Abstract
Congenital stationary night blindness (CSNB) is a heterogeneous group of non-progressive inherited retinal disorders with characteristic electroretinogram (ERG) abnormalities. Riggs and Schubert-Bornschein are subtypes of CSNB and demonstrate distinct ERG features. Riggs CSNB demonstrates selective rod photoreceptor dysfunction and occurs due to mutations in genes encoding proteins involved in rod phototransduction cascade; night blindness is the only symptom and eye examination is otherwise normal. Schubert-Bornschein CSNB is a consequence of impaired signal transmission between the photoreceptors and bipolar cells. Schubert-Bornschein CSNB is subdivided into complete CSNB with an ON bipolar signaling defect and incomplete CSNB with both ON and OFF pathway involvement. Both subtypes are associated with variable degrees of night blindness or photophobia, reduced visual acuity, high myopia, and nystagmus. Whole-exome sequencing of a family screened negative for mutations in genes associated with CSNB identified biallelic mutations in the guanine nucleotide-binding protein subunit beta-3 gene (GNB3). Two siblings were compound heterozygous for a deletion (c.170_172delAGA [p.Lys57del]) and a nonsense mutation (c.1017G>A [p.Trp339∗]). The maternal aunt was homozygous for the nonsense mutation (c.1017G>A [p.Trp339∗]). Mutational analysis of GNB3 in a cohort of 58 subjects with CSNB identified a sporadic case individual with a homozygous GNB3 mutation (c.200C>T [p.Ser67Phe]). GNB3 encodes the β subunit of G protein heterotrimer (Gαβγ) and is known to modulate ON bipolar cell signaling and cone transducin function in mice. Affected human subjects showed an unusual CSNB phenotype with variable degrees of ON bipolar dysfunction and reduced cone sensitivity. This unique retinal disorder with dual anomaly in visual processing expands our knowledge about retinal signaling.
Keywords: night blindness, congenital stationary, retinal dystrophies, G-protein beta 3 subunit, human GNB3, exome, light signal transduction
Main Text
Congenital stationary night blindness (CSNB) is a heterogeneous group of non-progressive inherited retinal disorders that follow autosomal-dominant, autosomal-recessive, or X-linked patterns of inheritance.1 CSNB results from defects in visual signal transduction either within rod photoreceptors or in rod and cone bipolar pathways.1 Full-field electroretinogram (ERG) testing is an essential tool to diagnose CSNB and helps to localize the functional deficit to photoreceptor or bipolar-cell signaling.1, 2 In brief, a bright flash (3.0 and 10.0 cd.s.m−2 [candela second per square meter] as defined by the International Society for Clinical Electrophysiology of Vision [ISCEV]) under dark-adapted (DA) conditions normally elicits an initial negative wave (a-wave) that reflects phototransduction-related photoreceptor hyperpolarization predominated by the rods; the subsequent positive wave (b-wave) mostly reflects ON bipolar cell depolarization. Under light-adapted (LA) conditions, a standard flash (3.0 cd.s.m−2) elicits an initial a-wave generated by cone photoreceptor hyperpolarization with an additional contribution from cone OFF bipolar cells; the subsequent b-wave is generated from within ON and OFF bipolar cells. The LA 30 Hz stimuli (3.0 cd.s.m−2) lead to a sinusoidal response generated in the inner retina that is driven by cones.3, 4, 5, 6, 7
The most frequent forms of CSNB have a normal fundus and can be subdivided into Riggs (CSNBAD1 [MIM: 610445], CSNBAD2 [MIM: 163500], CSNBAD3 [MIM: 610444], CSNB1D [MIM: 613830], and CSNB1G [MIM: 616389]) and Schubert-Bornschein (CSNB1A [MIM: 310500], CSNB1B [MIM: 257270], CSNB1C [MIM: 613216], CSNB1E [MIM: 614565], CSNB1F [MIM: 615058], CSNB2A [MIM: 300071], and CRSD [MIM: 610427]) types on the basis of distinct ERG abnormalities.8, 9 Riggs CSNB causes reduced rod ERG amplitudes (a- and b-wave) consistent with primary rod dysfunction; cone function is preserved. Night blindness might be the only symptom and affected individuals usually have normal distance visual acuity. Riggs CSNB is linked to mutations in proteins involved in rod phototransduction and can be inherited as autosomal-dominant (RHO [MIM: 180380], PDE6B [MIM: 180072], and GNAT1 [MIM: 139330]) and recessive (GNAT1 and SLC24A1 [MIM: 603617]) traits.10, 11, 12, 13, 14, 15 Schubert-Bornschein CSNB is characterized by normal DA a-wave and reduced b-wave (electronegative configuration: b/a < 1) in response to a bright flash (3.0 or 10.0 cd.s.m−2), consistent with impaired signal transmission between the photoreceptors and bipolar cells. Subjects with Schubert-Bornschein CSNB commonly present with some degree of night blindness, variable photophobia, nystagmus, reduced visual acuity, and myopia.1 Schubert-Bornschein CSNB is subcategorized into complete (c) and incomplete (ic) CSNB.16 cCSNB is characterized by selective ON bipolar cell dysfunction, and icCSNB is due to a signaling defect involving both ON and OFF bipolar pathways. cCSNB is linked to mutations in NYX (MIM: 300278), GRM6 (MIM: 604096), TRPM1 (MIM: 603576), GPR179 (MIM: 614515), and LRIT3 (MIM: 615004); these genes encode proteins localized at the dendritic tip of ON bipolar cells.17, 18, 19, 20, 21, 22, 23, 24, 25 icCSNB is linked to mutations in CACNA1F (MIM: 300110), CABP4 (MIM: 608965), and CACNA2D4 (MIM: 608171), coding for proteins localized to the photoreceptor synaptic terminal.26, 27, 28, 29 Mutations in known genes do not account for all cases of CSNB, and more gene defects are yet to be discovered.1 Identification of new gene defects will enable better understanding of visual signal processing within the photoreceptors and from photoreceptors to bipolar cells.
The study protocol followed the tenets of the Declaration of Helsinki and was approved by the institutional ethics review board of each participating hospital or university. Prior informed consent was obtained from all participating members and parents (on behalf of children). Family A, with three affected individuals, was identified at the Hospital for Sick Children, Toronto (Figure 1A). Affected members had mildly reduced vision and normal fundus appearance; childhood-onset night blindness was present in the proband (III-2) and maternal aunt (II-5). Mutational analysis of the proband did not identify any pathogenic variant in 17 genes known to be mutated in CSNB or in 114 additional genes known to be mutated in retinal dystrophies. 11 members of the family were recruited. Detailed eye examinations and ISCEV standard ERG testing was performed in ten members (except I-1).2, 30 LA ERG with long-duration white flashes (150 ms and 200 ms; 250 cd.m−2; to analyze cone ON and OFF pathways) was performed in the three affected members.31 Whole-exome capture and sequencing (WES) was performed in two trios (two affected subjects and their unaffected parents; Figure 1A) at The Center for Applied Genomics, Toronto as previously described (see the Supplemental Material and Methods).32 The filtering steps used in the WES analysis in the pedigree are summarized in Table S1. Under the assumption of autosomal-recessive inheritance, genes carrying ≥two rare non-synonymous coding variants, splicing variants, or indels were prioritized; only 13 genes were shared among III-2 and II-5. WES data from the unaffected parents and sibling were sequentially used to filter through shared rare variants. A single variant in guanine nucleotide-binding protein subunit beta-3 (GNB3 [GenBank: NM_002075.3] or Gβ3 [MIM: 139130]) was shared between III-2 and II-5. This variant, c.1017G>A in exon 10 and predicted to lead to nonsense mutation p.Trp339∗, was homozygous in II-5 and heterozygous in III-2. Individual III-2 also carried a second heterozygous variant in GNB3; this variant, c.170_172delAGA in exon 4, was predicted to cause an in-frame deletion, p.Lys57del. Both the c.170_172delAGA and c.1017G>A variants were confirmed by Sanger sequencing (conditions available on request) and segregated with disease phenotype in the family (Figure 1A). Neither variant was reported in any public databases, including the Exome Aggregation Consortium (ExAC) Browser, the NHLBI ESP Exome Variant Server, and the 1000 Genomes Browser (release 14).33
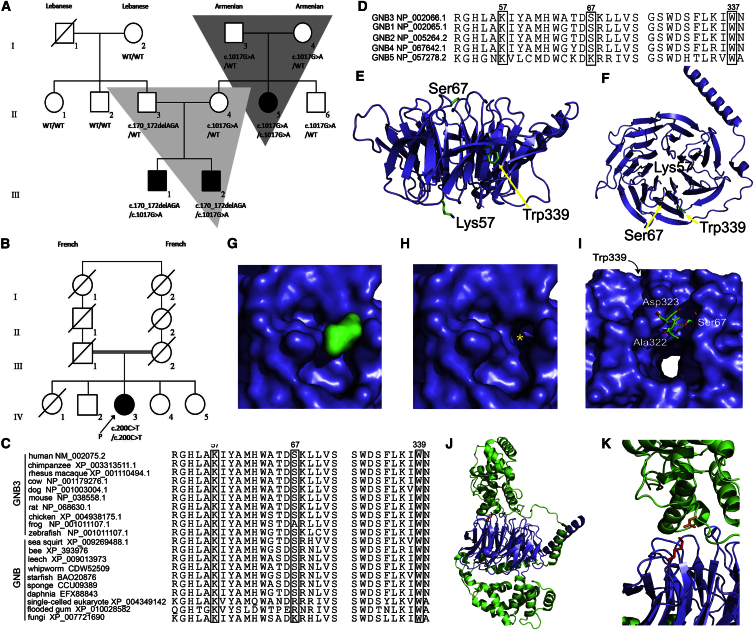
Figure 1.
Genetic Pedigree of Both Families and Protein Conservation Maps and Protein Modeling Results for the Detected GNB3 Variants
(A) A three generation pedigree (family A) with three affected members.
(B) Pedigree of a sporadic case subject (family B) born to distantly consanguineous parents.
(C) Conservation map of GNB3 in vertebrates and in GNB homologs across a range of taxonomic groups; Lys57 and Trp339 were the most conserved amino-acid residues.
(D) Conservation of amino-acid residues across GNB paralogs.
(E and F) Homology models of GNB3 generated with Phyre2. The structure of the GNB3 homology model based on the 1TBG structure of GNB1 (chain A [PDB: 1TBG]) and mutated residues are labeled.
(G and H) Surface representation of GNB3 illustrates how termination of the protein at Trp339 would create a significant loss of structure and the exposure of normally buried residues. The Trp surface (green) is shown in (G), and the loss of structure with removal of Trp339 and Asn340 (highlighted by yellow asterisk) is shown in (H).
(I) Illustration of the location of Ser67 showing the two potential hydrogen bonds to Ala322 and Asp323; the p.Ser67Phe mutation would disrupt both of these interactions. Trp339 is located in the same region of the β-barrel structure, potentially indicating an important biological role for this region of GNB3.
(J and K) The loss of Lys57 (p.Lys57del) would cause disruption of Gβ5:RGS interactions. GNB3 is shown in blue, RGS in green, Lys57 as red sticks, and the binding aspartate as orange sticks.
GNB3 mutational analysis was performed with Sanger sequencing (conditions available on request) in 58 additional CSNB cases; most individuals were excluded for mutations in genes associated with CSNB. The samples originated from different centers in Europe, the United States, Canada, and Israel. A presumably homozygous missense variant, c.200C>T in exon 4 of GNB3 and leading to p.Ser67Phe, was identified in a sporadic case subject (IV-3, family B; Figure 1B) clinically investigated at the Centre National de Références Maladies Sensorielles Génétiques in Montpellier (this individual was excluded for mutations in all known genes associated with CSNB). This extremely rare variant was found at a low frequency in the ExAC (MAF = 0.00004) and ESP (MAF = 0.0002) databases, but never in a homozygous state. In view of consanguineous ancestry, we determined this mutation to be most likely homozygous in the subject. Co-segregation could not be performed because the parents were deceased and no other family members were available for testing. Hence, the possibility of a heterozygous deletion involving the entirety of GNB3 cannot be completely excluded.
Geneious v.8.1.8 was used to align the amino acid sequences of GNB3 and GNB proteins in available species.34 The amino acid residues p.Lys57 and p.Trp339 in GNB3 are highly conserved across vertebrates, including zebrafish; p.Ser67Phe is conserved in most vertebrates (except frogs; Figure 1C). Residues p.Lys57 and p.Trp339 are conserved in all human GNB paralogs and in GNB proteins across a wide range of taxonomic groups (fungi, plants, and single-celled eukaryotes; Figures 1C and 1D). The p.Ser67 residue is conserved in all human paralogs, except GNB5, and is moderately conserved in GNB proteins present in other taxonomic groups. The mutant p.Phe67 is not seen in any paralog or homolog. All three variants, p.Lys57del, p.Trp339∗, and p.Ser67Phe, were predicted by MutationTaster to be disease causing with probabilities >0.99. Residues p.Lys57, p.Trp339, and p.Ser67 had PhylopPvert-100 average scores of 7.3, 9.6, and 4.1, respectively (strong conservation). The PhyloPMam average conservation score was 2.83 for p.Trp339 (strong conservation); p.Lys57 and p.Ser67 had scores of 2.22 and 2.46, respectively (moderate conservation).
GNB3 located on chromosome 12p13.31 encodes the β subunit of the G protein heterotrimer (Gαβγ); it is expressed in the retina and has been previously associated with a partial form of cCSNB in mice (Gnb3−/−).35, 36 GNB3 is a WD40 protein made up of seven highly conserved repeating units (∼40 amino acid motifs) usually ending with Trp-Asp (WD).37 WD40 repeat motifs act as sites for protein-protein interaction, and WD40 proteins regulate cellular function, including transmembrane signaling.38 Residues Lys57 and Ser67 involve WD40 repeat 1, and Trp339 involves WD40 repeat 7. To determine the pathological basis of the identified mutations, we created a homology model of GNB3 from the known GNB1 (Gβ1) amino-acid sequence (shares 83% identity and 97% amino acid similarity with GNB3) structure by using Phyre2 software with a confidence score of 100 (Figures 1E and 1F).39, 40 The Gβ1 subunit forms a seven-bladed β propeller, typical of WD40 repeat proteins, and the entire sequence of GNB3 was fit onto this structure. Despite being at the extreme C terminus of the GNB3 protein chain, Trp339 occupies an important position in the β-propeller structure, buried between the first and last β sheets that constitute the propeller. If a stop codon is introduced in place of Trp339, both Trp339 and Asn340 would be removed, creating a significant distortion of the circular propeller structure and exposing normally buried residues (Figures 1G and 1H). The β-propeller fold is commonly used to facilitate protein-protein interactions and the identified p.Trp339∗ would pathologically affect the ability of GNB3 to form important interactions within the G protein signaling complex. The residue Ser67 lies at the top ridge of the overall β-barrel structure of GNB3 with two potential hydrogen-bonding partners Ala322 and Asp323 (Figure 1I). The p.Ser67Phe mutation would remove the ability to participate in both hydrogen bonds, disrupting the structure of the top of the barrel where other protein-protein interactions occur. Interestingly, the region of the β barrel disrupted by p.Ser67Phe is the same region disrupted by p.Trp339∗, potentially indicating that this ridge or geographic region of GNB3 forms interactions important for cellular function. The other variant, p.Lys57del, lies within a surface loop on the GNB3 structure. To determine how this mutation might affect Gβ protein interactions with regulators of G protein signaling (RGS), we used the X-ray crystal structure of a Gβ5:RGS complex (PDB: 2PBI).41 Lys57 forms a conserved interaction with an aspartate in the RGS protein (Figures 1J and 1K), and loss of the Lys57 residue would cause disruption of bonding between GNB3 and cognate Gβ binding partners, inhibiting the formation of effective G protein complexes.
The ocular phenotype of the four affected individuals is summarized in Table S2. Childhood-onset night blindness was observed in three subjects. Photophobia and color vision deficits were noted only in middle aged subjects (II-5, family A and IV-3, family B). None had nystagmus and all subjects had a best-corrected visual acuity of 20/30 or better. Mild myopia was observed in one; two had hyperopia. All had normal fundus and visual fields. Central retinal thickness and retinal lamination were normal (spectral domain-optical coherence tomography) in all subjects; Gnb3-null mice also showed normal retinal morphology.35, 42 The visual parameters remained stable on follow-up in all cases; the proband from family B had 47 years of follow-up. These findings are consistent with the diagnosis of CSNB.43 All seven unaffected members in family A had normal eye examinations and normal ERGs (similar to the control eye data provided in Figure 2E).
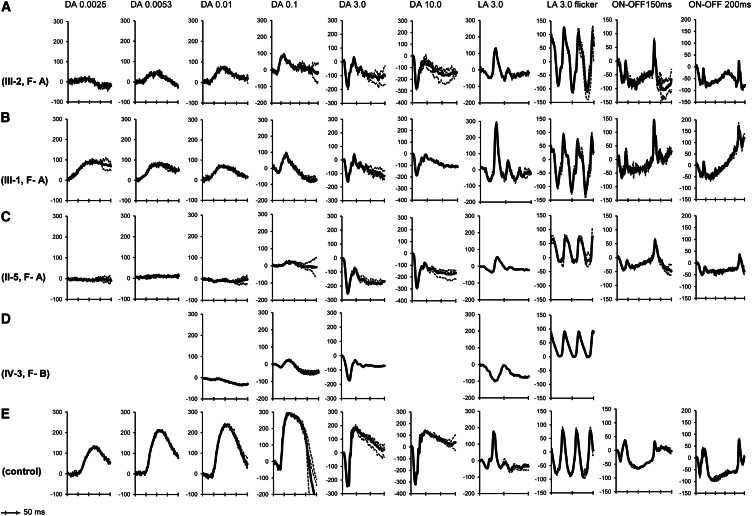
Figure 2.
ERG Traces from the Right Eye of All Four Affected Individuals and a Control Subject
ERG traces from family A (A, III-2; B, III-1; C, II-5), the family B proband (D; it is noted that only standard ERG protocols were tested in the subject), and a control subject (E). The nomenclature of the responses relates to the adaptive state of the eye (DA, dark adapted; LA, light adapted) and stimulus intensity in cd.s.m−2 (DA 0.01 relates to dark adapted responses to 0.01 cd.s.m−2 flash). The DA b-wave was either reduced or non-detectable between stimulus intensities of 0.0025 and 0.1 cd.s.m−2. On bright flash stimulation in the dark (3.0 and 10.0 cd.s.m−2), the a-wave is of normal amplitude; b-wave is markedly reduced at both intensities. The DA ERG results (DA 0.0025 to DA 10.0) are consistent with rod ON bipolar dysfunction in all affected subjects (A–D). The LA 3.0 single flash ERG showed normal a-wave amplitudes in all subjects, but the slope of the a-wave was reduced, and implicit times were markedly delayed in two subjects (C and D); the same subjects showed reduced b-wave. The 30 Hz flicker responses were completely normal in two subjects (represented in A and B), and showed marked delay in two subjects (represented in C and D). These findings suggest reduced cone sensitivity in two subjects (C and D). Long duration stimulation (ON-OFF ERG) showed selective reduction of b-wave in two subjects (A and C), consistent with cone ON bipolar dysfunction; d-wave (OFF response) was normal in all three tested (A–C).
The ERG phenotype in all affected subjects is shown in Figure 2 and the Supplemental Data (including Table S3). Rod ERG findings suggested partial (sibling pair, p.Lys57del and p.Trp339∗; Figures 2A and 2B) or severe rod ON bipolar dysfunction (II-5, family A, p.Trp339∗ homozygous and IV-3, family B, p.Ser67Phe homozygous; Figures 2C and 2D). The single flash and 30 Hz LA ERGs were normal in the sibling pair. The single flash LA 3.0 ERG a-wave showed normal amplitudes, but implicit times were markedly delayed in two subjects (24 ms in II-5, family A and 31 ms in IV-3, family B [normal range: 14–17 ms]; Figures 2C and 2D). The subsequent b-wave showed reduced amplitudes and delayed implicit times (36 ms in II-5, family A and 49 ms in IV-3, family B [normal range 27–30 ms]). The LA 30 Hz flicker ERG implicit times were also delayed in these two individuals. The LA ERG findings in these two subjects are peculiar and reminiscent of the functional abnormalities seen in Gnb3-null mice; this observation is suggestive of reduced cone photoreceptor sensitivity, atypical of CSNB.42 Further testing in individual II-5 (family A) confirmed this reduced cone sensitivity ([log(Sc)] = 0.83 [normal range: 1.59–2.17]); however, cone photoreceptor maximal amplitude (Rmp3 = −97μV) was normal in this individual (Supplemental Data). The normal or borderline LA 30 Hz flicker amplitudes in the two subjects (II-5, family A and IV-3, family B) suggest relative preservation of cone OFF-pathway. Long duration stimulus testing on family A confirmed cone OFF-pathway preservation (normal d-wave; Figures 2A–2C); two subjects showed selective cone ON bipolar dysfunction (reduced b-wave; Figures 2A and 2C).
The GNB3 sequence is extremely conserved across species and has a homology of 100%, 98%, and 92% with macaques, mice, and zebrafish respectively.44 In the mammalian retina, GNB3 immuno-localizes to the ON bipolar cells and cone photoreceptors in multiple species, including mice, macaques, and dogs.35, 44, 45, 46 The Gnb3-null mice show partial preservation of ON bipolar pathway signaling as seen in the sibling pair (III-1 and III-2, family A).35 In addition to the ON bipolar defect, the Gnb3-null mice show reduced cone sensitivity as seen in two subjects in the study (II-5, family A, p.Trp339∗ homozygous and IV-3, family B, p.Ser67Phe homozygous).42 The coexistence of cone sensitivity deficit and ON bipolar signaling deficit defines a unique retinal disorder with cone-receptoral and rod and cone post-receptoral phenotypes.
G proteins are heterotrimeric proteins consisting of single α, β, and γ subunits. There are at least 20 different Gα, 6 different Gβ, and 13 different Gγ subunits that associate together to form specific heterotrimer combinations and perform unique functions.35, 37, 42, 47 In the mammalian photoreceptors, light-activated opsins stimulate G-protein transducins. Rods have Gαt1β1γ1 as their G protein transducin, whereas cones have Gαt2β3γt2 as their G protein transducin.45, 46, 47, 48, 49 The G protein transducin activates phosphodiesterase, leading to a reduction in the levels of cGMP, closure of cGMP-gated channels, and membrane hyperpolarization. Hyperpolarization of the photoreceptors leads to decreased release of glutamate into the synapse, which deactivates the metabotropic glutamate receptor 6 (GRM6) on the ON bipolar cells (rods and cones). This activates the heterotrimeric G protein Gαoβ3γ13 to release the Gαo subunit to open the non-selective channel TRPM1, leading to ON bipolar cell depolarization.50, 51, 52, 53, 54, 55, 56, 57, 58 Hence, the Gβ3 subunit is specifically involved in modulating cone transducin function as well as cone and rod ON bipolar cell signaling. Consequently, severe biallelic mutations in GNB3 likely cause dysfunction of the G protein in cones (Gαt2β3γt2) and in rod and cone ON bipolar cells (Gαoβ3γ13); this causes the unique dual phenotype of CSNB with reduced cone sensitivity. The in-frame deletion (p.Lys57del) might confer some residual binding capability of the Gβ3 to the γ13 and γt2 subunits in the ON bipolar cells and cones, respectively; this might account for the residual ON bipolar signaling and relatively preserved cone function in the compound heterozygote individuals (sibling pair in family A, p.Lys57del and p.Trp339∗). The partial preservation of cone ON bipolar signaling could explain the absence of a broadened trough of LA 3.0 single flash and flicker ERGs in the sibling pair.
In any G protein heterotrimer, Gβ and Gγ subunits functionally form a monomer that cannot be dissociated under normal conditions. Truncating mutations at any point in Gβ3 would prevent correct assembly of the Gβγ functional monomer, thereby compromising G protein function, which would be the case for null alleles in GNB3 (p.Trp339∗ homozygous).37, 59 The ON bipolar cells in Gnb3-null mice showed reduced immuno-localization of other G protein subunits (Gαo and Gγ13), glutamate receptor (GRM6), and ion channel (TRPM1).35 Mutations in TRPM1 and GRM6 cause ON bipolar signaling dysfunction (cCSNB) in humans; animal models of Trpm1, Grm6, Gαo, and Gβ3 also cause an ON bipolar signaling defect.19, 20, 21, 22, 35, 53, 60, 61, 62, 63, 64 The human GNB3-mutant phenotype showed consistent ON bipolar involvement, but the severity was variable, which seems to correlate with the type of mutations. The cone photoreceptors in Gnb3-null mice showed reduced expression of other G protein subunits (Gαt2 and Gγt2), but members of phototransduction cascade (opsins, CNGA3, CNGB3, and RGS9BP) were normally expressed and localized. Hence, the Gnb3-null mice showed reduced cone sensitivity, but saturated cone response was normal.42 The human phenotype variably demonstrated cone sensitivity deficit; the maximal cone photoreceptor response was normal. Unlike in mammals, Gnb3 is expressed in both rods and cones in chicken retinas; a progressive retinal degeneration has been reported in aged Gnb3-null chicken.35, 44, 65 Of note, a prior study that investigated a wide range of generalized photoreceptor dystrophies and macular dystrophy (including MIM: 248200) identified no mutations in GNB3.66
A single nucleotide polymorphism in GNB3 (c.825C>T, rs5443, allele frequency: 35.98%) has been associated with an increased risk of multifactorial disease, such as hypertension (MIM: 145500], depression (MIM: 608516), obesity (MIM: 601665), and Alzheimer disease (MIM: 104300).67, 68, 69, 70, 71 An unbalanced translocation between chromosomes 8 and 12, der(8)t(8;12) (p23.1;p13.31), has been associated with childhood obesity syndrome; GNB3 is one of the 107 genes involved, and transgenic mouse models carrying duplicate copies of GNB3 were obese.72 A gain of function of GNB3 is the proposed mechanism conferring risk in these disorders.67, 72 The retinal phenotype discussed in the present study is autosomal recessive in inheritance and functional loss of GNB3 is the disease mechanism; all heterozygous carriers in family A were unaffected. None of the six individuals who underwent WES (family A) or the proband in family B had the c.825C>T variant in GNB3. None of the ten members in Family A or the proband in Family B had hypertension, depression, obesity, or Alzheimer disease.
To summarize, we identify an unusual and unique stationary retinal disorder with dual anomaly in visual processing associated with biallelic mutations in GNB3. The anomalies include partial or severe defects in ON bipolar signaling and variably reduced cone sensitivity. This GNB3-mutant phenotype commonly presents with childhood-onset night blindness and middle-age-onset photophobia, but affected individuals have near normal vision, absent nystagmus, and no high myopia. The rod system always shows evidence of an ON signaling defect; this can range from severe (as in cCSNB) to mild deficits. The cone ON bipolar signaling deficit is variable (severe, mild, or none); specialized ERG techniques (long-duration stimulus) are needed to uncover this deficit. Cone photoreceptor sensitivity is reduced in a subset of cases. Some features of the disease are reminiscent of cCSNB; these include night-blindness, the non-progressive nature of the disease, and selective involvement of the ON bipolar signaling pathway. G proteins are cardinal second messengers in signal transduction; GNB3 is the third G protein subunit to be associated with a Mendelian eye disease, after GNAT1 (Gαt1, rod α transducin; CSNB) and GNAT2 (Gαt2, cone α transducin; achromatopsia). This report enhances our knowledge about visual signal transduction.
Consortia
The members of the GNB3 Consortium are Eyal Banin, Beatrice Bocquet, Elfride De Baere, Ingele Casteels, Sabine Defoort-Dhellemmes, Isabelle Drumare, Christoph Friedburg, Irene Gottlob, Samuel G. Jacobson, Ulrich Kellner, Robert Koenekoop, Susanne Kohl, Bart P. Leroy, Birgit Lorenz, Rebecca McLean, Francoise Meire, Isabelle Meunier, Francis Munier, Thomy de Ravel, Charlotte M. Reiff, Saddek Mohand-Saïd, Dror Sharon, Daniel Schorderet, Sharon Schwartz, and Xavier Zanlonghi.
Acknowledgments
This work was supported by the Mira Godard Research fund (E.H.) and the Ophthalmology Research Fund (The Hospital for Sick Children). Funding for whole-exome sequencing was provided by the University of Toronto McLaughlin Centre. The authors acknowledge the contribution of the high-throughput sequencing platform of The Centre for Applied Genomics, Toronto, Canada. Some of the DNA samples included in this study originate from the NeuroSensCol DNA bank, part of the BioCollections network for research in neuroscience (principal investigator J.A. Sahel, co-principal investigator I. Audo, partner with Centre Hospitalier National d’Ophtalmologie [CHNO] des Quinze-Vingts, INSERM, and CNRS). The study was supported by the Fondation Voir et Entendre (C.Z.), Prix Dalloz for “La recherche en ophtalmologie” (C.Z.), LABEX LIFESENSES (reference ANR-10-LABX-65) supported by French state funds managed by the Agence Nationale de la Recherche within the Investissements d’Avenir program (ANR-11-IDEX-0004-0), a Foundation Fighting Blindness center grant (C-CMM-0907-0428-INSERM04), and Prix de la Fondation de l’Œil (I.A.). The authors acknowledge the families for their active participation in the study.
Published: April 7, 2016
Footnotes
Supplemental Data include Supplemental Material and Methods and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.03.021.
Contributor Information
Elise Héon, Email: elise.heon@sickkids.ca.
GNB3 Consortium:
Eyal Banin, Beatrice Bocquet, Elfride De Baere, Ingele Casteels, Sabine Defoort-Dhellemmes, Isabelle Drumare, Christoph Friedburg, Irene Gottlob, Samuel G. Jacobson, Ulrich Kellner, Robert Koenekoop, Susanne Kohl, Bart P. Leroy, Birgit Lorenz, Rebecca McLean, Francoise Meire, Isabelle Meunier, Francis Munier, Thomy de Ravel, Charlotte M. Reiff, Saddek Mohand-Saïd, Dror Sharon, Daniel Schorderet, Sharon Schwartz, and Xavier Zanlonghi
Web Resources
The URLs for data presented herein are as follows:
ExAC Browser, http://exac.broadinstitute.org/
GATK v.3.2.2, https://www.broadinstitute.org/gatk/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
MutationTaster, http://www.mutationtaster.org/
NCBI HomoloGene, http://www.ncbi.nlm.nih.gov/homologene
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
Picard tools v.1.112, MarkDuplicates, http://broadinstitute.github.io/picard/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Zeitz C., Robson A.G., Audo I. Congenital stationary night blindness: an analysis and update of genotype-phenotype correlations and pathogenic mechanisms. Prog. Retin. Eye Res. 2015;45:58–110. doi: 10.1016/j.preteyeres.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 2.McCulloch D.L., Marmor M.F., Brigell M.G., Hamilton R., Holder G.E., Tzekov R., Bach M. ISCEV Standard for full-field clinical electroretinography (2015 update) Doc. Ophthalmol. 2015;130:1–12. doi: 10.1007/s10633-014-9473-7. [DOI] [PubMed] [Google Scholar]
- 3.Frishman L.J. Origins of the Electroretinogram. In: Heckenlively J., Arden G.B., editors. Principles and Practice of Clinical Electrophysiology of Vision. Second Edition. MIT Press; 2006. pp. 139–185. [Google Scholar]
- 4.Bush R.A., Sieving P.A. A proximal retinal component in the primate photopic ERG a-wave. Invest. Ophthalmol. Vis. Sci. 1994;35:635–645. [PubMed] [Google Scholar]
- 5.Bush R.A., Sieving P.A. Inner retinal contributions to the primate photopic fast flicker electroretinogram. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 1996;13:557–565. doi: 10.1364/josaa.13.000557. [DOI] [PubMed] [Google Scholar]
- 6.Hood D.C., Birch D.G. Beta wave of the scotopic (rod) electroretinogram as a measure of the activity of human on-bipolar cells. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 1996;13:623–633. doi: 10.1364/josaa.13.000623. [DOI] [PubMed] [Google Scholar]
- 7.Sieving P.A., Murayama K., Naarendorp F. Push-pull model of the primate photopic electroretinogram: a role for hyperpolarizing neurons in shaping the b-wave. Vis. Neurosci. 1994;11:519–532. doi: 10.1017/s0952523800002431. [DOI] [PubMed] [Google Scholar]
- 8.Riggs L.A. Electroretinography in cases of night blindness. Am. J. Ophthalmol. 1954;38:70–78. doi: 10.1016/0002-9394(54)90011-2. [DOI] [PubMed] [Google Scholar]
- 9.Schubert G., Bornschein H. Analysis of the human electroretinogram. Ophthalmologica. 1952;123:396–413. doi: 10.1159/000301211. [DOI] [PubMed] [Google Scholar]
- 10.Dryja T.P., Berson E.L., Rao V.R., Oprian D.D. Heterozygous missense mutation in the rhodopsin gene as a cause of congenital stationary night blindness. Nat. Genet. 1993;4:280–283. doi: 10.1038/ng0793-280. [DOI] [PubMed] [Google Scholar]
- 11.Gal A., Orth U., Baehr W., Schwinger E., Rosenberg T. Heterozygous missense mutation in the rod cGMP phosphodiesterase beta-subunit gene in autosomal dominant stationary night blindness. Nat. Genet. 1994;7:551. doi: 10.1038/ng0894-551a. [DOI] [PubMed] [Google Scholar]
- 12.Dryja T.P., Hahn L.B., Reboul T., Arnaud B. Missense mutation in the gene encoding the alpha subunit of rod transducin in the Nougaret form of congenital stationary night blindness. Nat. Genet. 1996;13:358–360. doi: 10.1038/ng0796-358. [DOI] [PubMed] [Google Scholar]
- 13.Riazuddin S.A., Shahzadi A., Zeitz C., Ahmed Z.M., Ayyagari R., Chavali V.R., Ponferrada V.G., Audo I., Michiels C., Lancelot M.E. A mutation in SLC24A1 implicated in autosomal-recessive congenital stationary night blindness. Am. J. Hum. Genet. 2010;87:523–531. doi: 10.1016/j.ajhg.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naeem M.A., Chavali V.R., Ali S., Iqbal M., Riazuddin S., Khan S.N., Husnain T., Sieving P.A., Ayyagari R., Riazuddin S. GNAT1 associated with autosomal recessive congenital stationary night blindness. Invest. Ophthalmol. Vis. Sci. 2012;53:1353–1361. doi: 10.1167/iovs.11-8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neuillé M., Malaichamy S., Vadalà M., Michiels C., Condroyer C., Sachidanandam R., Srilekha S., Arokiasamy T., Letexier M., Démontant V. Next-generation sequencing confirms the implication of SLC24A1 in autosomal-recessive congenital stationary night blindness (CSNB) Clin. Genet. 2016 doi: 10.1111/cge.12746. Published online January 29, 2016. [DOI] [PubMed] [Google Scholar]
- 16.Miyake Y., Yagasaki K., Horiguchi M., Kawase Y., Kanda T. Congenital stationary night blindness with negative electroretinogram. A new classification. Arch. Ophthalmol. 1986;104:1013–1020. doi: 10.1001/archopht.1986.01050190071042. [DOI] [PubMed] [Google Scholar]
- 17.Bech-Hansen N.T., Naylor M.J., Maybaum T.A., Sparkes R.L., Koop B., Birch D.G., Bergen A.A., Prinsen C.F., Polomeno R.C., Gal A. Mutations in NYX, encoding the leucine-rich proteoglycan nyctalopin, cause X-linked complete congenital stationary night blindness. Nat. Genet. 2000;26:319–323. doi: 10.1038/81619. [DOI] [PubMed] [Google Scholar]
- 18.Pusch C.M., Zeitz C., Brandau O., Pesch K., Achatz H., Feil S., Scharfe C., Maurer J., Jacobi F.K., Pinckers A. The complete form of X-linked congenital stationary night blindness is caused by mutations in a gene encoding a leucine-rich repeat protein. Nat. Genet. 2000;26:324–327. doi: 10.1038/81627. [DOI] [PubMed] [Google Scholar]
- 19.Dryja T.P., McGee T.L., Berson E.L., Fishman G.A., Sandberg M.A., Alexander K.R., Derlacki D.J., Rajagopalan A.S. Night blindness and abnormal cone electroretinogram ON responses in patients with mutations in the GRM6 gene encoding mGluR6. Proc. Natl. Acad. Sci. USA. 2005;102:4884–4889. doi: 10.1073/pnas.0501233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z., Sergouniotis P.I., Michaelides M., Mackay D.S., Wright G.A., Devery S., Moore A.T., Holder G.E., Robson A.G., Webster A.R. Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am. J. Hum. Genet. 2009;85:711–719. doi: 10.1016/j.ajhg.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Genderen M.M., Bijveld M.M., Claassen Y.B., Florijn R.J., Pearring J.N., Meire F.M., McCall M.A., Riemslag F.C., Gregg R.G., Bergen A.A., Kamermans M. Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. Am. J. Hum. Genet. 2009;85:730–736. doi: 10.1016/j.ajhg.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Audo I., Kohl S., Leroy B.P., Munier F.L., Guillonneau X., Mohand-Saïd S., Bujakowska K., Nandrot E.F., Lorenz B., Preising M. TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2009;85:720–729. doi: 10.1016/j.ajhg.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Audo I., Bujakowska K., Orhan E., Poloschek C.M., Defoort-Dhellemmes S., Drumare I., Kohl S., Luu T.D., Lecompte O., Zrenner E. Whole-exome sequencing identifies mutations in GPR179 leading to autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2012;90:321–330. doi: 10.1016/j.ajhg.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peachey N.S., Ray T.A., Florijn R., Rowe L.B., Sjoerdsma T., Contreras-Alcantara S., Baba K., Tosini G., Pozdeyev N., Iuvone P.M. GPR179 is required for depolarizing bipolar cell function and is mutated in autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2012;90:331–339. doi: 10.1016/j.ajhg.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeitz C., Jacobson S.G., Hamel C.P., Bujakowska K., Neuillé M., Orhan E., Zanlonghi X., Lancelot M.E., Michiels C., Schwartz S.B., Congenital Stationary Night Blindness Consortium Whole-exome sequencing identifies LRIT3 mutations as a cause of autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2013;92:67–75. doi: 10.1016/j.ajhg.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strom T.M., Nyakatura G., Apfelstedt-Sylla E., Hellebrand H., Lorenz B., Weber B.H., Wutz K., Gutwillinger N., Rüther K., Drescher B. An L-type calcium-channel gene mutated in incomplete X-linked congenital stationary night blindness. Nat. Genet. 1998;19:260–263. doi: 10.1038/940. [DOI] [PubMed] [Google Scholar]
- 27.Bech-Hansen N.T., Naylor M.J., Maybaum T.A., Pearce W.G., Koop B., Fishman G.A., Mets M., Musarella M.A., Boycott K.M. Loss-of-function mutations in a calcium-channel alpha1-subunit gene in Xp11.23 cause incomplete X-linked congenital stationary night blindness. Nat. Genet. 1998;19:264–267. doi: 10.1038/947. [DOI] [PubMed] [Google Scholar]
- 28.Zeitz C., Kloeckener-Gruissem B., Forster U., Kohl S., Magyar I., Wissinger B., Mátyás G., Borruat F.X., Schorderet D.F., Zrenner E. Mutations in CABP4, the gene encoding the Ca2+-binding protein 4, cause autosomal recessive night blindness. Am. J. Hum. Genet. 2006;79:657–667. doi: 10.1086/508067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wycisk K.A., Zeitz C., Feil S., Wittmer M., Forster U., Neidhardt J., Wissinger B., Zrenner E., Wilke R., Kohl S., Berger W. Mutation in the auxiliary calcium-channel subunit CACNA2D4 causes autosomal recessive cone dystrophy. Am. J. Hum. Genet. 2006;79:973–977. doi: 10.1086/508944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marmor M.F., Fulton A.B., Holder G.E., Miyake Y., Brigell M., Bach M., International Society for Clinical Electrophysiology of Vision ISCEV Standard for full-field clinical electroretinography (2008 update) Doc. Ophthalmol. 2009;118:69–77. doi: 10.1007/s10633-008-9155-4. [DOI] [PubMed] [Google Scholar]
- 31.Dragas R., Westall C., Wright T. Changes in the ERG d-wave with vigabatrin treatment in a pediatric cohort. Doc. Ophthalmol. 2014;129:97–104. doi: 10.1007/s10633-014-9453-y. [DOI] [PubMed] [Google Scholar]
- 32.Vincent A., Forster N., Maynes J.T., Paton T.A., Billingsley G., Roslin N.M., Ali A., Sutherland J., Wright T., Westall C.A., FORGE Canada Consortium OTX2 mutations cause autosomal dominant pattern dystrophy of the retinal pigment epithelium. J. Med. Genet. 2014;51:797–805. doi: 10.1136/jmedgenet-2014-102620. [DOI] [PubMed] [Google Scholar]
- 33.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhingra A., Ramakrishnan H., Neinstein A., Fina M.E., Xu Y., Li J., Chung D.C., Lyubarsky A., Vardi N. Gβ3 is required for normal light ON responses and synaptic maintenance. J. Neurosci. 2012;32:11343–11355. doi: 10.1523/JNEUROSCI.1436-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ansari-Lari M.A., Muzny D.M., Lu J., Lu F., Lilley C.E., Spanos S., Malley T., Gibbs R.A. A gene-rich cluster between the CD4 and triosephosphate isomerase genes at human chromosome 12p13. Genome Res. 1996;6:314–326. doi: 10.1101/gr.6.4.314. [DOI] [PubMed] [Google Scholar]
- 37.Clapham D.E., Neer E.J. G protein beta gamma subunits. Annu. Rev. Pharmacol. Toxicol. 1997;37:167–203. doi: 10.1146/annurev.pharmtox.37.1.167. [DOI] [PubMed] [Google Scholar]
- 38.Neer E.J., Schmidt C.J., Nambudripad R., Smith T.F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 39.Sondek J., Bohm A., Lambright D.G., Hamm H.E., Sigler P.B. Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 40.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheever M.L., Snyder J.T., Gershburg S., Siderovski D.P., Harden T.K., Sondek J. Crystal structure of the multifunctional Gbeta5-RGS9 complex. Nat. Struct. Mol. Biol. 2008;15:155–162. doi: 10.1038/nsmb.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikonov S.S., Lyubarsky A., Fina M.E., Nikonova E.S., Sengupta A., Chinniah C., Ding X.Q., Smith R.G., Pugh E.N., Jr., Vardi N., Dhingra A. Cones respond to light in the absence of transducin β subunit. J. Neurosci. 2013;33:5182–5194. doi: 10.1523/JNEUROSCI.5204-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bijveld M.M., Florijn R.J., Bergen A.A., van den Born L.I., Kamermans M., Prick L., Riemslag F.C., van Schooneveld M.J., Kappers A.M., van Genderen M.M. Genotype and phenotype of 101 dutch patients with congenital stationary night blindness. Ophthalmology. 2013;120:2072–2081. doi: 10.1016/j.ophtha.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Ritchey E.R., Bongini R.E., Code K.A., Zelinka C., Petersen-Jones S., Fischer A.J. The pattern of expression of guanine nucleotide-binding protein beta3 in the retina is conserved across vertebrate species. Neuroscience. 2010;169:1376–1391. doi: 10.1016/j.neuroscience.2010.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng Y.W., Robishaw J.D., Levine M.A., Yau K.W. Retinal rods and cones have distinct G protein beta and gamma subunits. Proc. Natl. Acad. Sci. USA. 1992;89:10882–10886. doi: 10.1073/pnas.89.22.10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee R.H., Lieberman B.S., Yamane H.K., Bok D., Fung B.K. A third form of the G protein beta subunit. 1. Immunochemical identification and localization to cone photoreceptors. J. Biol. Chem. 1992;267:24776–24781. [PubMed] [Google Scholar]
- 47.Larhammar D., Nordström K., Larsson T.A. Evolution of vertebrate rod and cone phototransduction genes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:2867–2880. doi: 10.1098/rstb.2009.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lerea C.L., Somers D.E., Hurley J.B., Klock I.B., Bunt-Milam A.H. Identification of specific transducin alpha subunits in retinal rod and cone photoreceptors. Science. 1986;234:77–80. doi: 10.1126/science.3529395. [DOI] [PubMed] [Google Scholar]
- 49.Ong O.C., Yamane H.K., Phan K.B., Fong H.K., Bok D., Lee R.H., Fung B.K. Molecular cloning and characterization of the G protein gamma subunit of cone photoreceptors. J. Biol. Chem. 1995;270:8495–8500. doi: 10.1074/jbc.270.15.8495. [DOI] [PubMed] [Google Scholar]
- 50.Schneider F.M., Mohr F., Behrendt M., Oberwinkler J. Properties and functions of TRPM1 channels in the dendritic tips of retinal ON-bipolar cells. Eur. J. Cell Biol. 2015;94:420–427. doi: 10.1016/j.ejcb.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Euler T., Haverkamp S., Schubert T., Baden T. Retinal bipolar cells: elementary building blocks of vision. Nat. Rev. Neurosci. 2014;15:507–519. doi: 10.1038/nrn3783. [DOI] [PubMed] [Google Scholar]
- 52.Koike C., Numata T., Ueda H., Mori Y., Furukawa T. TRPM1: a vertebrate TRP channel responsible for retinal ON bipolar function. Cell Calcium. 2010;48:95–101. doi: 10.1016/j.ceca.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 53.Dhingra A., Lyubarsky A., Jiang M., Pugh E.N., Jr., Birnbaumer L., Sterling P., Vardi N. The light response of ON bipolar neurons requires G[alpha]o. J. Neurosci. 2000;20:9053–9058. doi: 10.1523/JNEUROSCI.20-24-09053.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dhingra A., Jiang M., Wang T.L., Lyubarsky A., Savchenko A., Bar-Yehuda T., Sterling P., Birnbaumer L., Vardi N. Light response of retinal ON bipolar cells requires a specific splice variant of Galpha(o) J. Neurosci. 2002;22:4878–4884. doi: 10.1523/JNEUROSCI.22-12-04878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okawa H., Pahlberg J., Rieke F., Birnbaumer L., Sampath A.P. Coordinated control of sensitivity by two splice variants of Gα(o) in retinal ON bipolar cells. J. Gen. Physiol. 2010;136:443–454. doi: 10.1085/jgp.201010477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koike C., Obara T., Uriu Y., Numata T., Sanuki R., Miyata K., Koyasu T., Ueno S., Funabiki K., Tani A. TRPM1 is a component of the retinal ON bipolar cell transduction channel in the mGluR6 cascade. Proc. Natl. Acad. Sci. USA. 2010;107:332–337. doi: 10.1073/pnas.0912730107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morgans C.W., Brown R.L., Duvoisin R.M. TRPM1: the endpoint of the mGluR6 signal transduction cascade in retinal ON-bipolar cells. BioEssays. 2010;32:609–614. doi: 10.1002/bies.200900198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang L., Max M., Margolskee R.F., Su H., Masland R.H., Euler T. G protein subunit G gamma 13 is coexpressed with G alpha o, G beta 3, and G beta 4 in retinal ON bipolar cells. J. Comp. Neurol. 2003;455:1–10. doi: 10.1002/cne.10396. [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Higuera I., Fenoglio J., Li Y., Lewis C., Panchenko M.P., Reiner O., Smith T.F., Neer E.J. Folding of proteins with WD-repeats: comparison of six members of the WD-repeat superfamily to the G protein beta subunit. Biochemistry. 1996;35:13985–13994. doi: 10.1021/bi9612879. [DOI] [PubMed] [Google Scholar]
- 60.Bellone R.R., Brooks S.A., Sandmeyer L., Murphy B.A., Forsyth G., Archer S., Bailey E., Grahn B. Differential gene expression of TRPM1, the potential cause of congenital stationary night blindness and coat spotting patterns (LP) in the Appaloosa horse (Equus caballus) Genetics. 2008;179:1861–1870. doi: 10.1534/genetics.108.088807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morgans C.W., Zhang J., Jeffrey B.G., Nelson S.M., Burke N.S., Duvoisin R.M., Brown R.L. TRPM1 is required for the depolarizing light response in retinal ON-bipolar cells. Proc. Natl. Acad. Sci. USA. 2009;106:19174–19178. doi: 10.1073/pnas.0908711106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen Y., Heimel J.A., Kamermans M., Peachey N.S., Gregg R.G., Nawy S. A transient receptor potential-like channel mediates synaptic transmission in rod bipolar cells. J. Neurosci. 2009;29:6088–6093. doi: 10.1523/JNEUROSCI.0132-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Masu M., Iwakabe H., Tagawa Y., Miyoshi T., Yamashita M., Fukuda Y., Sasaki H., Hiroi K., Nakamura Y., Shigemoto R. Specific deficit of the ON response in visual transmission by targeted disruption of the mGluR6 gene. Cell. 1995;80:757–765. doi: 10.1016/0092-8674(95)90354-2. [DOI] [PubMed] [Google Scholar]
- 64.Koyasu T., Kondo M., Miyata K., Ueno S., Miyata T., Nishizawa Y., Terasaki H. Photopic electroretinograms of mGluR6-deficient mice. Curr. Eye Res. 2008;33:91–99. doi: 10.1080/02713680701823232. [DOI] [PubMed] [Google Scholar]
- 65.Tummala H., Ali M., Getty P., Hocking P.M., Burt D.W., Inglehearn C.F., Lester D.H. Mutation in the guanine nucleotide-binding protein beta-3 causes retinal degeneration and embryonic mortality in chickens. Invest. Ophthalmol. Vis. Sci. 2006;47:4714–4718. doi: 10.1167/iovs.06-0292. [DOI] [PubMed] [Google Scholar]
- 66.Gao Y.Q., Danciger M., Akhmedov N.B., Zhao D.Y., Heckenlively J.R., Fishman G.A., Weleber R.G., Jacobson S.G., Farber D.B. Exon screening of the genes encoding the beta- and gamma-subunits of cone transducin in patients with inherited retinal disease. Mol. Vis. 1998;4:16. [PubMed] [Google Scholar]
- 67.Siffert W., Rosskopf D., Siffert G., Busch S., Moritz A., Erbel R., Sharma A.M., Ritz E., Wichmann H.E., Jakobs K.H., Horsthemke B. Association of a human G-protein beta3 subunit variant with hypertension. Nat. Genet. 1998;18:45–48. doi: 10.1038/ng0198-45. [DOI] [PubMed] [Google Scholar]
- 68.Zill P., Baghai T.C., Zwanzger P., Schüle C., Minov C., Riedel M., Neumeier K., Rupprecht R., Bondy B. Evidence for an association between a G-protein beta3-gene variant with depression and response to antidepressant treatment. Neuroreport. 2000;11:1893–1897. doi: 10.1097/00001756-200006260-00018. [DOI] [PubMed] [Google Scholar]
- 69.Siffert W., Forster P., Jöckel K.H., Mvere D.A., Brinkmann B., Naber C., Crookes R., Du P Heyns A., Epplen J.T., Fridey J. Worldwide ethnic distribution of the G protein beta3 subunit 825T allele and its association with obesity in Caucasian, Chinese, and Black African individuals. J. Am. Soc. Nephrol. 1999;10:1921–1930. doi: 10.1681/ASN.V1091921. [DOI] [PubMed] [Google Scholar]
- 70.Bullido M.J., Ramos M.C., Ruiz-Gómez A., Tutor A.S., Sastre I., Frank A., Coria F., Gil P., Mayor F., Jr., Valdivieso F. Polymorphism in genes involved in adrenergic signaling associated with Alzheimer’s. Neurobiol. Aging. 2004;25:853–859. doi: 10.1016/j.neurobiolaging.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 71.Weinstein L.S., Chen M., Xie T., Liu J. Genetic diseases associated with heterotrimeric G proteins. Trends Pharmacol. Sci. 2006;27:260–266. doi: 10.1016/j.tips.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 72.Goldlust I.S., Hermetz K.E., Catalano L.M., Barfield R.T., Cozad R., Wynn G., Ozdemir A.C., Conneely K.N., Mulle J.G., Dharamrup S., Unique Rare Chromosome Disorder Support Group Mouse model implicates GNB3 duplication in a childhood obesity syndrome. Proc. Natl. Acad. Sci. USA. 2013;110:14990–14994. doi: 10.1073/pnas.1305999110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.