Abstract
The Environment for Tree Exploration (ETE) is a computational framework that simplifies the reconstruction, analysis, and visualization of phylogenetic trees and multiple sequence alignments. Here, we present ETE v3, featuring numerous improvements in the underlying library of methods, and providing a novel set of standalone tools to perform common tasks in comparative genomics and phylogenetics. The new features include (i) building gene-based and supermatrix-based phylogenies using a single command, (ii) testing and visualizing evolutionary models, (iii) calculating distances between trees of different size or including duplications, and (iv) providing seamless integration with the NCBI taxonomy database. ETE is freely available at http://etetoolkit.org
Keywords: phylogenomics, tree visualization, tree comparison, NCBI taxonomy, hypothesis testing, phylogenetics.
The Environment for Tree Exploration (ETE) is a toolkit developed to facilitate the computation, analysis and visualization of phylogenetic data. ETE provides a comprehensive Python programming library (API) that allows researchers to automate common tasks in comparative genomics. Since its first release (Huerta-Cepas et al. 2010), ETE has been widely used as a computational framework to perform numerous phylogenomic analyses, including characterizing newly sequenced genomes (Richards et al. 2010; Wang et al. 2014), extracting information from large sets of phylogenetic trees (Derelle and Lang 2012; Chiapello et al. 2015; Marcet-Houben and Gabaldón 2015) and developing third party tools and databases (Zhang et al. 2013; Huerta-Cepas et al. 2014; Szitenberg et al. 2015). Here, we describe the latest version of the software (ETE v3), featuring a significantly improved API library and a novel collection of standalone tools. While the API continues to offer full programmatic control on data analysis and visualization, the new standalone tools facilitate the use of common phylogenetic methods at the genomic scale. We here describe the most notable additions.
Tree Building
The ete-build tool provides a unified interface to wrap the execution of reproducible phylogenetic workflows, comprising the reconstruction of gene–trees and supermatrix-based species trees. To do so, ETE relies on a versioned collection of external tools that are transparently installed and executed upon request. A single command is used to configure and launch complex phylogenetic pipelines, covering sequence alignment, trimming, substitution-model testing, tree inference, and image rendering (fig. 1A). In addition, the supermatrix-based reconstruction mode permits to build and concatenate multiple sequence alignments with ease, simplifying the inference of species trees based on multiple genes. Advanced options allow to automatically switch from amino-acid to nucleotide alignments based on sequence identity, resuming the execution of workflows, or even testing multiple strategies in parallel. As an example, a single command line can be used to test several alignment methodologies or phylogenetic inference programs simultaneously, making the tool particularly suitable to run phylogenomic pipelines. Notably, ETE-build was recently used to compute over one million phylogenetic trees for the EggNOG v4.5 database (Huerta-Cepas et al. 2016).
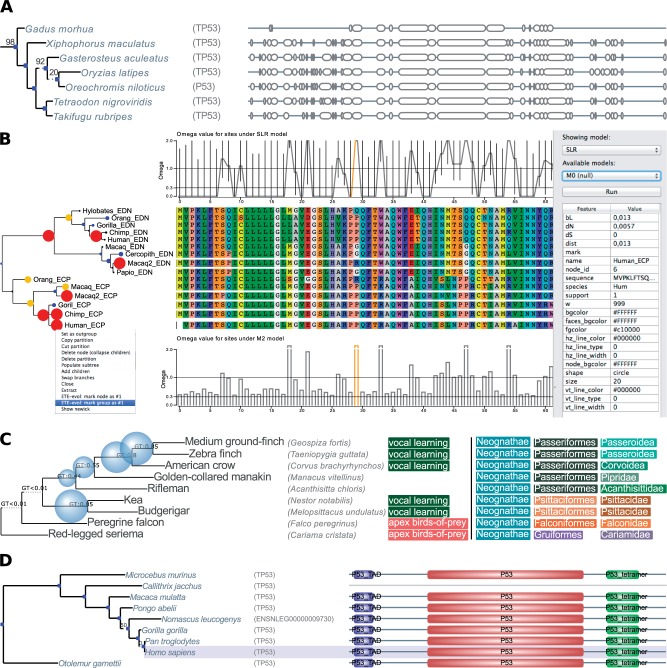
Fig. 1.
Several phylogenetic tree images generated using the ETE toolkit. (A) Gene tree reconstructed using ete-build. The figure shows the relationships between several P53 genes together with their aligned sequences visualized in condensed format. (B) Tree image generated by ete-evol for three models fitted to a classical example (Bielawski and Yang 2004). (i) The line chart on top of the alignment indicates the omega estimates for sites as calculated by the SLR software. (ii) The bar chart at the bottom part shows the dn/ds ratio for each site under the M2 site-model from CodeML. Line colors in both charts indicate the significance of assigning a site to a given class of positive selection (i.e., red for P-value <0.01 and orange for P-value <0.05). (iii) The color and size of tree nodes represent the dn/ds ratio estimated for tree branches using the free-ratio model from CodeML. Blue small circles indicate a ratio between 0.2 and 1, medium yellow nodes indicate a ratio >1, and big red nodes for infinite values. Note that the right side panel allows users to select the models to be displayed, and even starting new runs using predefined models. (C) Portion of a recently published bird species tree (Jarvis et al. 2014) annotated with gene–tree support values (blue spheres), custom node labeling (first aligned column) and taxonomic information (next aligned columns). (D) Example of a phylogenetic tree visualized with a sequence alignment and domain composition as used in the eggNOG database (Huerta-Cepas et al. 2016).
Testing Evolutionary Hypotheses
Measuring selective pressures on molecular sequences is a common task in evolutionary biology. Softwares such as CodeML (Yang 2007) or SLR (Massingham and Goldman 2005) provide the statistical and computational framework to perform these analyses. However, the use of such tools at the genomic scale requires substantial work on data preparation, on experimental design, and on results interpretation. To aid in these tasks, the ete-evol tool automates CodeML/SLR-based analyses by using pre-configured evolutionary models and directly producing a graphical representation of the results. These pre-configured models include site (Yang et al. 2000; Massingham and Goldman 2005), branch (Yang and Nielsen 2002), branch-site (Zhang et al. 2005), and clade (Yang and Nielsen 2002; Bielawski and Yang 2004) models. For instance, ete-evol can test, in parallel, and with a single call, the differential selective pressures along each branch in a given phylogeny. Importantly, fitted models are compared using a built-in likelihood ratio test. Evolutionary measures from the best-fitting models are then plotted (or interactively visualized) by mapping the predicted selective pressures acting on sites and branches into the tested topology, as well as on the multiple sequence alignment (fig. 1B). For convenience, raw output files produced by CodeML and SLR can also be visualized using ete-evol.
Comparing Trees
ETE v3 provides three measures to compute distances between trees, namely the Robinson–Foulds distance (Robinson and Foulds 1981), a branch congruence measure (%) and the TreeKO Speciation distance (Marcet-Houben and Gabaldón 2011). In contrast to existing software (Felsenstein 2005; Soria-Carrasco et al. 2007), ete-compare calculates all three distances at the same time; it accepts trees varying in size and containing duplication events; it allows filtering branches with low support; and it is optimized for comparing large datasets. In addition, ete-compare can provide a detailed list of the differences and coincidences among the compared trees for further analysis. Conveniently, the TreeKO method for splitting gene trees into duplication-free subtrees has been optimized and integrated into ETE’s API library, thereby enabling its use for other tests. For instance, ETE allows summarizing the phylogenetic signal (i.e., gene tree support) from an heterogenous sample of gene trees using a species tree topology as reference (fig. 1C).
Taxonomy Databases
Efficient queries to the NCBI-taxonomy database (Benson et al. 2014) are now available through the ete-ncbiquery tool or the relevant methods in the API. Extracting pruned subtrees, converting NCBI taxids into their corresponding scientific names, obtaining full lineage tracks, or annotating user-trees with taxonomic data, are common tasks that can be easily performed with the ete-ncbiquery tool. Importantly, all queries are carried out locally, avoiding unnecessary lags and permitting the integration of the tool into genomic and metagenomic pipelines.
Finally, other ETE-tools and methods are available that aid in routine tasks such as format conversion, topology manipulation, and custom visualization of trees linked to multiple sequence alignments (fig. 1D).
Conclusions
Although several software packages are available for the standalone exploration of trees (Letunic and Bork 2007; Huson and Scornavacca 2012; Asnicar et al. 2015) and the programmatic manipulation of data (Paradis et al. 2004; Knight et al. 2007; Sukumaran and Holder 2010; Vos et al. 2011; Talevich et al. 2012), ETE offers a unified framework to compute and analyze genome-wide collections of evolutionary data while providing unique visualization capabilities. Moreover, with the recent addition of the command line tools, ETE has significantly broadened its scope, simplifying many common tasks in phylogenomics for both expert and casual users.
Acknowledgments
We wish to thank Toni Gabaldón, Renato Alves, Falk Hildebrand, and Gabriela Aguileta for valuable feedback, as well as the GitHub community for contributions. This study was supported by the European Molecular Biology Laboratory (EMBL). Funding for open access charge: European Molecular Biology Laboratory (EMBL).
References
- Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N. 2015. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. Peer J. 3:e1029.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2014. GenBank. Nucleic Acids Res. 42:D32–D37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski JP, Yang Z. 2004. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J. Mol. Evol. 59:121–132. [DOI] [PubMed] [Google Scholar]
- Chiapello H, Mallet L, Guérin C, Aguileta G, Amselem J, Kroj T, Ortega-Abboud E, Lebrun M-H, Henrissat B, Gendrault A, et al. 2015. Deciphering genome content and evolutionary relationships of isolates from the fungus magnaporthe oryzae attacking different host plants. Genome. Biol. Evol. 7:2896–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, Lang BF. 2012. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol. Biol. Evol. 29:1277–1289. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package). Cladistics 5:164–166. [Google Scholar]
- Huerta-Cepas J, Capella-Gutiérrez S, Pryszcz LP, Marcet-Houben M, Gabaldón T. 2014. PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res. 42(D1):D897–D902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo J, Gabaldón T. 2010. ETE: a python Environment for Tree Exploration. BMC Bioinformatics 11:24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, et al. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44(D1):D286–D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C. 2012. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61:1061–1067. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SYW, Faircloth BC, Nabholz B, Howard JT, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R, Maxwell P, Birmingham A, Carnes J, Caporaso JG, Easton BC, Eaton M, Hamady M, Lindsay H, Liu Z, et al. 2007. PyCogent: a toolkit for making sense from sequence. Genome Biol. 8:R171.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. [DOI] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldón T. 2011. TreeKO: A duplication-aware algorithm for the comparison of phylogenetic trees. Nucleic Acids Res. 39:e66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldón T. 2015. Beyond the whole-genome duplication: phylogenetic evidence for an ancient interspecies hybridization in the baker’s yeast lineage. PLoS Biol. 13(8):e1002220.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massingham T, Goldman N. 2005. Detecting amino acid sites under positive selection and purifying selection. Genetics 169:1753–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. [DOI] [PubMed] [Google Scholar]
- Richards S, Gibbs RA, Gerardo NM, Moran N, Nakabachi A, Stern D, Tagu D, Wilson ACC, Muzny D, Kovar C, et al. 2010. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DF, Foulds LR. 1981. Comparison of phylogenetic trees. Math. Biosci. 53:131–147. [Google Scholar]
- Soria-Carrasco V, Talavera G, Igea J, Castresana J. 2007. The K tree score: quantification of differences in the relative branch length and topology of phylogenetic trees. Bioinformatics 23:2954–2956. [DOI] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT. 2010. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26:1569–1571. [DOI] [PubMed] [Google Scholar]
- Szitenberg A, John M, Blaxter ML, Lunt DH. 2015. ReproPhylo: an environment for reproducible phylogenomics. PLoS Comput. Biol. 11:e1004447.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talevich E, Invergo B, Cock P, Chapman B. 2012. Bio.Phylo: a unified toolkit for processing, analyzing and visualizing phylogenetic trees in Biopython. BMC Bioinformatics 13:209.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos RA, Caravas J, Hartmann K, Jensen MA, Miller C. 2011. BIO::Phylo-phyloinformatic analysis using perl. BMC Bioinformatics 12:63.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Haberer G, Gundlach H, Gläßer C, Nussbaumer T, Luo MC, Lomsadze A, Borodovsky M, Kerstetter RA, Shanklin J, et al. 2014. The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat. Commun. 5:3311.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19:908–917. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kapli P, Pavlidis P, Stamatakis A. 2013. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29:2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22:2472–2479. [DOI] [PubMed] [Google Scholar]