Abstract
The highly pathogenic avian influenza strain H5N1 was first detected in Europe in 2005, and has since been documented continent-wide in wild birds and poultry. However, the relative roles of each host group in transmission remain contentious. Using recently developed tools for analysis of ecological niches and geographic distributions of species, we compared ecological niche requirements for H5N1 between paired host groups (poultry versus wild birds, Anseriformes versus Falconiformes, swans versus non-swan Anseriformes). If environmental signals of different host groups are significantly different, the groups are likely to be involved in distinct transmission cycles. In contrast, models for which similarity cannot be rejected imply no unique ecological niches and no potential linkage of transmission cycles. In 24 similarity tests, we found significant similarity (13/24) or no significant differences (9/24). Although 2 of the 24 analyses showed significant differences, neither was unequivocal, so we conclude an overall signal of niche similarity among groups. We thus could not document distinct ecological niches for H5N1 occurrences in different host groups and conclude that the transmission cycles are broadly interwoven.
Keywords: ecological niche, highly pathogenic avian influenza strain H5N1, host group, niche similarity, Europe
Introduction
The highly-pathogenic avian influenza strain H5N1 spread from Southeast Asia and East Asia into Europe and Africa in 2005 (Gilbert et al., 2008). H5N1 has now been detected in 62 countries, including 26 in Europe (OIE, 2009). European H5N1 outbreaks appear to differ from those in other regions in that numbers of distinct outbreaks in wild birds and poultry are roughly equal: 379 reports for domestic birds against 294 in wild birds for 2006. This apparent leap in numbers of wild birds infected may be explained in part by more intensive surveillance than elsewhere, as nearly 121,000 wild birds were sampled in the European Union (EU) in 2006 (Hesterberg et al., 2009). Alternatively, these contrasts may point to viral evolution after 2005 (Whitworth et al., 2007), and consequent increased pathogenicity to wild bird species. For this reason, ecological or environmental differences among H5N1 strains transmitted in different host groups are of considerable interest.
Questions regarding the relative importance and linkage of transmission among wild birds and poultry remain controversial (Weber and Stilianakis, 2007; Keawcharoen et al., 2008) although both are likely to play roles (Kilpatrick et al., 2006). Avian influenza strains are known to be maintained in wild aquatic bird reservoirs (Swayne and Suarez, 2000). For example, Anas platyrhynchos has been identified as a possible H5N1 reservoir (Brown et al., 2006). Other factors, implicated in the transmission cycle include illegal wild animal-smuggling (Van Borm et al., 2005), infected poultry feed (Harder et al., 2009), and undocumented poultry trade.
Previous studies have attempted to understand regional-scale H5N1 ecology and geography, implicating various factors as correlates of H5N1 occurrence, e.g. duck abundance and rice cropping intensity in Southeast Asia (Gilbert et al., 2008), seasonal patterns of migration of wild birds (Si et al., 2009) and high seasonal variation in the values of the normalized difference vegetation index (NDVI) in studies in Africa and the Middle East (Williams et al., 2008; Williams and Peterson, 2009). Factors important to H5N1 transmission probably vary among regions. For example, Germany has suffered annual outbreaks since 2006 despite negligible rice cultivation and few free-grazing domestic ducks (Eurostat, 2008). Ecological niche models (ENMs) and associated techniques offer unique opportunities to study ecological associations of biological phenomena across broad regions. ENMs reconstruct coarse-resolution environmental and ecological requirements that determine geographic distributions (Soberón and Peterson, 2005), and have been used to explore diverse topics in distributional ecology, including disease distributions and transmission risks (Williams et al., 2008). Here, we use ENM-based niche-comparison approaches (Warren et al., 2008) with the objective to develop detailed comparisons of ecological niches among different potential host groups for H5N1 and to produce a first quantitative test of environmental connectivity of H5N1 transmission between wild and domestic birds.
Material and methods
Input data
Occurrence data were drawn from OIE (2009), which compiled 494 poultry (including chickens, ducks, geese, quail and turkeys, kept at all biosecurity levels) and 605 wild birds (of at least 21 species) with laboratory-confirmed highly pathogenic H5N1 detections across our study region (0°-58°E, 34°-60°N) during 2005–2008. The vast majority of the cases were detected due to host death, though a few cases were detected through surveillance of poultry or wild birds. We discarded points duplicated spatially at 0.01° resolution. To minimise problems caused by the non-independence of cases, we also rarefied points within spatial clusters to densities similar to those of surrounding areas, leaving a total of 89 poultry and 90 wild bird occurrence points for analysis (Table 1). Separately, we explored subsets of the wild bird data, to permit comparisons of wild Anseriformes (n = 102) versus wild Falconiformes (n = 21), and wild Cygnus spp. (n = 63) versus wild non-swan Anseriformes (n = 39). Because these latter data sets did not appear to manifest clumped distributions, we included them in their entirety.
Table 1.
Numbers of H5N1 cases used for generating, and area under the curve (AUC) values generated by each ecological niche models (ENM).
| Test group | Sample size | AUC
|
|
|---|---|---|---|
| Climate | Remotely sensed | ||
| Poultry | 89 | 0.841 | 0.898 |
| Wild birds | 90 | 0.907 | 0.926 |
| Anseriformes | 102 | 0.943 | 0.938 |
| Falconiformes | 21 | 0.977 | 0.960 |
| Cygnus | 63 | 0.907 | 0.875 |
| non-Cygnus Anseriformes | 39 | 0.936 | 0.908 |
ENMs were developed using two distinct and comparable suites of environmental data layers. The first was based on multi-temporal remotely sensed vegetation indices derived from the Moderate Resolution Imaging Spectroradiometer (MODIS) sensor (http://www.modis.gsfc.nasa.gov/) onboard the Terra satellite: NDVI (Tucker, 1979), enhanced vegetation index (EVI) (Huete et al., 2002), and land surface water index (LSWI) (Xiao et al., 2002). The NDVI, used to estimate vegetation growth and biomass production, is calculated as the following ratio of near infrared (NIR) and red spectral bands:
| (1) |
The EVI provides a similar vegetation measure, but corrects additionally for error due to aerosol reflectance and canopy background signals:
| (2) |
while the LSWI is calculated as the following ratio of near infrared and shortwave infrared (SWIR) bands:
| (3) |
In each case, we used maximum, mean, minimum and range for the year 2005, supplemented with information on topographic features, including aspect, compound topographic index and slope from the Hydro-1K digital elevation model data set (USGS, 2001), and elevation from GTOPO30 (all resampled to a spatial resolution of 0.01°) (USGS, 2006). The second data set consisted of 7 “bioclimatic” variables from the 10’ WorldClim data set (Hijmans et al., 2005): annual mean temperature, mean diurnal range, maximum temperature of warmest month, minimum temperature of coldest month, annual precipitation, precipitation of wettest month and precipitation of driest month (all resampled to a spatial resolution of 0.0083°).
Ecological niche models
ENMs have been developed via diverse methodological approaches, which have different strengths and weaknesses. For these analyses, we chose the algorithm Maxent, an approach that is known to perform particularly well in interpolation challenges (Elith et al., 2006). Maxent is a method for characterising probability distributions from incomplete information that has been applied as a method for estimating ecological niches and inferring species distributions from presence data (Phillips et al., 2006). Maxent has seen considerable success in model-comparison studies (Elith et al., 2006; Phillips et al., 2006). Although Maxent encounters challenges in estimating niches and geographic distributions across broad, unsampled regions (Peterson et al., 2007), it is excellent for relatively densely-sampled landscapes and interpolation challenges, as was the challenge in this study. A further benefit of using the Maxent algorithm is that it generates area under the curve (AUC) values as a measure of the predictive power of the model. Finally, Maxent is integral to the software available for background similarity testing that is central to this study (Warren et al., 2008). Maxent fits a probability distribution for occurrence of the biological phenomenon in question to the set of pixels across the study region, assuming that the best model will maximise the entropy of the probability distribution, subject to the constraint that pixel values where the biological phenomenon was detected should reflect presence at higher probability values. We used default parameters for compatibility with tools for testing niche similarity (see below), choosing logistic output format. Outputs were imported into ArcView, 3.3 (ESRI; Redlands, CA, USA) as floating-point grids, and then thresholded to the lowest predicted value associated with any known detection locality (Pearson et al., 2007).
Quantifying niche similarity
We followed Soberón (2007) and Warren et al. (2008) in considering the overlap between maps of habitat suitability in the environmental space as a measure of ecological niche similarity, using the D and I similarity metrics to quantify the similarity between two probability distributions. These statistics assume probability distributions defined over the geographic space, in which pX,i (or pY,i) denotes the probability assigned by the ENM for species X (or Y) to cell i. The D metric, Schoener’s statistic for niche overlap (Schoener, 1968), is calculated as:
| (4) |
The I metric was modified from the Hellinger distance (Van der Vaart, 1998) to be comparable to more conventional measures of niche overlap, and is calculated as:
| (5) |
The values for each metric range from 0 (no overlap) to 1 (identical). Following Warren et al. (2008), we used randomisation tests that ask whether two species’ modeled niches are more similar or more different than random expectations. Importantly, this allows specification of an area of analysis (“the background”), which we equated with M, i.e. the area accessible to a species over relevant time periods (Soberón and Peterson, 2005). Hence, using ENM Tools (Warren et al., 2008), we compared D and I values for paired ENMs for each of the paired groups of H5N1 hosts (poultry versus wild birds, Anseriformes versus Falconiformes and non-swans versus swans). We assumed a 300 km buffer around known case occurrences as a hypothesis of M. Analyses were conducted bi-directionally, so we compared the niche model for one host group to the background of the other, and vice versa. Numbers of points sampled from the background were set at observed sample sizes. In each test, 100 replicate randomisations were conducted to estimate probabilities for each test to estimate probabilities to the nearest 1%.
Given the unknown nature of the associations (positive or negative) between influenza occurrences between these groups, we used a two-tailed null hypothesis, i.e. niche overlap observed among host groups is not real difference, but rather explicable by differences in the background landscapes for each group. The hypothesis is rejected if observed similarity between models falls outside the 95% confidence limits of the null distribution, with greater than expected niche difference defined as <2.5%, and greater than expected similarity as >97.5%, of the distribution.
Results
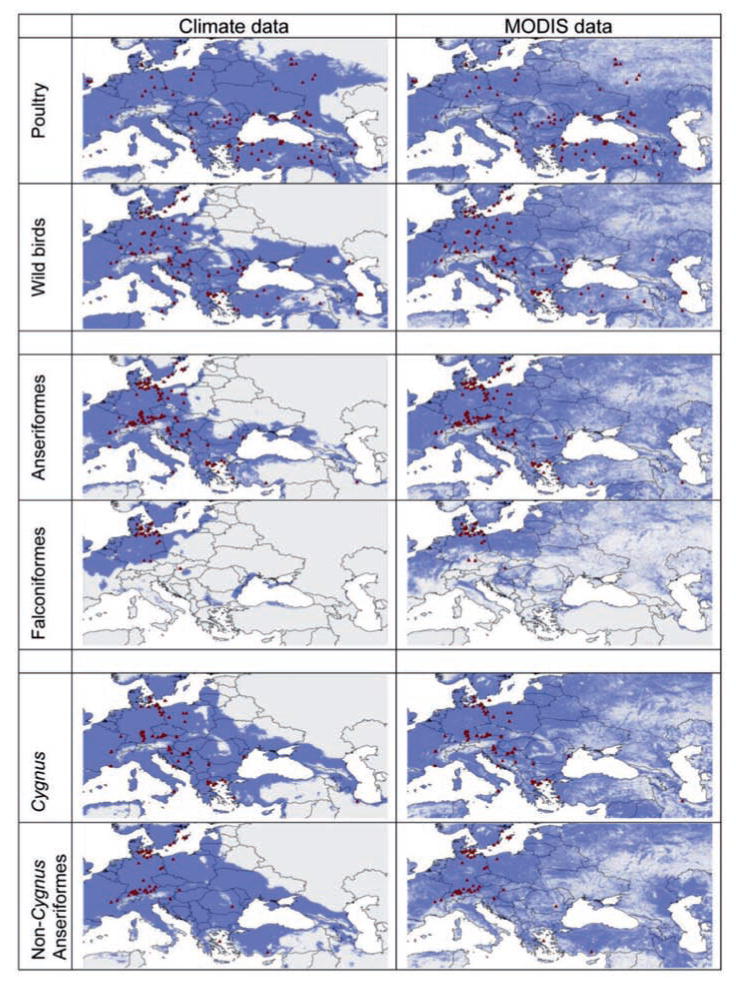
Niche models for H5N1 in poultry versus wild birds and swans versus other Anseriformes produced similar maps covering much of our study area, save for high-elevation regions. Models of occurrences in Anseriformes versus Falconiformes were less similar, with the latter omitting much of southern Europe. Generally, models based on climatic data were more restricted in areas predicted suitable than models based on remotely sensed data (Fig. 1). The predictive power of all models, as measured by AUC values, was high (0.840–0.970, see Table 1), although this approach has limitations (Lobo et al., 2008). Jackknife evaluations of variable contributions in each model are presented in Supplementary Material.
Fig. 1.
Ecological niche models (ENMs) for H5N1 detections in distinct avian groups across Europe. Dark grey indicates potential H5N1 presence (based on a least training presence threshold); light grey indicates absence. Triangles show cases used for model training. Each dataset was modeled using climatic and remotely sensed environmental datasets.
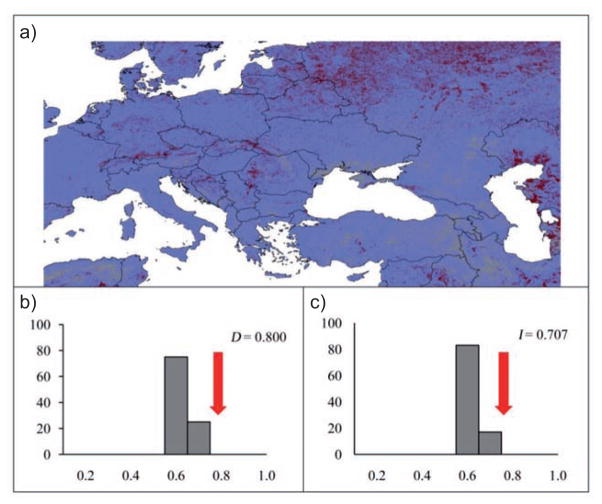
The background similarity tests, based on remotely sensed layers, used to assess whether two sets of occurrences were drawn from the same environmental niche, taking into account the availability of conditions across the region inhabited, indicated no ecological niche difference: seven of 12 pairs under comparison were more similar than random expectations, and in five of 12 pairs the null hypothesis could not be rejected (Table 2). Thus, in remotely sensed environmental dimensions, comparisons of ecological niches of poultry and wild birds, Anseriformes and Falconiformes, and Cygnus and non-Cygnus Anseriformes were unable to reject the null hypothesis of niche similarity. The similarity of niches between paired comparisons is shown by subtracting the remotely sensed ENM developed using H5N1 points detected in poultry from the remotely sensed wild bird model (Fig. 2a). Both background similarity tests assessing the H5N1 poultry versus wild bird niches were more similar than random expectations (Figs. 2b and 2c).
Table 2.
Background similarity tests using Schoener’s D and the Hellinger distance I for climatic and remotely sensed data. The null hypothesis of niche similarity is rejected if test statistic values are below 0.025 (indicated with asterisks); reciprocal tests are presented (A–B, B–A).
| Test caomarise | Climate
|
Remote sensing
|
|||||
|---|---|---|---|---|---|---|---|
| Overlap value | Background
|
Overlap value | Background
|
||||
| A – B | B – A | A – B | B – A | ||||
| APoultry versus | D | 0.676 | 0.00* | 0.90 | 0.800 | 1.00 | 1.00 |
| BWild birds | I | 0.649 | 0.00* | 0.31 | 0.707 | 1.00 | 0.99 |
| AAnseriformes versus | D | 0.476 | 0.21 | 1.00 | 0.357 | 0.19 | 0.94 |
| BFalconiformes | I | 0.661 | 0.94 | 1.00 | 0.538 | 0.19 | 1.00 |
| ACygnus versus | D | 0.785 | 0.95 | 0.78 | 0.754 | 0.73 | 1.00 |
| Bnon-Cygnus Anseriformes | I | 0.842 | 1.00 | 1.00 | 0.681 | 0.74 | 1.00 |
Fig. 2.
(a) Difference between suitability predicted for wild birds and poultry, based on remotely sensed environmental data. Grey indicates agreed prediction between models, white indicates areas predicted by poultry but not wild bird, and black indicates the reverse. (b) Histograms illustrating measured overlap (arrow) and distribution of background similarity among random replicate models for D, and (c) I metrics, for the wild bird remotely sensed model.
Background tests of niche similarity based on climate layers yielded similar, though more mixed, results (Table 2). The null hypothesis of similarity could not be rejected in comparisons of Falconiformes and Anseriformes, and swans and non-swan Anseriformes. However, although environments associated with wild bird and poultry H5N1 occurrences were indistinguishable from random similarity in two comparisons, they differed significantly in climatic dimensions in the remaining two comparisons.
In summary, in all but one randomisation test (that based on poultry data and climate layers), all pairs of model comparisons were found to be significantly similar (13/24), or not significant from random expectations (9/24). Although some climate-based comparisons of case occurrences in poultry and wild birds detected significant differences, they were contradicted by comparisons based on remotely sensed data. Hence, overall, the signal among our 24 comparisons was one of similarity among environments in which H5N1 was detected among host groups.
Discussion
Throughout our analyses, no clear signal emerged to suggest distinct ecological niches for H5N1 case occurrences in different host groups. Among wild bird groups, the picture was completely consistent: H5N1 cases in Falconiformes and Anseriformes, and cases in swans and non-swan Anseriformes, occurred under a single set of environmental conditions. This result points to functional linkage of transmission cycles and suggests that wild bird H5N1 case occurrences represent a single, coherent biological phenomenon. The picture was less clear for H5N1 in poultry and wild birds. Here, remotely sensed assessments detected greater-than-expected niche similarity but the climate-based tests were more complex: comparisons of poultry H5N1 cases against the background of wild bird cases were significantly non-similar, while the converse test was indistinguishable from random. Hence, most evidence (6/8 tests) point towards environmental similarity of wild bird and poultry H5N1 cases.
Occurrence data drawn from OIE (2009) did not systematically record the biosecurity level under which the hosts were raised, therefore, all poultry cases were grouped together to create a poultry model, rather than, for example, creating a “backyard bird” model and a “high biosecurity” model. Sometimes information on the affected population, and the number of susceptible individuals implied the biosecurity level under which the hosts were raised but frequently they did not. We felt that creating models based on the biosecurity level of host husbandry would be unreliably subjective. Moreover, the fact that H5N1 was detected in “high biosecurity” establishments, suggests failure of biosecurity measures at some level. Similarly, the OIE data did not record whether the affected population was raised under environmentally controlled conditions or not. Some industrial poultry facilities control the environment under which poultry are raised and this could conceivably be the case for up to 24/90 cases used for the poultry model (though probably fewer). It is possible that the effect of environmental (NDVI, EVI and LSWI) and bioclimatic (WorldClim) factors to transmission is reduced at these environmentally controlled transmission locations. However, it is unlikely that the analysis would be much affected by removing these cases except in a handful of cases when the locations were close to detection locations that (we assumed) were not environmentally controlled.
Kilpatrick et al. (2006) established likely pathways for H5N1 introductions into 52 countries by mapping phylogenetic data for H5N1 isolates to wild bird movements and trade in poultry and wild birds. They determined that 26 introductions (including 20/23 European introductions) were probable wild bird introductions, 11 were probable poultry introductions, whereas the remainder could not be assigned. Genomic analysis of H5N1 surface proteins (hemag-glutinin and neuraminidase) found no association between genotype and host (Janies et al., 2007), suggesting that the strain is not transmitted selectively to specific host groups.
Conclusions
No consistent signal of distinct ecological niches for H5N1 occurrences among the host bird groups tested was found. Our findings, though they cannot establish the transmission event introducing an isolate into a specific country, support the idea that host-specific transmission pathways do not exist, and that H5N1 circulates freely with respect to host group. Within Europe, we found no consistent evidence indicating distinct transmission cycles in different avian hosts.
Supplementary Material
Acknowledgments
Thanks to Chandrashekhar Biradar for help with remotely sensed data, and to Monica Pape Yoshinori Nakazawa and Sean Maher for expert advice on analyses. Thanks to the Wildlife Conservation Society, the National Biological Information Infrastructure of the U.S. Geological Survey and the Centers for Disease Control and Prevention/National Institutes of Health for funding this work.
References
- Brown JD, Stallknecht DE, Beck JR, Suarez DL, Swayne DE. Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg Infect Dis. 2006;12:1663–1670. doi: 10.3201/eid1211.060652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elith J, Graham CH, Anderson RP, Dudik M, Ferrier S, Guisan A, Hijmans RJ, Huettmann F, Leathwick JR, Lehmann A, Li J, Lohmann LG, Loiselle BA, Manion G, Moritz C, Nakamura M, Nakazawa Y, Overton JM, Peterson AT, Phillips SJ, Richardson K, Scachetti-Pereira R, Schapire RE, Soberon J, Williams S, Wisz MS, Zimmermann NE. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 2006;29:129–151. [Google Scholar]
- Eurostat. [accessed May 2011];Farm structure in Germany - Statistics Explained. 2008 http://epp.eurostat.ec.europa.eu/statistics_explained/index.php/Farm_structure_in_Germany.
- Gilbert M, Xiao X, Pfeiffer DU, Epprecht M, Boles S, Czarnecki C, Chaitaweesub P, Kalpravidh W, Minh PQ, Otte MJ, Martin V, Slingenbergh J. Mapping H5N1 highly pathogenic avian influenza risk in Southeast Asia. Proc Natl Acad Sci USA. 2008;105:4769–4774. doi: 10.1073/pnas.0710581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder TC, Teuffert J, Starick E, Gethmann J, Grund C, Fereidouni S, Durban M, Bogner KH, Neubauer-Juric A, Repper R, Hlinak A, Engelhardt A, Nockler A, Smietanka K, Minta Z, Kramer M, Globig A, Mettenleiter TC, Conraths FJ, Beer M. Highly pathogenic avian influenza virus (H5N1) in frozen duck carcasses, Germany, 2007. Emerg Infect Dis. 2009;15:272–279. doi: 10.3201/eid1502.080949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesterberg U, Harris K, Stroud D, Guberti V, Busani L, Pittman M, Piazza V, Cook A, Brown I. Avian influenza surveillance in wild birds in the European Union in 2006. Influenza Other Respir Viruses. 2009;3:1–14. doi: 10.1111/j.1750-2659.2008.00058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- Huete A, Didan K, Miura T, Rodriguez EP, Gao X, Ferreira LG. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens Environ. 2002;83:195–213. [Google Scholar]
- Janies D, Hill AW, Guralnick R, Habib F, Waltari E, Wheeler WC. Genomic analysis and geographic visualization of the spread of avian influenza (H5N1) Syst Biol. 2007;56:321–329. doi: 10.1080/10635150701266848. [DOI] [PubMed] [Google Scholar]
- Keawcharoen J, van Riel D, van Amerongen G, Bestebroer T, Beyer WE, van Lavieren R, Osterhaus A, Fouchier RAM, Kuiken T. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1) Emerg Infect Dis. 2008;14:600–607. doi: 10.3201/eid1404.071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM, Chmura AA, Gibbons DW, Fleischer RC, Marra PP, Daszak P. Predicting the global spread of H5N1 avian influenza. Proc Natl Acad Sci USA. 2006;103:19368–19373. doi: 10.1073/pnas.0609227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo JM, Jimenez-Valverde A, Real R. AUC: a misleading measure of the performance of predictive distribution models. Glob Ecol Biogeogr. 2008;17:145–151. [Google Scholar]
- OIE. [accessed on September 2009];Update on highly pathogenic avian influenza in animals (Type H5 and H7) 2009 http://www.oie.int/downld/avian%20influenza/A_AI-Asia.htm.
- Pearson RG, Raxworthy CJ, Nakamura M, Peterson AT. Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. J Biogeogr. 2007;34:102–117. [Google Scholar]
- Peterson AT, Papes M, Eaton M. Transferability and model evaluation in ecological niche modeling: a comparison of GARP and Maxent. Ecography. 2007;30:550–560. [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecol Model. 2006;190:231–259. [Google Scholar]
- Schoener TW. Anolis lizards of Bimini - resource partitioning in a complex fauna. Ecology. 1968;49:704–726. [Google Scholar]
- Si Y, Skidmore AK, Wang T, de Boer WF, Debba P, Toxopeus AG, Li L, Prius HH. Spatio-temporal dynamics of global H5N1 outbreaks match bird migration patterns. Geospat Health. 2009;4:65–78. doi: 10.4081/gh.2009.211. [DOI] [PubMed] [Google Scholar]
- Soberón J. Grinnellian and Eltonian niches and geographic distributions of species. Ecol Lett. 2007;10:1115–1123. doi: 10.1111/j.1461-0248.2007.01107.x. [DOI] [PubMed] [Google Scholar]
- Soberón J, Peterson AT. Interpretation of models of fundamental ecological niches and species’ distributional areas. Biodivers Inform. 2005;2:1–10. [Google Scholar]
- Swayne DE, Suarez DL. Highly pathogenic avian influenza. Rev Sci Tech Off Int Epiz. 2000;19:463–482. doi: 10.20506/rst.19.2.1230. [DOI] [PubMed] [Google Scholar]
- Tucker CJ. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens Environ. 1979;8:127–150. [Google Scholar]
- USGS. [accessed on June 2001];HYDRO1k Elevation Derivative Database. 2001 http://edc.usgs.gov/products/elevation/gtopo30/hydro/index.html.
- USGS. GTOPO30. [accessed on June 2006];Global 30 arc second elevation data. 2006 http://edc.usgs.gov/products/elevation/gtopo30/gtopo30.html.
- Van Borm S, Thomas I, Hanquet G, Lambrecht N, Boschmans M, Dupont G, Decaestecker M, Snacken R, van den Berg T. Highly pathogenic H5N1 influenza virus in smuggled Thai eagles, Belgium. Emerg Infect Dis. 2005;11:702–705. doi: 10.3201/eid1105.050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Vaart AW. Asymptotic statistics. Cambridge University Press; Cambridge, UK: 1998. [Google Scholar]
- Warren DL, Glor RE, Turelli M. Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution. 2008;62:2868–2883. doi: 10.1111/j.1558-5646.2008.00482.x. [DOI] [PubMed] [Google Scholar]
- Weber TP, Stilianakis NI. Ecologic immunology of avian influenza (H5N1) in migratory birds. Emerg Infect Dis. 2007;13:1139–1143. doi: 10.3201/eid1308.070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth D, Newman S, Mundkur T, Harris P, editors. Wild birds and avian influenza: an introduction to applied field research and disease sampling techniques. FAO; Rome, Italy: 2007. [Google Scholar]
- Williams RAJ, Fasina FO, Peterson AT. Predictable ecology and geography of avian influenza (H5N1) transmission in Nigeria and West Africa. Trans R Soc Trop Med Hyg. 2008;102:471–479. doi: 10.1016/j.trstmh.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Williams RAJ, Peterson AT. Ecology and geography of avian influenza (HPAI H5N1) transmission in the Middle East and northeastern Africa. Int J Health Geogr. 2009;8:e47. doi: 10.1186/1476-072X-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao XM, Boles S, Liu JY, Zhuang DF, Liu ML. Characterization of forest types in northeastern China, using multi-temporal SPOT-4 VEGETATION sensor data. Remote Sens Environ. 2002;82:335–348. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.