Abstract
Hydrogen sulfide (H2S) is a potent toxicant interfering with oxidative phosphorylation in mitochondria and creating extreme environmental conditions in aquatic ecosystems. The mechanistic basis of adaptation to perpetual exposure to H2S remains poorly understood. We investigated evolutionarily independent lineages of livebearing fishes that have colonized and adapted to springs rich in H2S and compared their genome-wide gene expression patterns with closely related lineages from adjacent, nonsulfidic streams. Significant differences in gene expression were uncovered between all sulfidic and nonsulfidic population pairs. Variation in the number of differentially expressed genes among population pairs corresponded to differences in divergence times and rates of gene flow, which is consistent with neutral drift driving a substantial portion of gene expression variation among populations. Accordingly, there was little evidence for convergent evolution shaping large-scale gene expression patterns among independent sulfide spring populations. Nonetheless, we identified a small number of genes that was consistently differentially expressed in the same direction in all sulfidic and nonsulfidic population pairs. Functional annotation of shared differentially expressed genes indicated upregulation of genes associated with enzymatic H2S detoxification and transport of oxidized sulfur species, oxidative phosphorylation, energy metabolism, and pathways involved in responses to oxidative stress. Overall, our results suggest that modification of processes associated with H2S detoxification and toxicity likely complement each other to mediate elevated H2S tolerance in sulfide spring fishes. Our analyses allow for the development of novel hypotheses about biochemical and physiological mechanisms of adaptation to extreme environments.
Keywords: ecological physiology, evolution, extreme environments, gene expression, H2S, Poecilia mexicana (Poeciliidae), RNA-sequencing.
Introduction
Extreme environments are characterized by physiochemical stressors lethal to most organisms, and their clearly defined and replicated selective regimes enable hypothesis-driven tests of organismal responses at all levels of biological organization (Waterman 1999; Bell 2012). Extremophiles can withstand such stressful conditions and provide ideal systems to study mechanisms underlying physiological adaptation (Storey KB and Storey JM 2005; Nevo 2011). In addition, they shed light into life’s capacities and limitations to adapt to novel environmental conditions, and understanding the ways organisms function in the context of natural stressors provides basic insights into the biochemical, physiological, and developmental processes that govern life (Waterman 1999; Tobler et al. 2015). Hydrogen sulfide (H2S) is a physiochemical stressor that is produced in many aquatic environments through geochemical or biological processes (Muyzer and Stams 2008). H2S has been hypothesized to have played a critical role in the origin of life and caused mass extinctions; accordingly, it has influenced basic physiological properties of organisms and shaped evolutionary diversification on Earth (Kump et al. 2005; Olson and Straub 2015). H2S has a wide variety of cytotoxic effects (Beauchamp et al. 1984; Reiffenstein et al. 1992), but its toxicity primarily unfolds through the inhibition of cytochrome c oxidase (COX) in the mitochondrial respiratory chain, which halts ATP production (Cooper and Brown 2008). Exposure to environmental H2S can consequently limit an organism's ability to survive and reproduce (Bagarinao 1992). Although environmental concentrations of H2S are typically low or highly transient, high and sustained concentrations create extreme environments that are lethal for most life (Tobler and Plath 2011; Riesch et al. 2015). Few metazoans have colonized H2S-rich habitats and evolved strategies to cope with the continuous exposure to this respiratory toxicant, giving rise to unique ecological communities in deep-sea hydrothermal vents, cold seeps, and freshwater sulfide springs (Van Dover 2000; Levin 2005; Greenway et al. 2014).
The mechanistic basis of physiological adaptation to high levels of H2S remains poorly understood. Previous studies have primarily focused on organisms with endosymbiotic sulfide oxidizing bacteria, which occur in some polychaete worms and mytilid mussels (Childress and Fisher 1992; Roeselers and Newton 2012). Other studies have compared physiological processes between H2S tolerant and nontolerant species across taxonomically disparate groups with vastly different body organizations and genomic architectures (rats vs. lugworms) and suggested that H2S-tolerant organisms may exhibit detoxification enzymes with higher activity rates (Hildebrandt and Grieshaber 2008). Nonetheless, the diversity of H2S’s physiological effects and molecular targets (Li et al. 2011; Olson 2011) suggests that adaptation to sulfide-rich environments could include a variety of alternative or complementary modifications. The identification of such adaptive modifications largely hinges on the comparison between closely related species and populations from sulfidic and nonsulfidic environments.
Recent progress in our understanding of the physiological effects and processing of H2S, driven primarily by biomedical research investigating the role of H2S in disease formation and potential biomedical applications (Szabo 2007; Li et al. 2011), enables hypothesis-driven tests of the potential molecular basis underlying adaptation to H2S-rich environments. Elevated tolerance to environmental H2S could be driven by three, nonmutually exclusive mechanisms. First, organisms perpetually exposed to high ambient H2S concentrations may minimize its flux from the environment into the body (Vismann 1991). H2S is lipid soluble and accordingly able to readily pass through biological membranes and invade organismal systems (Reiffenstein et al. 1992). Adaptation to environmental H2S may thus include modifications of the integumentary system and respiratory surfaces directly exposed to the environment. Such modifications could include changes in the structure and composition of biological membranes and molecular components involved in intercellular interactions, as well as excretion of compounds that create a barrier to H2S diffusion.
Second, increased tolerance could be mediated by an increased capability to maintain H2S homeostasis despite continuous influx from the environment. This would involve modification of mechanisms involved in the active elimination of H2S. All animals are able to detoxify low levels of H2S through enzymatic oxidation associated with the highly conserved sulfide:quinone oxidoreductase (SQR) pathway and subsequent excretion of oxidized sulfur species (typically sulfate; Hildebrandt and Grieshaber 2008; Shahak and Hauska 2008; Lagoutte et al. 2010). H2S can also be immobilized and sequestered by methylation, sulfhydration, or reaction with methemoglobin (Bagarinao and Vetter 1992; Levitt et al. 1999; Paul and Snyder 2012). Alternatively, increased capacity for the regulation of H2S homeostasis could be facilitated by reducing endogenous H2S production. Sulfide is endogenously produced through reactions associated with the transsulfuration pathway and the processing of sulfur-containing amino acids (Stipanuk 2004; Stipanuk and Ueki 2011).
Third, tolerance to H2S could be achieved through the modification of toxicity targets that make them less sensitive to adverse consequences caused by elevated endogenous concentration in the face of continuous influx from the environment. For example, COX—the primary target of H2S toxicity—in sulfide-tolerant organisms has been documented to exhibit structural changes that reduce blocking by H2S (Degn and Kristensen 1981; Pfenninger et al. 2014). Considering recent biomedical research that has found H2S to play a critical role in cell signaling and to have an increasing number of putative molecular targets, including ion channels and receptors, cell signaling proteins, and enzymes involved in metabolism (Li and Moore 2008; Li et al. 2011; Olson 2011; Whiteman et al. 2011), it is not straightforward to establish a comprehensive list of potential targets. Nonetheless, mitochondria in general—and the mitochondrial respiratory chain in particular—are expected to provide primary targets of selection mediated by the presence of environmental sulfide (fig. 1A), because they include both primary targets for H2S toxicity (COX, Cooper and Brown 2008) and are directly linked to H2S detoxification through SQR (Hildebrandt and Grieshaber 2008). To identify molecular mechanisms potentially underlying high H2S tolerance, we leveraged a natural system with closely related lineages that occur in environments with perpetually elevated concentrations of naturally occurring H2S as well as in adjacent nonsulfidic habitats.
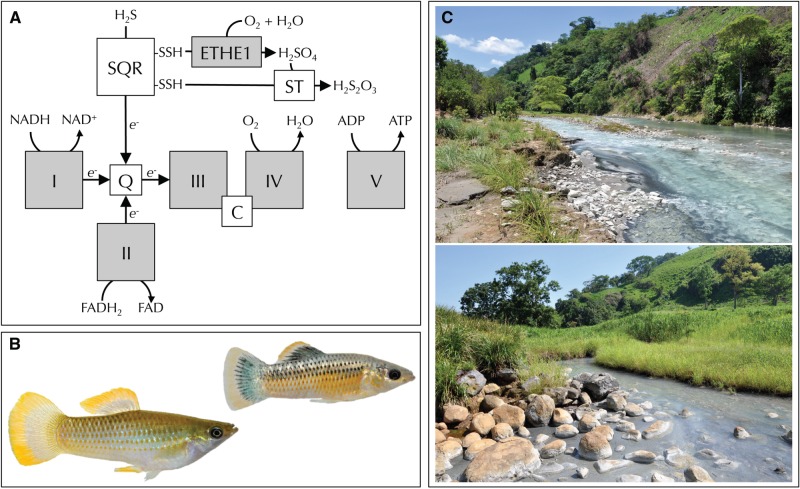
Fig. 1.
(A) Primary enzymes involved in the oxidative phosphorylation (OXPHOS) pathway of the mitochondrial respiratory chain, which represents a nexus of H2S toxicity and detoxification. H2S is toxic because it blocks COX (complex IV) of the respiratory chain. H2S is detoxified in mitochondria through pathways linked to the respiratory chain. Specifically, sulfide oxidation to thiosulfate is mediated by SQR, a sulfur dioxygenase (ETHE1), and a sulfur transferase (ST). SQR feeds electrons into the respiratory chain, which are ultimately transferred to oxygen by complex IV. (B) Representative specimens of the Poecilia mexicana species complex, including a male from a nonsulfidic population (left) and a male from a sulfide spring populations (right). (C) Representative sulfide springs in the Rio Puyacatengo (top) and Rio Pichucalco (bottom) drainages.
In southern Mexico, small livebearing fish of the Poecilia mexicana species complex (Poeciliidae) have repeatedly colonized H2S-rich freshwater springs in at least three river drainages (fig. 1B and C; Palacios et al. 2013). Sulfide spring populations are locally adapted and characterized by phenotypic modifications, including physiological, morphological, and life history traits that have largely evolved in convergence (Tobler and Hastings 2011; Tobler et al. 2011; Pfenninger et al. 2014; Riesch et al. 2014). In addition, sulfide spring populations are undergoing ecological speciation, and there is significant—albeit varying—genetic differentiation between sulfidic and adjacent nonsulfidic populations despite small geographic distances and a lack of physical barriers preventing fish movement (Plath et al. 2013; Pfenninger et al. 2015). Reproductive isolation between sulfidic and nonsulfidic populations is primarily mediated by natural and sexual selection against maladapted migrant individuals (Plath et al. 2010 , 2013).
The availability of evolutionarily replicated pairs of closely related populations in sulfidic and nonsulfidic environments provides a unique opportunity for comparative studies of organismal responses to continuous disruptions of H2S homeostasis. We contrasted variation in gene expression to test hypotheses about the potential molecular underpinnings of H2S tolerance. We focused on gill tissues, because they are in direct contact with the toxic environment, mediate a variety of physiological processes involved in the maintenance of homeostasis (Evans and Claiborne 2006; Evans et al. 2011), and exhibit strong transcriptional responses upon exposure to H2S (Tobler et al. 2014). We addressed four key questions: 1) Do levels of gene expression vary between proximate population pairs residing in sulfidic and nonsulfidic habitats? 2) Have gene expression patterns in replicated sulfide spring populations changed in convergence? 3) Is there any evidence for potentially adaptive changes in gene expression? 4) Do shared differentially expressed genes provide any insights about potential mechanisms underlying sulfide tolerance?
Results and Discussion
Transcriptome Assembly
Sequencing the gill tissue transcriptomes on an Illumina Hiseq platform yielded over 265 million reads for 35 individual fish collected from sulfidic and nonsulfidic habitats in the Tacotalpa, Puyacatengo, and Pichucalco river drainages (supplementary table S1 and fig. S1, Supplementary Material online). Nonsulfidic populations and sulfide spring populations in the Tacotalpa and Puyacatengo drainages nominally belong to the widespread species P. mexicana; the sulfide spring population in the Pichucalco drainage has been described as a highly endemic, distinct species (P. sulphuraria) (Palacios et al. 2013). Mapping against the reference genome of Xiphophorus maculatus (Schartl et al. 2013), a closely related species in the same family (Poeciliidae), resulted in a P. mexicana gill transcriptome that was composed of 35,468 transcripts from 29,143 unique loci (supplementary table S2, Supplementary Material online). Based on comparison with the X. maculatus reference annotation set, 8,503 loci were putatively unique to P. mexicana. When limiting the analyses to the longest transcript from each locus, the transcriptome size was 71,518,404 bp, with an N50 of 3,694 bp and a genome size of approximately 860 Mbp based on C-value estimates (Rasch et al. 1970). The longest transcript was 66,752 bp in length, stemming from the gene coding for the largest known protein (Titin; Opitz et al. 2003). This indicated that our analysis effectively captured even long transcripts present in the transcriptome. A BLAST search found that 22,566 of the 29,143 unique loci (77.4%) had matches in the SwissProt database (see supplementary table S3, Supplementary Material online, for complete annotation individual transcripts). These represented 17,021 unique SwissProt records, 13,127 of which were associated with a Gene Ontology (GO) term (Gene Ontology Consortium 2004).
Variation in Gene Expression
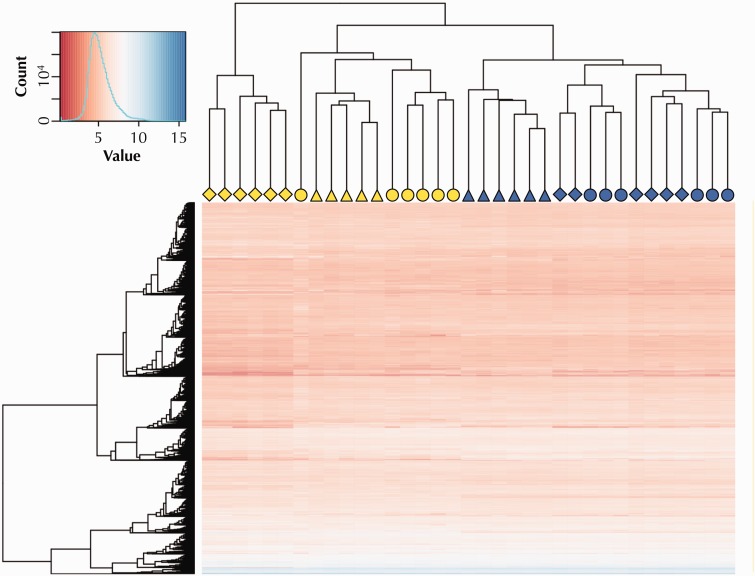
Hierarchical cluster analysis of the top 10,000 expressed genes recovered three major clusters, which did not primarily correspond to a geographical pattern in gene expression that could be related to our sampling scheme (i.e., clustering by river drainage). Instead, individuals clustered largely by habitat type (fig. 2). Individuals from nonsulfidic habitats appeared in a single cluster. Although there was some geographic structuring among nonsulfidic fish (Puyacatengo individuals were contained in a single subcluster), there was no apparent differentiation between fish from the Tacotalpa and Pichucalco drainages. This indicated that gene expression patterns in nonsulfidic habitats were relatively similar irrespective of river drainage, and considering that the Tacotalpa and Pichucalco drainages are most distant physically, geographic isolation alone likely played a minor role in driving population differences. In contrast, individuals from sulfide spring populations appeared in two separate clusters. One cluster only included individuals from the Pichucalco drainage (P. sulphuraria), which exhibited the most distinct gene expression patterns. The other cluster consisted of individuals from the Tacotalpa and Puyacatengo drainages, which—with the exception of an outlier individual from the Tacotalpa drainage—were arranged in distinct subclusters. General patterns recovered by the hierarchical cluster analysis were also evident in results from ordination of gene expression data (supplementary fig. S2, Supplementary Material online).
Fig. 2.
Results of a hierarchical cluster analysis and heat map showing expression variation in the 10,000 top expressed genes. Symbol shapes designate the population of origin for each individual in the analysis (○: Tacotalpa drainage; ▵: Puyacatengo drainage; ⋄: Pichucalco drainage). Yellow symbols represent individuals from sulfide springs, blue symbols those from nonsulfidic reference habitats.
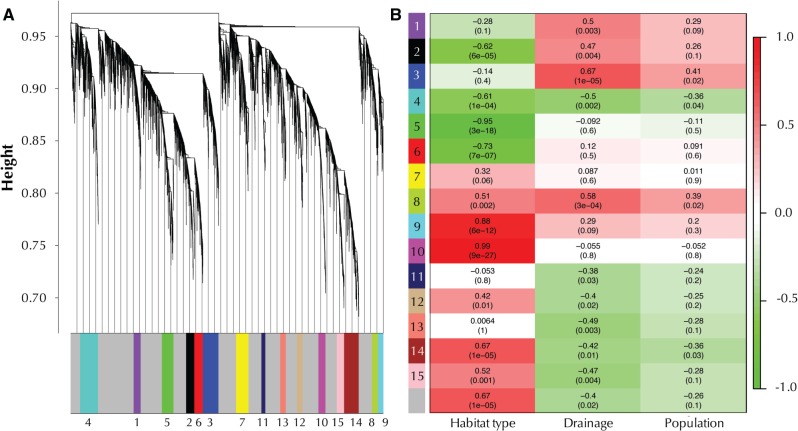
Weighted gene correlation network analysis (WGCNA) of the top 10,000 expressed genes revealed 15 modules of coexpressed genes (fig. 3A). Ten of the 15 modules were significantly correlated with habitat type (presence or absence of H2S), with modules 5 and 10 exhibiting correlation coefficients >0.9 (fig. 3B). These modules primarily contained genes associated with enzymatic H2S detoxification and processing of sulfur compounds (positively correlated with the presence of H2S, module 10), or with genes associated with immune function as well as cell migration and chemotaxis (negatively correlated with the presence of H2S, module 5). In contrast, there were much fewer significant associations between any modules and specific river drainages or specific populations, and correlation coefficients were considerably <0.9. This again supports that geographic distance per se plays a minor role in shaping population differences in gene expression and indicates that the presence or absence of H2S is a main driver of gene expression networks as well as connectivity among genes.
Fig. 3.
Results of WGCNA of the top 10,000 expressed genes. (A) Average linkage clustering tree based on topological overlap distances in gene expression patterns of sulfidic and nonsulfidic fish. Branches of the dendrogram correspond to modules, as shown in the color bar below. Note branches associated with the gray color represent genes that are not associated with any module. (B) Correlation between module eignenvalues and habitat type (presence or absence of H2S), river drainage, and population (collection site). Each row represents a module corresponding to color and identification number. Reported are Pearson correlation coefficients (on the top of each cell) and P values (on the bottom in parentheses). Cell coloration represents the correlation value according to the scale bar on the right.
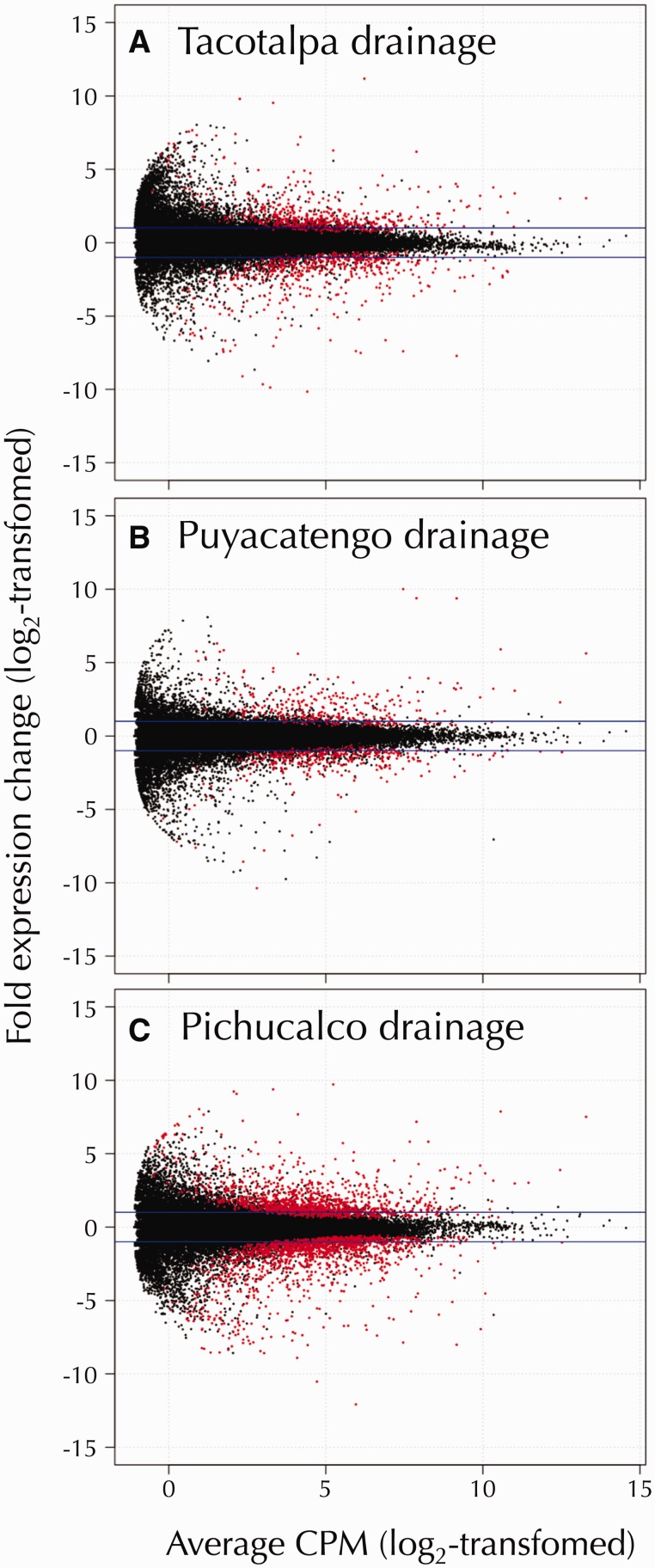
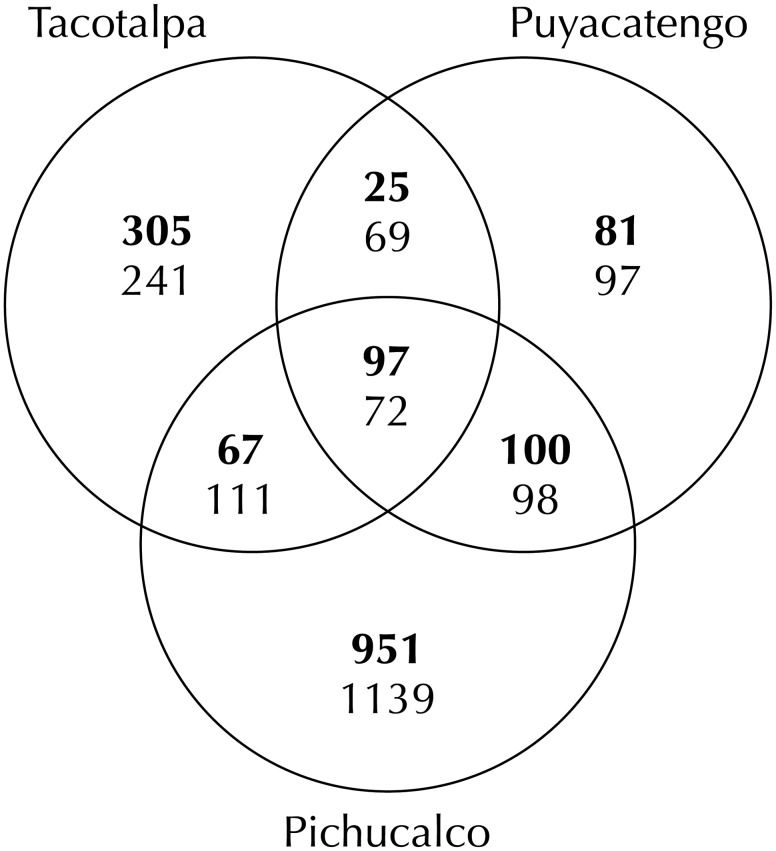
Comparison of gene expression between individuals from the three paired sulfidic and nonsulfidic sites revealed significant evidence for differential expression in each of the river drainages (fig. 4). Overall, 1,626 transcripts were significantly upregulated and 1,827 downregulated in sulfide spring fish in at least one of the river drainages (supplementary table S4, Supplementary Material online). Nonetheless, the number of differentially expressed genes was not equally distributed across different river drainages. Gene expression differences were least pronounced in the Puyacatengo (303 upregulated and 336 downregulated genes) and the Tacotalpa drainages (494 upregulated and 493 downregulated), and most pronounced in the Pichucalco drainage (1,215 upregulated and 1,420 downregulated). The vast majority of gene expression differences between sulfidic and nonsulfidic populations was unique to a particular drainage; 82% of differentially expressed transcripts were upregulated and 81% downregulated in only one of the three population pairs (fig. 5). Populations in the Pichucalco drainage exhibited the highest number of unique gene expression differences (951 unique upregulated and 1,139 unique downregulated transcripts), followed by the populations in the Tacotalpa (305 upregulated and 241 downregulated) and the Puyacatengo (81 upregulated and 97 downregulated) drainages.
Fig. 4.
Volcano plots depicting the fold change in gene expression (log2-transformed) between the sulfidic and nonsulfidic populations as a function of the concentration of each transcript (in counts per million [CPM], log2-transformed). Transcripts with evidence for significant differential expression (FDR ≤ 0.001) are colored in red. The horizontal blue lines indicate a 4-fold difference in transcript abundance. Results are shown separately for the Tacotalpa (A), Puyacatengo (B), and Pichucalco (C) river drainages each of the river drainages investigated (panels A–C).
Fig. 5.
Venn diagram depicting shared and unique variation in gene expression between sulfidic and nonsulfidic populations across three river drainages. Numbers in each section correspond to the number of differentially expressed transcripts derived from estimations of gene expression in eXpress. The number of upregulated transcripts are listed on top (in bold font) and the number of downregulated transcript below.
The differences in the number of unique differentially expressed genes in each of the drainages largely corresponded to previously documented differences in the divergence times between different pairs of sulfidic and nonsulfidic populations, which is consistent with neutral drift driving a large portion of gene expression variation among populations (Whitehead and Crawford 2006). The sulfide spring population in the Pichucalco drainage represents the oldest (∼300,000 years) and most distinct lineage and shares little gene flow with populations from adjacent nonsulfidic habitats (Plath et al. 2013; Pfenninger et al. 2014). Unlike sulfide spring populations in the other river drainages, they are not closely related to P. mexicana in adjacent nonsulfidic habitats, but rather to populations in Northern Mexico (Tobler et al. 2011; Palacios et al. 2013). Hence, distinctness of gene expression patterns in the sulfide spring population of the Pichucalco river drainage likely reflects phylogenetic divergence. In contrast, populations in the Tacotalpa and Puyacatengo drainages have colonized sulfide springs more recently (<50,000 years ago; Pfenninger et al. 2014). Sulfide springs in these drainages have been colonized independently (Tobler et al. 2011; Palacios et al. 2013), and sulfidic populations remain connected to adjacent nonsulfidic populations by low rates of gene flow (Plath et al. 2013).
Overall, these results suggest that the presence of H2S and historical patterns of population connectivity have interacted to shape population differences in gene expression patterns across small spatial scales. Accordingly, there was little evidence that convergent evolution has shaped genome-wide gene expression patterns in different sulfide spring populations. The absence of convergence at the transcriptome level maybe due to several, nonmutually exclusive reasons (Kaeuffer et al. 2012). First, the documented variation in gene expression may be selectively neutral and not affect fitness along the environmental gradient from sulfide springs to regular freshwater habitats. Variation in gene expression hence could reflect the effects of genetic drift that causes random differences among populations (Whitehead and Crawford 2006). Second, variation in gene expression may be related to population attributes and sources of selection that are unrelated to the presence or absence of H2S per se. For example, variation in age structure, reproductive status, parasitization, and the presence or absence of other abiotic and biotic environmental factors could all affect transcription of genes (Naumova et al. 2012; Choi et al. 2014; Yu et al. 2014; McTaggart et al. 2015; Whittington et al. 2015), potentially causing unique patterns of gene expression in individual populations. Finally, absence of convergence could be a consequence of different mechanisms underlying adaptation in independent lineages of sulfide spring fishes (Wilkens and Strecker 2003; Hoekstra et al. 2006; Losos 2011). Although field-based studies of broad gene expression patterns are inadequate to differentiate how these factors may shape convergent evolution, identification of shared differentially expressed genes and vetting of functional annotations against a priori predictions can nonetheless provide insights about mechanisms of adaptation.
Shared Differentially Expressed Genes and Transcripts
Even though genome-wide variation in gene expression patterns showed little evidence for convergence, there was a small, but significant number of transcripts that were consistently differentially expressed in the same direction across two or more sulfidic and nonsulfidic population pairs. Of the 1,626 transcripts that showed evidence for upregulation in at least one of the drainages, 289 (18%) were shared between two drainages and 97 (6%) among all three drainages (table 1). Of the 1,827 transcripts that showed evidence for downregulation in at least one of the drainages, 350 (19%) were shared between two drainages and 71 (4%) among all drainages (table 2). Transcripts that are consistently differentially expressed among all three population pairs represent primary candidates for playing a role in adaptation to sulfide spring environments (Ghalambor et al. 2015), and the following discussion of the potential functional consequences of variation in gene expression—unless noted otherwise—refers only to differentially expressed genes shared among all population pairs.
Table 1.
List of Genes that Were Consistently Upregulated in Sulfide Spring Fishes of All Drainages.
| Gene ID | Accession Number | Gene Name | Xmac Annotation | Number of Transcripts |
|---|---|---|---|---|
| TCONS_00000049 | O75452 | Retinol dehydrogenase 16 | rdh1 | 1 |
| TCONS_00002627 | Q16878 | Cysteine dioxygenase type 1 | CDO1 | 1 |
| TCONS_00002704 | Q00610 | Clathrin heavy chain 1 | CLTCL1 | 2 |
| TCONS_00002706 | P53007 | Tricarboxylate transport protein | si:dkey-178e17.1 | 1 |
| TCONS_00002887 | n/a | XLOC_002625 | 1 | |
| TCONS_00004013 | Q9Y6N5 | Sulfide:quinone oxidoreductase | sqrdl | 1 |
| TCONS_00004833 | Q5BKX6 | Solute carrier family 45 member 4 | SLC45A4 (2 of 2) | 1 |
| TCONS_00005215 | Q9BRT3 | Migration and invasion enhancer 1 | XLOC_004459 | 1 |
| TCONS_00006194 | P02144 | Myoglobin | Mb | 1 |
| TCONS_00006595 | Q8NHV1 | GTPase IMAP family member 7 | zgc:152658 | 1 |
| TCONS_00006596 | P10620 | Microsomal glutathione S-transferase 1 | mgst1.2 | 1 |
| TCONS_00006640 | P52294 | Importin subunit alpha-5 | kpna1 | 1 |
| TCONS_00006715 | Q5T6X4 | Protein FAM162B | fam162a | 3 |
| TCONS_00007401 | B7ZAP0 | RAB GTPase activating protein 1-like | rabgap1l2 | 1 |
| TCONS_00008470 | P15121 | Aldose reductase | akr1b1 | 1 |
| TCONS_00010008 | O00483 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex | ndufa4l2 | 1 |
| TCONS_00010278 | P41236 | Protein phosphatase inhibitor 2 | ppp1r2 | 1 |
| TCONS_00010396 | Q7KZN9 | Cytochrome c oxidase assembly protein COX15 | cox15 | 1 |
| TCONS_00011484 | Q96PR1 | Potassium voltage-gated channel subfamily C member 2 | kcnc2 | 1 |
| TCONS_00011526 | P58743 | Prestin | slc26a5 | 1 |
| TCONS_00011778 | P21266 | Glutathione S-transferase Mu 3 | ENSXMAG00000009961 | 1 |
| TCONS_00012045 | Q12805 | EGF-containing fibulin-like extracellular matrix protein 1 | EFEMP1 | 1 |
| TCONS_00012336 | Q9H628 | Ras-related and estrogen-regulated growth inhibitor-like protein | zgc:171704 | 2 |
| TCONS_00012508 | P53985 | Monocarboxylate transporter 1 | slc16a1 | 1 |
| TCONS_00012770 | P00390 | Glutathione reductase | Gsr | 1 |
| TCONS_00013706 | P99999 | Cytochrome c | XLOC_011171 | 1 |
| TCONS_00014743 | Q6ZQY3 | Acidic amino acid decarboxylase GADL1 | GADL1 (1 of 2) | 1 |
| TCONS_00014769 | Q6ZQY3 | Acidic amino acid decarboxylase GADL1 | GADL1 (2 of 2) | 1 |
| TCONS_00014831 | Q96P48 | Arf-GAP with Rho-GAP domain, ANK repeat, and PH domain-containing protein 1 | ENSXMAG00000006436 | 1 |
| TCONS_00014991 | Q16822 | Phosphoenolpyruvate carboxykinase | pck2 | 1 |
| TCONS_00015284 | P27144 | Adenylate kinase 4 | ak4 | 2 |
| TCONS_00015294 | Q9BYD5 | Cornifelin | ponzr10 (4 of 5) | 1 |
| TCONS_00015352 | Q8TDN7 | Alkaline ceramidase 1 | acer1 | 1 |
| TCONS_00015381 | Q96B33 | Claudin-23 | cldn23 | 2 |
| TCONS_00016368 | Q16831 | Uridine phosphorylase 1 | upp1 | 1 |
| TCONS_00016470 | P60174 | Triosephosphate isomerase | tpi1a | 1 |
| TCONS_00017022 | Q9UBX3 | Mitochondrial dicarboxylate carrier | SLC25A10 | 2 |
| TCONS_00017053 | Q86WA9 | Sodium-independent sulfate anion transporter | slc26a11 | 1 |
| TCONS_00017057 | O15525 | Transcription factor MafG | MAFG (2 of 2) | 1 |
| TCONS_00017983 | P09972 | Fructose-bisphosphate aldolase C | aldocb | 2 |
| TCONS_00018345 | P25325 | 3-mercaptopyruvate sulfurtransferase | zgc:162544 (1 of 2) | 1 |
| TCONS_00018346 | P25325 | 3-mercaptopyruvate sulfurtransferase | zgc:162544 (2 of 2) | 1 |
| TCONS_00018398 | Q9UQK1 | Protein phosphatase 1 regulatory subunit 3C | ppp1r3ca | 2 |
| TCONS_00019672 | Q9UPQ4 | Tripartite motif-containing protein 35 | TRIM35 (2 of 15) | 1 |
| TCONS_00020083 | P10599 | Thioredoxin | ENSXMAG00000001739 | 1 |
| TCONS_00020502 | P35558 | Phosphoenolpyruvate carboxykinase | pck1 | 1 |
| TCONS_00021089 | Q684P5 | Rap1 GTPase-activating protein 2 | rap1gap2a | 1 |
| TCONS_00021347 | P07339 | Cathepsin D | ctsd | 1 |
| TCONS_00021997 | O95571 | Persulfide dioxygenase ETHE1 | ETHE1 | 2 |
| TCONS_00022001 | O95571 | Persulfide dioxygenase ETHE1 | ETHE1 | 3 |
| TCONS_00022084 | Q86YN6 | Peroxisome proliferator-activated receptor gamma coactivator 1-beta | ENSXMAG00000019179 | 1 |
| TCONS_00022105 | Q5VV67 | Peroxisome proliferator-activated receptor gamma coactivator-related protein 1 | XLOC_017890 | 1 |
| TCONS_00022352 | n/a | XLOC_018095 | 1 | |
| TCONS_00022678 | P09105 | Hemoglobin subunit theta-1 | ENSXMAG00000000181 | 2 |
| TCONS_00022913 | Q7L5N7 | Lysophosphatidylcholine acyltransferase 2 | LPCAT4 (1 of 2) | 1 |
| TCONS_00022971 | P00918 | Carbonic anhydrase | cahz | 2 |
| TCONS_00023196 | Q53RY4 | Keratinocyte-associated protein 3 | tmem54 | 1 |
| TCONS_00023347 | Q9Y241 | HIG1 domain family member 1A | hig1 | 1 |
| TCONS_00023958 | Q15120 | Pyruvate dehydrogenase kinase, isozyme 3 | pdk3a | 1 |
| TCONS_00024060 | Q56VL3 | OCIA domain-containing protein 2 | ociad2 | 1 |
| TCONS_00025513 | P25325 | 3-mercaptopyruvate sulfurtransferase | ENSXMAG00000008042 | 1 |
| TCONS_00025981 | P06733 | Alpha-enolase | eno1a | 1 |
| TCONS_00026107 | n/a | abch1 | 1 | |
| TCONS_00026499 | O60675 | Transcription factor MafK | maff | 1 |
| TCONS_00026767 | P0CG29 | Glutathione S-transferase theta-2 | ENSXMAG00000006919 | 4 |
| TCONS_00026929 | P48506 | Glutamate-cysteine ligase catalytic subunit | gclc | 1 |
| TCONS_00028224 | n/a | XLOC_022849 | 1 | |
| TCONS_00028623 | n/a | XLOC_023166 | 1 | |
| TCONS_00028908 | n/a | XLOC_023394 | 1 | |
| TCONS_00030449 | Q9Y6M7 | Sodium bicarbonate cotransporter 3 | SLC4A7 (1 of 2) | 2 |
| TCONS_00030815 | Q96FC7 | Phytanoyl-CoA hydroxylase-interacting protein-like | ENSXMAG00000007363 | 1 |
| TCONS_00033175 | Q06830 | Peroxiredoxin 1 | prdx1 | 3 |
| TCONS_00034020 | Q8N4X5 | Actin filament-associated protein 1-like 2 | si:dkey-220o5.5 | 1 |
| TCONS_00034143 | O15394 | Neural cell adhesion molecule 2 | ENSXMAG00000017303 | 1 |
| TCONS_00034516 | n/a | XLOC_028149 | 1 | |
| TCONS_00034517 | P07911 | Uromodulin | XLOC_028150 | 1 |
Note.—Provided are the GenBank accession number for the top BLAST hit, the gene name, the corresponding identifier in the Xiphophorus maculatus (Xmac) reference genome, and the number of transcripts with evidence for upregulation for each gene. For additional information, see supplementary table S4, Supplementary Material online.
n/a, not applicable.
Table 2.
List of Genes that Were Consistently Downregulated in Sulfide Spring Fishes of All Drainages.
| Gene ID | Accession Number | Gene Name | Xmac Annotation | Number of Transcripts |
|---|---|---|---|---|
| TCONS_00000141 | Q9BQR3 | Serine protease 27 | zgc:165423 | 3 |
| TCONS_00000307 | P05787 | Keratin, type II cytoskeletal 8 | wu:fd11e11 | 1 |
| TCONS_00000716 | O43865 | Adenosylhomocysteinase 2 | ahcyl1 (2 of 3) | 2 |
| TCONS_00001614 | n/a | XLOC_001505 | 1 | |
| TCONS_00003728 | O43520 | Phospholipid-transporting ATPase IC | atp8b3 | 1 |
| TCONS_00003846 | P51589 | Cytochrome P450 2J2 | ENSXMAG00000005929 | 1 |
| TCONS_00004097 | n/a | XLOC_003586 | 1 | |
| TCONS_00004170 | Q13823 | Nucleolar GTP-binding protein 2 | gnl2 | 1 |
| TCONS_00004688 | n/a | XLOC_004052 | 1 | |
| TCONS_00005037 | O00483 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 | NDUFA4 (1 of 2) | 2 |
| TCONS_00005679 | O94900 | Thymocyte selection-associated high mobility group box protein TOX | Tox | 1 |
| TCONS_00005785 | P51857 | 3-oxo-5-beta-steroid 4-dehydrogenase | AKR1D1 (1 of 2) | 1 |
| TCONS_00006859 | P05787 | Keratin, type II cytoskeletal 8 | krt4 | 1 |
| TCONS_00007568 | Q96IP4 | Protein FAM46A | FAM46B (2 of 2) | 1 |
| TCONS_00007657 | Q92539 | Phosphatidate phosphatase LPIN2 | LPIN2 (1 of 3) | 1 |
| TCONS_00008606 | n/a | XLOC_007101 | 1 | |
| TCONS_00010147 | P62508 | Estrogen-related receptor gamma | Esrrgb | 1 |
| TCONS_00010450 | O43490 | Prominin-1 | prom2 | 2 |
| TCONS_00010764 | Q8IXH8 | Cadherin-like protein 26 | CDH26 (2 of 4) | 1 |
| TCONS_00011212 | Q5FVE4 | Long-chain-fatty-acid-CoA ligase ACSBG2 | acsbg1 | 1 |
| TCONS_00011532 | Q9HBU6 | Ethanolamine kinase 1 | etnk1 | 1 |
| TCONS_00011703 | Q96HN2 | Adenosylhomocysteinase 3 | ahcyl1 (3 of 3) | 1 |
| TCONS_00011898 | Q9UPQ4 | Tripartite motif-containing protein 35 | trim35-12 | 1 |
| TCONS_00012317 | O60825 | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 | pfkfb2a | 1 |
| TCONS_00013668 | Q9NZW5 | MAGUK p55 subfamily member 6 | mpp6b | 1 |
| TCONS_00014144 | P08779 | Keratin, type I cytoskeletal 16 | krt97 | 3 |
| TCONS_00014168 | Q9GZV3 | High affinity choline transporter 1 | ENSXMAG00000017837 | 1 |
| TCONS_00015517 | Q9HBA0 | Transient receptor potential cation channel subfamily V member 4 | trpv4 | 1 |
| TCONS_00015752 | Q96QD5 | DEP domain-containing protein 7 | depdc7 | 1 |
| TCONS_00016045 | Q9NXI6 | RING finger protein 186 | im:7152348 | 1 |
| TCONS_00016260 | Q99259 | Glutamate decarboxylase 1 | gad1b | 1 |
| TCONS_00017126 | n/a | XLOC_013895 | 1 | |
| TCONS_00018061 | Q6UXB0 | Protein FAM131A | FAM131A | 1 |
| TCONS_00018951 | P52943 | Cysteine-rich protein 2 | CRIP3 | 1 |
| TCONS_00019566 | Q02218 | 2-oxoglutarate dehydrogenase | OGDH (2 of 2) | 2 |
| TCONS_00019615 | Q8WVX9 | Fatty acyl-CoA reductase 1 | FAR2 | 1 |
| TCONS_00019789 | O14745 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | SLC9A3R1 (2 of 2) | 1 |
| TCONS_00019956 | Q86UD5 | Mitochondrial sodium/hydrogen exchanger 9B2 | si:dkey-162b23.4 | 2 |
| TCONS_00020193 | Q9NSE2 | Cytokine-inducible SH2-containing protein | Cishb | 1 |
| TCONS_00020392 | Q96S86 | Hyaluronan and proteoglycan link protein 3 | hapln3 | 1 |
| TCONS_00020515 | Q8N5B7 | Ceramide synthase 5 | cers5 | 1 |
| TCONS_00021208 | Q02779 | Mitogen-activated protein kinase 10 | map3k10 | 1 |
| TCONS_00021855 | Q04912 | Macrophage-stimulating protein receptor | mst1rb | 1 |
| TCONS_00022897 | P55017 | Solute carrier family 12 member 3 | ENSXMAG00000000951 | 2 |
| TCONS_00023777 | Q9NUV9 | GTPase IMAP family member 4 | ENSXMAG00000019889 | 2 |
| TCONS_00024008 | Q0VF96 | Cingulin-like protein 1 | CGNL1 | 1 |
| TCONS_00024654 | O76013 | Keratin, type I cuticular Ha6 | zgc:171226 (2 of 2) | 1 |
| TCONS_00024675 | P08727 | Keratin, type I cytoskeletal 19 | zgc:171226 (1 of 2) | 2 |
| TCONS_00024983 | O95267 | RAS guanyl-releasing protein 1 | rasgrp4 | 2 |
| TCONS_00025014 | Q9P0J1 | [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1 | pdp1 | 1 |
| TCONS_00025959 | Q8TD43 | Transient receptor potential cation channel subfamily M member 4 | trpm4a | 1 |
| TCONS_00027843 | Q6NUK1 | Calcium-binding mitochondrial carrier protein SCaMC-1 | SLC25A24 (2 of 2) | 1 |
| TCONS_00028646 | O14493 | Claudin-4 | Cldne | 2 |
| TCONS_00028684 | Q9Y5Z4 | Heme-binding protein 2 | soul5 | 2 |
| TCONS_00030067 | Q8N1W1 | Rho guanine nucleotide exchange factor 28 | arhgef28 | 1 |
| TCONS_00031911 | P58107 | Epiplakin | XLOC_025876 | 1 |
| TCONS_00032188 | P36269 | Gamma-glutamyltransferase 5 | GGT5 (3 of 3) | 1 |
Note.—Provided are the GenBank accession number for the top BLAST hit, the gene name, the corresponding identifier in the Xiphophorus maculatus (Xmac) reference genome, and the number of transcripts with evidence for downregulation for each gene. For additional information, see supplementary table S4, Supplementary Material online.
GO enrichment analysis indicated significant overrepresentation of certain GO terms associated with shared differentially expressed genes relative to the background set (all 13,127 transcripts associated with a GO term). Specifically, shared upregulated genes were enriched for 60 GO terms associated with biological processes, 16 associated with molecular function, and 12 associated with cellular components (supplementary table S5 and figs. S3–S5, Supplementary Material online); shared downregulated genes were enriched for 13 terms associated with biological processes and 3 associated with cellular components (supplementary table S6 and figs. S6 and S7, Supplementary Material online). As predicted, the mitochondrial respiratory chain and other parts of mitochondria (GO:0070469, as well as GO:0044429 and associated terms) belonged to the cellular components with evidence for significant enrichment in shared differentially expressed genes, reflecting the fact that these components represent a nexus in H2S toxicity and detoxification (Cooper and Brown 2008; Hildebrandt and Grieshaber 2008). In the following sections, we focus on the discussion of enriched GO terms associated with biological processes in the context of the three general mechanisms potentially driving elevated tolerance to environmental H2S.
Evidence for Differential Expression of Genes that Could Mediate H2S Exclusion
Increased tolerance to high environmental concentrations of H2S may be mediated by the formation of barriers that minimize diffusion into the body (Vismann 1991). Our analyses have provided no evidence suggesting that sulfide spring fishes differentially express genes that could be associated with decreasing the flux of H2S. In fact, genes encoding for claudins and keratins (table 2), which are associated with the establishment of skin barriers (Fuchs and Weber 1994; Günzel and Yu 2013) and enriched relative to the background set (GO:0061436), were consistently downregulated and not upregulated in sulfide spring populations. In addition, shared differentially expressed genes associated with the function of cellular membranes (basolateral and apical plasma membranes; GO:0016323 and GO:0016324) were primarily involved in ion transport during osmoregulatory processes (see below).
The apparent lack of modifications to the integumentary system and respiratory surfaces is not entirely unexpected, because there is likely a trade-off between H2S exclusion and oxygen acquisition. Barriers that reduce diffusion of H2S likely reduce the diffusion of oxygen across gill epithelia. This trade-off is also reflected in the expression of a morphological trait, where sulfide spring fishes exhibit higher gill surface areas per unit body mass than fish from nonsulfidic environments (Tobler et al. 2011). Hence, effective exclusion of H2S may be constrained by organisms’ need for oxygen acquisition in the hypoxic waters of sulfide springs. Evidence for structural modifications that mediate H2S exclusion from the body has also been scarce in other systems (see Bagarinao 1992 for a review), and behavioral adaptations, including alternative respiratory strategies that minimize the contact of respiratory surfaces with H2S-rich water, may be more important to minimize the influx of H2S into the body (Abel et al. 1987; Brauner et al. 1995; Greenway et al. 2014).
Evidence for Differential Expression of Genes that Could Mediate Maintenance of H2S Homeostasis
Increased tolerance to high environmental concentrations of H2S may be mediated by an increased ability of sulfide spring fishes to maintain endogenous H2S homeostasis despite its continuous influx from the environment. Indeed, we found consistent upregulation of genes involved in enzymatic H2S oxidation (GO:0019418 and associated terms), glutathione metabolism (GO:0006749 and associated terms), as well the transport of oxidized sulfur species (GO:0008272 and associated terms). Genes associated with H2S oxidation primarily belonged to the SQR pathway (including SQR, persulfide dioxygenase [ETHE1], and a mitochondrial dicarboxylate carrier; table 1), which represents the primary route of enzymatic H2S detoxification in metazoans (Hildebrandt and Grieshaber 2008; Shahak and Hauska 2008). Upregulation of enzymes involved in glutathione metabolism (e.g., glutathione s-transferases and glutathione reductase) is also consistent with increased H2S detoxification capability, because sulfur molecules sequestered by SQR may be transferred to glutathione (Jackson et al. 2012). Upregulation of genes associated with enzymatic H2S oxidation has been documented in other animals (Ma et al. 2012; Liu et al. 2015) and human tissues (Lagoutte et al. 2010; Mimoun et al. 2012) exposed to elevated H2S concentrations. A recent laboratory study also indicated that sulfide spring P. mexicana from one river drainage investigated here (Tacotalpa) retained higher constitutive SQR expression than reference populations even in the absence of H2S (Tobler et al. 2014), and we verified the presence of differential amounts of protein of SQR using western blot (supplementary fig. S8, Supplementary Material online). Furthermore, a population genomic study has also uncovered significant signatures of positive selection on SQR in some sulfide spring populations of P. mexixana (Pfenninger et al. 2015), perhaps suggesting that there are both transcriptional and structural modifications to components of this pathway. Collectively, these results suggested that an increased expression of genes involved in H2S detoxification likely contribute to the maintenance of low endogenous concentrations, and adaptation in sulfide spring fishes likely involves a higher regulatory capacity during environmental H2S exposure.
In contrast, we found no evidence that an increased ability to maintain low endogenous H2S concentration may be linked to lower rates of endogenous production, which is primarily mediated by the processing of sulfur-containing amino acids (Kabil et al. 2014). Several genes associated with the metabolic processing of sulfur amino acids (GO:0000096 and associated terms) were consistently upregulated in sulfide spring fishes. These included cysteine dioxygenase and mercaptopyruvate sulfurtransferase (table 1), the latter of which has been directly implicated in the endogenous production of H2S (Stipanuk 2004; Stipanuk and Ueki 2011; Módis et al. 2012). Although the enzymatic processing of sulfur amino acids can result in the production of a wide variety of metabolites and may not precipitate in differences in endogenous H2S production between sulfidic and nonsulfidic fish populations, these findings suggested that endogenous production of H2S in sulfide spring fish is probably higher and certainly not lower than in close relatives from nonsulfidic habitats. This is consistent with a previous study that documented an increase in the expression of cystathionine γ lyase, a primary producer of endogenous H2S (Stipanuk and Ueki 2011), upon short-term exposure to H2S both in sulfidic and nonsulfidic populations (Tobler et al. 2014). It remains to be investigated whether upregulation of genes associated with the processing of sulfur-containing amino acids and endogenous H2S production represents a maladaptive response to environmental H2S exposure, or whether it contributes to H2S processing and adaptation in ways not currently understood.
Evidence for Differential Expression of Genes that Could Reduce Side Effects of H2S Toxicity
Increased tolerance to high environmental concentrations of H2S may be mediated by modification of toxicity targets that allow for proper organismal function, even endogenous H2S concentrations are elevated. H2S toxicity is primarily mediated through its binding to COX, effectively halting oxidative phosphorylation (OXPHOS) in the mitochondrial respiratory chain. Our analysis indicated significant enrichment and upregulation of genes associated with the transfer of electrons from cytochrome c to oxygen (GO:0006123), which is mediated by COX. This included upregulation of cytochrome c (table 1), which receives electrons from coenzyme Q through complex III of the respiratory chain (Vedel et al. 1999). Although most proteins associated with the respiratory chain are organized in complexes, anchored in the inner membrane of mitochondria, and can only transfer electrons, cytochrome c is concentrated in the intermembrane space, only loosely associated with the inner membrane, and can permanently hold electrons until being processed by COX (Hatefi 1985; Saraste 1999). Upregulation of cytochrome c may be adaptive in sulfidic environments by buffering against deviations in the stoichiometric balance between different protein complexes of the respiratory chain (Lemos et al. 2004). Deviations in the stoichiometric balance are expected during exposure to environmental H2S both because of COX blockage and because SQR—in addition to complexes I and II—transfers electrons to coenzyme Q (Lagoutte et al. 2010). Supplementary electron flow has been linked to oxidative stress (see below), and upregulation of cytochrome c could alleviate bottlenecks in electron flow. In addition, enrichment of genes associated with the transfer of electrons from cytochrome c to oxygen include proteins that are critical for the assembly of the COX enzyme (Glerum et al. 1997), although the adaptive significance of this transcriptional change remains unclear. A previous study already indicated that two lineages of sulfide spring fish in the P. mexicana complex (Puyacatengo and Pichucalco drainages) have evolved a COX that is resistant to the toxic effects of H2S and allows for the retention of high enzyme activity in the presence of H2S (Pfenninger et al. 2014). Consequently, there is evidence for both structural and transcriptional variation between sulfidic and nonsulfidic populations that are likely related to the maintenance of electron flow in the respiratory chain and the minimization of oxidative stress in the presence of H2S.
Through its interference with COX function and OXPHOS, H2S also has profound impacts on other aspects of energy metabolism. Even though there was no evidence for enrichment in other genes involved in the mitochondrial respiratory chain, it is important to note that there was evidence for consistent differential expression in other genes associated with OXPHOS and mitochondrial bioenergetics (TCONS_00005037 and TCONS_00005038, which are part of a subunit of NADH dehydrogenase [complex I in fig. 1A]; TCONS_00019566 and TCONS_00019567, which encode for oxoglutarate dehydrogenase, an enzyme of the citric acid cycle). Inspection of metabolic pathway maps generated with iPath indicated that components of complexes I and II of the respiratory chain (fig. 1A), which harvest electrons from organic substrates, were consistently downregulated in all sulfide spring populations (supplementary fig. S9, Supplementary Material online). This is consistent with a potential shift in the relative importance of H2S and organic substrates as electron donors for OXPHOS (Völkel and Grieshaber 1997; Goubern et al. 2007). In addition, fish from sulfidic habitats upregulated and showed enrichment for a variety of genes involved in carbohydrate metabolism, particularly glycolysis (GO:0061621 and associated terms) and gluconeogenesis (GO:0006094 and associated terms). Glycolysis is a primary pathway producing ATP in an oxygen (OXPHOS)-independent manner through the processing of glucose, and gluconeogenesis generates glucose from a variety of carbon substrates (Richards 2009). Increases in glycolysis are commonly observed in organisms exposed to hypoxic conditions (Soengas and Aldegunde 2002; Richards 2009; Speers-Roesch et al. 2013). Potential upregulation of anaerobic metabolism in sulfide spring fish may be linked to hypoxia in sulfidic environments (reduced aerobic ATP production due to shortages on oxygen supply; see below) and/or to the interruption of OXPHOS by H2S (reduced aerobic ATP production due to blocking of COX). Hence, future studies will need to unravel how COX blockage and oxygen availability interact to modulate anaerobic metabolism in sulfide spring fish.
Finally, H2S has also been shown to be a potent generator of reactive oxygen species and oxidative stress (Eghbal et al. 2004). Sulfide spring fishes exhibited a consistent upregulation of genes involved in responses to reactive oxygen species (GO:0000302) and oxidative stress in general (GO:0006979). This does not only include the genes associated with the glutathione pathway discussed above, but also peroxiredoxin and thioredoxin (table 1), which act as antioxidants (Immenschuh and Baumgart-Vogt 2005; Koharyova and Kollarova 2008). Consequently, our analyses of gene expression variation across different population of sulfide spring fishes provided evidence for modification of genes that are directly or indirectly related to the toxic effects of H2S.
Evidence for Differential Expression of Genes Associated with Environmental Factors that Correlate with the Presence of H2S
Sulfide springs and adjacent nonsulfidic habitats not only differ in the presence and absence of H2S, but they vary in a suite of abiotic and biotic environmental characteristics that may act as a source of selection and shape population differences in trait expression (Tobler and Plath 2011; Greenway et al. 2014). Sulfide springs are typically characterized by having lower dissolved oxygen concentrations, a lower pH, and higher concentrations of dissolved salts than adjacent nonsulfidic habitats (Tobler et al. 2006; Rosales 2012). Differences in gene expression both between sulfidic and nonsulfidic populations as well as among different sulfidic populations could therefore be driven by a variety of environmental factors that were not measured in this study. For example, we found enrichment in genes that have been associated with responses to pH (GO:0009268), sodium transport (GO:0006814), water loss via skin (GO:0033561), and metabolic processes (GO:0061621, GO:0006094, and associated terms), which may be related to environmental factors correlated with the presence of H2S. Furthermore, there was evidence for enrichment in a variety of other GO terms (supplementary tables S5 and 6, Supplementary Material online), for which the putative functional links to the presence of H2S and other environmental factors remain elusive. Future studies will have to investigate how exposure to different environmental factors separately and jointly shapes patterns of gene expression in organisms that are exposed to multifarious selective regimes.
Conclusions
Transcriptome analyses in replicated lineages of sulfide spring fishes and corresponding reference populations provided evidence for substantial variation in gene expression across small spatial scales. Genes that were consistently differentially expressed across sulfide spring populations provide promising candidates for the identification of molecular mechanisms underlying adaptation to these extreme environments, although future work will also need to scrutinize how genes that were only differentially regulated in a subset of lineages may contribute to coping with H2S toxicity. There was clear evidence for the upregulation of genes associated with the detoxification and subsequent excretion of H2S. At the same time, differential expression was evident for genes associated with energy metabolism and oxidative stress, two primary physiological processes affected by H2S. Hence, modification of processes associated with detoxification and toxicity likely complement each other to mediate elevated H2S tolerance in sulfide spring fishes. Furthermore, the mitochondrial respiratory chain appears to represent a center of adaptation to the perpetual exposure to H2S, which is consistent with its joint role in both H2S toxicity and detoxification. Our analyses allow for the development of testable hypotheses about biochemical and physiological mechanisms of adaptation to H2S-rich environments. Future studies will have to focus their efforts on three key problems: 1) Identifying the drivers underlying variation in gene regulation in natural populations will require controlled laboratory experiments. Rearing fish from replicated populations under standardized conditions and exposing individuals to varying H2S concentrations will help disentangle plastic and genetic contributions to gene expression patterns observed in the wild (Whitehead 2012). 2) Quantifying genetic variation in protein-coding genes and identifying variants that have likely been shaped by natural selection will allow testing how variation in protein structure and protein regulation interact to mediate adaptation (Hoekstra and Coyne 2007; Wray 2007). 3) Quantifying the joint functional consequences of variation in gene expression and genetic variation will provide a mechanistic understanding of the biochemical and physiological processes that govern differences in H2S tolerance.
Our study contributes to a broader effort to understand molecular genetic mechanisms underlying life in extreme environment (Dassanayake et al. 2011; Teets et al. 2012; Kelley et al. 2014; Kavembe et al. 2015). The findings suggest that adaptation to physiochemical stressors involves the modification of multiple, complex physiological pathways that likely complement each other. Similar results have been found in other systems, where resistance to pesticides involves modifications to the integument that slows down diffusion of toxins into the body as well as alterations of toxicity targets that reduce their binding activity (Zhu et al. 2013). This illustrates the phenomenal challenges of relating variation in specific genes and their expression to physiological performance and ultimately organismal fitness (Storz and Wheat 2010). Acknowledging the complexity of organisms conceptually and empirically during investigations of adaptation will be paramount to understand the multifarious, layered strategies organisms use to cope with stressful environmental conditions. Although a wide variety of biochemical, physiological, structural, and behavioral coping strategies have been discovered for specific physicochemical stressors (including the one studied here; Riesch et al. 2015), disentangling the relative contributions of different traits to overall organismal performance remains a key challenge that should reveal potential synergies and redundancies among strategies. Ultimately, elucidating how organisms respond and adapt to physiochemical stressors that govern life in extreme environments will be instrumental for predicting the resilience and response of species to environmental stress in the face of global environmental change (Tobler et al. 2015).
Materials and Methods
Study Sites and Sample Collection
Samples of Poecilia for transcriptome analyses were collected in the area around the city of Teapa (Tabasco, Mexico). Sulfide spring complexes inhabited by Poecilia are located in the foothills of the Sierra Madre and are distributed across three major tributaries of the Río Grijalva (from east to west: Tacotalpa, Puyacatengo, and Pichucalco drainages; supplementary fig. S1, Supplementary Material online). Sulfide springs in each drainage were colonized independently by P. mexicana–like ancestors (Tobler et al. 2011; Palacios et al. 2013). All populations investigated here taxonomically belong to P. mexicana, except for the sulfide spring population in the Río Pichucalco drainage, which has been described as a distinct and highly endemic species (P. sulphuraria) (Alvarez del Villar 1948; Palacios et al. 2013). In each of the drainages, we collected individual fish from one sulfide spring complex and one proximate, nonsulfidic habitat of similar size and structure (N = 5–6 per site; see supplementary table S7, Supplementary Material online, for details). Fish were caught with a seine (2 × 4 meters). Immediately after capture, individual fish were sacrificed, measured and weighed, and gill tissues were extracted from both sides of the body using previously sterilized scissors and forceps. Tissues were preserved in 2 ml of RNAlater (Ambion, Inc.). All experiments were approved by the Institutional Animal Care and Use Committee of Kansas State University (ACUP #3473).
RNA Isolation and RNAseq Library Construction
RNA was isolated from gills by pulverizing 50–100 mg of tissue frozen in liquid nitrogen with a Covaris Cryoprep at setting 3. RNA was then extracted with Qiagen’s RNeasy Plus mini kit. PolyA+ mRNA was prepared from 50 μg total RNA using Invitrogen’s Dynabeads mRNA purification kit. RNA was bound to and eluted twice from Dynabeads to minimize ribosomal RNA contamination. mRNA was fragmented to an average size of 400 nt using NEB’s mRNA Fragmentation Module by incubation at 94 °C for 4 min. Fragmented mRNA was purified using Agencourt RNAClean XP beads and eluted in 12 μl ddH2O. First-strand cDNA was synthesized in a 20 μl reaction using Invitrogen’s double-stranded cDNA kit, primed with 1 μl of a mix of random hexamers:oligo dT primers (2 μg:1 μg), and incubated with Superscript II at 45 °C for 1 h. Double-stranded cDNA was synthesized directly from the first-strand cDNA using the NEBNext mRNA Second Strand Synthesis kit. After second-strand cDNA synthesis, the reaction was purified with Agencourt Ampure XP beads and eluted in 25 μl ddH2O.
Double-stranded cDNA was used as input for Illumina sequencing library preparation with end-repair using the NEBNext end-repair kit, A-tailing with Taq polymerase, ligation with Truseq barcoded adapters, and amplification with Kapa Library Amplification Readymix. All steps were cleaned up with Ampure XP beads. RNAseq libraries were quantified on an Agilent 2100 Bioanalyzer High Sensitivity DNA chip and pooled in sets of 12 based on nanomolar (nM) concentration. Individuals were split into pools such that samples from different habitat types and drainages were sequenced together, and there was no evidence for significant lane effects. Libraries were sequenced on an Illumina HiSeq 2000 with paired-end 101 bp reads at the Stanford Center for Genomics and Personalized Medicine. Sequence data were deposited in the Sequence Read Archive on GenBank (study accession ID: PRJNA290391).
Transcriptome Assembly and Annotation
Raw RNA-seq reads from Illumina sequencing were sorted by barcode and dynamically trimmed to quality 20 using TrimGalore! (Krueger 2014). All reads shorter than 50 bases were removed, and only paired reads were used for the analysis, which resulted in removal of ∼10% of sequenced bases (supplementary table S1, Supplementary Material online). Trimmed reads for each of the 35 individuals were mapped to the platyfish (X. maculatus) genome (version 4.4.2 release-75; Schartl et al. 2013). Mapping to the reference genome was accomplished using Stampy (version 1.0.23), which is designed for mapping reads to a divergent reference genome (Lunter and Goodson 2011). Cufflinks (version 2.2.1) was then used to extract putatively expressed regions for each individual, with the platyfish reference gene set as a guide (Trapnell et al. 2010). The number of unique loci to the P. mexicana data set was queried using cuffcompare. Putatively expressed regions for each individual were merged with cuffmerge, and a FASTA file of all expressed loci was extracted using gffread, as implemented in Cufflinks. Platyfish mitochondrial transcripts were removed from the FASTA file and replaced with the appropriate Poecilia mitochondrial reference sequences, which included 13 protein-coding genes and the 16S ribosomal RNA sequence (GenBank: KC992995). Note that mapping reads to a de novo reference transcriptome assembly of P. mexicana using Trinity (see Kelley et al. 2012 for general approaches) yielded in qualitatively similar results. Mapping to the platyfish genome, however, led to fewer unannotated transcripts, which facilitated the functional interpretation of results.
To annotate transcripts, we first conducted a BLAST search of the longest transcript for each locus against the SwissProt database (http://ca.expasy.org/sprot/; BLASTX, critical e-value = 0.001; database accessed November 1, 2014). For each locus, we retained the top BLAST hit for subsequent analyses, and all transcripts for each locus were given the same SwissProt annotation. Sequences with a match in the SwissProt database were subsequently annotated with GO IDs (Gene Ontology Consortium 2004). GO IDs describe gene product characteristics and are hierarchically organized in terms of biological processes, molecular functions, and cellular components (Gene Ontology Consortium 2004).
Quantifying and Comparing Gene Expression Patterns
Transcript abundance was estimated by remapping the RNA-seq reads to the extracted transcriptome using Stampy (Lunter and Goodson 2011). Overall, transcript abundance (expression values) was estimated using eXpress (Roberts and Pachter 2013). We used the edgeR function calcNormFactors (Robinson et al. 2010; Robinson and Oshlack 2010) to estimate an effective library size, which accounts for differences in the number of total reads (total library size) among individuals. We modeled the common dispersion using the estimateCommonDisp function. We also modeled tagwise (gene-wise) dispersion using the estimateTagwiseDisp function to prevent outliers from driving any signal of differential expression between comparisons. We tested for differential expression between pairs of sulfidic and nonsulfidic ecotypes within the same drainage using an exact test implemented in edgeR. To control for multiple testing, we used a genome-wide false discovery rate (FDR) of 0.001 (Benjamini and Hochberg 1995).
To describe whether gene expression patterns in replicated sulfide spring populations changed in convergence, we conducted hierarchical cluster analysis. If gene expression patterns have changed in convergence, individuals from lineages living under the same environmental conditions (sulfidic vs. nonsulfidic) should cluster together. The hierarchical cluster analysis was performed on the top 10,000 expressed genes using the log-counts-per-million with prior count set to 5 to avoid taking the log of zero counts. We then used the heatmap.2 function from the GPLOTS package in R to generate hierarchical clusters. In addition, we conducted a WGCNA using the top 10,000 expressed genes. We used the variance-stabilizing transformation function in DESeq2 (Love et al. 2014) and constructed networks with the R package WGCNA (Langfelder and Horvath 2008). Clustering of gene expression profiles for the retained genes was consistent with hierarchical clustering based on the most expressed genes (fig. 2). The soft threshold (power) for the combined network was 6, which was the lowest value that optimized the scale free topology.
Identifying Potentially Adaptive Changes and Testing Hypotheses about the Function of Differentially Expressed Genes
The sets of differentially expressed genes with FDR < 0.001 for each pair of sulfidic and nonsulfidic ecotypes within the same drainage were intersected to identify genes with shared patterns of differential expression. GO annotations were then used in an enrichment analysis for biological processes, molecular function, and cellular components as implemented in GOrilla with a P value threshold of 0.001 (Eden et al. 2009). Furthermore, we visualized metabolic pathways associated with shared differentially expressed genes through annotation with KEGG orthologs (Kyoto Encyclopedia of Genes and Genomes; Kanehisa et al. 2012) as implemented in iPath v2 (Letunic et al. 2008).
Supplementary Material
Supplementary figures S1–S9 and tables S1–S7 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank the communities of Teapa and Tapijulapa, as well as local landowners and Villa Luz Nature Park for providing access to study sites. We are indebted to Centro de Investigación e Innovación para la Enseñanza y Aprendizaje and Universidad Juárez Autónoma de Tabasco for their hospitality and support over many years of research. O. E. Cornejo, Z. W. Culumber, K. K. Dhillon, M. E. Palacios-Mejia, and C. N. Passow assisted during work in the field, helped in the laboratory, provided technical advice, and/or commented on previous versions of his manuscript. Permits were kindly provided by the Mexican Federal Agencies SEMARNAT and CONAPESCA (DGOPA.09004.041111.3088, SGPA/DGVS/04315/11, PRMN/DGOPA-003/2014, PRMN/DGOPA-009/2015). This work was funded through grants from the National Science Foundation (IOS-1121832, IOS-1463720) and the Army Research Office (W911NF-15-1-0175) to M.T. and J.L.K., a L’Oreal Fellowship for Women in Science to J.L.K., and a Ralph E. Powe Junior Faculty Enhancement Award from Oak Ridge Associated Universities to M.T.
References
- Abel DC, Koenig CC, Davis WP. 1987. Emersion in the mangrove forest fish Rivulus marmoratus: a unique response to hydrogen sulfide. Environ Biol Fish. 18:67–72. [Google Scholar]
- Alvarez del Villar J. 1948. Descripción de una nueva especie de Mollienisia capturada en Baños del Azufre, Tabasco (Pisces, Poeciliidae). An Esc Nac Cienc Biol. 5:275–281. [Google Scholar]
- Bagarinao T. 1992. Sulfide as an environmental factor and toxicant: tolerance and adaptations in aquatic organisms. Aquat Toxicol. 24:21–62. [Google Scholar]
- Bagarinao T, Vetter RD. 1992. Sulfide-hemoglobin interactions in the sulfide-tolerant salt marsh resident, the California killifish Fundulus parvipinnis. J Comp Physiol B. 162:614–624. [Google Scholar]
- Beauchamp RO, Bus JS, Popp JA, Boreiko CJ, Andjelkovich DA. 1984. A critical review of the literature on hydrogen sulfide toxicity. Crit Rev Toxicol 13:25–97. [DOI] [PubMed] [Google Scholar]
- Bell EM. 2012. Life at extremes: environments, organisms and strategies for survival. Oxfordshire (UK): CABI. [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 57:289–300. [Google Scholar]
- Brauner CJ, Ballantyne CL, Randall DJ, Val AL. 1995. Air-breathing in the armored catfish (Hoplosternum littorale) as an adaptation to hypoxic, acidic, and hydrogen sulfide-rich waters. Can J Zool. 73:739–744. [Google Scholar]
- Childress JJ, Fisher CR. 1992. The biology of hydrothermal vent animals: physiology, biochemistry, and autotrophic symbioses. Oceanogr Mar Biol Ann Rev. 30:337–441. [Google Scholar]
- Choi YJ, Aliota MT, Mayhew GF, Erickson SM, Christensen BM. 2014. Dual RNA-seq of parasite and host reveals gene expression dynamics during filarial worm-mosquito interactions. PLoS Negl Trop Dis. 8:e2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CE, Brown GC. 2008. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. J Bioenerg Biomembr. 40:533–539. [DOI] [PubMed] [Google Scholar]
- Dassanayake M, Oh DH, Haas JS, Hernandez A, Hong H, Ali S, Yun DJ, Bressan RA, Zhu JK, Bohnert HJ, et al. 2011. The genome of the extremophile crucifer Thellungiella parvula. Nat Genet. 43:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degn H, Kristensen B. 1981. Low sensitivity of Tubifex sp. respiration to hydrogen sulfide and other inhibitors. Comp Biochem Physiol. 69B:809–817. [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eghbal MA, Pennefather PS, O'Brien PJ. 2004. H2S cytotoxicity mechanism involves reactive oxygen species formation and mitochondrial depolarisation. Toxicology 203:69–76. [DOI] [PubMed] [Google Scholar]
- Evans DH, Claiborne JB. 2006. The physiology of fishes. Boca Raton (FL): Taylor & Francis. [Google Scholar]
- Evans TG, Hammill E, Kaukinen K, Schulze AD, Patterson DA, English KK, Curtis JMR, Miller KM. 2011. Transcriptomics of environmental acclimatization and survival in wild adult Pacific sockeye salmon (Oncorhynchus nerka) during spawning migration. Mol Ecol. 20:4472–4489. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Weber K. 1994. Intermediate filaments: structure, dynamics, function, and disease. Ann Rev Biochem. 63:345–382. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32:D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghalambor CK, Hoke KL, Ruell EW, Fischer EK, Reznick D, Hughes KA. 2015. Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature 525:372–375. [DOI] [PubMed] [Google Scholar]
- Glerum DM, Muroff I, Jin C, Tzagoloff A. 1997. COX15 codes for a mitochondrial protein essential for the assembly of yeast cytochrome oxidase. J Biol Chem. 272:19088–19094. [DOI] [PubMed] [Google Scholar]
- Goubern M, Andriamihaja M, Nubel T, Blachier F, Bouillaud F. 2007. Sulfide, the first inorganic substrate for human cells. FASEB J. 21:1699–1706. [DOI] [PubMed] [Google Scholar]
- Greenway R, Arias-Rodriguez L, Diaz P, Tobler M. 2014. Patterns of macroinvertebrate and fish diversity in freshwater sulphide springs. Diversity 6:597–632. [Google Scholar]
- Günzel D, Yu ASL. 2013. Claudins and the modulation of tight junction permeability. Physiol Rev. 93:525–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatefi Y. 1985. The mitochondrial electron transport and oxidative phosphorylation system. Ann Rev Biochem. 54:1015–1069. [DOI] [PubMed] [Google Scholar]
- Hildebrandt TM, Grieshaber M. 2008. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J. 275:3352–3361. [DOI] [PubMed] [Google Scholar]
- Hoekstra H, Coyne JA. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61:995–1016. [DOI] [PubMed] [Google Scholar]
- Hoekstra H, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. 2006. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 313:101–104. [DOI] [PubMed] [Google Scholar]
- Immenschuh S, Baumgart-Vogt E. 2005. Peroxiredoxins, oxidative stress, and cell proliferation. Antioxid Redox Signal. 7:768–777. [DOI] [PubMed] [Google Scholar]
- Jackson MR, Melideo SL, Jorns M. 2012. Human sulfide:quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry 51:6804–6815. [DOI] [PubMed] [Google Scholar]
- Kabil O, Vitvitsky V, Banerjee R. 2014. Sulfur as a signaling nutrient through hydrogen sulfide. Ann Rev Nutr. 34:171–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeuffer R, Peichel CL, Bolnick DI, Hendry AP. 2012. Parallel and nonparallel aspects of ecological, phenotypic, and genetic divergence across replicate population pairs of lake and stream stickleback. Evolution 66:402–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavembe GD, Franchini P, Irisarri I, Machado-Schiaffino G, Meyer A. 2015. Genomics of adaptation to multiple concurrent stresses: insights from comparative transcriptomics of a cichlid fish from one of Earth’s most extreme environment, the hypersaline Soda Lake Magadi in Kenya, East Africa. J Mol Evol. 81:90–109. [DOI] [PubMed] [Google Scholar]
- Kelley JL, Passow C, Plath M, Arias-Rodriguez L, Yee MC, Tobler M. 2012. Genomic resources for a model in adaptation and speciation research: the characterization of the Poecilia mexicana transcriptome. BMC Genomics 13:652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL, Peyton JT, Fiston-Lavier AS, Teets NM, Yee MC, Bustamante C, Lee RE, Denlinger DL. 2014. Insights into evolution of the small genome of the Antarctic midge. Nat Commun. 5:4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koharyova M, Kollarova M. 2008. Oxidative stress and thioredoxin system. Gen Physiol Biophys. 27:71–84. [PubMed] [Google Scholar]
- Krueger F. 2014. Trim Galore! version 0.3.7. Available from: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
- Kump LR, Pavlov A, Arthur MA. 2005. Massive release of hydrogen sulfide to the surface ocean and atmosphere during intervals of oceanic anoxia. Geology 33:397–400. [Google Scholar]
- Lagoutte E, Mimoun S, Andriamihaja M, Chaumontet C, Blachier F, Bouillaud F. 2010. Oxidation of hydrogen sulfide remains a priority in mammalian cells and causes reverse electron transfer in colonocytes. Biochim Biophys Acta. 1797:1500–1511. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Meiklejohn CD, Hartl DL. 2004. Regulatory evolution across the protein interaction nextwork. Nat Genet. 36:1059–1060. [DOI] [PubMed] [Google Scholar]
- Letunic I, Yamada T, Kanehisa M, Bork P. 2008. iPath: interactive exploration of biochemical pathways and networks. Trend Biochem Sci. 33:101–103. [DOI] [PubMed] [Google Scholar]
- Levin LA. 2005. Ecology of cold seep sediments: interactions of fauna with flow, chemistry and microbes. Oceanogr Mar Biol Ann Rev. 43:1–46. [Google Scholar]
- Levitt MD, Furne J, Springfield J, Suarez F, DeMaster E. 1999. Detoxification of hydrogen sulfide and methanethiol in the cecal mucosa. J Clin Invest. 104:1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Moore PK. 2008. Putative biological roles of hydrogen sulfide in health and disease: a breath of not so fresh air? Trend Pharmacol Sci. 29:84–90. [DOI] [PubMed] [Google Scholar]
- Li L, Rose P, Moore PK. 2011. Hydrogen sulfide and cell signaling. Ann Rev Pharmacol Toxicol. 51:169–187. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ma X, Li X, Zhou D, Gao B, Bai Y. 2015. Sulfide exposure results in enhanced sqr transcription through upregulating the expression and activation of HSF1 in echiuran worm Urechis unicinctus. Aquat Toxicol. 170:229–239. [DOI] [PubMed] [Google Scholar]
- Losos JB. 2011. Convergence, adaptation, and constraint. Evolution 65:1827–1840. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Goodson M. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21:936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YB, Zhang ZF, Shao MY, Kang KH, Shi XL, Dong YP, Li JJ. 2012. Response of sulfide-quinone oxidoreductase to sulfide exposure in the echiuran worm Urechis unicinctus. Mar Biotechnol 14:245–251. [DOI] [PubMed] [Google Scholar]
- McTaggart SJ, Cézard T, Garbutt JS, Wilson PJ, Little TJ. 2015. Transcriptome profiling during a natural host-parasite interaction. BMC Genomics 16:643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimoun S, Andriamihaja M, Chaumontet C, Atanasiu C, Benamouzig R, Blouin JM, Tome D, Bouillaud F, Blachier F. 2012. Detoxification of H2S by differentiated colonic epithelial cells: implications of the sulfide oxidizing unit and of the cell respiratory capacity. Antioxid Redox Signal 17:1–10. [DOI] [PubMed] [Google Scholar]
- Módis K, Coletta C, Erdélyi K, Papapetropoulos A, Szabo C. 2012. Intramitochondrial hydrogen sulfide production by 3-mercaptopyruvate sulfurtransferase maintains mitochondrial electron flow and supports cellular bioenergetics. FASEB J. 27:601–611. [DOI] [PubMed] [Google Scholar]
- Muyzer G, Stams AJM. 2008. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol. 6:441–454. [DOI] [PubMed] [Google Scholar]
- Naumova OY, Palejev D, Vlasova NV, Lee M, Rychkov SY, Babich ON, Vaccarino M, Grigorenko EL. 2012. Age-related changes of gene expression in the neocortex: preliminary data on RNA-Seq of the transcriptome in three functionally distinct cortical areas. Dev Psychopathol. 24:1427–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo E. 2011. Evolution under environmental stress at macro and microscales. Genome Biol Evol. 3:1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson KR. 2011. The therapeutic potential of hydrogen sulfide: separating hype from hope. Am J Physiol Regul Integr Comp Physiol. 301:R297–R312. [DOI] [PubMed] [Google Scholar]
- Olson KR, Straub KD. 2015. The role of hydrogen sulfide in evolution and the evolution of hydrogen sulfide in metabolism and signaling. Physiology 31:60–72. [DOI] [PubMed] [Google Scholar]
- Opitz CA, Kulke M, Leake MC, Neagoe C, Hinssen H, Hajjar RJ, Linke WA. 2003. Damped elastic recoil of the titin spring in myofibrils of human myocardium. Proc Natl Acad Sci U S A. 100:12688–12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios M, Arias-Rodriguez L, Plath M, Eifert C, Lerp H, Lamboj A, Voelker G, Tobler M. 2013. The redescovery of a long described species reveals additional complexity in speciation patterns of poeciliid fishes in sulfide springs. PLoS One 8:e71069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BD, Snyder SH. 2012. H2S signalling through protein sulfhydration and beyond. Nat Rev Mol Cell Biol. 13:499–507. [DOI] [PubMed] [Google Scholar]
- Pfenninger M, Lerp H, Tobler M, Passow C, Kelley JL, Funke E, Greshake B, Erkoc UK, Berberich T, Plath M. 2014. Parallel evolution of cox genes in H2S-tolerant fish as key adaptation to a toxic environment. Nat Commun. 5:3873. [DOI] [PubMed] [Google Scholar]
- Pfenninger M, Patel S, Arias-Rodriguez L, Feldmeyer B, Riesch R, Plath M. 2015. Unique evolutionary trajectories in repeated adaptation to hydrogen sulphide-toxic habitats of a neotropical fish (Poecilia mexicana). Mol Ecol. 24:5446–5459. [DOI] [PubMed] [Google Scholar]
- Plath M, Pfenninger M, Lerp H, Riesch R, Eschenbrenner C, Slattery P, Bierbach D, Herrmann N, Schulte M, Arias-Rodriguez L, et al. 2013. Genetic differentiation and selection against migrants in evolutionarily replicated extreme environments. Evolution 67:2647–2661. [DOI] [PubMed] [Google Scholar]
- Plath M, Riesch R, Oranth A, Dzienko J, Karau N, Schiessl A, Stadler S, Wigh A, Zimmer C, Arias-Rodriguez L, et al. 2010. Complementary effects of natural and sexual selection against immigrants maintains differentiation between locally adapted fish. Naturwissenschaften 97:769–774. [DOI] [PubMed] [Google Scholar]
- Rasch EM, Prehn LM, Rasch RW. 1970. Cytogenetic studies of Poecilia (Pisces): 2. Triploidy and DNA levels in naturally occurring populations associated with the gynogenetic teleost, Poecilia formosa (Girard). Chromosoma 31:18–40. [DOI] [PubMed] [Google Scholar]
- Reiffenstein R, Hulbert W, Roth S. 1992. Toxicology of hydrogen sulfide. Ann Rev Pharmacol Toxicol. 1992:109–134. [DOI] [PubMed] [Google Scholar]
- Richards JG. 2009. Metabolic and molecular responses of fish to hypoxia In: Richards JG, Farrell AP, Brauner CJ, editors. Fish physiology: hypoxia. New York: Academic Press; p. 443–485. [Google Scholar]
- Riesch R, Plath M, Schlupp I, Tobler M, Langerhans RB. 2014. Colonization of toxic springs drives predictable life-history shift in livebearing fishes (Poeciliidae). Ecol Lett. 17:65–71. [DOI] [PubMed] [Google Scholar]
- Riesch R, Tobler M, Plath M. 2015. Hydrogen sulfide-toxic habitats In: Riesch R, Tobler M, Plath M, editors. Extremophile fishes: ecology, evolution, and physiology of teleosts in extreme environments. Heidelberg (Germany): Springer; p. 137–159. [Google Scholar]
- Roberts A, Pachter L. 2013. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 10:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeselers G, Newton ILG. 2012. On the evolutionary ecology of symbioses between chemosynthetic bacteria and bivalves. Appl Microbiol Biotechnol. 94:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales LL. 2012. Investigation of karst brackish-sulfidic springs and their role in the hydrogeology, subsurface water-rock interactions, and speleogenesis at northern Sierra de Chiapas, Mexico [dissertation]. [Socorro (NM)]: New Mexico Institute of Mining and Technology. [Google Scholar]
- Saraste M. 1999. Oxidative phosphorylation at the fin de siècle. Science 283:1488–1493. [DOI] [PubMed] [Google Scholar]
- Schartl M, Walter RB, Shen Y, Garcia T, Catchen J, Amores A, Braasch I, Chalopin D, Volff JN, Lesch KP, et al. 2013. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat Genet. 45:567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahak Y, Hauska G. 2008. Sulfide oxidation from cyanobacteria to humans: sulfide-quinone oxidoreductase (SQR) In: Hell R, Dahl C, Knaff DB, Leustek T, editors. Advances in photosynthesis and respiration. Heidelberg (Germany): Springer; p. 319–335. [Google Scholar]
- Soengas JL, Aldegunde M. 2002. Energy metabolism of fish brain. Comp Biochem Physiol B. 131:271–296. [DOI] [PubMed] [Google Scholar]
- Speers-Roesch B, Mandich M, Groom DJE, Richards JG. 2013. Critical oxygen tensions as predictors of hypoxia tolerance and tissue metabolic responses during hypoxia exposure in fishes. J Exp Mar Biol Ecol. 449:239–249. [Google Scholar]
- Stipanuk MH. 2004. Sulfur amino acid metabolism: pathways for production and removal of homocysteine amd cysteine. Ann Rev Nutr. 24:539–577. [DOI] [PubMed] [Google Scholar]
- Stipanuk MH, Ueki I. 2011. Dealing with methionine/homocysteine sulfur: cysteine metabolism to taurine and inorganic sulfur. J Inherit Metab Dis. 34:17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey KB, Storey JM. 2005. Biochemical adaptation to extreme environments In: Walz W, editor. Integrative physiology in the proteomics and post-genomics age. Totowa (NJ): Humana Press; p. 169–200. [Google Scholar]
- Storz JF, Wheat CW. 2010. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution 64:2489–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo C. 2007. Hydrogen sulphide and its therapeutic potential. Nat Rev Drug Discov. 6:917–935. [DOI] [PubMed] [Google Scholar]
- Teets NM, Peyton JT, Colinet H, Renault D, Kelley JL, Kawarasaki Y, Lee RE, Jr, Denlinger DL. 2012. Gene expression changes governing extreme dehydration tolerance in an Antarctic insect. Proc Natl Acad Sci U S A. 109:20744–20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler M, Hastings L. 2011. Convergent patterns of body shape differentiation in four different clades of poeciliid fishes inhabiting sulfide springs. Evol Biol. 38:412–421. [Google Scholar]
- Tobler M, Henpita C, Bassett B, Kelley JL, Shaw J. 2014. H2S exposure elicits differential expression of candidate genes in fish adapted to sulfidic and non-sulfidic environments. Comp Biochem Physiol A Mol Integr Physiol. 175:7–14. [DOI] [PubMed] [Google Scholar]
- Tobler M, Palacios M, Chapman LJ, Mitrofanov I, Bierbach D, Plath M, Arias-Rodriguez L, Garcia de Leon FJ, Mateos M. 2011. Evolution in extreme environments: replicated phenotypic differentiation in livebearing fish inhabiting sulfidic springs. Evolution 65:2213–2228. [DOI] [PubMed] [Google Scholar]
- Tobler M, Plath M. 2011. Living in extreme habitats In: Evans J, Pilastro A, Schlupp I, editors. Ecology and evolution of poeciliid fishes. Chicago (IL): University of Chicago Press; p. 120–127. [Google Scholar]
- Tobler M, Riesch R, Plath M. 2015. Extremophile fishes: an integrative synthesis In: Riesch R, Tobler M, Plath M, editors. Extremophile fishes: ecology, evolution, and physiology of teleosts in extreme environments. Heidelberg (Germany): Springer; p. 279–296. [Google Scholar]
- Tobler M, Schlupp I, Heubel K, Riesch R, Garcia de Leon FJ, Giere O, Plath M. 2006. Life on the edge: hydrogen sulfide and the fish communities of a Mexican cave and surrounding waters. Extremophiles 10:577–585. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dover CL. 2000. The ecology of deep-sea hydrothermal vents. Princeton (NJ): Princeton University Press. [Google Scholar]
- Vedel F, Lalanne E, Sabar M, Chétrit P, De Paepe R. 1999. The mitochondrial respiratory chain and ATP synthase complexes: composition, structure, and mutational studies. Plant Physiol Biochem. 37:629–643. [Google Scholar]
- Vismann B. 1991. Sulfide tomerance: physiological mechanisms and ecological implications. Ophelia 34:1–27. [Google Scholar]
- Völkel S, Grieshaber MK. 1997. Sulfide oxidation and oxidative phosphorylation in the mitochondria of the lugworm Arenicola marina. J Exp Biol. 200:83–92. [DOI] [PubMed] [Google Scholar]
- Waterman TH. 1999. The evolutionary challenges of extreme environments (part 1). J Exp Zool. 285:326–359. [PubMed] [Google Scholar]
- Whitehead A. 2012. Comparative genomics in ecological physiology: toward a more nuanced understanding of acclimation and adaptation. J Exp Zool. 215:884–891. [DOI] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. 2006. Neutral and adaptive variation in gene expression. Proc Natl Acad Sci U S A. 103:5425–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman M, Le Trionnaire SLE, Chopra M, Fox BW, Whatmore J. 2011. Emerging role of hydrogen sulfide in health and disease: critical appraisal of biomarkers and pharmacological tools. Clinic Sci. 121:459–488. [DOI] [PubMed] [Google Scholar]
- Whittington CM, Griffith OW, Qi W, Thompson MB, Wilson AB. 2015. Seahorse brood pouch transcriptome reveals common genes associated with vertebrate pregnancy. Mol Biol Evol. 32:3114–3131. [DOI] [PubMed] [Google Scholar]
- Wilkens H, Strecker U. 2003. Convergent evolution of the cavefish Astyanax (Characidae, Teleostei): genetic evidence from reduced eye-size and pigmentation. Biol J Linnean Soc. 80:545–554. [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 8:206–216. [DOI] [PubMed] [Google Scholar]
- Yu Y, Zhao C, Su Z, Wang C, Fuscoe JC, Tong W, Shi L. 2014. Comprehensive RNA-Seq transcriptomic profiling across 11 organs, 4 ages, and 2 sexes of Fischer 344 rats. Sci Data. 1:140013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Gujar H, Gordon JR, Haynes KF, Potter MF, Palli SR. 2013. Bed bugs evolved unique adaptive strategy to resist pyrethroid insecticides. Sci Rep. 3:1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.