Highlight
A clear interconnection between photorespiration and primary nitrogen assimilation is established in Lotus japonicus, and key transcription factors connected to both routes are identified using transcriptomics and gene co-expression networks.
Key words: Co-expression networks, Lotus, japonicus, nitrogen metabolism, nitrogen nutrition, photorespiration, transcriptomics.
Abstract
Nitrogen is one of the most important nutrients for plants and, in natural soils, its availability is often a major limiting factor for plant growth. Here we examine the effect of different forms of nitrogen nutrition and of photorespiration on gene expression in the model legume Lotus japonicus with the aim of identifying regulatory candidate genes co-ordinating primary nitrogen assimilation and photorespiration. The transcriptomic changes produced by the use of different nitrogen sources in leaves of L. japonicus plants combined with the transcriptomic changes produced in the same tissue by different photorespiratory conditions were examined. The results obtained provide novel information on the possible role of plastidic glutamine synthetase in the response to different nitrogen sources and in the C/N balance of L. japonicus plants. The use of gene co-expression networks establishes a clear relationship between photorespiration and primary nitrogen assimilation and identifies possible transcription factors connected to the genes of both routes.
Introduction
Nitrogen is one of the most important nutrients for plants, and, in natural soils, its availability is often a major limiting factor for plant growth. The use of nitrogen by plants involves several steps, including uptake, assimilation, translocation, and different forms of recycling and remobilization processes, all of them of crucial importance in terms of nitrogen utilization efficiency (Hirel et al., 2007; Masclaux-Daubresse et al., 2010). Primary nitrogen assimilation by plants involves the use of different forms of inorganic nitrogen (NO3 − and/or NH4 +), depending on nitrogen availability, plant species, and adaptations. Alternatively, symbiosis with bacteria enables some plant species, most notably legumes, to use atmospheric N2 that is reduced to NH4 + in the nodules by the action of bacterial nitrogenase. In addition, efficient secondary NH4 + assimilation must also exist in plants in order to reassimilate the NH4 + ions that can be produced endogenously in the plants from processes such as photorespiration, phenylpropanoid biosynthesis, or amino acid catabolism (Betti et al., 2014).
The NH4 + generated by the photorespiratory nitrogen cycle can be up to 20 times that resulting from the reduction of NO3 − (Canvin, 1990; Guo et al., 2007). The large amounts of NH4 + generated by the photorespiratory cycle are produced in the mitochondria as a result of the conversion of two molecules of glycine into one molecule of serine by the combined action of glycine decarboxylase (GDC; EC 2.1.2.10) and serine hydroxymethyl transferase (SHMT; EC 2.1.2.1). NH4 + is transported to the chloroplast where it is reassimilated by the plastidic glutamine synthetase and ferredoxin-glutamate synthase cycle (GS2/Fd-GOGAT). Several important pathways such as nitrogen assimilation, respiration, one-carbon metabolism, purine biosynthesis (Bauwe et al., 2010), and redox signaling (Foyer et al., 2009) interact in different ways with photorespiration. Moreover, it has been described that the conditions that inhibited photorespiration (elevated CO2) also strongly inhibited NO3 − assimilation in C3 plants (Rachmilevitch et al., 2004; Bloom, 2015). Photorespiration serves as a mechanism for plants to use NO3 − as a nitrogen source without diverting energy from CO2 fixation. The linkage between photorespiration and NO3 − assimilation provides higher plants with a relatively abundant nitrogen source that other organisms cannot afford to use, but that C3 plants can use with little additional energy cost (Bloom, 2015).
Furthermore, the form of nitrogen available to plants can affect their time and rate of seed germination, leaf expansion and function, dry matter partitioning between shoot and root, and root architecture (Andrews et al., 2013). Despite the fact that more energy is needed for the assimilation of NO3 −, most plants prefer NO3 − over NH4 +. With the exception of NH4 +-tolerant species, NH4 + as a sole nitrogen source, in addition to internal production of NH4 + by processes such as photorespiration (Keys et al., 1978), may prove toxic to the plant. Indeed, in comparison with NO3 − uptake, much less is known about the regulatory mechanisms controlling either NH4 + or N2 acquisition (Ruffel et al., 2008). Notably, the toxic effect of external NH4 + can be relieved by co-provision of NO3 −, so-called mixed nutrition (NH4NO3) (Forde and Clarkson, 1999). Also, a fascinating and still poorly understood aspect of nitrogen nutrition is that in most cases the growth of a plant on NH4NO3 can surpass the maximal growth compared with either NO3 − or NH4 + alone. This relief of NH4 + toxicity by NO3 − may be related to a synergism between the signaling routes of NH4 + and NO3 − (Britto and Kronzuker, 2002).
Lotus japonicus is a temperate legume that can grow using the atmospheric N2 fixed in the nodules or by using external sources of nitrogen, such as NO3 − and/or NH4 +. The utilization of NO3 − requires its reduction to NH4 + from the consecutive action of nitrate reductase (NR; EC 1.7.1.1/2) and nitrite reductase (NiR; EC 1.7.7.1) enzymes. Then, the NH4 + synthesized as a result of both primary and secondary assimilation is assimilated into glutamine and then into glutamate by the enzymes glutamine synthetase (GS; EC 6.3.1.2) and glutamate synthase (GOGAT; EC 1.4.7.1 or EC 1.4.1.14) (Márquez et al., 2005). Different sets of GS and GOGAT isoforms exist in plants which are specifically associated with different processes such as primary NO3 − or NH4 + assimilation, N2 fixation, and/or secondary NH4 + reassimilation. In particular, plastidic GS2 and Fd-GOGAT have been reported to be the crucial enzymes for the reassimilation of photorespiratory NH4 + thanks to the isolation of photorespiratory mutants deficient in GS2 (Wallsgrove et al., 1987) or Fd-GOGAT (Somerville and Ogren, 1980).
The first GS photorespiratory mutants isolated from legume plants were identified several years ago in our laboratory from the model legume L. japonicus (Orea et al., 2002; Márquez et al., 2005). These mutants were shown to be specifically deficient in GS2 and have been substantially characterized at the molecular and physiological levels (Orea et al., 2002; Márquez et al., 2005; Betti et al., 2006, 2012a , 2014; García-Calderón et al., 2012), including recent transcriptomics and metabolomics studies (Díaz et al., 2010; Betti et al., 2012a; Pérez-Delgado et al., 2013). Under a CO2-enriched atmosphere, where photorespiration is suppressed, the mutants did not show any visible phenotype (Orea et al., 2002).
The objective of this study is to determine the effect of different forms of nitrogen nutrition and the effect of photorespiration on gene expression in order to analyze the possible interconnections among these processes in L. japonicus plants. For this purpose, a comparative transcriptomic study was carried out using wild-type (WT) plants and plants with a deficiency in GS2 (Ljgln2-2) grown with different nitrogen regimes (NO3 −, NH4 +, NH4NO3, or under conditions of biological nitrogen fixation) and in different photorespiratory conditions. Furthermore, a set of gene co-expression networks was built to study the connection between photorespiratory genes and genes of primary nitrogen assimilation, with the ultimate goal of identifying regulatory candidate genes co-ordinating these processes.
We first demonstrate that several important transcriptomic changes occurred in leaves of L. japonicus when plants were cultivated with different nitrogen sources, including genes involved in nitrogen, carbon, and secondary metabolism. To study the possible interconnections between primary nitrogen assimilation and photorespiration, WT and Ljgln2-2 mutant plants were examined under different forms of nitrogen nutrition and different photorespiratory conditions. The data obtained provide novel information on the possible role of plastidic GS2 in the response to different nitrogen sources and on the C/N balance of L. japonicus plants. Finally, co-expression networks were built using the nitrogen- and photorespiration-responsive genes previously identified. A clear interconnection between nitrogen assimilation and photorespiration was established in L. japonicus, and several key transcription factors that could be involved in the co-ordinate regulation of these metabolic routes were identified.
Materials and methods
Growth conditions and harvesting of plant material
Lotus japonicus (Regel) Larsen cv. Gifu (B-129-S9) was initially obtained from Professor Jens Stougaard (Aarhus University, Aarhus, Denmark) and then self-propagated at the University of Seville. The Ljgln2-2 mutant, which lacks GS2 protein and activity (Betti et al., 2006), was isolated from the photorespiratory mutant screening carried out using ethyl methanesulfonate as described previously (Orea et al., 2002). The mutant progeny of two consecutive backcrosses into the WT background were used. WT and mutant seeds were scarified and surface-sterilized, then germinated in 1% agar in Petri dishes and transferred to pots using vermiculite as solid support. Five seedlings were planted in each pot and grown until plants had seven trefoils in a growth chamber under 16 h:8h day:night, 20:18 °C, with a photosynthetic photon flux density of 250 μmol m−2 s−1 and a constant humidity of 70%. When required, CO2 was automatically injected to a final concentration of 0.7% (v/v) (high CO2 atmosphere).
Nodulated plants were inoculated with Mesorhizobium loti and watered with nitrogen-free ‘Hornum’ medium supplemented with 3mM KCl (Handberg and Stougaard, 1992). Mesorhizobium loti TONO JA76 (Kawaguchi et al., 2002) was grown in YM liquid medium (Vincent, 1970) at 28 °C to an optical density at 600 nm=1, and then collected by centrifugation for 30min at 2408 g and resuspended in 0.75% (w/v) NaCl. Once sown in the pots, the plants were inoculated by the addition of 2ml of this bacterial suspension.
Plants under different forms of nitrogen nutrition were watered with ‘Hornum’ nutrient solution containing 10mM KNO3 (NO3 − plants), 10mM NH4Cl supplemented with 3mM KCl (NH4 + plants), or 5mM NH4NO3 supplemented with 3mM KNO3 (NH4NO3 plants). The nutrient solutions were renewed every 3 d. These nutritional conditions were used taking into consideration the recommended growth conditions for L. japonicus (Handberg and Stougaard, 1992; Orea et al,. 2005). Moreover, previous works have shown that the concentrations of the different nitrogen sources used here are not toxic for L. japonicus (Orea et al., 2005). After all the plants had reached the same size (an average of seven trefoils), leaf tissue was harvested. All the leaf samples used in this work were harvested 4h after the beginning of the light period. The stage of seven trefoils was reached after ~38, 35, 36, or 42 d in the case of plants growing under symbiotic conditions or with NH4NO3, NO3 −, or NH4 + alone, respectively. Every harvest involved at least three independent biological replicates for each genotype and treatment. A biological replicate consisted of tissue pooled from five plants grown in the same pot.
Plants were grown continuously either under normal CO2 atmosphere (0.04% v/v), to permit normal rates of photorespiration, or under high CO2 (0.7% v/v) atmosphere in order to suppress photorespiration and to permit the normal growth of the Ljgln2-2 mutant.
RNA extraction and qRT–PCR
Leaf material was flash-frozen in liquid nitrogen, homogenized with a mortar and pestle, and kept at −80 °C until use. Three independent biological replicates were used for the quantitative real-time reverse transcription–PCR (qRT–PCR) analysis. Total RNA was isolated using the hot borate method (Sánchez et al., 2008). The integrity and concentration of the RNA preparations were checked using an Experion bioanalyzer (Bio-Rad) with RNA StdSens chips and a Nano-Drop ND-1000 (Nano-Drop Technologies), respectively.
For qRT–PCR analysis, total RNA was treated with the TURBO DNA-free DNase (Ambion). Reverse transcription was carried out using SuperScript III reverse transcriptase (Invitrogen), oligo(dT), and RNAsin RNase inhibitor (Ambion). DNA contamination and RNA integrity were checked by carrying out qRT–PCRs with oligonucleotides that amplified an intron in the LjHAR1 gene and the 3' and 5' ends of L. japonicus glyceraldehyde-3-phosphate dehydrogenase, respectively. qRT–PCRs were carried out in 10 μl in a Lightcycler 480 thermal cycler (Roche) using a SensiFAST SYBR No-ROX Kit (Bioline). Expression data were normalized using the geometric mean of four housekeeping genes: LjGPI-anchored protein (probeset chr3.CM0047.42), LjPp2A (probeset chr2.CM0310.22), LjUBC10 (probeset chr1.TM0487.4), and LjUBQ4 (probeset chr5.CM0956.27) that were selected amongst the most stably expressed genes in plants (Czechowski et al., 2004). A list of all the oligonucleotides used is provided in Supplementary Table S1 at JXB online.
DNA chip hybridization and data analysis
Two independent biological replicates were used for the transcriptomic analysis of plants grown in different nitrogen sources. Microarray slides were designed and produced using Agilent eArray (Agilent Technologies; http://www.agilent.com) specifically developed for L. japonicus, and hybridized with total leaf RNA according to the manufacturer’s instructions.
The microarrays were scanned, the raw image files were processed, and data analysis was performed by Agilent Corporation. MIAME compliant data are deposited at Array Express (http://www.ebi.ac.uk/arrayexpress) as E-MTAB-4177. Data were normalized using the function rma of the limma package (Linear Models for Microarray Data, v. 2.10.5) (Smyth, 2004) of bioconductor.
The differentially expressed genes of plants grown under different forms of nitrogen nutrition were determined with a one-way ANOVA using a false discovery rate (FDR) threshold of P<0.01 based on the 10 000 permutation test with the MeV module of the TM4 package (http://www.TM4.org) (Saeed et al., 2003) followed by a post-hoc Tukey test with R (http://www.R-project.org/). Hierarchical clustering of transcriptomic data was carried out with Expander software version 7.1 (Shamir et al., 2005; Ulitsky et al., 2010) using average linkage.
Differentially expressed genes between WT and Ljgln2-2 mutant plants and between photorespiratory and non-photorespiratory conditions were determined using Rank products (Breitling et al., 2004) passing an FDR threshold of P<0.1. The differentially expressed genes were visualized using the MapMan program (Usadel et al., 2005) and analyzed according to the corresponding metabolic pathways or functional categories using Pathexpress (Goffard and Weiller, 2007). The default threshold of P<0.1 and FDR rate correction was used for Pathexpress.
Gene sequences were obtained at the Kazusa database (http://www.kazusa.or.jp/lotus/).
Co-expression network construction
Microarray data collection and pre-processing
Microarray data used in this work were obtained from the experiments published by Sánchez et al. (2008, 2011), Høgslund et al. (2009), Díaz et al. (2010), Betti et al. (2012b), and Pérez-Delgado et al. (2013). These experiments have a total of 240 hybridizations. CEL files of these experiments are available in the public microarrays database EBI (https://www.ebi.ac.uk/arrayexpress/). Code numbers of experiments are: E-MEXP-1204, E-TABM-715, E-MEXP-2344, E-MEXP-2690, E-MEXP-1726, E-MEXP-3710, and E-MEXP-3603. Background correction and normalization of the raw data sets were performed using Robust MultiChip Analysis (RMA) implemented in the ‘affy’ R package (Gautier et al., 2004).
Identification of differentially expressed genes
The non-parametric Rank product method (Breitling et al., 2004) was used to identify differentially expressed genes between treatment and control conditions using an FDR threshold of P<0.05.
Network construction
The network construction was developed according to Canales et al. (2014). A weighted gene co-expression network was constructed using the WGCNA R package version 1.27.1 (Langfelder and Horvath, 2008) with differentially expressed genes. First, the Pearson correlation matrix was weighted by raising it to a power (β). To choose the appropriate power, the network topology for various soft-thresholding powers was evaluated using pickSoftThreshold function and β=6 was chosen because this ensured an approximate scale-free topology of the resulting network. Next, the pairwise measure of gene co-expression of the resulting weighted network was transformed into a topological overlap (TO) similarity measure, which is a robust measure of pairwise interconnectedness (Yip and Horvath, 2007). A TO similarity measure between two genes (ij) is defined as: T0ij=Σu aiuauj+aij min (ki,kj)+1–aij where ki=u aiu was the node connectivity and a is the network adjacency. Finally, the co-expression network was visualized using Cytoscape v 3.0 and analyzed using the NetworkAnalyser plugin (Doncheva et al., 2012). In order to simplify the display of the network and to focus on relevant relationships, only edges in this network of the corresponding TO similarity measure above a threshold of 0.11 are presented in this work. Furthermore, the NetworkAnalyser plugin was used to assess which genes in the network form hubs.
Results and Discussion
Transcriptomics of L. japonicus leaves from plants grown in different nitrogen sources
Previous studies indicated that the expression levels of different key genes for nitrogen metabolism and photorespiration were affected by a defect in the photorespiratory cycle (Pérez-Delgado et al., 2013, 2015). In addition, it has been shown that the rate of photorespiration strongly influences the efficiency with which plants can use different nitrogen sources such as NO3 − or NH4 + (Bloom, 2015). Therefore, in the present study, we examined the transcriptomic response of L. japonicus plants to different forms of nitrogen nutrition, as a first step to determine if the regulation of primary nitrogen assimilation and of photorespiration may be interconnected in legumes. The study was carried out in leaves because this is where the photorespiratory cycle is active. Special attention was paid to the genes encoding transcription factors whose expression was altered under the different conditions considered.
Lotus japonicus plants were grown with different mineral nitrogen sources (NO3 −, NH4 +, or NH4NO3) or under conditions of biological nitrogen fixation (Nod). A comparative transcriptomic analysis was carried out in leaves of plants grown under the four different nitrogen regimes. Genes that showed significantly different levels of expression among treatments were identified by performing a one-way ANOVA together with a Tukey test (further details are given in the Materials and methods). The P-values were corrected for multiple testing using an FDR method with a cut-off of 0.01. A total of 542 probesets were found to be differentially expressed in plants cultivated with the different nitrogen treatments. A global clustering analysis of the whole data set indicated that the transcriptional responses to NO3 − or NH4 + were very similar (Fig. 1). Moreover, plants growing with either NO3 − or NH4 + clustered more closely to the plants grown under purely symbiotic conditions (Nod) than to the plants grown with NH4NO3, indicating that the co-provision of NO3 − and NH4 + triggers a unique transcriptional response compared with the other nitrogen regimes. A hierarchical clustering of the genes that showed significantly different levels of expression between treatments was carried out based on their levels of expression under each nitrogen condition. Five different clusters were obtained according to this analysis (Fig. 2) and are analyzed hereafter. A complete list of the genes and of the transcription factors present in each cluster can be found in Supplementary Tables S2 and S3, respectively.
Fig. 1.
Hierarchical clustering of the different nitrogen nutrition treatments according to the probesets differentially expressed in these conditions as determined by ANOVA using an FDR threshold of P<0.01. Plants were cultivated under four different nitrogen regimens: under symbiotic conditions (Nod) or with NH4NO3 (Mix), NO3 − only, or NH4 + only. The clustering analysis was carried out with the Expander software using average linkage. a and b indicate the biological replicates of the samples in the microarray (E-MTAB-4177).
Fig. 2.
Hierarchical clustering of the probesets differentially expressed among plants cultivated with different forms of nitrogen nutrition according to the ANOVA carried out. The different nitrogen conditions used were: purely symbiotic conditions (Nod), NH4NO3 (Mix), NO3 − only, or NH4 + only. The analysis was carried out with the Expander software and the clusters were determined using average linkage and a distance threshold of 0.3. For each cluster, the average log2 of the fold change in expression of all the differentially expressed probesets under each form of nitrogen nutrition is represented. The number of probesets in each cluster is indicated in parentheses. a and b indicate the biological replicates of the samples in the microarray (E-MTAB-4177). (This figure is available in colour at JXB online.)
Cluster 1
This first cluster contains genes that were more expressed in nodulated plants compared with the other nutritional conditions. Several genes for the biosynthesis of phenolic compounds were found among this group. 4-Coumarate:CoA ligase (probeset chr4.CM0061.26), which catalyzes one of the first common steps for the biosynthesis of all phenolic compounds, was more expressed in nodulated plants. Genes for the synthesis and modification of lignin precursors such as cinnamoyl-CoA reductase (probesets Ljwgs_051770.2 and chr5.CM0200.51) and caffeoyl CoAO-methyltransferase (probeset Ljwgs_030453.1) were also present in this cluster together with a gene for isoflavonoid biosynthesis (isoflavone reductase, probeset chr2.CM0249.94). Phenolic compounds are a large family of secondary metabolites involved in different processes, including plant–pathogen interactions, pollination, light screening, seed development, and allelopathy. Moreover, several of these compounds show an important antioxidant capacity (Hernández et al., 2008). Different genes for phenolic metabolism were found to be altered in their expression levels under different growing conditions in L. japonicus plants, some of which were also connected to photorespiration (García-Calderón et al., 2015). The higher expression of genes for the biosynthesis of phenolic compounds in nodulated plants confirms previous results that showed the existence of important differences in redox metabolism in nodulated plants compared with those cultivated with NO3 − (M. García-Calderón et al., unpublished). Also, it is well known that flavonoids and isoflavonoids have a key role in nodulation (Falcone Ferreyra et al., 2012) and this may also explain the higher expression levels of these genes in nodulated plants. Finally, only one transcription factor belonging to the ERF (ethylene response factor) subfamily was found in this group of genes (probeset TM1489.5) (see Supplementary Table S3).
Cluster 2
In a similar way to cluster 1, the 89 probesets of this group corresponded to genes showing the highest levels of expression in nodulated plants; but, in addition to that, their expression levels were lowest under NH4NO3 nutrition. In agreement with the analysis of cluster 1, several genes involved in the biosynthesis of phenolic compounds were more expressed in nodulated plants, such as the genes encoding a 4-coumarate:CoA ligase isoform (probeset Ljwgs_064187.1) and enzymes involved in flavonoid biosynthesis such as flavanone 4-hydroxylase (probeset Ljwgs_024122.1) and anthocyanidin reductase (chr1.CM0579.2), again suggesting an increased requirement for this class of secondary metabolites in nodulated plants. The only transcription factor present in this group is a zinc finger protein of the C2H2 type (probeset TM0489.21.1) of unknown function (see Supplementary Table S3).
Cluster 3
The 173 probesets in this group corresponded to genes with higher expression in plants growing under NH4NO3 nutrition compared with the other three nutritional conditions. This group included genes encoding enzymes of both nitrogen and carbon metabolism. Key enzymes for amino acid metabolism such as lysine-ketoglutarate dehydrogenase (probeset chr4.CM0343.10) and asparagine synthetase (probesets gi897772 and TM1307.9.1) were more expressed under NH4NO3 nutrition. On the other hand, genes encoding enzymes of central carbon metabolism were also present in this cluster, such as phosphoenolpyruvate carboxykinase (probeset Ljwgs_107668.1), phosphoenolpyruvate carboxylase (probeset chr5.CM0311.22), and enolase (probeset chr1.TM0356.3), together with genes involved in the metabolism of storage polysaccharides such as fructans (β-fructofuranosidase, probesets Ljwgs_038389.1.1 and Ljwgs_012880.1). Eleven transcription factors were highly expressed in NH4NO3-fed plants compared with the other nutritional forms (Supplementary Table S3). While none of these factors has been characterized in L. japonicus, some interesting information can be obtained by searching for their orthologous genes in Arabidopsis. Probesets chr1.CM0088.102 and chr1.CM0375.38 are orthologs to the Arabidopsis transcription factors ATML1, a HUA-like transcription factor family member, and to AXR2, respectively, all of them involved in embryo and shoot development (Takada et al., 2013; Jali et al., 2014; Sato et al., 2015). Two NAC transcription factors (probesets Ljwgs_045720.1 and Ljwgs_055404.1) correspond to Arabidopsis VNI2 and NTL9, which regulate leaf longevity and senescence (Yoon et al., 2008; Yang et al., 2011). These transcription factors may be related to the fact that the growth of plants cultivated with NH4NO3 can surpass the maximal growth compared with either NO3 − or NH4 + alone (Orea et al., 2005).
Cluster 4
The 213 probesets in this group corresponded to genes with higher levels of expression under NH4 + nutrition (and, to a minor extent, under NO3 − conditions) compared with NH4NO3 nutrition and purely symbiotic conditions. Very few genes coding for enzymes of central metabolic pathways, with the exception of an NO3 − transporter and a potassium transporter, were found in this cluster. Ten genes related to the response to different kinds of abiotic or biotic stress were induced by NH4 + nutrition, which may be justified by the higher levels of oxidative stress that are normally observed in plants growing with NH4 + as the sole nitrogen source (Britto and Kronzuker, 2002). On the other hand, a lot of genes related to the modification of chromatin, RNA transcription and translation, and protein post-translational modification showed higher expression mainly in plants grown with NH4 +. These included several genes coding for histones, DNA helicases, RNA helicases, RNases, and several RNA-binding proteins, proteins involved in the regulation of translation, post-translational modification of proteins, protein degradation (proteases, ubiquitin-conjugation enzymes and components of the proteasome), and protein targeting different organelles. Moreover, several transcription factors clustered in this group (see Supplementary Table S3). Some of them were related to cell and tissue development, such as the orthologs of Arabidopsis LZF1 (probeset Ljwgs_049567.1), BSM (probeset Ljwgs_142668.1), and ATRNJ (probeset Ljwgs_035337.1) that are required for the biogenesis of mitochondria and chloroplasts (Chang et al., 2008; Quesada et al., 2011; Chen et al., 2015) or the orthologs of Arabidopsis RSM1, AGL62, PIF3, and CDF4 (probesets TM1490.11, chr5.CM0180.16.1, chr5.CM0048.61, and chr4.CM0399.23, respectively) that are involved in the morphogenesis of different plants tissues (see Hamaguchi et al., 2008; Roszak and Köhler, 2011; Wang et al., 2014; Pi et al., 2015, respectively, for more information on these transcription factors). In addition, some of the transcription factors found in this cluster are involved in the generation of circadian rhythms. This included the ortholog to the core clock protein TOC1 (probeset Ljwgs_147347.1) (Strayer et al., 2000) as well as other proteins involved in the circadian system such as the orthologs to the JMJD5 protein (probeset Ljwgs_035693.2) (Jones et al., 2010) and the ARR3 protein (probeset chr2.CM0028.12) (Salomé et al., 2006). Interestingly, probeset chr5.CM0909.32 encodes the transcription factor OBF4 (also called TGA4), whose ortholog gene (At5g10030) has been associated with the response to NO3 − (Álvarez et al., 2014) and NH4 + (Patterson et al., 2010) in Arabidopsis. These data suggest that the leaf response to NH4 + nutrition is mediated mainly by reorganization of gene expression and cell function rather than through the modification of central metabolic routes.
Cluster 5
The expression of the genes corresponding to the eight probesets of this group was mainly different between NO3 −-fed and NH4 +-fed plants. With the exception of a serine protease, these genes encoded proteins of unknown function. This is in good agreement with the data from Fig. 1 that indicate that there are few transcriptional differences in the leaves of plants grown with these two nitrogen sources.
In summary, transcriptomic data analysis showed that different groups of genes were differentially expressed in leaves of L. japonicus depending on the nitrogen source provided. Under purely symbiotic conditions, a higher expression of genes related to the biosynthesis of phenolic compounds, flavonoids, and isoflavonoids was observed, probably reflecting the importance of these secondary metabolites in the rhizobial symbiosis process. On the other hand, plants growing under NH4NO3 nutrition showed changes in genes of primary metabolism. An induction of genes involved in carbon and nitrogen metabolism was observed in plants growing with NH4NO3 (cluster 3). It is well known that NO3 − functions as a signal molecule to induce the transcripts and activities of enzymes related to NH4 + assimilation and organic acid synthesis providing carbon skeletons for amino acid synthesis (Scheible et al., 1997; Wang et al., 2004). These results indicate the existence of important differences in the link between carbon and nitrogen metabolism under the different forms of nitrogen nutrition examined. Therefore, it was of great interest to explore further the possible interconnections of processes mainly related to carbon metabolism such as photorespiration and nitrogen assimilation. Interestingly, 40 transcription factors responsive to the different nitrogen regimes were identified, suggesting that they could have a relevant function in the different nitrogen signaling or metabolic processes involved.
Interconnection of photorespiration and nitrogen assimilation: studies with Ljgln2-2 mutants
In order to analyze the possible interactions between the assimilation of different nitrogen sources and photorespiration, we first compared the levels of expression of key genes for nitrogen assimilation in plants grown under different nitrogen regimes, maintained either under normal photorespiratory conditions (normal CO2 atmosphere) or under photorespiratory-suppressed conditions (high CO2 atmosphere). For reasons of simplicity, the results obtained for some representative genes (LjASN1, LjGLN2, and LjGLU1) are presented in Fig. 3A, but the whole data set can be found in Supplementary Fig. S1. The levels of atmospheric CO2 had a significant influence on the levels of expression of several nitrogen-assimilatory genes (Fig. 3A; Supplementary Fig. S1). It has to be taken into consideration that the impact of atmospheric CO2 on gene expression may be due to its effects on the photorespiratory cycle and/or to the high availability of carbon under high CO2 conditions. In addition, the patterns of expression of the nitrogen-assimilatory genes in the different nitrogen sources were different in plants cultivated in photorespiratory-suppressed conditions and in plants cultivated in active photorespiratory conditions. These results indicated that the atmospheric CO2 concentration/photorespiration affect the response of L. japonicus plants to different nitrogen sources. A good agreement was found between the gene expression levels measured by qRT–PCR and the expression levels measured by microarray for the WT plants under normal CO2 conditions and under nutrition with different nitrogen forms (Supplementary Table S4).
Fig. 3.
(A) Expression levels of some key genes of nitrogen metabolism in WT plants under a CO2-enriched atmosphere (CO2, gray bars) or normal air (A, white bars) and under different nitrogen conditions: purely symbiotic conditions (Nod), NH4NO3, NO3 − only, or NH4 + only. (B) Expression levels of the same genes in WT (gray bars) and Ljgln2-2 plants (M, gray striped bars) grown under the same different forms of nitrogen nutrition and CO2-enriched atmosphere. LjASN1, asparagine synthetase 1; LjGLN2, plastidic glutamine synthetase; LjGLU1, ferredoxin-dependent GOGAT. Data are the mean ±SD of three independent biological replicates. *Indicates a significant difference between high CO2 and normal air conditions in (A) and between the WT and Ljgln2-2 in (B) as determined by Student’s test (P<0.05).
The key enzyme at the interface between carbon and nitrogen metabolism is GS since this enzyme incorporates the NH4 + resulting from primary and secondary nitrogen assimilation processes to yield glutamine. In particular, the plastidic GS2 isoform is the enzyme in charge of photorespiratory NH4 + reassimilation in plants, and it is therefore the main point of connection between nitrogen assimilation and photorespiratory metabolism. For this reason, the influence of the nitrogen source on the levels of expression of different nitrogen-assimilatory genes was also studied in the photorespiratory mutant Ljgln2-2 (that lacks the plastidic GS2 enzyme) in comparison with the WT. Both WT and mutant plants were grown under high CO2 conditions in order to permit the normal growth of the mutant. Again, for reasons of simplicity, the results obtained for some representative genes (LjASN1, LjGLN2, and LjGLU1) are presented in Fig. 3B, but the whole data set can be found in Supplementary Fig. S2. The patterns of expression of the nitrogen-assimilatory genes in mutant plants in the different nitrogen sources were different from the patterns of expression of these genes in the WT plants cultivated in the same nitrogen sources. These results indicated that the lack of GS2 affects the response of L. japonicus plants to different nitrogen sources.
On the other hand, a more integrative analysis of all the expression data of nitrogen assimilation genes obtained in WT and Ljgln2-2 mutant plants in different photorespiratory conditions shows that there was a close similarity in the differential levels of gene expression of WT plants in high CO2 versus air treatments (Fig. 3A) compared with WT versus Ljgln2-2 mutant plants in high CO2 (Fig. 3B) under the different nitrogen forms of nutrition examined. This suggests that the deficiency in GS2 has a similar effect on the transcript levels of nitrogen-assimilatory genes as the decrease of CO2 concentration and the presence of active photorespiratory conditions in the WT plants under the different forms of nitrogen nutrition studied.
It has been examined whether other genes, in addition to key genes for nitrogen assimilation, could be modulated by changes in CO2 concentration/photorespiratory conditions and/or the lack of GS2. For this purpose, a complete transcriptomic analysis was carried out using the available microarray data from the experiments published by Díaz et al. (2010) and Betti et al. (2012b ). The transcriptional changes produced in the WT by the decrease of the CO2 atmospheric concentration were compared with the transcriptional changes produced by plastidic GS deficiency in plants grown under non-photorespiratory conditions (high CO2).
A Rank product analysis with FDR correction (P<0.1) identified 1668 differentially expressed probesets when comparing WT plants grown in high CO2 or in normal air conditions, which represent genes that can be modulated by carbon and/or active photorespiratory conditions. On the other hand, 1200 differentially expressed probesets as a result of the lack of GS2 under non-photorespiratory conditions were identified using the same statistical cut-off. Interestingly, 24% of these probesets (288) were common to the two transcriptome comparisons carried out (Fig. 4A; Supplementary Table S5). Moreover, these probesets showed a strong linear correlation (r 2=0.91) between the levels of their corresponding fold change in gene expression in the two transcriptomic analyses, with a slope of the regression line of 0.9 that indicates that the transcriptional changes were always very similar between the two transcriptomic comparisons carried out (Fig. 4B). Only 18 of the 288 common probesets showed an opposite response to the decrease of CO2 concentration and to the lack of GS2. All these results indicate the existence of an important number of genes whose expression levels changed in the same way in response to the lack of GS2 and to the decrease of the atmospheric CO2 concentration in addition to the genes for nitrogen metabolism that were studied previously in this work.
Fig. 4.
(A) Venn diagram showing the number of probesets modulated by the decrease of CO2 concentration and/or the absence of plastidic GS (P<0.1 and FDR correction). (B) Comparison of the fold change values for the probesets that are significantly elicited by both conditions.
The MapMan and PathExpress tools (Usadel et al., 2005; Goffard and Weiller, 2007) were utilized to analyze in more detail the above-mentioned group of 288 probesets (Supplementary Fig. S3). The MapMan program allows the visualization of the changes observed in transcriptomic data by providing an overview of metabolic pathways. Visualization of the 288 probesets using this software indicated that the corresponding genes were apparently evenly distributed among the different routes of primary and secondary metabolism (Supplementary Fig. S3). However, PathExpress permitted the identification of four significantly over-represented metabolic pathways within this group of genes. The routes for flavonoid biosynthesis, histidine metabolism, d-arginine and d-ornithine metabolism, and starch and sucrose metabolism were significantly over-represented within the 288 probesets. Moreover, pyruvate decarboxylase (probeset Ljwgs_007060.2), a key gene of central carbon metabolism that has also been reported to have a role in stress tolerance (Pinhero et al., 2011), was found in this group together with other genes related to carbon metabolism encoding a sugar transporter (probeset Ljwgs_136027.1), a diacylglycerol kinase (probeset Ljwgs_067758.1), and an α-galactosidase (probeset Ljwgs_025958.1), amongst others. In addition, several genes related to redox function were modulated such as glutathione S-transferase (probesets Ljwgs_037557.1 and Ljwgs_027688.1), cytochrome P450 (probesets chr1.CM0591.58 and Ljwgs_099719.2), and glutaredoxin (probeset chr1.CM0109.23.1), suggesting that there is an interconnection of carbon, photorespiration, and the lack of GS2 with redox metabolism or stress responses. It is very interesting to note that there was a modulation of a glycolate oxidase (LjGO1) gene (probeset Ljwgs_013523.1). This gene may have a role in stress responses in L. japonicus in relation to photorespiratory NH4 + accumulation (Pérez-Delgado et al., 2013). Previous studies showed that several genes differentially expressed in Ljgln2-2 and WT plants under non-photorespiratory conditions were also elicited by drought stress specifically in Ljgln2-2, thus confirming the existence of a relationship between the lack of GS2 and the stress-responsive machinery in L. japonicus (Díaz et al., 2010).
The present study has shown first that the lack of GS2 affects the response of L. japonicus plants to different nitrogen sources. Secondly, a novel link between the lack of plastidic GS2 and the decrease in external CO2 provided to the plants was observed, which is supported by the finding of 288 commonly modulated probesets in both types of situations. Therefore, it appears that a relationship between the lack of GS2 and carbon metabolism must exist in L. japonicus plants. Previous work from our group has established how a GS2 defect in nitrogen assimilation affects carbon metabolism in this plant (Betti et al., 2012a; García-Calderón et al., 2012; Pérez-Delgado et al., 2013). Finally, six transcription factors were also found in the group of 288 commonly modulated probesets (Supplementary Table S6). These transcription factors may be important for the C/N balance and for the photorespiratory metabolism of L. japonicus plants in relation to GS2 and were further investigated together with the nitrogen-responsive transcription factors that were previously identified in the first section of the paper using co-expression networks as is shown below.
Use of co-expression networks to study the interconnection between photorespiration and nitrogen assimilation
To explore further the interaction between photorespiration and primary nitrogen assimilation, a gene co-expression network was constructed using the WGCNA R package and the transcriptomic data available for L. japonicus. The use of gene co-expression networks was proved to be a very useful tool for the prediction of network driver genes and for the identification of transcription factors with a role in the control of gene expression in nitrogen metabolism (Vidal et al., 2010; Canales et al., 2014). This new analysis integrates the data obtained in the previous experiments of this study with the aim of determining if the genes for nitrogen assimilation and photorespiratory metabolism are interconnected and to search for regulatory genes that may co-ordinate these processes. The co-expression networks generated were visualized using the Cytoscape software and analyzed using the NetworkAnalyser plugin (Doncheva et al., 2012).
A first co-expression network was constructed using the genes for primary nitrogen assimilation and for photorespiratory metabolism (Supplementary Table S7). This analysis identifies the most connected genes between the two groups across multiple experiments (Fig. 5; Supplementary Table S8). Eighteen genes for primary nitrogen assimilation were clearly connected to 15 photorespiratory genes. Among the genes of nitrogen metabolism, we found seven low affinity NO3 − transporters (LjNPF), two high affinity NO3 − transporters (LjNRT2), two NH4 + transporters (LjAMT and LjAMT1), one asparagine synthetase (LjASN2), one glutamate dehydrogenase (LjGDH4), one cytosolic glutamine synthetase (LjGLN1.2), two NADH-dependent glutamate synthases (LjGLT1 and LjGLT2), and two aspartate aminotransferases (LjAAT). This result suggests that primary nitrogen assimilation is connected to photorespiration through NO3 − and NH4 + transporters and NH4 + assimilatory genes. Among the photorespiratory genes that were connected to genes of primary nitrogen assimilation, we found two phosphoglycolate phosphatases (LjPglP1 and LjPglP2), four glycine decarboxylases (LjGDC-H1, LjGDC-P1, LjGDC-P2, and LjGDC-T), one glycerate kinase (LjGlyK2), two aminotransferases (LjGGT and LjSGAT2), one serine hydroxymethyltransferase (LjSHMT1), one glycolate oxidase (LjGO2), hydroxypyruvate reductase (LjHPR), plastidic glutamine synthetase (LjGLN2), and two chloroplast envelope transporters (LjDiT1 and LjDiT2.1). The vast majority of the different photorespiratory genes were connected to some extent to primary nitrogen assimilation. These results confirm the existence of a clear connection between primary nitrogen assimilation and photorespiration, as suggested also by different authors using other types of approaches (Rachmilevitch et al., 2004; Pérez-Delgado et al., 2013, 2015; Bloom, 2015). Other co-expression networks were generated as a control in order to evaluate the connection between photorespiratory genes and genes of other pathways. The connection between photorespiratory genes and genes of nucleotide synthesis, cellulose synthesis, and genes for DNA repair and cell division was very low, indicating that the connection between photorespiration and nitrogen metabolism was significant (Supplementary Table S9).
Fig. 5.
Co-expression network analysis of the connections detected among photorespiratory genes (rectangles in the left-hand column) and genes of primary nitrogen assimilation (rectangles in the right-hand column). Edges represent predicted regulatory interactions between target genes. The genes present in the network image are: phosphoglycolate phosphatase (LjPglP1 and LjPglP2); glycine decarboxylase (LjGDC-H1, LjGDC-P1, LjGDC-P2, and LjGDC-T); glycerate kinase (LjGlyK2); glutamate:glyoxylate aminotransferase (LjGGT); serine:glyoxylate aminotransferase (LjSGAT2); serine hydroxymethyltransferase (LjSHMT1); glycolate oxidase (LjGO2); hydroxypyruvate reductase (LjHPR); plastidic glutamine synthetase (LjGLN2); plastidic dicarboxylate transporter (LjDiT1 and LjDiT2.1); NO3 − transporter (LjNPF); NH4 + transporter (LjAMT); asparagine synthetase (LjASN2); glutamate dehydrogenase (LjGDH4); cytosolic glutamine synthetase (LjGLN1.2); NADH-dependent glutamate synthase (LjGLT1 and LjGLT2); and aspartate aminotransferase (LjAAT). (This figure is available in colour at JXB online.)
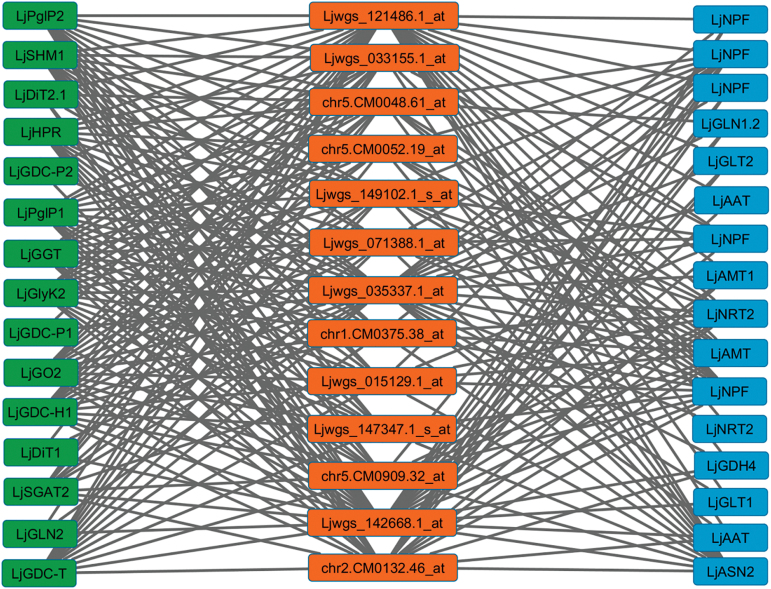
A second co-expression network was constructed using the genes of primary nitrogen assimilation and photorespiratory metabolism and also different transcription factor genes from L. japonicus, obtained in the available databases (Jin et al., 2014) (see Supplementary Table S10). A total of 370 transcription factors were connected to at least one gene for primary nitrogen assimilation and to at least one photorespiratory gene (Supplementary Table S11). A high percentage of these genes belong to the families ERF, basic helix–loop–helix (bHLH), and Myb. To distill from this analysis the candidate regulatory genes that may co-ordinate primary nitrogen assimilation and photorespiration, a third co-expression network was built from the previous one using only the transcription factors that were modulated by either the nitrogen source (Supplementary Table S3) or the GS2 deficiency and the variation in atmospheric CO2 concentration (Supplementary Table S6). Thirteen transcription factors connected to both photorespiration and nitrogen assimilation were identified in this analysis (Table 1; Fig. 6; Supplementary Table S12).
Table 1.
Transcription factors (TFs) connected to genes of primary nitrogen assimilation and to photorespiratory genes using a co-expression networks analysis
Transcription factors highlighted with an asterisk were also modulated by the transfer from photorespiratory suppressed conditions to active photorespiratory conditions.
| Probeset | TF family | Number of connections | Gene (Kazusa 3.0) | Ortholog gene in Arabidopsis |
|---|---|---|---|---|
| Ljwgs_121486.1_at | Unknown | 31 | Lj2g3v1984810.1 | At4g17800 |
| Ljwgs_035337.1_at* | Trihelix | 31 | Lj0g3v0261399.1 | At5g63420 |
| Ljwgs_142668.1_at* | mTERF | 30 | Lj2g3v2197630.1 | At4g02990 |
| chr2.CM0132.46_at | bHLH | 23 | Lj2g3v1984450.1 | At4g37850 |
| Ljwgs_149102.1_s_at | Unknown | 17 | Lj4g3v3015070.1 | At4g12750 |
| chr5.CM0048.61_at | bHLH | 17 | Lj5g3v1533330.1 | At1g09530 |
| Ljwgs_015129.1_at* | bHLH | 16 | Lj3g3v0028580.1 | At2g22770 |
| chr5.CM0909.32_at* | bZIP | 15 | Lj5g3v1697630.1 | At5g10030 |
| Ljwgs_071388.1_at* | Myb-related | 15 | Lj4g3v0973380.1 | At4g39250 |
| chr1.CM0375.38_at* | Unknown | 14 | Lj1g3v4450520.1 | At3g23050 |
| chr5.CM0052.19_at* | ERF | 14 | Lj5g3v1937400.1 | At5g64750 |
| Ljwgs_033155.1_at | bHLH | 13 | Lj0g3v0136069.1 | At1g32640 |
| Ljwgs_147347.1_s_at* | ARR | 11 | Lj4g3v1658890.2 | At5g61380 |
Fig. 6.
Co-expression network analysis of the connections detected among photorespiratory genes (rectangles in the left-hand column) and genes of primary nitrogen assimilation (rectangles in the right-hand column) with transcription factor genes (rectangles in the central column). Edges represent predicted regulatory interactions between transcription factors and target genes. The photorespiratory genes and genes of primary nitrogen assimilation present in the network image are: phosphoglycolate phosphatase (LjPglP1 and LjPglP2); glycine decarboxylase (LjGDC-H1, LjGDC-P1, LjGDC-P2, and LjGDC-T); glycerate kinase (LjGlyK2); glutamate:glyoxylate aminotransferase (LjGGT); serine:glyoxylate aminotransferase (LjSGAT2); serine hydroxymethyltransferase (LjSHMT1); glycolate oxidase (LjGO2); hydroxypyruvate reductase (LjHPR); plastidic glutamine synthetase (LjGLN2); plastidic dicarboxylate transporter (LjDiT1 and LjDiT2.1); NO3 – transporter (LjNPF); NH4 + transporter (LjAMT); asparagine synthetase (LjASN2); glutamate dehydrogenase (LjGDH4); cytosolic glutamine synthetase (LjGLN1.2); NADH-dependent glutamate synthase (LjGLT1 and LjGLT2); and aspartate aminotransferase (LjAAT). Transcription factors are represented by their probeset. (This figure is available in colour at JXB online.)
Two of the transcription factors identified (probesets Ljwgs_121486.1_at and Ljwgs_035337.1_at) are of particular interest because they are connected to all the photorespiratory genes and primary nitrogen assimilation genes in the co-expression network. The probeset Ljwgs_035337.1_at encodes a transcription factor whose ortholog gene in Arabidopsis (At5g63420) plays a vital role in embryo morphogenesis and apical-basal pattern formation by regulating chloroplast development (Chen et al., 2015), whereas the function of the gene encoded by the probeset Ljwgs_121486.1_at is unknown. The genes encoding both transcription factors were more expressed in plants growing with NH4 + (cluster 4 from Fig. 2). Probeset Ljwgs_142668.1_at is connected to all the photorespiratory genes and almost all the nitrogen genes, and encodes a transcription factor whose ortholog gene in Arabidopsis is required for the biogenesis of mitochondria and chloroplasts (Quesada et al., 2011). The corresponding gene was also more expressed in plants growing with NH4 + (cluster 4). In addition, the probesets chr2.CM0132.46_at, Ljwgs_149102.1_s_at, and chr5.CM0048.61_at also showed a high number of connections to both photorespiratory and primary nitrogen assimilation genes. chr2.CM0132.46_ at and Ljwgs_149102.1_s_at encode transcription factors of unknown function, while chr5.CM0048.61_at encodes PIF3, a bHLH transcription factor whose ortholog gene in Arabidopsis (At1g09530) is involved in morphogenesis (Wang et al., 2014). Probesets Ljwgs_015129.1_at, chr5.CM0909.32_at, and Ljwgs_071388.1_at are connected to different NO3 − transporters, one NH4 + transporter, and one asparagine synthetase, and to 10 different photorespiratory genes. Ljwgs_015129.1_at corresponds to a bHLH transcription factor whose ortholog gene in Arabidopsis (At2g22760) is involved in the regulation of the formation of an endoplasmic reticulum-derived structure, the ER body (Matsushima et al., 2004). The probeset chr5.CM0909.32_at encodes the transcription factor OBF4 (also named TGA4 transcription factor), whose ortholog gene in A. thaliana (At5g10030) has been associated with the NO3 − response (Álvarez et al., 2014) and NH4 + response (Patterson et al., 2010). Ljwgs_071388.1_at corresponds to a Myb-related transcription factor whose ortholog gene in Arabidopsis (At4g39250) is involved in plant development (Zhang et al., 2013). Finally, it was also found that another four transcription factors were connected to several photorespiratory genes as well as to NO3 − transporters and to an asparagine synthetase (probesets Ljwgs_033155.1_at and Ljwgs_147347.1_s_at), and also to one NH4 + transporter (chr1.CM0375.38_at and chr5.CM0052.19_at). chr1.CM0375.38_at is a transcription factor whose ortholog gene in Arabidopsis (AXR2, At3g23050) is involved in embryo and shoot development (Sato et al., 2015). The probesets chr5.CM0052.19_at and Ljwgs_033155.1_at encode an ERF and a bHLH factor, respectively, whose orthologous genes in Arabidopsis (At5g64750 and At1g32640, respectively) are involved in the response to abscisic acid and abiotic stress (Pandey et al., 2005; De Ollas et al., 2015). The ortholog to probeset Ljwgs_147347.1_s_at is the core clock protein TOC1, involved in the generation of circadian rhythms (Shimizu et al., 2015).
The functional significance of the transcription factors identified in relation to photorespiration was further confirmed by the fact that eight of the 13 genes described here (highlighted with an asterisk in Table 1) were also modulated by the transfer from photorespiratory-suppressed conditions to active photorespiratory conditions (as can be revealed by analyzing the data from Pérez-Delgado et al., 2013). Of particular interest is the probeset chr5.CM0052.19_at, whose expression levels were modulated up to 8-fold when plants were transferred to active photorespiratory conditions. Furthermore, two of the transcription factors detected with this co-expression analysis (probesets Ljwgs_015129.1_at and Ljwgs_033155.1_at) were also identified among the transcription factors that were commonly affected by the decrease of CO2 concentration and the lack of GS2 (Supplementary Table S6).
The results obtained in this work demonstrate the existence of multiple interconnections between primary nitrogen assimilation and photorespiration in L. japonicus plants, and allowed us to identify several transcription factors that could have a crucial role in the joint regulation of these processes. Further work would still be required to obtain mutants from the transcription factors identified and to determine their exact physiological significance.
Conclusions
In this work it has been shown that several important transcriptomic changes occur in leaves of L. japonicus when plants are cultivated with different nitrogen sources. This includes genes involved in nitrogen, carbon, and secondary metabolism, as well as several transcription factors that may play an important role in nitrogen signaling or metabolism. In addition, the study of the Ljgln2-2 mutants provides novel insights into the effects of the lack of plastidic GS in the response to different nitrogen sources and also shows the existence of a common transcriptomic response due to the lack of GS2 and to the changes in external CO2/photorespiratory activity, thus emphasizing the importance of GS2 in the C/N balance in L. japonicus plants. Finally, the use of co-expression networks allowed us to establish a clear interconnection between photorespiration and primary nitrogen assimilation and to identify several candidate genes for the co-ordination of these metabolic routes.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primer sequences used for qRT–PCR measurements.
Table S2. Differentially expressed genes in plants grown with different nitrogen sources (NO3 −, NH4 +, NH4NO3, and nodulated plants) divided into different clusters.
Table S3. Differentially expressed transcription factor genes in plants grown with different nitrogen sources (NO3 −, NH4 +, NH4NO3, and nodulated plants) divided into different clusters.
Table S4. Validation of microarray and qRT–PCR data.
Table S5. Genes elicited by both the decrease of CO2 concentration and the lack of GS2.
Table S6. Transcription factors elicited by both the decrease of CO2 concentration and the lack of GS2.
Table S7. List of the genes for primary nitrogen assimilation and photorespiration.
Table S8. Co-expression network analysis of photorespiratory genes and genes of primary nitrogen assimilation.
Table S9. Control co-expression analysis of photorespiratory genes and genes of nitrogen metabolism, nucleotide synthesis, cellulose synthesis, and genes for DNA repair and cell division. Groups of genes were taken from MapMan.
Table S10. List of Lotus japonicus transcription factors available in the current databases.
Table S11. Co-expression network analysis of photorespiratory genes, genes for primary nitrogen assimilation, and transcription factors.
Table S12. Co-expression network analysis of photorespiratory genes, genes for primary nitrogen assimilation, and the transcription factors that were previously detected as responsive to nitrogen nutrition or changes in the photorespiratory conditions (from Supplementary Tables S3 and S6).
Figure S1. Expression levels of some key genes for nitrogen metabolism in WT plants under a CO2-enriched atmosphere (CO2) or normal air (A) and different nitrogen sources.
Figure S2. Expression levels of some key genes of nitrogen metabolism in WT and Ljgln2-2 mutant plants (M) grown with different forms of nitrogen nutrition and under a CO2-enriched atmosphere.
Figure S3. MapMan overview of general metabolism and Pathexpress analysis of over-represented pathways for the 288 probesets that were modulated by both the decrease of CO2 concentration and the lack of plastidic GS.
Acknowledgements
We acknowledge financial support given by projects AGL2014-54413-R from FEDER-Ministerio de Economía y Competitividad, Spain, as well as P10-CVI-6368 and BIO-163 from Consejería de Economía, Innovación y Ciencia, Junta de Andalucía (Spain). The authors would like to thank V Plan Propio de Investigación for a fellowship to CMP, the CITIUS Biology facilities of the University of Seville for qRT–PCR measurements, and M.J. Cubas and A. Gómez for technical and secretarial assistance.
References
- Álvarez J, Riveras E, Vidal EA, et al. 2014. System approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. The Plant Journal 80, 1–13. [DOI] [PubMed] [Google Scholar]
- Andrews M, Raven JA, Lea PJ. 2013. Do plants need nitrate? The mechanisms by which nitrogen form affects plants. Annals of Applied Biology 163, 174–199. [Google Scholar]
- Bauwe H, Hageman M, Fernie AR. 2010. Photorespiration: players, partners and origin. Trends in Plant Science 15, 330–336. [DOI] [PubMed] [Google Scholar]
- Betti M, Arcondéguy T, Márquez AJ. 2006. Molecular analysis of two mutants from Lotus japonicus deficient in plastidic glutamine synthetase. Functional properties of purified GLN2 enzymes. Planta 224, 1068–1079. [DOI] [PubMed] [Google Scholar]
- Betti M, García-Calderón M, Pérez-Delgado CM, Credali A, Estivill G, Galván F, Vega JM, Márquez AJ. 2012. a Glutamine synthetase in legumes: recent advances in enzyme structure and functional genomics. International Journal of Molecular Sciences 13, 7994–8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betti M, García-Calderón M, Pérez-Delgado CM, Credali A, Pal’ove-Balang P, Estivill G, Repçkák M, Vega JM, Galván F, Márquez AJ. 2014. Reassimilation of ammonium in Lotus japonicus . Journal of Experimental Botany 65, 5557–5566. [DOI] [PubMed] [Google Scholar]
- Betti M, Pérez-Delgado C, García-Calderón M, Díaz P, Monza J, Márquez AJ. 2012. b Cellular stress following water deprivation in the model legume Lotus japonicus . Cells 1, 1089–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom AJ. 2015. Photorespiration and nitrate assimilation: a major intersection between plant carbon and nitrogen. Photosynthesis Research 123, 117–128. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Letters 573, 83–92. [DOI] [PubMed] [Google Scholar]
- Britto DT, Kronzucker HJ. 2002. NH4 + toxicity in higher plants: a critical review. Journal of Plant Physiology 159, 567–584. [Google Scholar]
- Canales J, Moyano TC, Villarroel E, Gutiérrez RA. 2014. System analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Frontiers in Plant Science 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canvin DT. 1990. Photorespiration and CO2-concentrating mechanisms. In: Dennis DT, Turpin DH, eds. Plant physiology, biochemistry, and molecular biology , Singapore: Langman Scientific-Technical, 253–273. [Google Scholar]
- Chang CS, Li YH, Chen LT, et al. 2008. LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. The Plant Journal 54, 205–219. [DOI] [PubMed] [Google Scholar]
- Chen H, Zou W, Zhao J. 2015. Ribonuclease J is required for chloroplast and embryo development in Arabidopsis. Journal of Experimental Botany 66, 2079–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK. 2004. Real-time RT–PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. The Plant Journal 38, 366–379. [DOI] [PubMed] [Google Scholar]
- De Ollas C, Arbona V, Gómez-Cadenas A. 2015. Jasmonic acid interacts with abscisic acid to regulate plant responses to water stress conditions. Plant Signaling and Behavior 10, e1078953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz P, Betti M, Sánchez DH, Udvardi MK, Monza J, Márquez AJ. 2010. Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytologist 188, 1001–1013. [DOI] [PubMed] [Google Scholar]
- Doncheva NT, Assenov Y, Domingues FS, Albrecht M. 2012. Topological analysis and interactive visualization of biological networks and protein structures. Nature Protocols 7, 670–685. [DOI] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Rius SP, Casati P. 2012. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Frontiers in Plant Science 3, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Clarkson DT. 1999. Nitrate and ammonium nutrition of plants: physiological and molecular perspectives. Advances in Botanical Research 30, 1–90. [Google Scholar]
- Foyer CH, Bloom A, Queval G, Noctor G. 2009. Photorespiratory metabolism: genes, mutants, energetics and redox signalling. Annual Review of Plant Biology 60, 455–484. [DOI] [PubMed] [Google Scholar]
- García-Calderón M, Chiurazzi M, Espuny MR, Márquez AJ. 2012. Photorespiratory metabolism and nodule function: behaviour of Lotus japonicus mutants deficient in plastid glutamine synthetase. Molecular Plant-Microbe Interactions 25, 211–219. [DOI] [PubMed] [Google Scholar]
- García-Calderón M, Pons-Ferrer T, Mrázova A, et al. 2015. Modulation of phenolic metabolism under stress conditions in a Lotus japonicus mutant lacking plastidic glutamine synthetase. Frontiers in Plant Science 6, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. 2004. Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315. [DOI] [PubMed] [Google Scholar]
- Goffard N, Weiller G. 2007. Pathexpress: a web-based tool to identify relevant pathways in gene expression data. Nucleic Acids Research 35, W176–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Zhou Y, Shen Q, Zhang F. 2007. Effect of ammonium and nitrate nutrition on some physiological processes in higher plants—growth, photosynthesis, photorespiration, and water relations. Plant Biology 9, 21–29. [DOI] [PubMed] [Google Scholar]
- Hamaguchi A, Yamashino T, Koizumi N, Kiba T, Kojima M, Sakakibara H, Mizuno T. 2008. A small subfamily of Arabidopsis RADIALIS-LIKE SANT/MYB genes: a link to HOOKLESS1-mediated signal transduction during early morphogenesis. Bioscience, Biotechnology, and Biochemistry 72, 2687–2696. [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J. 1992. Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. The Plant Journal 2, 487–496. [Google Scholar]
- Hernández I, Alegre L, van Breusegem F, Munné-Bosch S. 2008. How relevant are flavonoids as antioxidants in plants. Trends in Plant Science 14, 125–132. [DOI] [PubMed] [Google Scholar]
- Hirel B, Le Gouis J, Gallais A. 2007. The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. Journal of Experimental Botany 58, 2369–2387. [DOI] [PubMed] [Google Scholar]
- Høgslund N, Radutoiu S, Krusell L, et al. 2009. Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. PLoS One 4, e6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jali SS, Rosloski SM, Janakirama P, Steffen JG, Zhurov V, Berleth T, Clark RM, Grbic V. 2014. A plant-specific HUA2-LIKE (HULK) gene family in Arabidopsis thaliana is essential for development. The Plant Journal 80, 242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JP, Zhang H, Kong L, Gao G, Luo JC. 2014. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Research 42, D1182–D1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Covington MF, DiTacchio L, Vollmers C, Panda S, Harmer SL. 2010. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proceedings of the National Academy of Sciences, USA 107, 21623–21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi M, Imaizumi-Anraku H, Koiwa H, Niwa S, Ikuta A, Syono K, Akao S. 2002. Root, root hair and symbiotic mutants of the model legume Lotus japonicus . Molecular Plant-Microbe Interactions 15, 17–26. [DOI] [PubMed] [Google Scholar]
- Keys AJ, Bird IF, Cornelius MJ, Lea PJ, Wallsgrove RM, Miflin BJ. 1978. Photorespiratory nitrogen cycle. Nature 275, 741–743. [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez AJ, Betti M, García-Calderón M, Pal’ove-Balang P, Díaz P, Monza J. 2005. Nitrate assimilation in Lotus japonicus . Journal of Experimental Botany 56, 1741–1749. [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. 2010. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Annals of Botany 105, 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Fukao Y, Nishimura M, Hara-Nishimura I. 2004. NAI1 gene encodes a basic-helix–loop–helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. The Plant Cell 16, 1536–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orea A, Pajuelo P, Pajuelo E, Quidiello C, Romero JM, Márquez AJ. 2002. Isolation of photorespiratory mutants from Lotus japonicus deficient in glutamine synthtetase. Physiologia Plantarum 115, 352–361. [DOI] [PubMed] [Google Scholar]
- Orea A, Pajuelo P, Romero JM, Márquez AJ. 2005. Nitrate assimilation: influence of nitrogen supply. In: Márquez AJ, ed. Lotus japonicus handbook . Dordrecht, The Netherlands: Springer, 295–313. [Google Scholar]
- Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S. 2005. ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiology 139, 1185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson K, Cakmak T, Cooper A, Lager I, Rasmusson AG, Escobar MA. 2010. Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant, Cell and Environment 33, 1486–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Delgado CM, García-Calderón M, Márquez AJ, Betti M. 2015. Reassimilation of photorespiratory ammonium in Lotus japonicus plants deficient in plastidic glutamine synthetase. PLoS One 10, e0130438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Delgado CM, García-Calderón M, Sánchez DH, Udvardi MK, Kopka J, Márquez AJ, Betti M. 2013. Transcriptomic and metabolic changes associated to photorespiratory ammonium accumulation in the model legume Lotus japonicus . Plant Physiology 162, 1834–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T. 2015. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Developmental Cell 33, 576–588. [DOI] [PubMed] [Google Scholar]
- Pinhero R, Pazhekattu R, Marangoni AG, Liu Q, Yada RY. 2011. Alleviation of low temperature sweetening in potato by expressing Arabidopsis pyruvate decarboxylase gene and stress-inducible rd29A: a preliminary study. Physiology and Molecular Biology of Plants 17, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Sarmiento-Mañús R, González-Bayón R, Hricová A, Pérez-Marcos R, Graciá-Martínez E, Medina-Ruiz L, Leyva-Díaz E, Ponce MR, Micol JL. 2011. Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. The Plant Journal 68, 738–753. [DOI] [PubMed] [Google Scholar]
- Rachmilevitch S, Cousins AB, Bloom AJ. 2004. Nitrate assimilation in plant shoots depends on photorespiration. Proceedings of the National Academy of Sciences, USA 101, 11506–11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak P, Köhler C. 2011. Polycomb group proteins are required to couple seed coat initiation to fertilization. Proceedings of the National Academy of Sciences, USA 108, 20829–20831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Freixes S, Balzergue S, et al. 2008. Signaling of the plant nitrogen status triggers specific transcriptome responses depending on the nitrogen source in Medicago truncatula . Plant Physiology 146, 2020–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, et al. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–8. [DOI] [PubMed] [Google Scholar]
- Salomé PA, To JP, Kieber JJ, McClung CR. 2006. Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. The Plant Cell 18, 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez DH, Lippold F, Redestig H, Hannah MA, Erban A, Krämer U, Kopka J, Udvardi MK. 2008. Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus . The Plant Journal 53, 973–987. [DOI] [PubMed] [Google Scholar]
- Sánchez DH, Pieckenstain FL, Szymanski J, Erban A, Bromke M, Hannah MA, Kraemer U, Kopka J, Udvardi MK. 2011. Comparative functional genomics of salt stress in related model and cultivated plants identifies and overcomes limitations to translational genomics. PLoS One 6, e17094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Sasaki S, Matsuzaki J, Yamamoto KT. 2015. Negative phototropism is seen in Arabidopsis inflorescences when auxin signaling is reduced to a minimal level by an Aux/IAA dominant mutation, axr2. Plant Signaling and Behavior 10, e990838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M. 1997. Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. The Plant Cell 9, 783–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir R, Maron-Katz A, Tanay A, Linhart C, Steinfeld I, Sharan R, Shiloh Y, Elkon R. 2005. EXPANDER—an integrative program suite for microarray data analysis. BMC Bioinformatics 6, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H, Araki T, Endo M. 2015. Photoperiod sensitivity of the Arabidopsis circadian clock is tissue-specific. Plant Signaling and Behavior 10, e1010933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology 3, 3. [DOI] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL. 1980. The inhibition of photosynthesis in Arabidopsis mutants lacking glutamate synthase activity. Nature 286, 257–259. [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. 2000. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. [DOI] [PubMed] [Google Scholar]
- Takada S, Takada N, Yoshida A. 2013. ATML1 promotes epidermal cell differentiation in Arabidopsis shoots. Development 140, 1919–1923. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Maron-Katz A, Shavit S, Sagir D, Linhart C, Elkon R, Tanay A, Sharan R, Shiloh Y, Shamir R. 2010. Expander: from expression microarrays to networks and functions. Nature Protocols 5, 303–322. [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, et al. 2005. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiology 138, 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Tamayo KP, Gutiérrez RA. 2010. Gene networks for N-sensing, signaling and response in Arabidopsis thaliana. Wiley Interdisciplinary Reviews: System Biology and Medicine 2, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JM. 1970. The cultivation, isolation and maintenance of rhizobia. In: Vincent JM, ed. A manual for the practical study of the root-nodule bacteria . Oxford: Blackwell Scientific Publications, 1–13. [Google Scholar]
- Wallsgrove RM, Turner JC, Hall NP, Kendall AC, Bright SW. 1987. Barley mutants lacking chloroplast glutamine synthetase. Biochemical and genetic analysis. Plant Physiology 83, 155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM. 2004. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis . Plant Physiology 136, 2512–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fan X, Lin F, He G, Terzaghi W, Zhu D, Deng XW. 2014. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proceedings of the National Academy of Sciences, USA 111, 10359–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SD, Seo PJ, Yoon HK, Park CM. 2011. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. The Plant Cell 23, 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip AM, Horvath S. 2007. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics 8, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HK, Kim SG, Kim SY, Park CM. 2008. Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Molecules and Cells 25, 438–445. [PubMed] [Google Scholar]
- Zhang M, Wu F, Shi J, Zhu Y, Zhu Z, Gong Q, Hu J. 2013. ROOT HAIR DEFECTIVE3 family of dynamin-like GTPases mediates homotypic endoplasmic reticulum fusion and is essential for Arabidopsis development. Plant Physiology 163, 713–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.