Abstract
Contrary to previous understanding, recent evidence indicates that synonymous codon changes may sometimes face strong selection. However, it remains difficult to generalize the nature, strength, and mechanism(s) of such selection. Previously, we showed that synonymous variants of a key enzyme-coding gene (fae) of Methylobacterium extorquens AM1 decreased enzyme production and reduced fitness dramatically. We now show that during laboratory evolution, these variants rapidly regained fitness via parallel yet variant-specific, highly beneficial point mutations in the N-terminal region of fae. These mutations (including four synonymous mutations) had weak but consistently positive impacts on transcript levels, enzyme production, or enzyme activity. However, none of the proposed mechanisms (including internal ribosome pause sites or mRNA structure) predicted the fitness impact of evolved or additional, engineered point mutations. This study shows that synonymous mutations can be fixed through strong positive selection, but the mechanism for their benefit varies depending on the local sequence context.
Keywords: fitness, codon use, epistasis, Shine–Dalgarno sequence, mRNA folding.
Introduction
Relative to nonsynonymous mutations, synonymous mutations are thought to face at most modest selection, and have thus been used to estimate neutral mutation rates for many decades. However, a growing body of evidence suggests that synonymous mutations—either alone or in conjunction with other mutations—have nonnegligible impacts on cells. These effects range from altering mRNA structure and stability (Kudla et al. 2009; Goodman et al. 2013; Firnberg et al. 2014), protein or nucleic acid binding sites (Li et al. 2012), translation rate (Sørensen and Pedersen 1991), protein folding (Zhang et al. 2009), protein solubility (Rosano and Ceccarelli 2009), and fitness (Carlini 2004; Novella et al. 2004; Coleman et al. 2008; Amoros-Moya et al. 2010; Agashe et al. 2013; Firnberg et al. 2014; Kashiwagi et al. 2014). Genome-wide features such as codon usage bias (i.e., nonrandom use of synonymous codons) are also thought to evolve under selection (Sharp and Li 1987), supporting the idea that synonymous mutations may play important roles in shaping evolutionary dynamics of populations. For instance, altering codon pair bias in the poliovirus drastically reduces its pathogenicity, suggesting that synonymous mutations may be an effective way to develop live virus vaccines (Coleman et al. 2008). Although most studies have reported deleterious effects of synonymous mutations, a fraction of such mutations are predicted to be advantageous, as indicated by the distribution of fitness effects of point mutations in ribosomal genes of Salmonella enterica Serovar typhimurium (Lind et al. 2010). However, the potential fitness benefit is not sufficient to guarantee fixation through positive selection, because synonymous mutations also have to outcompete beneficial nonsynonymous and noncoding mutations during evolution. We know of only one such case, where two beneficial synonymous mutations were fixed during laboratory evolution of Pseudomonas fluorescens (Bailey et al. 2014). However, the mechanism(s) responsible for the beneficial effects of synonymous mutations, the degree of evolutionary repeatability, and the generality of such events remain unclear.
Previously, we demonstrated large deleterious effects of synonymous mutations in a key enzyme-coding gene (fae, encoding the formaldehyde activating enzyme) (Vorholt et al. 2000) in the bacterium Methylobacterium extorquens AM1 (Agashe et al. 2013). Extending earlier ideas by Akashi (1994), we had calculated the genome-wide frequency of each codon at evolutionarily conserved versus variable amino acid residues, comparing with a sister strain M. extorquens DM4. We designated codons enriched at conserved residues (putatively under strong selection) as “frequent”, and codons depleted at such sites as “rare” (likely under weak selection). The resulting codon use is strongly correlated with the output of standard codon use metrics (Agashe et al. 2013), with the added benefit of avoiding biases that arise from using a small subset of reference genes such as ribosomal genes. Using this table of frequent and rare codons, we generated seven synonymous fae alleles (WT: Wild Type [8% rare codons]; AF: All frequent codons [0% rare]; AR: All rare codons [99% rare]; RN: Random residues rare [50% rare]; VA: Variable residues rare [50% rare]; CO: Conserved residues rare [50% rare]; AC: Active sites rare [11% rare]). We replaced the wild type fae with each allele in turn, maintaining the native regulatory region upstream of the gene. We found that the fitness effects of these alleles arose as a direct result of low enzyme production, but could not be consistently explained by either relative codon use or mRNA structure and folding. Instead, fitness was negatively correlated with the number of internal Shine–Dalgarno (SD)-like motifs in transcripts, which are proposed to decrease elongation speed (Li et al. 2012) and reduce protein production. Therefore, the exact physiological cause of low protein production could vary depending on the specific nucleotide sequence of each allele. To gain insight into the fitness effects of these codon changes, we asked: what are the evolutionary solution(s) to overcome the deleterious synonymous mutations; what is the mechanism underlying the solution(s); and is it a repeatable phenomenon?
We allowed eight to ten replicate populations of each of our synonymous mutants to evolve in the laboratory in growth media that contained methylamine as the sole carbon source. This regime imposed selection on the focal mutated gene, fae. Formaldehyde activating enzyme (FAE) catalyzes the condensation of formaldehyde with tetrahydromethanopterin, which is critical for single-carbon (C1) metabolism. Thus, FAE is essential during growth on substrates such as methanol and methylamine (Vorholt et al. 2000). Each mutant carried a synonymous variant of fae (described above) that was not expressed in sufficient quantities to permit wild-type (WT) fitness on methanol (Agashe et al. 2013) or methylamine (fig. 1). To facilitate adaptation, four replicate populations of each mutant were also evolved at a higher effective population size (for AF and AC, ∼5-fold increase in Ne; for other strains ∼50-fold increase; see supplementary table S1, Supplementary Material online). We terminated the evolution experiments after population growth rate stabilized for a few generations (∼60–80 generations for RN, AF and AC, and ∼250 generations for the rest; see supplementary fig. S1, Supplementary Material online). We found that all mutants rapidly evolved increased fitness on methylamine. Despite beginning with highly recoded versions of fae containing 46–150 synonymous mutations (across 186 total codons), we found that single, highly beneficial point mutations in fae could often alleviate these fitness defects. Most of these mutations were unique to a single ancestral variant. Four of these nine mutations were synonymous, demonstrating that synonymous mutations can play a major role in adaptive evolution.
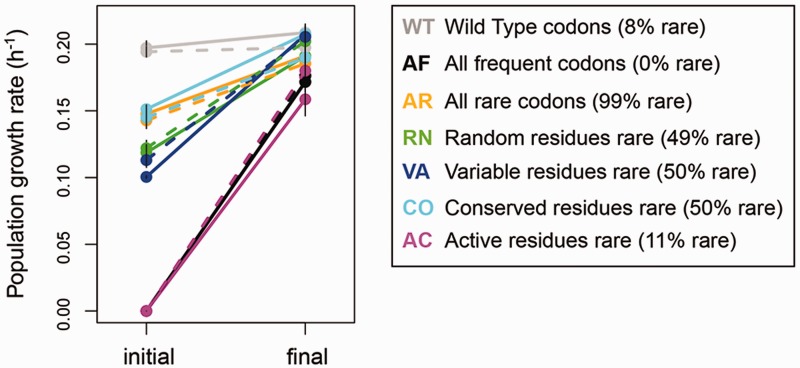
Fig. 1.
Populations rapidly evolve increased fitness on methylamine. Summary of population level initial and final growth rate of fae synonymous variants in methylamine (±SD). Dashed lines = large population size (n = 3–4 replicate populations per strain, with three technical replicates each); solid lines = small population size (n = 5–6 populations per strain, with three technical replicates each).
Results
Rapid Increase in Fitness Is Associated with Repeatable fae-Associated Point Mutations
During laboratory evolution, populations of the mutant ancestors quickly overcame the large fitness defects caused by the initial set of synonymous substitutions at 26–85% of fae codons. All evolving populations showed rapid and parallel increases in growth rate in <100 generations (paired t-test, P < 0.05 for populations of both sizes), with populations of four of the mutants attaining almost WT-level fitness (fig. 1; supplementary fig. S1, Supplementary Material online). Simultaneously evolved WT populations did not show a significant fitness gain over this short timescale (paired t-test, P > 0.3 for populations of both sizes; supplementary fig. S1, Supplementary Material online). The fitness gains were especially striking for AF and AC populations, because the ancestors could not grow on methylamine at all and the growth medium had to be supplemented with succinate (a, multi-carbon substrate for which fae is dispensable) for the first few generations of experimental evolution (see supplementary methods, Supplementary Material online). The large increase in population level growth rate was mirrored by the growth rate of isolated clones from evolved populations (supplementary fig. S2A, Supplementary Material online).
Since we expected fae to be a critical target of selection during growth on methylamine, we tested whether mutations in this gene were responsible for the observed adaptation. We found that nearly all evolved populations had acquired a point mutation either within the fae coding sequence or in the intergenic region directly upstream of fae (henceforth “fae-associated mutations”). Of the nine coding point mutations in fae segregating in populations of the synonymous variants AF, AC, and VA, four were synonymous (fig. 2A). All replicate populations of these three strains carried exactly one of these coding mutations, apart from one AC population that did not acquire a mutation within or near fae (fig. 2B). None of the control WT evolved lines had any mutations associated with the fae gene. Codon bias at specific sites was not important for selection: none of the coding mutations resulted in a WT codon, and different mutations occurred at the same site in a given ancestor (e.g., three AC mutations at bp 12 and two VA mutations at bp 40; fig. 2A). Only one mutation (eVA2) affected a known active residue of FAE, but it was synonymous and hence unlikely to directly impact enzyme catalytic activity. Populations of the other three mutants (AR, CO, and RN) did not gain any coding mutations in fae. However, some AR populations had point mutations in the intergenic region upstream of fae, and all replicate populations of CO and RN mutants acquired a substitution just upstream of the fae start site (fig. 2). Out of 55 evolved populations, only seven (six AR and one AC population) improved fitness without acquiring fae-associated mutations. Thus, as expected, fae was the most important target of selection during laboratory evolution, and hence we focus here on fae-associated evolved mutations.
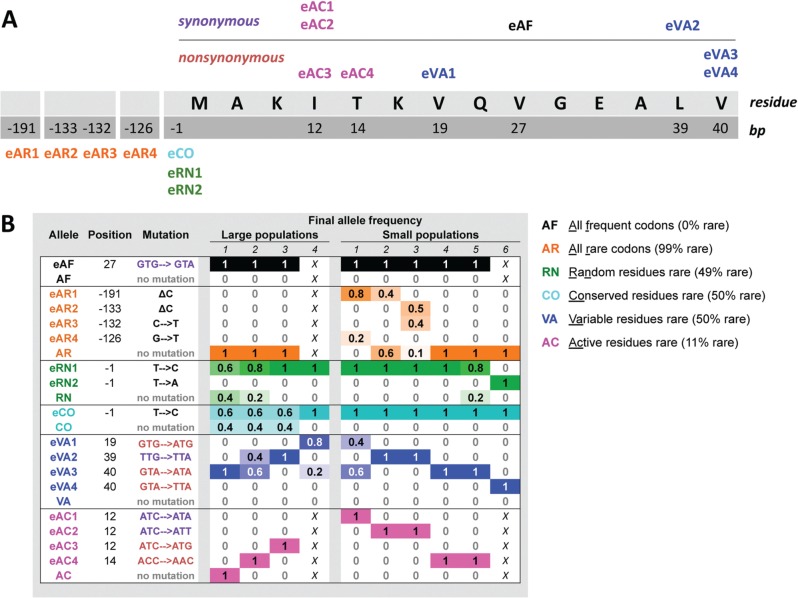
Fig 2.
Evolved coding and noncoding mutations associated with fae. (A) The first 14 amino acid residues of the fae coding and relevant upstream sequence are shown, indicating nucleotide positions that acquired mutations during experimental evolution. Coding mutations are shown above the gene sequence and noncoding mutations are shown below. No mutations were observed in the rest of the fae gene, and each tested clone had a maximum of one mutation. Alleles are named as follows: “e” indicates evolved allele, letters identify the ancestral fae strain, and the number differentiates multiple evolved alleles for each ancestor. (B) Each evolved mutation is described, along with its final frequency in replicate evolved populations (n = 5 isolated clones per large population; n = 10 clones for small populations). The frequency of clones without a fae-associated mutation is also indicated for each ancestor. Each column represents an independent population, with the cell value and color intensity indicating the final frequency of the corresponding allele in that population (maximum color intensity indicates a fixed mutation with frequency 1). In some cases, there were fewer replicate populations, and these “missing” populations are marked with an X.
Remarkably, most of the point mutations (henceforth “SNPs”) seem to have fixed repeatedly across replicate populations of a given strain (fig. 2B), with the exception of mutations in AR populations. Sequencing the fae gene from a few colonies from intermediate time steps suggested that most of these mutations rose to high frequencies relatively early during experimental evolution, and in rare cases displaced other fae-associated mutations (supplementary fig. S3, Supplementary Material online). At the end of the evolution experiment, we also observed some polymorphism in three replicate populations of AR, VA, CO, and RN strains (fig. 2B). Most of these coexisting alleles were fixed in at least one other population, and hence it is quite likely that further evolution would resolve these polymorphisms as we have observed in other experiments with M. extorquens AM1 (Lee and Marx 2013). None of the SNPs were present in the original independent colony used to initiate each population, indicating that they evolved independently in replicate populations. An extreme case in point is the same synonymous mutation that was fixed in each of the eight replicate AF populations. Similarly, a noncoding mutation just upstream of the fae gene was fixed in 7/10 CO populations and 6/10 RN populations, and rose to ≥40% frequency in the remaining replicate populations of both strains. The large and small populations were evolved in two different laboratories on different continents at different times, arguing strongly against cross-contamination across replicates being responsible for the high repeatability. Such a high degree of parallelism at the level of the exact point mutation suggests that each of these mutations was both easily accessible and of particularly large benefit to strains bearing the specific fae allele. On the other hand, different populations of VA and AC sampled four distinct mutations, suggesting a more rugged fitness landscape for these two allele backgrounds. Notably, there was no obvious difference between the pattern of fixation of evolved synonymous and nonsynonymous mutations (fig. 2B).
Physiological Impacts of Beneficial Mutations Vary across Strains
To test the fitness and physiological consequences of the evolved mutations, we replaced the ancestral fae allele (or upstream region) of each mutant with the corresponding evolved allele (or upstream region) carrying the SNP. We successfully generated such strains (denoted “snp”) for ten of the 15 fae-associated mutations (including eight of nine coding mutations); we were unable to obtain the remaining strains. In each case, the evolved SNP significantly increased fitness (fig. 3A). For AF and AC, the fitness gains were extremely large, allowing nearly WT-level growth for strains that were previously unable to grow on methylamine. Even the relatively modest fitness gains in other strains represented a growth advantage of at least 20%. In most cases, the SNP explained all of the fitness increase in the evolved clone carrying the mutation (i.e., “snp” fitness was indistinguishable from that of an evolved isolate carrying the SNP; supplementary fig. S4A, Supplementary Material online). Furthermore, we found that potential background mutations in the rest of the genome rarely improved fitness (tested by replacing the evolved fae allele in evolved isolates with the respective ancestral fae; supplementary fig. S4A, Supplementary Material online). Together, these data suggest that all evolved fae mutations arose under very strong positive selection.
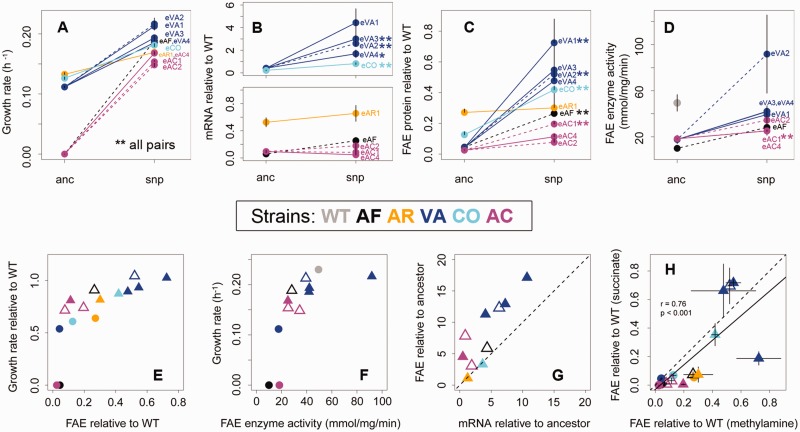
Fig 3.
Impact of evolved mutations on fitness, gene expression, protein production and enzyme activity. Average (±SEM) values of (A) growth rate, (B) mRNA, (C) protein, and (D) enzyme activity per milligram total cell protein are shown for the ancestor (“anc”) and an ancestor carrying only the evolved SNP (“snp”). For panels A and B, n = 3–10 biological replicates per strain; for panel C, n = 3–6 biological replicates per strain; for panel D, n = 3 biological replicates with three technical replicates each. In panel B, the split y-axis allows visualization of the high impact of SNPs in strain VA as well as the lower impact of SNPs in other strains. In panels B and C, error bars on only one side are shown for clarity. Asterisks mark significant differences between “anc” – “snp” pairs after correcting for multiple comparisons using the Benjamini–Hochberg method that controls for false discovery rate (*P = 0.05; **P < 0.05). (E and F) Growth rate as a function of (E) FAE protein production and (F) enzyme activity. (G) Impact of evolved SNPs on mean enzyme levels versus mean mRNA levels. The dashed line indicates y = x. (H) Amount of FAE protein produced during growth on succinate versus methylamine. The dashed line indicates y = x, and the solid line indicates the best-fit linear regression with associated correlation strength and P value. In panels E–H, circles = “anc” and triangles = “snp” strains; open triangles = synonymous SNPs. mRNA and protein values were calculated relative to WT (e.g., mRNAanc/mRNAWT). In all panels, strains are colored as in figure 1 and dashed lines and open triangles indicate synonymous mutations.
Surprisingly, the large fitness gains in evolved populations were not always associated with large increases in fae mRNA or protein levels. Compared with their ancestors, evolved isolates rarely showed a significant increase in fae mRNA levels (supplementary fig. S2B, Supplementary Material online), but some isolates exhibited a significant increase in FAE enzyme levels (supplementary fig. S2C, Supplementary Material online). Furthermore, although SNPs tended to increase fae mRNA, protein or enzyme activity, the increase was not significant in all cases and the magnitude of the effect and their impact on fitness also varied substantially across alleles (fig. 3B–D). In general, the average fitness effect of evolved alleles was associated with increased protein production (fig. 3E) and enzyme activity per milligram total cell protein in cell lysates (fig. 3F). As with growth rate, potential “background” mutations elsewhere in the genome had weak or no impacts on mRNA and protein production; instead, evolved fae-associated SNPs usually had a larger effect (supplementary fig S4B and C, Supplementary Material online). Notably, the magnitude of these phenotypic effects was not significantly different when comparing synonymous versus nonsynonymous mutations (Student’s t-test for each phenotype: P > 0.2; supplementary fig. S5, Supplementary Material online). Together, these results suggest that relatively minor increases in FAE enzyme were sufficient to impart large fitness gains during laboratory evolution.
Cellular enzyme levels depend on higher-level metabolic regulation, as well as the specific outcome of enzyme synthesis in response to this regulation (e.g., via intrinsic factors like the strength of the ribosome binding site). WT M. extorquens cells synthesize large amounts of FAE enzyme even during growth on succinate (when it is not essential), and further upregulate fae on switching to C1 substrates (Okubo et al. 2007). Thus, low FAE enzyme levels in the ancestral variants could result from a failure to appropriately upregulate enzyme synthesis during methylotrophy. Alternatively, abnormal transcript or protein production could change the outcome of otherwise normal upregulation in response to C1 substrates. We found that evolved SNPs had a much larger impact on protein production than predicted by their impact on transcript levels (fig. 3G). The production of more enzyme molecules per mRNA suggests that the SNPs were beneficial because they increased translation rate or protein folding and stability. We also observed that differences in enzyme production during growth on methylamine were well correlated with differences seen during growth on succinate (fig. 3H). Thus, FAE levels relative to WT were maintained regardless of metabolic condition. Together, these results indicate that evolved mutations rescued fitness by altering intrinsic properties of transcripts or proteins rather than restoring a disturbed metabolic regulation to a normal state.
As mentioned above, the physiological impact of evolved SNPs varied substantially, suggesting variation in the mechanisms responsible for their fitness effects. Evolved SNPs of strain VA were associated with over a 5-fold increase in mRNA levels and a 10-fold increase in protein levels relative to the ancestor (fig. 3G). In contrast, eAF, eAC, eCO, and eAR alleles caused relatively weak increases in mRNA and enzyme levels (fig. 3G). The lack of significant increases in transcript or enzyme levels for some of the eAC alleles is surprising in light of their enormous fitness benefits. Given the extremely steep response of growth to changes in FAE, it is possible that growth-dependent dilution could be masking differences in protein production. Such feedback was observed previously when using a regulated promoter to induce fae alleles (Agashe et al. 2013), and in empirical and theoretical work on lactose growth in Escherichia coli (Perfeito et al. 2011). Furthermore, although we never found detectable FAE protein in either AF or AC ancestors, we always detected minute quantities of enzyme in all evolved isolates of these strains. Thus, although the magnitude of the impact of evolved AF and AC SNPs on FAE protein was low (being at the limit of detection), it might have been a sufficiently large change to be biologically important.
The 5′ (N-Terminal) Region of fae Largely Determines the Fitness Effects of Alleles
Intriguingly, all coding mutations occurred in the N-terminal region of the gene encompassing the first 14 residues (out of a total of 186), suggesting that attributes of the N-terminal sequence might underlie the initial fitness defect and subsequent recovery. N-terminal regions of genes are typically enriched in rare codons and are associated with significantly lower mRNA secondary structure (Eyre-Walker and Bulmer 1993; Bentele et al. 2013; Goodman et al. 2013; Hockenberry et al. 2014). Thus, these regions may evolve under different selection pressures than the rest of the gene (e.g., Goodman et al. 2013; Firnberg et al. 2014), potentially because the 5′ transcript structure determines co-translational dynamics or mRNA degradation. Alternatively, internal SD-like motifs in the N-terminal region may promote ribosomal stalling and decrease translation speed (Li et al. 2012; Chevance et al. 2014) although it is not clear whether and why such motifs would be particularly deleterious at the 5′ end of the mRNA. All three alleles with coding SNPs (AF, AC, and VA) had stronger predicted 5′ mRNA structure than the WT allele, reflected by their higher minimum folding energy (MFE; supplementary fig. S6A, Supplementary Material online). Thus, evolved SNPs could be beneficial if they decreased 5′ mRNA structure and folding. On the other hand, ancestral fitness was negatively correlated with the number of internal SD-like motifs in the transcript (Agashe et al. 2013). Therefore, evolved SNPs could also be beneficial if they decreased affinity to the anti-SD sequence and increased protein production. Note that while translation initiation is generally thought to be rate-limiting for protein production, elongation rates can also impact protein production (Yu et al. 2015). For an essential enzyme such as FAE, increasing protein production is expected to directly increase growth rate in selective (C1) media.
To test the importance of the N-terminal region for fitness, we generated fae knockout strains of M. extorquens AM1 carrying plasmid-borne ancestral alleles containing an additional N-terminal insert (13 residues including a FLAG tag). The N-terminal region of the WT fae allele is enriched in frequent codons, with only two of the first 14 residues encoded by rare codons. Therefore, we encoded the N-terminal insert using frequent codons (see supplementary methods, Supplementary Material online). The N-terminal insert was predicted to equalize the anti-SD affinity of all alleles as well as their 5′ MFE, representing either an increase or a decrease in mRNA structure depending on the ancestral sequence. Remarkably, we found that for all gene versions, the N-terminal insert removed the deleterious effect of the synonymous changes in the rest of the allele, conferring growth levels near WT (fig. 4). Thus, adding a small stretch of amino acids reversed the large fitness defects of the synonymous fae alleles, supporting the hypothesis that the evolved SNPs were localized within a critical region of the fae allele. However, since the N-terminal insert affected both anti-SD affinity and MFE, we could not test their independent impacts on fitness. To do this, we next tested the fitness impact of point mutations in the N-terminal region of fae.
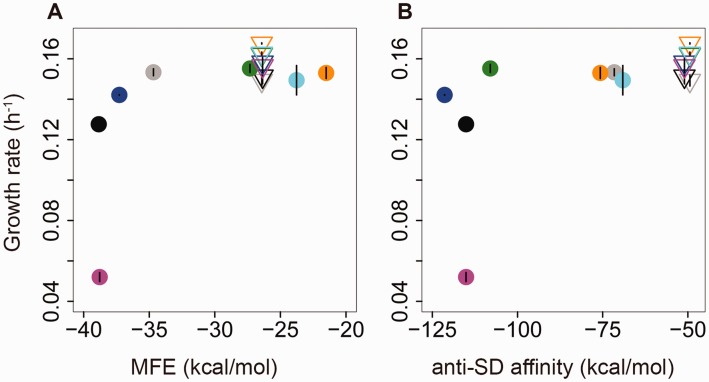
Fig. 4.
Effect of N-terminal insert on fitness. Mean Growth rate (±SEM; n = 3 biological replicates) of strains carrying plasmid-borne fae alleles induced with 1 μM cumate is shown as a function of (A) 5′ MFE and (B) anti-SD affinity. Filled circles indicate ancestral fae alleles, and open inverted triangles indicate alleles carrying the N-terminal insert rich in frequent codons. Points are colored as in figure 1. For MFE calculations we used 100 bp long sequences (−50 to +50 bp relative to the fae start site). For anti-SD affinity calculations, we used 55 bp long sequences (−5 to +50 bp relative to the fae start site).
Reduced 5′ mRNA Structure or Anti-SD Affinity Is Not Sufficient to Improve Fitness
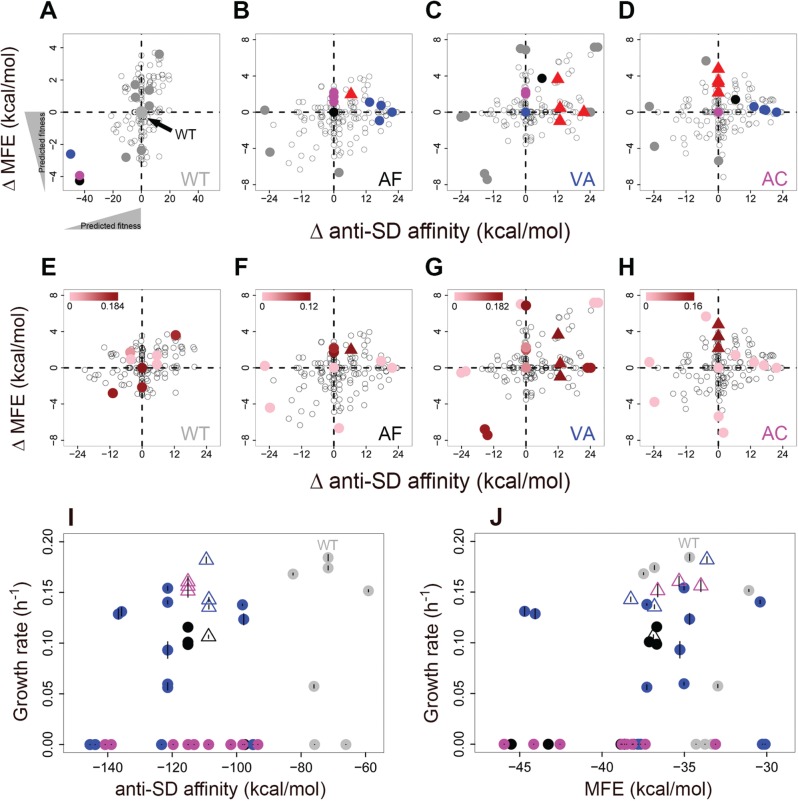
We first systematically analyzed the predicted effects of point mutations on MFE or anti-SD affinity, calculating the predicted impact of all possible point mutations in the first 50 coding nucleotides of each allele (fig. 5A–D and supplementary fig. S7, Supplementary Material online). We then compared the predicted impact of evolved mutations with the distribution of effects of all 5′ mutations that could have occurred in this region. Note that ancestral AF, VA, and AC had stronger mRNA structure as well as anti-SD affinity compared with WT (fig. 5A), leading us to predict that a decrease in either of these values should be beneficial. We found that each of the evolved coding SNPs in AF, VA, and AC are predicted to decrease either MFE or anti-SD affinity, potentially explaining their fitness benefit (fig. 5B–D). For instance, in strain VA, all evolved SNPs decreased anti-SD affinity and were among the largest-effect point mutations possible given the ancestral sequence (supplementary fig. S8A, Supplementary Material online). The VA ancestor had the highest number of SD-like hexamers (supplementary fig. S6B, Supplementary Material online), so a reduction in anti-SD binding may explain the large increase in protein production and consequently, fitness. In contrast, the noncoding SNPs upstream of the fae start codon in AR, CO, and RN did not have large effects on predicted N-terminal MFE or on anti-SD affinity, although several point mutations with potentially large beneficial effects were accessible to each ancestor (supplementary figs. S7 and S8, Supplementary Material online). It is plausible that these mutations were beneficial because they altered regulatory elements (e.g., promoter sequences) that increased protein production, or affected the structure of the 5′ UTR (untranslated region) to increase translation initiation (Espah Borujeni et al. 2014).
Fig. 5.
(A–D) Predicted effect of fae coding SNPs on MFE and anti-SD binding affinity of the ancestral alleles (mutant – ancestor; kcal/mol in both cases). Dashed lines indicate no effect on anti-SD affinity and MFE. In each panel, coordinates (0,0) refer to the respective ancestral strain (indicated with a filled circle colored according to the strain). Hence, the actual MFE and affinity values at this point differ across panels. In panel A, the difference in MFE and anti-SD affinity is also shown for ancestral AF, VA, and AC with respect to the WT, leading to the predicted fitness effects of point mutants in these strains. Open circles = all possible mutations in the first 50 bases of the coding sequence; red triangles = evolved mutations in each strain; colored circles = evolved mutations from other strains that could have (but did not) occur in the focal strain; grey points = engineered mutations (highlighted in subsequent panels). (E–H) Fitness impact of a subset of mutations from panels A–D. Growth rate (±SEM) conferred by plasmid-borne alleles induced with 100 ng/ml anhydrous tetracycline. Points are shaded according to growth rate, with darker color indicating faster growth as indicated by the bar in each panel. Triangles = evolved mutations; filled circles = tested mutations; open circles = untested possible mutations. For MFE calculations we used 100 bp long sequences (−50 to +50 bp relative to the fae start site). For anti-SD affinity calculations, we used 55 bp long sequences (−5 to +50 bp relative to the fae start site). (I and J) Growth rate (±SEM) of strains carrying point mutations, as a function of predicted (I) anti-SD affinity and (J) MFE. Triangles = evolved fae alleles; filled circles = engineered mutations (from panels D–F). Points are colored according to each ancestral allele as indicated in panels A–D.
Next, we experimentally tested whether an introduced mutation that was predicted to alter anti-SD affinity and/or mRNA structure would be sufficient to change fitness. For each ancestral allele (including WT), we identified mutations predicted to be most beneficial (top right quadrant of fig. 5A–D) or detrimental (bottom left quadrant of fig. 5A–D) with respect to one or both parameters. We expressed plasmid-borne fae alleles carrying each of these 37 mutations in a Δfae strain, inducing fae expression at a level where strains with plasmid-borne and chromosomal WT fae had similar growth rates in methylamine. Surprisingly, the fitness impact of these variants did not match the predicted impact based on the altered anti-SD affinity and/or MFE (fig. 5E–H; supplementary table S5, Supplementary Material online). For instance, for strain VA, two alleles that increase anti-SD affinity and decrease MFE (and are therefore predicted to be disadvantageous) turned out to be among the most beneficial alleles tested (bottom left quadrant in fig. 5F). In addition, of a pair of alleles with nearly identical predicted impacts on MFE, one conferred very high fitness while the other conferred very low fitness (topmost points along x = 0 in fig. 5F). Strikingly, there was no pattern even for mutants of the WT allele (fig. 5E), suggesting that the observed lack of predictability with respect to fitness effects of point mutations was not an outcome of the specific mutant ancestors. Combining all mutations across alleles, we did not observe an association with either anti-SD affinity or MFE (fig. 5I and 5J), showing that neither mechanism explains the observed fitness effects of point mutations. This was also reflected in the fact that change in fitness was not correlated with change in either MFE or anti-SD affinity (supplementary fig. S9, Supplementary Material online).
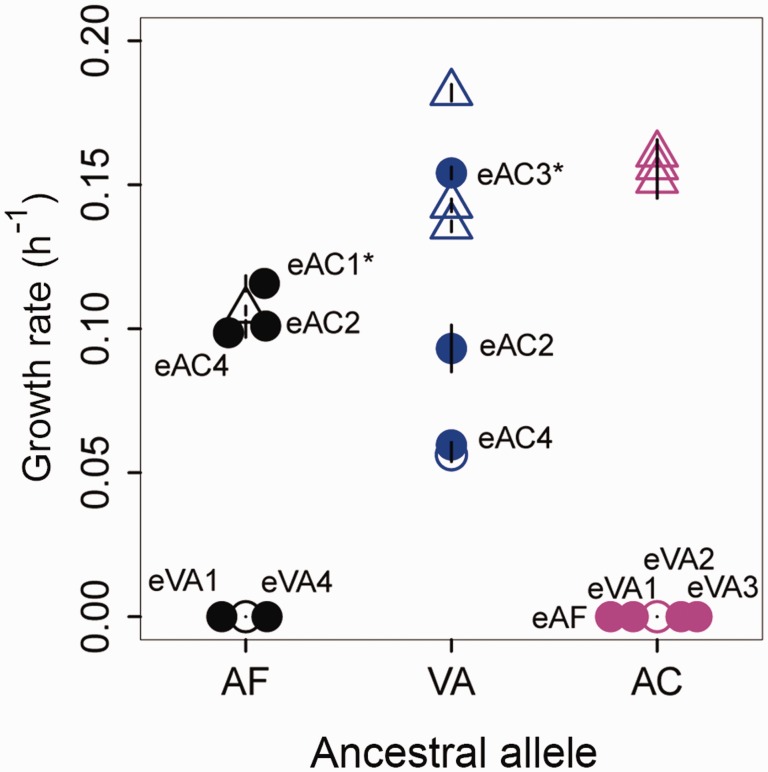
The Benefit of Evolved Mutations Depends upon Epistatic Interactions within the fae Allele
Although all coding SNPs occurred in the N-terminal region, we found that each ancestor acquired unique mutations. This is somewhat surprising because the 5′ sequences of AF, VA, and AC were similar enough that each evolved SNP could have occurred across all strains (fig. 5A–C). It is possible that different strains simply did not sample the same set of mutations, or they were sampled and subsequently lost because they were not as beneficial as other mutations that outcompeted them. Our panel of plasmid-borne point mutations (above) included instances where we could test the fitness effect of evolved mutations in alternate allelic backgrounds in which they were not observed. Of 12 such cross-swaps, only two were significantly more beneficial than mutations that evolved to high frequency in the specific ancestor (eAC1 moved into AF, and eAC3 moved into VA; fig. 6). The variable fitness effects of evolved mutations across ancestral alleles may potentially result from pleiotropic effects in each specific allelic context (e.g., due to epistasis). Note that this epistasis would necessarily involve interactions within the fae allele, since the rest of the genomic sequence was identical across all strains. Thus, in most cases even if the populations had sampled other mutations, they would have been quickly outcompeted by the mutations that eventually fixed or rose to high frequency. This is especially striking for variant AC, where none of the nine additional point mutations tested (including four that evolved in AF and VA) provided any fitness benefit (figs. 5F and 6). In contrast, multiple point mutations in VA conferred fitness benefits comparable to two of the evolved mutations (fig. 5E and 5H), and it is unclear why none of these alternative mutations were observed in any of the VA populations. In addition, the fitness impact did not differ systematically for synonymous and nonsynonymous mutations in the panel (supplementary fig. S10, Supplementary Material online), supporting our results for the original evolved mutations. Together, these patterns suggest that each ancestral variant fixed one of the best possible sequence-specific solutions to the common problem of low enzyme production.
Fig. 6.
Fitness effect of evolved mutations in different allelic backgrounds. Open circles = ancestral fae alleles; triangles = evolved fae alleles; filled circles = ancestral fae alleles with evolved mutations from a different strain (labeled). Cross-swaps conferring significantly higher fitness than actually evolved alleles are marked with an asterisk. Overlapping points are displaced slightly along the x-axis for visualization. Points are colored according to the ancestral fae allele as indicated in figure 1.
Discussion
We report the surprisingly parallel and frequent occurrence of large-effect beneficial synonymous mutations that overcome the deleterious effects of prior synonymous changes in a protein-coding bacterial gene. In our experiments, synonymous mutations in replicate populations across different ancestors showed a minimum benefit of 20%, and three of them elevated growth rate from undetectable to 75% of WT fitness. Another recent study also observed the emergence of two independent synonymous beneficial mutations during laboratory evolution of P. fluorescens (Bailey et al. 2014). Both these mutations occurred in a single population that had already fixed another beneficial mutation, increasing fitness by 12–15% relative to the immediate ancestor and 7–8% relative to the WT ancestor. In both studies, the fitness benefits of synonymous and nonsynonymous mutations were comparable and the benefit was derived from increased gene expression or protein production. Laboratory-evolved DNA and RNA viruses also show parallel synonymous mutations that were most likely fixed through positive selection (Novella et al. 2004; Bull et al. 2012; Kashiwagi et al. 2014), although the precise fitness contributions of individual mutations were not measured in these experiments. Together, these studies show that synonymous mutations can have enormous implications for adaptive evolution that cannot be attributed to the idiosyncrasies of a specific organism or selective pressure. Whereas previous evolution experiments have found large-effect beneficial point mutations, this study uncovered a surprising level of repeatability at the nucleotide level. This high repeatability is especially interesting given that many of these mutations were synonymous, and in contrast to their large fitness benefits, many had relatively weak impacts on expression of the focal gene. Finally, this study demonstrates that the proposed strategy of using multiple synonymous mutations to generate attenuated viruses (e.g., Coleman et al. 2008) may be overly simplistic, for it may be quite easy for the virus to rapidly fix beneficial mutations that can reverse the initial fitness defect.
What was the mechanism responsible for the initial fitness decline in synonymous variants, and for their subsequent recovery? Possibilities include erroneous transcriptional regulation, premature transcript degradation, transcript structure hindering translation, and low translation rate. Our data show that transcript levels were low independent of metabolic feedback, most likely due to intrinsic features of the mutant alleles. These may include transcript secondary structure, which alters in vitro expression (Allert et al. 2010); increased transcriptional pausing in synonymous variants (Larson et al. 2014); rapid mRNA degradation (Dressaire et al. 2013); or disrupted binding to regulatory elements such as sRNA or riboswitches (e.g., reviewed in Deana 2005). All these problems could potentially be alleviated by evolved point mutations. Our results for alleles carrying an additional N-terminal insert implicate both 5′ mRNA structure and internal SD-like motifs as underlying causes of the phenotypic effects of coding synonymous mutations. However, our experimental data on point mutations that were predicted to substantially alter anti-SD binding affinity and/or 5′ mRNA structure showed that neither mechanism was sufficient to predict fitness effects. This was also true for a set of mutations in the WT allele, indicating that our results are applicable in the context of a “normal” gene, and cannot be attributed to the initial set of variants that we had synthesized. Furthermore, a recent report found no evidence for ribosomal pausing due to internal SD-like sequences, either in in vitro “toeprinting” assays or in ribosome profiling experiments (Mohammad et al. 2016). Instead, the authors suggest that experimental artifacts may be responsible for earlier reports of ribosomal pausing at SD-like sequences. Thus, selection against internal SD-like motifs may be globally much weaker and more context-dependent than previously believed. As such, our evolved alleles may represent a subset of the possible alleles that improve anti-SD binding and mRNA structure, and likely affect fitness through alternative mechanisms such as sequence motifs for other forms of gene regulation. Therefore, just as the fitness impacts of nonsynonymous mutations are highly context-dependent and variable, a single mechanism cannot predict the fitness effects of synonymous mutations. Overall, this study indicates that different mechanisms were responsible for the low fitness of strains with different fae ancestral alleles, leading to parallelism between replicate populations and yet diverse evolutionary solutions across populations carrying distinct ancestral alleles.
Theory predicts that bacterial codon usage and tRNA gene copy number co-evolve due to translational selection acting on highly expressed genes (e.g., Higgs and Ran 2008). However, we observe that changing codon use in a key metabolic gene leads to large fitness defects that were not explained by relative codon usage. Further, evolutionary restoration of fitness most often involved single large-effect point mutations associated with the focal gene, rather than changes in codon frequencies or tRNA genes. Thus, this study indicates that not all deleterious codon changes will generate the sustained selection pressure necessary to generate co-evolutionary dynamics between tRNA genes and genome-wide codon use. Of course, codon changes in natural bacterial populations are more likely spread apart in time and along the genome (compared with our ancestral variants that had many mutations in a single gene). Such a combination of weak selection and larger evolutionary timeframes could allow multiple small-effect mutations to be tested, potentially generating the proposed co-evolutionary dynamics. On the other hand, even small tRNA gene copy number changes are likely to affect global rather than local translation, affecting fitness (Bloom-Ackermann et al. 2014). Hence, it is not clear whether slight changes in codon use would generate selection strong enough to allow the fixation of such mutations in tRNA genes. Our results do suggest, however, that whenever codon differences impose strong selection (e.g., with laterally transferred genes), populations may find diverse but local evolutionary solutions to the immediate physiological problem of regulating gene expression. Thus, co-evolutionary dynamics between tRNA gene copy number and codon use may be unlikely in the short term, and subsequent changes in tRNA gene copy number may instead serve to fine-tune global protein production in the long term.
One of our most interesting findings is the high repeatability of synonymous mutations. For mutant AF, each of the eight evolved populations fixed the same synonymous point mutation. What drives this repeatability? Theory suggests that depending on population size, clonal interference between competing mutations maximizes repeatability, while genetic drift and epistatic interactions between mutations decrease repeatability (Szendro et al. 2013). Thus, one possibility is that low population size and extremely strong selection for FAE function in AF (initial fitness relative to the WT was nearly 0) may have allowed the first beneficial mutation to sweep to fixation before competing mutations arose. On the other hand, despite similar population size and selection strength in AC (similar initial fitness), four different SNPs were fixed across replicate populations. Furthermore, even with substantially lower selection strength and higher population size (initial growth rate was ∼1/3rd the growth rate of the WT), all ten replicate populations of mutants CO and 9/10 populations of RN fixed the same noncoding mutation upstream of the fae start site. Thus, the relative strength of selection and genetic drift alone cannot explain the high repeatability of the genetic basis of adaptation. Instead, high repeatability may arise if very few beneficial mutations with comparable selective advantage were available to each allele. The number of beneficial mutations could be limited both by within- and between-gene negative epistasis. However, the lack of evidence for strong epistatic interactions between fae-associated and putative genomic background mutations suggests that between-gene epistasis was not an important factor, potentially because fae is essential for C1 metabolism. Indeed, we found that three of the four highly beneficial VA- or AC-evolved SNPs tested were not beneficial when placed in the AF allele (controlling for the genomic background). Thus, the most likely explanation for variable repeatability of evolved mutations is that epistatic interactions within the specific sequence of each allele determined the number of beneficial mutations that could be sampled in the experiment.
Such context dependence of mutational benefit could occur if interactions between the point mutation and other bases in the transcript altered mRNA folding or binding to regulatory elements. For instance, the fitness effect of N-terminal synonymous mutations in the TEM-1 beta lactamase gene in E. coli was strongly correlated with mRNA stability and structure, but this was not the case for synonymous mutations elsewhere in the coding sequence (Firnberg et al. 2014). In S. enterica, deleterious fitness effects of synonymous mutations that disrupted base pairing in the folded mRNA were successfully reversed by introducing mutations that restored base pairing (Lind and Andersson 2013). Interestingly, in the Lenski E. coli long-term experimental evolution lines, nearly twice as many mutations that alter predicted mRNA structure (folding energy) occurred in nonessential compared with essential genes, suggesting that such mutations more commonly evolve under purifying selection (Chursov et al. 2013). A recent analysis of E. coli and Bacillus subtilis also shows that N-terminal regions of membrane protein sequences are enriched in “programmed” (putatively selected) translational pauses arising from internal SD-like sequences; and such pauses may be necessary for proper targeting of these proteins to the cell membrane (Fluman et al. 2014). Thus, the N-terminal regions of bacterial proteins may often evolve under multiple selective pressures that determine the fate of point mutations in the region via local interactions.
Synonymous mutations may thus have numerous context-dependent physiological impacts, and our results show that none of the currently proposed mechanistic hypotheses can completely explain the observed range of fitness effects of point mutations. Moreover, we do not yet understand the relative importance of the distinct underlying mechanisms for a given allele and sequence context. Regardless, it is clear that synonymous mutations can repeatedly evolve under extremely strong positive selection, and their broader role in adaptive evolution deserves more attention.
Methods Summary
We used ancestral fae variants, strains, and growth media as described previously (Agashe et al. 2013) and detailed in the online supplementary material. In brief, all strains used in this experiment were generated from a laboratory strain of M. extorquens AM1, CM2563, that lacks both the WT fae (derived from CM2006, an isolate of CM198.1 described in Marx and Lidstrom 2002) and the cel operon for cellulose synthesis (to prevent flocculation; Delaney et al. 2013) (supplementary table S2, , Supplementary Material online, set “anc-orig”). All fae alleles carried a C-terminal FLAG tag that allowed us to quantify FAE protein using western blots. The WT strain was generated using the same cloning process used for other ancestral strains, and also carried an identical FLAG tag. We used a minimal growth medium (“Hypho”) supplemented with methylamine (C1), succinate (multi-carbon) or a mixture of both carbon sources as required. We initiated each replicate experimental evolution line of each ancestor using a distinct colony picked at random from a Hypho-succinate agar plate. We evolved populations on Hypho-methylamine in batch culture, initially supplementing AC and AF populations with succinate to prevent extinction. We periodically froze population samples at −80 °C to allow later resuscitation, and plated out an aliquot to check for contamination by other bacteria. We used aliquots of frozen samples to measure change in population-level growth rate during the experiment.
We plated out an aliquot of the final frozen sample of each evolved population on Hypho-succinate agar, and sequenced the fae gene and upstream region of five to ten randomly picked colonies per population. We also sequenced this region for each founding clone to confirm that evolved mutations arose during the evolution experiment. We used homologous recombination with a scar-less allelic exchange vector (Marx 2008) to move (a) evolved SNPs into the respective ancestral strains (“snp”), and (b) ancestral fae alleles into the respective evolved strains (“bkg”). We measured growth rate in Hypho-methylamine in 48-well microplates, using optical density of growing cultures at 600 nm. For other phenotypic measurements, we grew cultures to mid-log phase in Hypho-succinate and induced fae expression with methylamine; this was necessary since some ancestors (AF and AC) could not grow on methylamine alone. After 2 h, we harvested cells to quantify fae mRNA, FAE protein, and enzyme activity using fae allele-specific qPCR primers, anti-FLAG antibody and monitoring production of NADPH respectively.
We calculated predicted MFE of all alleles using RNAfold (Lorenz et al. 2011), and estimated anti-SD binding affinity as the ΔG value for binding between a given hexamer and the anti-SD sequence (Li et al. 2012). We tested the sensitivity of MFE calculations to changes in folding temperature and allowing dangling free ends. We found that MFE calculated at 30°C (growth temperature of M. extorquens) was strongly correlated with MFE at a wide range of temperatures (supplementary fig. S11, Supplementary Material online). Similarly, ignoring dangling-end free energy had little impact on MFE (supplementary fig. S12, Supplementary Material online). The anti-SD sequence of M. extorquens has not been experimentally demonstrated, and we used a computationally predicted ribosome binding sequence (GGAG/GAGG; see supplementary methods, Supplementary Material online) that is routinely used to express plasmid-borne genes in M. extorquens. We tested the fitness impact of an N-terminal insert and of point mutations predicted to alter anti-SD affinity and/or MFE using inducible plasmids pHC115 (Chou and Marx 2012) and pLC291 (Chubiz et al. 2013), respectively. We separately cloned the mutated, evolved, and ancestral alleles into the expression plasmid and transformed an fae knockout strain of M. extorquens AM1 with each plasmid. We induced fae expression with 1 μM cumate (pHC115-based plasmids) or 100 ng/ml anhydrotetracycline (pLC291-based plasmids) in Hypho-methylamine media; and measured the growth rates as described above. To confirm that the lack of correlation between growth rate and anti-SD affinity was independent of the specific anti-SD sequence that we used, we re-calculated the affinity using the entire 16S rRNA tail sequence of M. extorquens, or estimated the highest possible anti-SD affinity conferred by each point mutation (instead of cumulative affinity of hexamers in all frames, as done earlier). In both cases, growth rate remained uncorrelated with anti-SD affinity (supplementary fig. S13, Supplementary Material online).
Supplementary Material
Supplementary material, tables S1–S5, and figures S1–S13 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Swetha Kasetty, Agrim Saini, and Nilima Walunjkar for laboratory assistance and Aparna Agarwal, Jessica Lee, Saurabh Mahajan, and Laasya Samhita for critical comments on the manuscript. We acknowledge funding and support from the Department of Science and Technology’s INSPIRE Faculty program, India (award IFA-13 LSBM-64 to D.A.); National Center for Biological Sciences, India (D.A., M.S., K.P., G.D., V.S.); Council for Scientific and Industrial Research, India (M.S.); University Grants Commission, India (K.P., G.D., A.H.); and the National Institutes of Health, USA (award GM078209-03S1 to C.J.M.).
References
- Agashe D, Martinez-Gomez NC, Drummond DA, Marx CJ. 2013. Good codons, bad transcript: large reductions in gene expression and fitness arising from synonymous mutations in a key enzyme. Mol Biol Evol. 30:549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. 1994. Synonymous codon usage in Drosophila melanogaster—natural selection and translational accuracy. Genetics 136:927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allert M, Cox JC, Hellinga HW. 2010. Multifactorial determinants of protein expression in prokaryotic open reading frames. J Mol Biol. 402:905–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoros-Moya D, Bedhomme S, Hermann M, Bravo IG. 2010. Evolution in regulatory regions rapidly compensates the cost of nonoptimal codon usage. Mol Biol Evol. 27:2141–2151. [DOI] [PubMed] [Google Scholar]
- Bailey SF, Hinz A, Kassen R. 2014. Adaptive synonymous mutations in an experimentally evolved Pseudomonas fluorescens population. Nat Commun. 5:1–7. [DOI] [PubMed] [Google Scholar]
- Bentele K, Saffert P, Rauscher R, Ignatova Z, Blüthgen N. 2013. Efficient translation initiation dictates codon usage at gene start. Mol Syst Biol. 9:675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom-Ackermann Z, Navon S, Gingold H, Towers R, Pilpel Y, Dahan O. 2014. A comprehensive tRNA deletion library unravels the genetic architecture of the tRNA pool. PLos Genet. 10:e1004084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Molineux IJ, Wilke CO. 2012. Slow fitness recovery in a codon-modified viral genome. Mol Biol Evol. 29:2997–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlini DB. 2004. Experimental reduction of codon bias in the Drosophila alcohol dehydrogenase gene results in decreased ethanol tolerance of adult flies. J Evol Biol. 17:779–785. [DOI] [PubMed] [Google Scholar]
- Chevance FFV, Le Guyon S, Hughes KT. 2014. The effects of codon context on in vivo translation speed. PLoS Genet. 10:e1004392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H-H, Marx CJ. 2012. Optimization of gene expression through divergent mutational paths. Cell Reports. 1:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubiz LM, Purswani J, Carroll SM, Marx CJ. 2013. A novel pair of inducible expression vectors for use in Methylobacterium extorquens. BMC Res Notes. 6:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chursov A, Frishman D, Shneider A. 2013. Conservation of mRNA secondary structures may filter out mutations in Escherichia coli evolution. Nucleic Acids Res. 41:7854–7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. 2008. Virus attenuation by genome-scale changes in codon pair bias. Science 320:17842.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A. 2005. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 19:2526–2533. [DOI] [PubMed] [Google Scholar]
- Delaney NF, Kaczmarek ME, Ward LM, Swanson PK, Lee M-C, Marx CJ. 2013. Development of an optimized medium, strain and high-throughput culturing methods for Methylobacterium extorquens. PLoS One 8:e62957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressaire C, Picard F, Redon E, Loubière P, Queinnec I, Girbal L, Cocaign-Bousquet M. 2013. Role of mRNA stability during bacterial adaptation. PLoS One 8:e59059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espah Borujeni A, Channarasappa AS, Salis HM. 2014. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 42:2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Bulmer M. 1993. Reduced synonymous substitution rate at the start of Enterobacterial genes. Nucleic Acids Res. 21:4599–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firnberg E, Labonte JW, Gray JJ, Ostermeier M. 2014. A comprehensive, high-resolution map of a gene's fitness landscape. Mol Biol Evol. 31:1581–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluman N, Navon S, Bibi E, Pilpel Y. 2014. mRNA-programmed translation pauses in the targeting of E. coli membrane proteins. eLife 3:e03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman DB, Church GM, Kosuri S. 2013. Causes and effects of N-terminal codon bias in bacterial genes. Science 342:475–479. [DOI] [PubMed] [Google Scholar]
- Higgs PG, Ran W. 2008. Coevolution of codon usage and tRNA genes leads to alternative stable states of biased codon usage. Mol Biol Evol. 25:2279–2291. [DOI] [PubMed] [Google Scholar]
- Hockenberry AJ, Sirer MI, Amaral LAN, Jewett MC. 2014. Quantifying position-dependent codon usage bias. Mol Biol Evol. 31:1880–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi A, Sugawara R, Sano Tsushima F, Kumagai T, Yomo T. 2014. Contribution of silent mutations to thermal adaptation of RNA bacteriophage Qβ. J Virol. 88:11459–11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. 2014. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344:1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Marx CJ. 2013. Synchronous waves of failed soft sweeps in the laboratory: remarkably rampant clonal interference of alleles at a single locus. Genetics 193:943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-W, Oh E, Weissman JS. 2012. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484:538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Andersson DI. 2013. Fitness costs of synonymous mutations in the rpsT gene can be compensated by restoring mRNA base pairing. PLoS One 8:e63373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Berg OG, Andersson DI. 2010. Mutational robustness of ribosomal protein genes. Science 330:825–827. [DOI] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Hoener Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol. 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx CJ. 2008. Development of a broad-host-range sacB-based vector for unmarked allelic exchange. BMC Res Notes 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx CJ, Lidstrom ME. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. BioTechniques 33:1062–1067. [DOI] [PubMed] [Google Scholar]
- Mohammad F, Woolstenhulme CJ, Green R, Buskirk AR. 2016. Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep. 14:686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella IS, Zárate S, Metzgar D, Ebendick-Corpus BE. 2004. Positive selection of synonymous mutations in Vesicular Stomatitis Virus. J Mol Biol. 342:1415–1421. [DOI] [PubMed] [Google Scholar]
- Okubo Y, Skovran E, Guo X, Sivam D, Lidstrom ME. 2007. Implementation of microarrays for Methylobacterium extorquens AM1. OMICS 11:325–340. [DOI] [PubMed] [Google Scholar]
- Perfeito L, Ghozzi S, Berg J, Schnetz K, Lässig M. 2011. Nonlinear fitness landscape of a molecular pathway. PLos Genet. 7:e1002160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosano GL, Ceccarelli EA. 2009. Rare codon content affects the solubility of recombinant proteins in a codon bias-adjusted Escherichia coli strain. Microb Cell Fact. 8:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. 1987. The codon adaptation index: a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szendro IG, Franke J, de Visser JAGM, Krug J. 2013. Predictability of evolution depends nonmonotonically on population size. Proc. Natl Acad Sci. 110:571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen MA, Pedersen S. 1991. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J Mol Biol. 222:265–280. [DOI] [PubMed] [Google Scholar]
- Vorholt JA, Marx CJ, Lidstrom ME, Thauer RK. 2000. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J Bacteriol. 182:6645–6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C-H, Dang Y, Zhou Z, Wu C, Zhao F, Sachs MS, Liu Y. 2015. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol Cell 59:744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. 2009. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 16:274–280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.