mutations in order to improve diagnosis
Sequence analysis using the Ion Torrent Variant Caller (v4.0-r76860) plug-in revealed a total of 1,399 nucleotide variations in the 16 patient samples. These variations were positioned in the 5′-UTR, 3′-UTR and in both introns and exons, with an average of 87 variants per patient (Table II). The identified variations were annotated using online Ion Reporter software: 1,368 (97.8%) of them were predicted to be non-coding or synonymous, whereas 31 (2.2%) were non-synonymous and insertion or deletion variants that lead to alterations in 1 or more amino acids (Table II). Non-synonymous and frame shift variation sites were described (data available upon request).
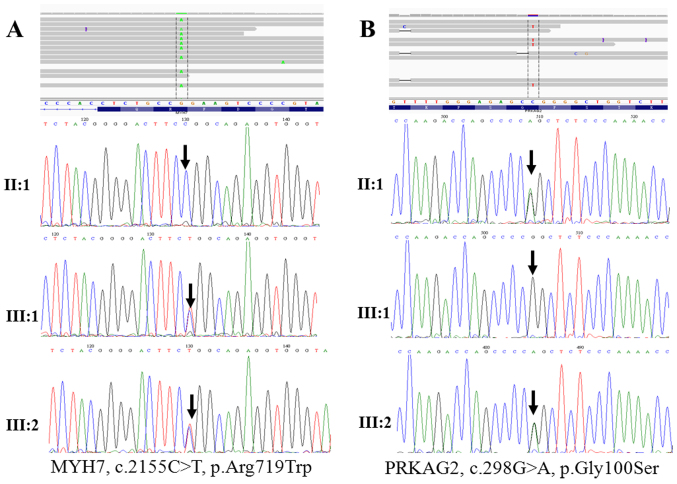
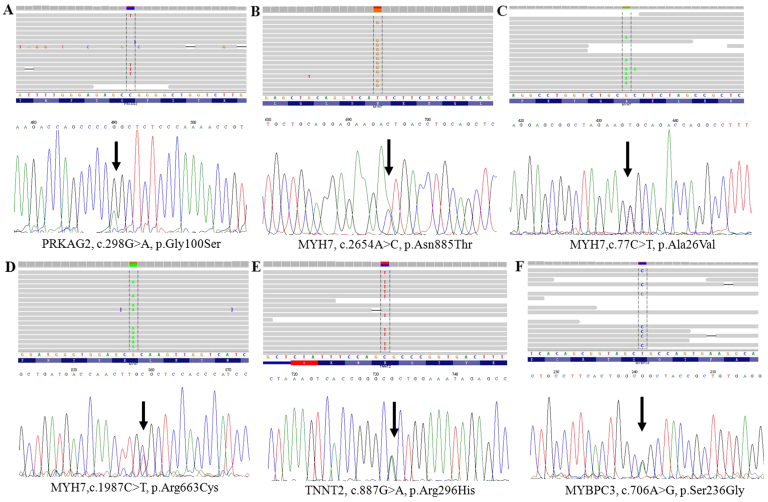
The entire potential non-synonymous and frameshift variation sites were filtered (24). Among these variants, 5 known heterozygous mutations (MYH7, p.Arg719Trp, p.Ala26Val and p.Arg663Cys; PRKAG2, p.Gly100Ser and MYBPC3, p.Ser236Gly) are already known to be associated with inherited cardiomyopathy, and one variant was of uncertain significance (TNNT2, p.Arg296His). Of note, we identified a novel A>C mutation located at nucleotide position c.2654 (according to the cDNA reference sequence, GenBank accession number NM_000257.3) within exon 22 of MYH7, which resulted in the replacement of asparagine at the 885th amino acid with threonine (p.Asn885Thr), as shown in Table IV. These HCM-and DCM-related pathogenic mutations were validated by conventional capillary-based sequencing (Figs. 2A and B and 3), and the PCR primers for the first generation sequencing are listed in Table III. All of the putative mutations were verified by Sanger sequencing (Figs. 1–3), demonstrating that Ion Torrent PGM sequencing achieves a high degree of accuracy with regard to detecting rare mutations.
Table IV.
Pathogenic mutations detected in subjects.
| Subject no. | Gene symbol | Ref Chr. | Transcript | Nucleotide changes | Amino acid changes | SIFT | Poly Phen-2 | Mutation Taster | MAF in 1000G | MAF in ExAC (EA) | Novel/known (Refs.) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A12 | MYH7 | chr14:23895180 | NM_000257.3 | c.2155C>T | p.Arg719Trp | N/A | N/A | N/A | 0.000 | 0.000 | Known (46) |
| A13 | MYH7 | chr14:23895180 | NM_000257.3 | c.2155C>T | p.Arg719Trp | N/A | N/A | N/A | 0.000 | 0.000 | Known (46) |
| A11 | PRKAG2 | chr7:151478406 | NM_016203.3 | c.298G>A | p.Gly100Ser | N/A | N/A | N/A | 0.071 | 0.035 | Known (47) |
| A13 | PRKAG2 | chr7:151478406 | NM_016203.3 | c.298G>A | p.Gly100Ser | N/A | N/A | N/A | 0.071 | 0.035 | Known (47) |
| A15 | PRKAG2 | chr7:151478406 | NM_016203.3 | c.298G>A | p.Gly100Ser | N/A | N/A | N/A | 0.071 | 0.035 | Known (47) |
| A37 | MYH7 | chr14:23902865 | NM_000257.3 | c.77C>T | p.Ala26Val | N/A | N/A | N/A | 0.008 | 0.006 | Known (35) |
| A94 | MYBPC3 | chr11:47370041 | NM_000256.3 | c.706A>G | p.Ser236Gly | N/A | N/A | N/A | 0.031 | 0.030 | Known (52) |
| A36 | MYH7 | chr14:23894003 | NM_000257.3 | c.2654A>C | p.Asn885Thr | NT | PD | DC | 0.000 | 0.000 | Novel |
| 64 | MYH7 | chr14:23896043 | NM_000257.3 | c.1987C>T | p.Arg663Cys | N/A | N/A | N/A | 0.000 | 0.000 | Known (53) |
| A86 | TNNT2 | chr1:201328348 | NM_001276345.1 | c.887G>A | p.Arg296His | NT | PD | DC | 0.000 | 0.000 | VSU |
N/A, not applicable; NT, not tolerated; PD, probably damaging; DC, disease causing; MAF, minor allele frequency; 1000G, 1000 Genomes Project; EA, East Asian; ExAC, Exome Aggregation Consortium; VUS, variant of uncertain significance.
Figure 2.
Pedigree of the family with hypertrophic cardiomyopathy (HCM) by Sanger sequencing analysis. (A) Sanger sequncing showing the results for the MYH7, p.Arg719Trp mutation in family members; the results were positive in III:1, III:2, and negative in II:1. (B) Sanger sequencing showing the results for the PRKAG2, p.Gly100Ser mutation in family members; the results were positive in II:1, III:2 and negative in III:1.
Figure 3.
(A–F) Sanger sequencing showing gene mutations observed in this study of cardiomyopathy patients from China. Mutation sites are indicated by arrows.
Table III.
PCR primers used for the validation of the gene sequence variants.
| Gene symbol | Nucleotide changes | Primers (5′→3′) | Fragment size (bp) |
|---|---|---|---|
| MYH7 | c.2155C>T | Sense: GCTAATCAGTGACAAAGCCAGGATC Antisense: AGGGTGGAAGAGCCAACAGTAGC |
1,434 |
| PRKAG2 | c.298G>A | Sense: CAGTCCTGTGTGGTCAGAACTTGG Antisense: GGACCAGAAGGATTACGCTTTGAT |
907 |
| MYH7 | c.77C>T | Sense: AGCCAGCTTCTGCTCACTCCAG Antisense: GCCACTTGTAAGGGTTGACGGT |
1,013 |
| MYBPC3 | c.706A>G | Sense: CACCATACTTGGCTAATTTTCGT Antisense: GGATGACTGTTGACGGGACATAATGT |
1,542 |
| MYH7 | c.2654A>C | Sense: GCTAATCAGTGACAAAGCCAGGATC Antisense: AGGGTGGAAGAGCCAACAGTAGC |
1,434 |
| MYH6 | c.5410C>A | Sense: TGATGGAGGAGGGAAAGGTGATT Antisense: GGGTGCCAGGTGAACGGTTAA |
2,286 |
| MYH7 | c.1987C>T | Sense: GCAGAATCCATGTCCACCTGT Antisense: TGTCCTAGGAGGTCCTGTTCC |
1,248 |
| TNNT2 | c.887G>A | Sense: AGGGTGATTGTGAGGGTTACAG Antisense: GAGGGTCAAGAGAATGTGTCGT |
2,007 |
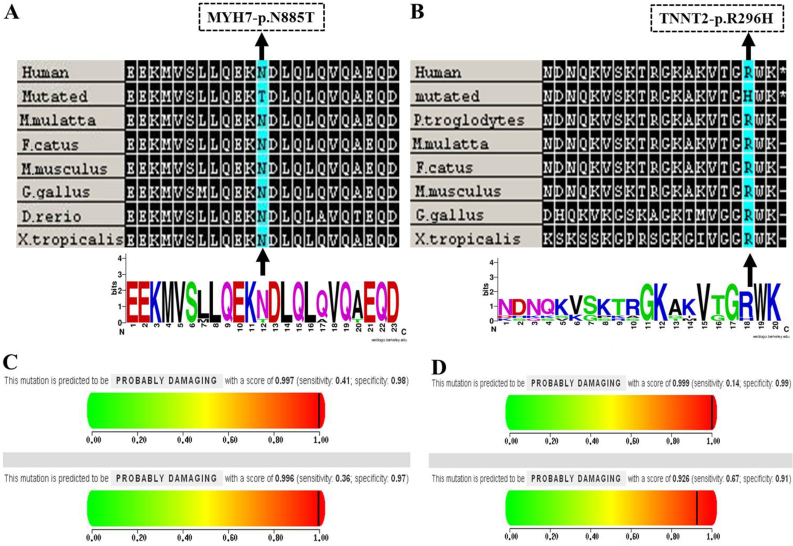
Notably, the novel MYH7, p.Asn885Thr mutation was consistently predicted to be deleterious by the PolyPhen-2 (25), SIFT (26), and MutationTaster (27) algorithms which showed that the mutation is probably damaging with scores of 0.997 and 0.996 on the HumDiv and HumVar models, respectively (Fig. 4C). Secondly, this mutation was not found in reference alleles from the 100 healthy controls, and was also absent from the HGMD database (www.hgmd.cf.ac.uk), NCBI dbSNP137 (http://www.ncbi.nlm.nih.gov/projects/SNP/) and 1000 Genomes project (http://www.1000genomes.org/). Finally, the mutation, p.Asn885Thr, occurs within a 3 helix bundle with the second helix interrupted and it was highly conserved across a number of species (Fig. 4A). As with the mutation of p.Asn885Thr in MYH7, notably, a variant of uncertain significance (TNNT2, p.Arg296His) was predicted to be probably damaging to amino acids using in silico programs, which is relevant for the pathogenicity of HCM (Fig. 4B and D).
Figure 4.
(A and B) Evolutionary conservation analyses of the MYH7, p.Asn885Thr and TNNT2, p.Arg296His mutations were performed, respectively. Clustal W was used to align sequences from Homo sapiens, Macaca mulatta, Felis catus, Mus musculus, Gallus gallus, Danio rerio and Xenopus tropicalis and WebLogo was used to generate sequence logos. The MYH7, p.Asn885Thr and TNNT2, p.Arg296His mutations are marked by a black arrow; (C and D) Results of the PolyPhen-2 analysis predicting the pathogenicity of the mutations of MYH7, p.Asn885Thr and TNNT2, p.Arg296His, respectively.
Discussion
The identification of pathogenic mutations is critical for understanding the molecular pathogenesis of inherited cardio-myopathy, as this in turn will aid the clinical diagnosis of this disease. In the present study, we established a method for the rapid detection of potentially pathogenic mutations in a panel of 12 major genes closely associated with the occurrence of HCM or DCM using the Ion Torrent PGM system. A novel heterozygous mutation (MYH7, p.Asn885Thr) and a variant of uncertain significance (TNNT2, p.Arg296His) were identified.
The Ion Torrent PGM technique is based on the PCR amplification of DNA obtained from subjects which is followed by pooling and barcode labeling of the fragments in each sample and high-throughput sequencing (30,31). The multiplex amplification primers were designed to amplify the principal HCM and DCM disease-related genes in a condensed format that required only 6 microtubes. This process avoids the need for multiple amplifications and reduces the reagent cost per patient. Approximately 90% of the obtained sequences were matched to the coding regions of the target genes using the Coverage Analysis plug-in; however, 10% of the coding region was not covered. This may be due to several factors. Firstly, as the primer pairs were mixed in one reaction, multiplex PCR has a low specific amplification. In addition, the sequence stretches of low complexity and as well as GC-rich regions are difficult to capture (30). Such uncovered regions must be completed using conventional capillary-based sequencing, although there is a risk of missing some mutations using this technique. Despite the drawback regarding lower sequence coverage, the Ion Torrent PGM-based approach using multiplex PCR remains a high-throughput, low-cost method for the detection of mutations.
Herein, we constructed a library to sequence the genomic DNA isolated from patients (19, A15, A36, A37 and A7). Similarly, the second (A13, A63, A64, A94, A12 and 38) and third (51, 64, A51, A86 and A88) pool were also used to construct libraries, respectively, and a total of 8 patients were identified as carriers of pathogenic mutations (Table IV). We detected mutations in 12 disease genes in 7 (7/8) patients with HCM and in 2 (2/8) of the 8 patients with DCM. Of these 7 mutations, 5 are known (MYH7, p.Arg719Trp, p.Ala26Val and p.Arg663Cys; PRKAG2, p.Gly100Ser and MYBPC3, p.Ser236Gly), one is a variant of uncertain significance (TNNT2, p.Arg296His) and one is a novel mutation (MYH7, p.Asn885Thr). Our DCM mutation detection rates are consistent with those of a previous study (32), but the HCM detection rate was higher than that in earlier studies (33,34). As shown in Table IV, 9 subjects [a total of 7 subjects with HCM, one subject (A37) with DCM, and one (A11) subject who was clinically asymptomatic] carried mutations; subject A13 carried a double heterozygous mutation (PRKAG2, p.Gly100Ser plus MYH7, p.Arg719Trp). Mutations were distributed mostly in MYH7 (50%, 5/10) and PRKAG2 (30%, 3/10), followed by MYBPC3 (10%, 1/10) and TNNT2 (10%, 1/10). Mutations were not found in the LMNA, MYH6, MYL2, TPM1, MYL3, SCN5A, ACTC1 and TNNI3 genes. No pathogenic mutations were detected in 87.5% (7/8) of the patients with DCM. This may be due to the fact that there are fewer known DCM-related genes compared with the number of HCM-related genes (3,35); thus, the likelihood of detecting a mutation using our limited gene panel is low. Alternatively, lifestyle and enviornmental factors may play a more important role in the etiology of DCM which is not taken into account when performing genetic screening alone (32).
To the best of our knowledge, this is the first study to describe the novel MYH7 mutation (p.Asn885Thr) in patients with HCM. Sequence conservation analysis revealed that this residue is highly conserved across a number of species (Fig. 4A), thereby impairing its contractile function. Our results suggest that this novel mutation may be functionally deleterious and thus, play an important role in the pathogenesis of HCM, and it expands the mutational spectrum of the MYH7 gene. Moreover, to the best of our knowledge we are the first to report a variant of uncertain significance, p.Arg296His in TNNT2 which is associated with HCM; this illustrated that the status of the variant TNNT2, p. Arg296His may be upgraded to pathogenic. Determining the pathogenicity of a mutation remains a major clinical challenge (36); further independent studies with in vitro or animal models (37,38) are essential in order to validate our results.
His558Arg polymorphism of the SCN5A gene is associated with dilated cardiomyopathy (39), atrial fibrillation (40), idiopathic cardiac conduction disorders (41) and Brugada syndrome (42). His558Arg is a common polymorphism that interacts with the other mutation, eventually modifying or modulating the disease phenotypes (39,42). We identified the common polymorphism of His558Arg in the SCN5A gene in 4 patients with DCM (subject nos. 19, A51, A7 and A37) and validated these findings using Sanger sequencing (data available upon request). Of note, the mutation MYH7, p.Ala26Val plus the common polymorphism of SCN5A, His558Arg were detected in one patient (A37) with DCM. Potentially, a polymorphism of His558Arg modifies or modulates the variant MYH7, p.Ala26Val which causes DCM, and may affect the age of onset, the severity and rate of progression of DCM.
Some patients with inherited cardiomyopathy may have more than one disease-causing mutation, which can occur in either the same gene (compound heterozygotes) or in different genes (double heterozygotes) (43,44). As a consequence of these complex mutations, the individual usually has a malignant phenotype of inherited cardiomyopathy (45). Notably, we have identified, for the first time to the best of our knowledge, a double heterozygous (MYH7, p.Arg719Trp plus PRKAG2, p.Gly100Ser) mutation in a proband (III:2) with familial HCM. He exhibited a malignant phenotype of HCM that manifested with increased interventricular septum thickness (Table I and Fig. 2A and B). His elder sister (III:1) was also diagnosed with HCM and carried MYH7, p.Arg719Trp, but not PRKAG2, p.Gly100Ser. Family screening revealed that the proband's 45-year-old mother (II:1), who was asymptomatic, was also affected, and carried the PRKAG2, p.Gly100Ser mutation (Fig. 2A and B). These results suggest that the pathogenic MYH7, Arg719Trp mutation probably originated in the proband's father and grandfather. The MYH7, p.Arg719Trp (46) and PRKAG2, p.Gly100Ser (47) mutations have previously been shown to be associated with malignant familial HCM and sporadic HCM, respectively. We suggest that PRKAG2, p.Gly100Ser exacerbates the clinical severity of HCM in individuals with the MYH7 p.Arg719Trp mutation and thus, have a 'double dose' gene mutation effect (48,49). This is associated with a poor prognosis, and explains why the proband exhibited an early onset malignant phenotype of HCM (50). As a mutation carrier, the proband's mother (A11) is a family member at risk who was clinically asymptomatic (51); long-term follow-up is therefore essential in this subject. However, her children are at high risk of developing HCM, and thus genetic testing may be particularly helpful in this group. Indeed, we have recommended genetic testing for all first-degree relatives of the proband, since it may facilitate clinical decisions that impact diagnosis and treatment strategies.
In conclusion, Ion Torrent PGM targeted sequencing is a rapid and cost-effective method for the clinical genetic screening of patients with inherited cardiomyopathy. Correct recognition of the responsible gene mutations is essential for providing optimal presymptomatic intervention and genetic counseling for probands and their family members. The gene panel testing of 12 major disease-related genes in patients with inherited cardiomyopathy reported in this study has the potential to assist in revealing the etiology of genetically heterogeneous HCM, and it is a highly reliable and effective method for the screening of pathogenic mutations in candidate genes. However, the coverage of these targeted regions must be further improved. Furthermore, we detected a novel (MYH7, p.Asn885Thr) mutation and a variant of uncertain significance (TNNT2, p.Arg296His) that we suggest be upgraded to pathogenic status; these add new data to the spectrum of mutations in HCM. Moreover, a double heterozygous (PRKAG2, p.Gly100Ser plus MYH7, p.Arg719Trp) mutation was found in a proband with familial HCM. This data has the potential to allow us to better facilitate risk stratification and guide familial management of the disease. Finally, we note that genes beyond this initial 12-gene panel should be included in future tests as their relevance to DCM becomes clear.
Acknowledgments
We would like to thank all the participants of this study from the inherited cardiomyopathy registry, and we would like to thank the staff of the First Hospital of Yunnan Province for their support. The present study was supported by the Major Program of Applied Basic Research of Yunnan Province, China (no. 2013FC007).
References
- 1.Callis TE, Jensen BC, Weck KE, Willis MS. Evolving molecular diagnostics for familial cardiomyopathies: at the heart of it all. Expert Rev Mol Diagn. 2010;10:329–351. doi: 10.1586/erm.10.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes SE, McKenna WJ. New insights into the pathology of inherited cardiomyopathy. Heart. 2005;91:257–264. doi: 10.1136/hrt.2004.040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 4.Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, et al. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Spaendonck-Zwarts KY, Posafalvi A, van den Berg MP, Hilfiker-Kleiner D, Bollen IA, Sliwa K, Alders M, Almomani R, van Langen IM, van der Meer P, et al. Titin gene mutations are common in families with both peripartum cardiomyopathy and dilated cardiomyopathy. Eur Heart J. 2014;35:2165–2173. doi: 10.1093/eurheartj/ehu050. [DOI] [PubMed] [Google Scholar]
- 6.Zhou YM, Dai XY, Qiu XB, Yuan F, Li RG, Xu YJ, Qu XK, Huang RT, Xue S, Yang YQ. HAND1 loss-of-function mutation associated with familial dilated cardiomyopathy. Clin Chem Lab Med. 2015 Nov 18; doi: 10.1515/cclm-2015-0766. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 7.Zhao CM, Bing-Sun, Song HM, Wang J, Xu WJ, Jiang JF, Qiu XB, Yuan F, Xu JH, Yang YQ. TBX20 loss-of-function mutation associated with familial dilated cardiomyopathy. Clin Chem Lab Med. 2016;54:325–332. doi: 10.1515/cclm-2015-0328. [DOI] [PubMed] [Google Scholar]
- 8.Qu XK, Yuan F, Li RG, Xu L, Jing WF, Liu H, Xu YJ, Zhang M, Liu X, Fang WY, et al. Prevalence and spectrum of LRRC10 mutations associated with idiopathic dilated cardiomyopathy. Mol Med Rep. 2015;12:3718–3724. doi: 10.3892/mmr.2015.3843. [DOI] [PubMed] [Google Scholar]
- 9.Zhang XL, Qiu XB, Yuan F, Wang J, Zhao CM, Li RG, Xu L, Xu YJ, Shi HY, Hou XM, et al. TBX5 loss-of-function mutation contributes to familial dilated cardiomyopathy. Biochem Biophys Res Commun. 2015;459:166–171. doi: 10.1016/j.bbrc.2015.02.094. [DOI] [PubMed] [Google Scholar]
- 10.Zhou W, Zhao L, Jiang JQ, Jiang WF, Yang YQ, Qiu XB. A novel TBX5 loss-of-function mutation associated with sporadic dilated cardiomyopathy. Int J Mol Med. 2015;36:282–288. doi: 10.3892/ijmm.2015.2206. [DOI] [PubMed] [Google Scholar]
- 11.Zhang XL, Dai N, Tang K, Chen YQ, Chen W, Wang J, Zhao CM, Yuan F, Qiu XB, Qu XK, et al. GATA5 loss-of-function mutation in familial dilated cardiomyopathy. Int J Mol Med. 2015;35:763–770. doi: 10.3892/ijmm.2014.2050. [DOI] [PubMed] [Google Scholar]
- 12.Xu L, Zhao L, Yuan F, Jiang WF, Liu H, Li RG, Xu YJ, Zhang M, Fang WY, Qu XK, et al. GATA6 loss-of-function mutations contribute to familial dilated cardiomyopathy. Int J Mol Med. 2014;34:1315–1322. doi: 10.3892/ijmm.2014.1896. [DOI] [PubMed] [Google Scholar]
- 13.Zhao L, Xu JH, Xu WJ, Yu H, Wang Q, Zheng HZ, Jiang WF, Jiang JF, Yang YQ. A novel GATA4 loss-of-function mutation responsible for familial dilated cardiomyopathy. Int J Mol Med. 2014;33:654–660. doi: 10.3892/ijmm.2013.1600. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Liu WD, Yang ZL, Yuan F, Xu L, Li RG, Yang YQ. Prevalence and spectrum of GATA4 mutations associated with sporadic dilated cardiomyopathy. Gene. 2014;548:174–181. doi: 10.1016/j.gene.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Reinstein E, Orvin K, Tayeb-Fligelman E, Stiebel-Kalish H, Tzur S, Pimienta AL, Bazak L, Bengal T, Cohen L, Gaton DD, et al. Mutations in TAX1BP3 cause dilated cardiomyopathy with septo-optic dysplasia. Hum Mutat. 2015;36:439–442. doi: 10.1002/humu.22759. [DOI] [PubMed] [Google Scholar]
- 16.Dhandapany PS, Razzaque MA, Muthusami U, Kunnoth S, Edwards JJ, Mulero-Navarro S, Riess I, Pardo S, Sheng J, Rani DS, et al. RAF1 mutations in childhood-onset dilated cardiomyopathy. Nat Genet. 2014;46:635–639. doi: 10.1038/ng.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou Y, Song L, Wang Z, Ma A, Liu T, Gu H, Lu S, Wu P, Zhang Y, Shen L, et al. Prevalence of idiopathic hypertrophic cardiomyopathy in China: A population-based echocardiographic analysis of 8080 adults. Am J Med. 2004;116:14–18. doi: 10.1016/j.amjmed.2003.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–789. doi: 10.1161/01.CIR.92.4.785. [DOI] [PubMed] [Google Scholar]
- 19.Yuan F, Qiu XB, Li RG, Qu XK, Wang J, Xu YJ, Liu X, Fang WY, Yang YQ, Liao DN. A novel NKX2-5 loss-of-function mutation predisposes to familial dilated cardiomyopathy and arrhythmias. Int J Mol Med. 2015;35:478–486. doi: 10.3892/ijmm.2014.2029. [DOI] [PubMed] [Google Scholar]
- 20.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 21.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, Naidu SS, Nishimura RA, Ommen SR, Rakowski H, American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines 2011 ACCF/AHA Guideline for the Diagnosis and Treatment of Hypertrophic Cardiomyopathy a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Developed in collaboration with the American Association for Thoracic Surgery, American Society of Echocardiography, American Society of Nuclear Cardiology, Heart Failure Society of America, Heart Rhythm Society, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J Am Coll Cardiol. 2011;58:e212–260. doi: 10.1016/j.jacc.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Mestroni L, Maisch B, McKenna WJ, Schwartz K, Charron P, Rocco C, Tesson F, Richter A, Wilke A, Komajda M. Collaborative Research Group of the European Human and Capital Mobility Project on Familial Dilated Cardiomyopathy: Guidelines for the study of familial dilated cardiomyopathies. Eur Heart J. 1999;20:93–102. doi: 10.1053/euhj.1998.1145. [DOI] [PubMed] [Google Scholar]
- 23.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE, Collaborative Research Group of the European Human and Capital Mobility Project on Familial Dilated Cardiomyopathy ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 25.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 28.Sikkema-Raddatz B, Johansson LF, de Boer EN, Almomani R, Boven LG, van den Berg MP, van Spaendonck-Zwarts KY, van Tintelen JP, Sijmons RH, Jongbloed JD, Sinke RJ. Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Hum Mutat. 2013;34:1035–1042. doi: 10.1002/humu.22332. [DOI] [PubMed] [Google Scholar]
- 29.Tarabeux J, Zeitouni B, Moncoutier V, Tenreiro H, Abidallah K, Lair S, Legoix-Né P, Leroy Q, Rouleau E, Golmard L, et al. Streamlined ion torrent PGM-based diagnostics: BRCA1 and BRCA2 genes as a model. Eur J Hum Genet. 2014;22:535–541. doi: 10.1038/ejhg.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa JL, Sousa S, Justino A, Kay T, Fernandes S, Cirnes L, Schmitt F, Machado JC. Nonoptical massive parallel DNA sequencing of BRCA1 and BRCA2 genes in a diagnostic setting. Hum Mutat. 2013;34:629–635. doi: 10.1002/humu.22272. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Buckton AJ, Wilkinson SL, John S, Walsh R, Novotny T, Valaskova I, Gupta M, Game L, Barton PJ, et al. Towards clinical molecular diagnosis of inherited cardiac conditions: a comparison of bench-top genome DNA sequencers. PLoS One. 2013;8:e67744. doi: 10.1371/journal.pone.0067744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakdawala NK, Funke BH, Baxter S, Cirino AL, Roberts AE, Judge DP, Johnson N, Mendelsohn NJ, Morel C, Care M, et al. Genetic testing for dilated cardiomyopathy in clinical practice. J Card Fail. 2012;18:296–303. doi: 10.1016/j.cardfail.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brito D, Miltenberger-Miltenyi G, Vale Pereira S, Silva D, Diogo AN, Madeira H. Sarcomeric hypertrophic cardio-myopathy: genetic profile in a Portuguese population. Rev Port Cardiol. 2012;31:577–587. doi: 10.1016/j.repc.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 34.Zou Y, Wang J, Liu X, Wang Y, Chen Y, Sun K, Gao S, Zhang C, Wang Z, Zhang Y, et al. Multiple gene mutations, not the type of mutation, are the modifier of left ventricle hypertrophy in patients with hypertrophic cardiomyopathy. Mol Biol Rep. 2013;40:3969–3976. doi: 10.1007/s11033-012-2474-2. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, Feng Y, Zhang YM, Ding XX, Song YZ, Zhang AM, Liu L, Zhang H, Ding JH, Xia XS. Targeted next-generation sequencing of candidate genes reveals novel mutations in patients with dilated cardiomyopathy. Int J Mol Med. 2015;36:1479–1486. doi: 10.3892/ijmm.2015.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das KJ, Ingles J, Bagnall RD, Semsarian C. Determining pathogenicity of genetic variants in hypertrophic cardiomyopathy: importance of periodic reassessment. Genet Med. 2014;16:286–293. doi: 10.1038/gim.2013.138. [DOI] [PubMed] [Google Scholar]
- 37.Hassel D, Dahme T, Erdmann J, Meder B, Huge A, Stoll M, Just S, Hess A, Ehlermann P, Weichenhan D, et al. Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat Med. 2009;15:1281–1288. doi: 10.1038/nm.2037. [DOI] [PubMed] [Google Scholar]
- 38.Geisterfer-Lowrance AA, Christe M, Conner DA, Ingwall JS, Schoen FJ, Seidman CE, Seidman JG. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272:731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- 39.Cheng J, Morales A, Siegfried JD, Li D, Norton N, Song J, Gonzalez-Quintana J, Makielski JC, Hershberger RE. SCN5A rare variants in familial dilated cardiomyopathy decrease peak sodium current depending on the common polymorphism H558R and common splice variant Q1077del. Clin Transl Sci. 2010;3:287–294. doi: 10.1111/j.1752-8062.2010.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L, Zhang W, Fang C, Jiang S, Shu C, Cheng H, Li F, Li H. Polymorphism H558R in the human cardiac sodium channel SCN5A gene is associated with atrial fibrillation. J Int Med Res. 2011;39:1908–1916. doi: 10.1177/147323001103900535. [DOI] [PubMed] [Google Scholar]
- 41.Nikulina SY, Chernova AA, Shulman VA, Maksimov VN, Gavrilyuk OA, Tretyakova SS, Marilovceva OV. An investigation of the association of the H558R polymorphism of the SCN5A gene with idiopathic cardiac conduction disorders. Genet Test Mol Biomarkers. 2015;19:288–294. doi: 10.1089/gtmb.2015.0012. [DOI] [PubMed] [Google Scholar]
- 42.Marangoni S, Di Resta C, Rocchetti M, Barile L, Rizzetto R, Summa A, Severi S, Sommariva E, Pappone C, Ferrari M, et al. A Brugada syndrome mutation (p.S216L) and its modulation by p.H558R polymorphism: standard and dynamic characterization. Cardiovasc Res. 2011;91:606–616. doi: 10.1093/cvr/cvr142. [DOI] [PubMed] [Google Scholar]
- 43.Roncarati R, Viviani Anselmi C, Krawitz P, Lattanzi G, von Kodolitsch Y, Perrot A, di Pasquale E, Papa L, Portararo P, Columbaro M, et al. Doubly heterozygous LMNA and TTN mutations revealed by exome sequencing in a severe form of dilated cardiomyopathy. Eur J Hum Genet. 2013;21:1105–1111. doi: 10.1038/ejhg.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lekanne Deprez RH, Muurling-Vlietman JJ, Hruda J, Baars MJ, Wijnaendts LC, Stolte-Dijkstra I, Alders M, van Hagen JM. Two cases of severe neonatal hypertrophic cardiomyopathy caused by compound heterozygous mutations in the MYBPC3 gene. J Med Genet. 2006;43:829–832. doi: 10.1136/jmg.2005.040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho CY. Genetics and clinical destiny: improving care in hyper-trophic cardiomyopathy. Circulation. 2010;122:2430–2440. doi: 10.1161/CIRCULATIONAHA.110.978924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anan R, Greve G, Thierfelder L, Watkins H, McKenna WJ, Solomon S, Vecchio C, Shono H, Nakao S, Tanaka H. Prognostic implications of novel beta cardiac myosin heavy chain gene mutations that cause familial hypertrophic cardiomyopathy. J Clin Invest. 1994;93:280–285. doi: 10.1172/JCI116957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang BL, Xu RL, Zhang J, Zhao XX, Wu H, Ma LP, Hu JQ, Zhang JL, Ye Z, Zheng X, Qin YW. Identification and functional analysis of a novel PRKAG2 mutation responsible for Chinese PRKAG2 cardiac syndrome reveal an important role of non-CBS domains in regulating the AMPK pathway. J Cardiol. 2013;62:241–248. doi: 10.1016/j.jjcc.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 48.Van Driest SL, Vasile VC, Ommen SR, Will ML, Tajik AJ, Gersh BJ, Ackerman MJ. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:1903–1910. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Y, Feng Y, Zhang YM, Ding XX, Song YZ, Zhang AM, Liu L, Zhang H, Ding JH, Xia XS. Targeted next-generation sequencing reveals hot spots and doubly heterozygous mutations in Chinese patients with familial cardiomyopathy. BioMed Res Int. 2015;2015:561819. doi: 10.1155/2015/561819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brisca G, Fiorillo C, Nesti C, Trucco F, Derchi M, Andaloro A, Assereto S, Morcaldi G, Pedemonte M, Minetti C, et al. Early onset cardiomyopathy associated with the mitochondrial tRNALeu((UUR)) 3271T>C MELAS mutation. Biochem Biophys Res Commun. 2015;458:601–604. doi: 10.1016/j.bbrc.2015.01.157. [DOI] [PubMed] [Google Scholar]
- 51.Wang AL, Kong DH, Chen DX, Wan J, Yu YX. Mutation of V896M in cardiac myosin binding protein-c gene in two Chinese families with hypertrophic cardiomyopathy. Mol Med Rep. 2010;3:759–763. doi: 10.3892/mmr.2010.333. [DOI] [PubMed] [Google Scholar]
- 52.Millat G, Chanavat V, Rousson R. Evaluation of a new NGS method based on a custom AmpliSeq library and Ion Torrent PGM sequencing for the fast detection of genetic variations in cardiomyopathies. Clin Chim Acta. 2014;433:266–271. doi: 10.1016/j.cca.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 53.Glotov AS, Kazakov SV, Zhukova EA, Alexandrov AV, Glotov OS, Pakin VS, Danilova MM, Poliakova IV, Niyazova SS, Chakova NN, et al. Targeted next-generation sequencing (NGS) of nine candidate genes with custom AmpliSeq in patients and a cardiomyopathy risk group. Clin Chim Acta. 2015;446:132–140. doi: 10.1016/j.cca.2015.04.014. [DOI] [PubMed] [Google Scholar]