Abstract
Genomewide scans for natural selection (GWSS) have become increasingly common over the last 15 years due to increased availability of genome-scale genetic data. Here, we report a representative survey of GWSS from 1999 to present and find that (i) between 1999 and 2009, 35 of 49 (71%) GWSS focused on human, while from 2010 to present, only 38 of 83 (46%) of GWSS focused on human, indicating increased focus on nonmodel organisms; (ii) the large majority of GWSS incorporate interpopulation or interspecific comparisons using, for example FST, cross-population extended haplotype homozygosity or the ratio of nonsynonymous to synonymous substitutions; (iii) most GWSS focus on detection of directional selection rather than other modes such as balancing selection; and (iv) in human GWSS, there is a clear shift after 2004 from microsatellite markers to dense SNP data. A survey of GWSS meant to identify loci positively selected in response to severe hypoxic conditions support an approach to GWSS in which a list of a priori candidate genes based on potential selective pressures are used to filter the list of significant hits a posteriori. We also discuss four frequently ignored determinants of genomic heterogeneity that complicate GWSS: mutation, recombination, selection and the genetic architecture of adaptive traits. We recommend that GWSS methodology should better incorporate aspects of genomewide heterogeneity using empirical estimates of relevant parameters and/or realistic, whole-chromosome simulations to improve interpretation of GWSS results. Finally, we argue that knowledge of potential selective agents improves interpretation of GWSS results and that new methods focused on correlations between environmental variables and genetic variation can help automate this approach.
Keywords: genetic architecture, genomewide scans for selection, mutation, natural selection, recombination
Introduction
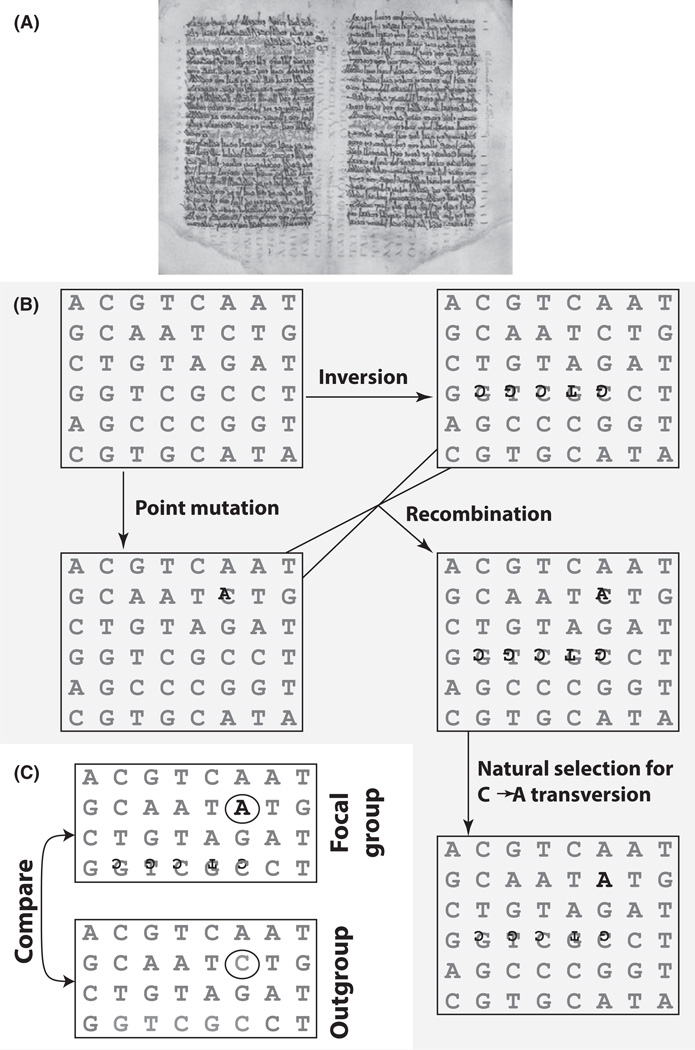
The genome provides an organic record of evolution that is frequently likened to a palimpsest (Delwiche 2004; Weiss & Kawasaki 2006)—a writing medium that is recycled, continuously written over and reoriented so as to partially or wholly obscure older text (Fig. 1A). By this metaphor, chromosomes are the parchment and DNA sequence the text. Mutation obfuscates older genetic text; recombination and chromosomal rearrangements change the content, sense and/or order of the text; and natural selection may secure permanent erasure and replacement of older text (Fig. 1B). In the latter case, reference to other copies of the genetic text—in closely related species or populations where the text has not been altered by natural selection—may enable inference of the original, ancestral genetic text (Fig. 1C).
Fig. 1.
A palimpsest as a metaphor for the genome and the population genetic processes that change it. (A) The Codex Nitriensis is a palimpsest. The lower, faded text is written in Greek and dates to the sixth century A.D., while the upper, bolder text is written in Syriac Aramaic and dates to several centuries later. (B) A genetic text in which the content and/or sense of the text is changed by mutation, chromosomal rearrangement and recombination. These events obscure or permanently alter the original text. Initially, an individual chromosome text is affected by a C-to-A point mutation, while another is affected by a chromosomal inversion of five nucleotides. Recombination can bring these separate mutations together in individual sequence texts. Mutations are in black, while the original sequence is in grey. Relative sizes of letters indicate their frequency in the population. If the A is advantageous, it may eventually fix in the population. At fixation, the A is most commonly found in combination with the noninverted sequence because that is the sequence it originally arose upon. (C) Comparison of the focal group’s sequence text to that of a closely related outgroup (population or species) can help with inference of the ancestral sequence text. While some methods for detecting natural selection (FST outlier, dN/dS, McDonald–Kreitman test, etc.) require such comparisons, other methods can potentially identify the targets of natural selection without comparison to outgroup sequences. The majority of studies documented here do incorporate a test that utilizes out-group comparison (see text).
The modern evolutionary biologist attempting to infer past events from the historical but palimpsest-like text of a species’ genome is therefore faced with an exciting though exacting task: identify regions of the genome critical to adaptation despite the muddled historical record encoded in the palimpsest-like genome. Increasingly, the task of identifying targets of natural selection is performed using genomewide, population-level data. Indeed, genomewide scans for natural selection (GWSS), in which anomalous patterns of genetic diversity are linked to selective events, have produced a number of important results. For example, in humans, frequency of a null variant of CYP3A5 is positively correlated with population distance from the equator; given that CYP3A5 functions in salt homoeostasis, it has been suggested that climatic environmental variables act as selective agents at this locus (Thompson et al. 2004). Subsequently, a number of GWSS corroborated this locus as a target of selection in Europeans and Asians (Carlson et al. 2005; Voight et al. 2006; Olesyk et al. 2008). As an interesting parallel, based on a comparison between the genomes of wild and domestic camels, Jirimutu et al. (2012) found that 11 copies of CYP2J (a member of the same cytochrome P450 family to which CYP3A5 belongs) are found in the wild camel; this far exceeds the copy number of this gene in other mammals (e.g. humans have only one copy). The selective pressure for maintenance of this high copy number is likely also related to salt homoeostasis, as camels are able to ingest large quantities of salt without developing hypertension (Jirimutu et al. 2012). As the number of species investigated using methods of GWSS increases, interspecific comparisons such as this that consider targets of selection and putative selective pressures will refine our understanding of a variety of evolutionary processes including convergent evolution.
The use of genomewide data, which, unlike candidate gene approaches, interrogates variation across the genome, is meant to identify selective targets unbiased by a priori expectations (Ellegren 2014). Yet, a number of factors may undermine this bias-free hope for GWSS. Even though the explicit bias of a candidate gene study is eliminated in GWSS, empirical and simulation studies have shown that some selective events are inherently more difficult to identify. For example, selection on standing variation (Hermisson & Pennings 2005; Przeworski et al. 2005) and selection targeting molecular variants with complex mutational properties (Zhang et al. 2012; Haasl & Paysuer 2013) involve population genetic dynamics that often differ from those underlying stereotypical signatures of selection such as extended haplotype homozygosity (Sabeti et al. 2002). Thus, GWSS based on standard summary statistics and methods may fail to identify a range of selective events, including soft sweeps, polygenic selection and selection targeting genetic variants such as microsatellites or copy number variants (Innan & Kim 2004; Pritchard & Di Rienzo 2010; Haasl et al. 2014). At the biological level, another potential bias derives from the fact that different taxa are characterized by a remarkable diversity of demographic and natural histories as well as a wide variety of environmental factors that may act as selective pressures. Frequently, GWSS are performed with the expectation that certain categories of genes are likely to stand out due to what is known of the focal species biology. When studying high-altitude populations, for example, the understandable tendency is to focus on selection targeting genes associated with adaptation to hypoxic conditions despite the fact that whole-genome sequences or dense genotypes are available (e.g. in yak, Qiu et al. 2012; in human, Tibetan and Andean populations, Bigham et al. 2010; Wuren et al. 2014; in pig, Dong et al. 2014). As we will argue, interpretation of GWSS results is improved by consideration of candidate genes determined a priori.
Here, we survey the findings of >100 GWSS to date. Taking a broad view of biodiversity and ecological circumstance, two extreme possibilities might be found among this catalogue of recent GWSS: (i) GWSS identify a disparate array of selective targets with little overlap between studies or (ii) within and among species, the targets and modes of selection identified by GWSS are largely similar. The latter case would signal something profound about evolution, as this would suggest a subset of the genome’s diversity is the primary source of evolutionary change at both micro- and macroscales. Indeed, previous authors have found some interspecific evidence that supports disproportionate targeting of certain DNA regions. For example, Marden (2013) showed that the results of candidate gene studies and GWSS in organisms as diverse as Clamydomonas, Drosophila mojavensis, the Red abalone snail, the Bactrian camel and humans are enriched for metabolic enzymes. Using a GWSS, Vernot et al. (2012) found that the number, although not effect size, of regulatory variants under selection far exceeded the number of selected variants in protein-coding genes. Similarly, a GWSS comparing variation in 2773 protein-coding genes between normal and dwarf forms of the whitefish Coregonus clupeaformis found very few divergence outliers that were protein-coding mutations, suggesting an abundance of regulatory mutations under selection (Hebert et al. 2013).
Yet, it is important to consider the possibility that convergence of natural selection on a subset of molecular targets might result from something other than a true biological bias towards a subset of critical proteins. For example, apparent biological bias may result from inability to detect unusual modes or targets of selection, failure to correct for complications such as variation in recombination rate or focus on a biased set of organisms and/or environments.
The goal of this perspective article was threefold. First, we briefly discuss major genetic factors that complicate GWSS and may lead to nonbiological biases in results. In particular, we discuss how variability in mutation, recombination, natural selection and the genetic architecture of adaptive traits affect the success of GWSS. Second, we survey recent GWSS that include a variety of methods and cover a broad taxonomic range. The nonstandardized nature of GWSS (still in its infancy) precludes us from performing a true, quantitative meta-analysis of this catalogue of GWSS. However, we discuss the most important genetic, evolutionary and methodological trends observed in this representative set of GWSS and discuss whether the data seem to conform to disparate or similar selective targets across studies and species. Furthermore, we perform a more detailed comparison of GWSS focused on the intense selective pressure of hypoxic conditions at high altitude. Finally, based on genetic complications discussed in the first section and early empirical trends identified in our survey of GWSS, we recommend solutions and best practices to improve the efficacy and impact of future GWSS.
Complicating genetic factors in GWSS
Genomewide scans for natural selection convert heterogeneity in patterns of variation across the genome into inferences about natural selection. All factors that cause variation to differ from one locus to the next therefore affect the success of GWSS. Here, we briefly describe challenges and predictions generated by four determinants of genomic heterogeneity: mutation, recombination, selection and the genetic architecture of adaptive traits.
Selection targets variants that arise through a wide spectrum of mutational events, including single-nucleotide substitutions, insertions, deletions, transpositions and inversions (Fig. 1). The mutational class of a variant affects the signature of selection. For example, microsatellites mutate by adding or subtracting repeats to a tandem array. With realistic mutation rates, this process recurrently generates the same adaptive allele on short timescales, violating the common assumption that beneficial alleles have single mutational origins. Additionally, microsatellites often harbour many alleles, leading to complex fitness surfaces (Haasl & Paysuer 2013). Collectively, these characteristics predict little power for standard approaches to find instances of positive selection that involve microsatellites (Haasl et al. 2014). Furthermore, the rates at which the full variety of mutational events occurs span several orders of magnitude. In humans, single-nucleotide mutations happen at a rate of 10−8–10−9/site/generation (Nachman & Crowell 2000; Roach et al. 2010), microsatellite mutation rates range from 10−2 to 10−6 (Weber & Wong 1993; Sun et al. 2012), and large-scale copy number variants arise at a genomewide rate of 10−2 (Itsara et al. 2010). There is heterogeneity even among single-nucleotide changes, including an order of magnitude elevation in rate at CpG dinucleotides (Campbell et al. 2012). Beneficial mutations appear at different rates across the genome and signatures of selection vary among classes of mutational variants.
Although it is possible to pinpoint specific mutations targeted by positive selection, most GWSS approaches look for the effects of selection on linked diversity. The length of sequence over which polymorphism is distorted (relative to neutral predictions) is inversely related to the local meiotic recombination rate (Maynard Smith & Haigh 1974; Kaplan et al. 1989). As a result, frequency increases in beneficial variants (‘selective sweeps’) with the same selective intensity will be easier to detect in regions with little recombination. Indeed, a positive correlation between nucleotide diversity and recombination rate across the Drosophila melanogaster genome provided the first general evidence for recurrent selective sweeps (Begun & Aquadro 1992). Genomic variation in the recombination rate assumes two forms. Broadscale rate differences among chromosomes or on megabase scales within chromosomes (Broman et al. 1998; Kong et al. 2002; Jensen-Seaman et al. 2004; Shifman et al. 2006; Backström et al. 2010; Wong et al. 2010) likely reflect meiotic constraints, including crossover interference, suppressed recombination near centromeres and requirements for at least one crossover per chromosome or per chromosome arm (Hassold & Hunt 2001; Pardo-Manuel de Villena & Sapienza 2001; Fledel-Alon et al. 2009). In multiple species, crossovers disproportionately occur at a subset of sites (‘hot spots’) interdigitated by stretches of sequence that rarely experience recombination (‘coldspots’); variation in the location and intensity of hot spots produces dramatic fluctuations in recombination rate on the fine scale (Gerton et al. 2000; Jeffreys et al. 2001; Myers et al. 2005; Coop et al. 2008; Kong et al. 2010; Comeron et al. 2012). The degree of recombination rate heterogeneity varies among species (Smukowski & Noor 2011; Kaur & Rockman 2014), suggesting caution when GWSS are applied to taxa without independent information about the rate of crossing over. Recombination rates also vary among individuals (Brooks & Marks 1986; Broman et al. 1998; Koehler et al. 2002; Kong et al. 2010; Comeron et al. 2012). Theory describing the effects of interindividual differences on signatures of selection is needed (Comeron et al. 2012).
Methods of GWSS usually assume that overall patterns of genomic diversity reflect neutral processes, including nonequilibrium demographic history. However, recurrent selection shapes linked variation. The effects of purifying selection (background selection) and selective sweeps on linked diversity depend on the intensity of selection, the local recombination rate and the mutation rate to selected alleles (Maynard Smith & Haigh 1974; Kaplan et al. 1989; Stephan et al. 1992; Charlesworth et al. 1993). Because these parameters vary along genomes, recurrent selection generates heterogeneous patterns of polymorphism. For example, nucleotide diversity covaries with local recombination rate in a variety of species (Cutter & Payseur 2013). Ignoring recurrent linked selection complicates GWSS in two ways. First, positive selection and purifying selection can be conflated. By reducing diversity, background selection also elevates relative measures of population differentiation (Charlesworth et al. 1997), which provide the basis of several common GWSS methods (such as FST-outlier approaches). Second, appropriate thresholds for identifying selective sweeps are unclear. Using patterns of variation at sites affected by linked selection to formulate baseline expectations (as in the commonly employed outlier strategy) violates the basic null model (neutrality) of GWSS and muddles comparisons among genomic windows. Species with large population sizes are especially susceptible to this problem (Leffler et al. 2012; Corbett-Detig et al. 2015). In one notable example, signs of linked selection seem to be pervasive across the Drosophila genome (Begun et al. 2007; Sella et al. 2009; Langley et al. 2012). Ironically, the ability to detect individual instances of selection can decrease as the fraction of the genome affected by linked selection grows.
Finally, genomic regions, genes or variants identified by GWSS are expected to control variation in an organismal trait that in turn affects fitness. How phenotypic selection is projected on to the genome is determined by the genetic architecture of the selected trait. Much of the theory underlying GWSS assumes that selection on individual variants is strong, a situation that arises when adaptive trait differences are conferred by one or a few mutations. Even in this simple scenario, the signature of selection depends on characteristics of adaptive mutations, including dominance (Teshima & Przeworski 2006) and starting allele frequencies (Hermisson & Pennings 2005; Przeworski et al. 2005). When selection targets complex phenotypes—at which variation reflects the action of many mutations—GWSS are less likely to succeed (Pritchard & Di Rienzo 2010). As the number of causative mutations grows, the intensity of selection experienced by each mutation decreases, and the resulting signature of selection is dampened. Selection on a highly polygenic trait generates minimal changes in the frequencies of causative variants; the response to selection mostly comes from covariances among variants (Latta 1998; McKay & Latta 2002; Le Corre & Kremer 2003). In this case, common GWSS approaches fail and alternative strategies are required (Le Corre & Kremer 2012; Berg & Coop 2014; Kemper et al. 2014). When adaptive trait differences are instead generated by a moderate number of substitutions, theory predicts an exponential distribution of phenotypic effects and selection coefficients among substitutions (Orr 1998, 2002). The key point is that the same selection differential applied to phenotypes with contrasting genetic architectures leaves distinct imprints on genomic patterns of variation (Le Corre & Kremer 2012). Because selection affects multiple phenotypes, differences in inheritance provide another source of genomic heterogeneity.
A survey of empirical GWSS
To identify a representative set of GWSS over the last 15 years, we queried the online database Web of Science using a number of different queries, including: ‘selection and genome*wide’, ‘selection and genome and scan’, ‘genome-wide scan’, and ‘genomic scan and selection’. These queries were deemed sufficiently vague to collect the majority of GWSS, while including key terms that would limit the number of query hits. In addition to query results that were clearly not relevant, we rejected a number of GWSS from inclusion in our study for a variety of reasons. We did not include genome scans that used amplified fragment length polymorphisms (AFLPs) as genetic markers. These ecological genomic studies represent an important first look at genomic level data in these nongenetic model organisms. However, AFLPs are usually dominant markers, which limits them to FST-outlier approaches (Luikart et al. 2003), and are plagued by fragment-size homoplasy that reduces power to detect natural selection by ~15% (Caballero et al. 2008). We excluded most studies that search for the genetic targets of artificial selection, including GWSS applied to different breeds of domesticated animals. Exceptions to this include cases where GWSS were used to identify selective targets associated with domestication from the wild (Vigouroux et al. 2002; Chapman et al. 2008) or adaptation to natural selective pressures, such as domestic pigs to high altitude (Dong et al. 2014). We included several GWSS with relatively low marker density—for example scans that only use several thousand SNPs or ~100 microsatellites. While these studies provide lower power to detect targets of natural selection, we included them to increase taxonomic diversity in the data set and because these genetic data span the full genome. We also included several instances of genomic scans that analyse exome or transcriptome sequences only and refer to these studies as exomic scans for natural selection (ESS). Finally, we note that the set of GWSS and ESS included here are meant to be representative rather than comprehensive. For example, although we include several studies that report the draft genome sequence of a species and use dN/dS to scan the newly obtained genome for positive selection, a complete accounting of such studies is beyond the scope of this review.
Qualitative trends
Table 1 lists details of 132 GWSS and ESS. Additional information, including marker number, focal population(s) and major findings, is included in Table S1 (Supporting information). Not surprisingly, the predominant subject species of GWSS is human. The primary driver of this trend is no doubt the abundance of publicly available SNP data from a diversity of natural human populations; sources include the HapMap project (International HapMap Consortium 2005), Human Genome Diversity Panel (Cann et al. 2002) and 1000 Genomes Project (1000 Genomes Project Consortium 2012). These data make it possible to perform GWSS of importance from the computational laboratory alone. Furthermore, these data also provide reference sets of human population genetic variation for studies in which newly sampled human populations are the focus [e.g. Oceanians (Kimura et al. 2008); Indian ethnic groups (Metspalu et al. 2011); Sardinians (Piras et al. 2012); and pygmy populations from the Philippines and Papua New Guinea (Migliano et al. 2013)]. From 1999 to 2009, 35 of 49 (71%) GWSS focused on human, while from 2010 to present, only 38 of 83 (46%) of GWSS focused on human; the decreasing percentage of human studies indicates that genomewide data are becoming easier to obtain in nonmodel organisms.
Table 1.
Summary of representative genomewide scans for natural selection (GWSS) to date
| References | Type | Species | Methodology | Marker type |
|---|---|---|---|---|
| Jaquiery et al. (2012) | gwss |
Acyrthosiphon pisum (pea aphid species complex) |
FST outlier | STR |
| Schubert et al. (2014) | GWSS | Ancient and extant horses | Comparative scans for selection | WGS |
| Gagnaire et al. (2012a) | ess | Anguilla rostrata (American eel) | FST outlier; logistic regression | SNP |
| Gagnaire et al. (2012b) | ess |
Anguilla rostrata and A. anguilla (eels) |
Extension of McDonald–Kreitman test | SNP |
| White et al. (2011) | GWSS | Anopheles gambiae (mosquito) | FST outlier; SFS | SNP |
| Wallberg et al. (2014) | GWSS | Apis mellifera (honeybee) | FST outlier | SNP |
| Chavez-Galarza et al.(2013) | gwss |
Apis mellifera iberiensis (honeybee in Iberia) |
FST outlier | SNP |
| Hancock et al. (2011b) | GWSS-GLMM | Arabadopsis thaliana | GLMM | SNP |
| Huber et al. (2014) | GWSS | Arabadopsis thaliana | SweepFinder and FST outlier | WGS |
| Lobreaux & Melodelima (2015) | GWSS-GLMM | Arabadopsis thaliana | GLMM (climatic variables) | SNP |
| Qiu et al. (2012) | GWSS |
Bos grunniens and Bos taurus (yak and cattle) |
Comparative genomics | WGS |
| Edea et al. (2014) | GWSS | Bos taurus (cattle) | LD outlier | SNP |
| Jirimutu et al. (2012) | GWSS |
Camelus bactrianus ferus (wild Bactrian camel) |
dN/dS | WGS |
| Akey et al. (2010) | gwss | Canis familiaris (dog) | FST outlier | SNP |
| Quilez et al. (2011) | gwss |
Canis familiaris (dog, breed: Boxer) |
Regions of homozygosity | SNP |
| Pollinger et al. (2005) | gwss |
Canis familiaris (dog, breed: Dauschund) |
FST outlier; regions of homozygosity | STR |
| Hagenblad et al. (2009) | gwss |
Canis lupus (Eurasian wolf in Scandinavia) |
Ewens–Watterson; lnRV/H; FST outlier | STR |
| Hebert et al. (2013) | ESS |
Coregonus clupeaformis (whitefish) |
FST outlier | SNP |
| Tsumura et al. (2014) | gwss |
Cryptomeria japonica (Japanese cedar) |
FST outlier | SNP |
| Pool et al. (2012) | GWSS | Drosophila melanogaster | Modified SweepFinder | WGS |
| Langley et al. (2012) | GWSS | Drosophila melanogaster | McDonald–Kreitman test; FST outlier | WGS |
| Reinhardt et al. (2014) | GWSS | Drosophila melanogaster | FST outlier | GWS |
| Begun et al. (2007) | GWSS | Drosophila simulans | Modified HKA test; SFS | GWS |
| Shapiro et al. (2007) | gwss | Drosophila spp. | Ka/Ks; SFS; McDonald–Kreitman test | SNP |
| Gu et al. (2009) | gwss |
Equus ferus caballus (Thoroughbred) |
Ewens–Watterson test; FST outlier | STR |
| Steane et al. (2014) | gwss-glmm |
Eucalyptus tricarpa (Red ironbark eucalyptus) |
Bayescan | DArT |
| Zhan et al. (2013) | GWSS |
Falco peregrinus and Falco cherrug (falcons) |
Comparative genomics | WGS |
| Star et al. (2011) | GWSS | Gadus morhua (Atlantic cod) | Comparative genomics | WGS |
| Makinen et al. (2008) | gwss |
Gasterosteus aculeatus (three-spined stickleback) |
FST outlier; lnRH | STR; indel |
| Kane & Rieseberg (2007) | gwss | Helianthus annuus (sunflower) | lnRV and lnRH; FST outlier | STR |
| Chapman et al. (2008) | gwss | Helianthus annuus (sunflower) | lnRV and lnRH | STR |
| Huttley et al. (1999) | GWSS | Homo sapiens | Extended LD | STR |
| Akey et al. (2002) | gwss | Homo sapiens | Variety of FST -based methods | SNP |
| Akey et al. (2002) | gwss | Homo sapiens | FST outlier | SNP |
| Payseur et al. (2002) | GWSS | Homo sapiens | SFS | STR |
| Kayser et al. (2003) | gwss | Homo sapiens | lnRV; RST outlier | STR |
| Storz et al. (2004) | gwss | Homo sapiens | FST outlier; SFS | STR |
| Shriver et al. (2004) | gwss | Homo sapiens | FST outlier | SNP |
| Bustamante et al. (2005) | ESS | Homo sapiens | dN/dS | WES |
| Carlson et al. (2005) | GWSS | Homo sapiens | SFS | SNP |
| International HapMap Consortium (2005) | GWSS | Homo sapiens | LRH; population differentiation | SNP |
| Weir et al. (2005) | GWSS | Homo sapiens | FST outlier | SNP |
| Voight et al. (2006) | GWSS | Homo sapiens | iHS | SNP |
| Wang et al. (2006) | GWSS | Homo sapiens | LD decay | SNP |
| Mattiangeli et al. (2006) | gwss | Homo sapiens | Ewens–Watterson test | STR |
| Kelley et al. (2006) | GWSS | Homo sapiens | SFS | SNP |
| Zhang et al. (2006) | GWSS | Homo sapiens | WGLRH | SNP |
| Bubb et al. (2006) | GWSS | Homo sapiens | High SNP density | WGS |
| Williamson et al. (2007) | GLMM | Homo sapiens | CLRT | SNP |
| International HapMap Consortium (2007) | GWSS | Homo sapiens | iHS; EHH | SNP |
| Sabeti et al. (2007) | GWSS | Homo sapiens | iHS; XP-EHH | SNP |
| Tang et al. (2007) | GWSS | Homo sapiens | Modified EHH; LRH | SNP |
| Kimura et al. (2007) | GWSS | Homo sapiens | Haplotype homozygosity (Rsb) | SNP |
| Haygood et al. (2007) | ESS | Homo sapiens | dN/dS | WGS |
| Hancock et al. (2008) | gwss-glmm | Homo sapiens | GLMM (climatic variables) | SNP |
| Olesyk et al. (2008) | GWSS | Homo sapiens | FST and heterozygosity outliers | SNP |
| Johansson & Gyllensten (2008) | GWSS | Homo sapiens | (Haplotype length + FST) outliers | SNP |
| Kimura et al. (2008) | GWSS | Homo sapiens | LRH; SFS | SNP |
| O’Reilly et al. (2008) | GWSS | Homo sapiens | Novel recombination rate-based test | SNP |
| Myles et al. (2008) | GWSS | Homo sapiens | FST outlier | SNP |
| Amato et al. (2009) | GWSS | Homo sapiens | FST outlier | SNP |
| Pickrell et al. (2009) | GWSS | Homo sapiens | iHS; XP-EHH; FST outlier | SNP |
| López Herráez et al. (2009) | GWSS | Homo sapiens | Modified Rsb | SNP |
| Chen et al. (2009) | GWSS | Homo sapiens | Modified McDonald–Kreitman test | Indel |
| Andres et al. (2009) | ESS | Homo sapiens | CLRT | WES |
| Hancock et al. (2010) | GWSS-GLMM | Homo sapiens | GLMM (four ecoregion variables) | SNP |
| Yi et al. (2010) | ESS | Homo sapiens | PBS | WES |
| Albrechtsen et al. (2010) | GWSS | Homo sapiens | Excessive Identity by descent | SNP |
| Bigham et al. (2010) | GWSS | Homo sapiens | lnRH; WGRLH; SFS | SNP; CNV |
| Simonson et al. (2010) | GWSS | Homo sapiens | iHS; XP-EHH | SNP |
| Beall et al. (2010) | GWSS | Homo sapiens | Allele frequency differences | SNP |
| Lappalainen et al. (2010) | GWSS | Homo sapiens | iHS, LRH, EHH; FST outlier | SNP |
| Chen et al. (2010) | GWSS | Homo sapiens | XP-CLR | SNP |
| Xu et al. (2010) | GWSS | Homo sapiens | iHS; XP-EHH; XP-CLR; FST outlier | SNP |
| Metspalu et al. (2011) | GWSS | Homo sapiens | XP-EHH; iHS | SNP |
| Fumagalli et al. (2011) | GWSS-GLMM | Homo sapiens | GLMM | SNP |
| Hancock et al. (2011a) | GWSS-GLMM | Homo sapiens | GLMM | SNP |
| Granka et al. (2012) | GWSS | Homo sapiens | iHS; XP-EHH | SNP |
| Piras et al. (2012) | GWSS | Homo sapiens | EHH and XP-EHH | SNP |
| Vernot et al. (2012) | GWSS | Homo sapiens | FST outlier vs. DNase I peak | WGS |
| Zhang et al. (2012) | GWSS | Homo sapiens | CNV frequency differentiation | SNP; CNV |
| Jarvis et al. (2012) | GWSS | Homo sapiens | FST outlier; XP-EHH; iHS | SNP |
| Andersen et al. (2012) | GWSS | Homo sapiens | Composite of multiple methods | SNP; WGS |
| Scheinfeldt et al. (2012) | GWSS | Homo sapiens | Locus-specific branch length | SNP |
| Suo et al. (2007) | GWSS | Homo sapiens | iHS; XP-EHH | SNP |
| Migliano et al. (2013) | GWSS | Homo sapiens | iHS; XP-EHH | SNP |
| Somel et al. (2013) | ESS | Homo sapiens | dN/dS | SNP |
| Hider et al. (2013) | GWSS | Homo sapiens | SFS; Rsb; PBS | WGS |
| Frichot et al. (2013) | GWSS-GLMM | Homo sapiens | Latent factor mixed models | SNP |
| Raj et al. (2013) | GWSS | Homo sapiens | iHS; FST outlier | SNP |
| Liu et al. (2013) | gwss | Homo sapiens | Long-range haplotype method | SNP |
| Bhatia et al. (2014) | GWSS | Homo sapiens | Deviations in local ancestry | SNP |
| Colonna et al. (2014) | GWSS | Homo sapiens | iHS; XP-EHH; FST outlier | SNP; indel |
| Eichstaedt et al. (2014) | GWSS | Homo sapiens | iHS; XP-EHH; FST outlier | SNP |
| Clemente et al. (2014) | GWSS | Homo sapiens | iHS; SFS | SNP |
| Haasl et al. (2014) | GWSS | Homo sapiens | Novel ksk2 test | WGS |
| Ali et al. (2014) | GWSS | Homo sapiens | iHS; XP-EHH | SNP |
| Wuren et al. (2014) | GWSS | Homo sapiens | iHS; XP-EHH | SNP |
| Fangy et al. (2014) | GWSS | Homo sapiens | iHS and Derived Intraallelic Nucleotide Diversity test |
WGS |
| Enard et al. (2014) | ESS | Homo sapiens | iHS; XP-EHH | WGS |
| Sjostrand et al. (2014) | GWSS | Homo sapiens | Novel Maximum Frequency of Private Haplotypes test |
SNP |
| Leffler et al. (2013) | GWSS | Homo sapiens, Pan troglodytes | Haplotype sharing between species | WGS |
| Nielsen et al. (2005) | ESS | Homo sapiens, Pan troglodytes | dN/dS likelihood ratio test | WES |
| Clark et al. (2003) | ESS |
Homo sapiens, Pan troglodytes, Mus musculus |
dN/dS in the human lineage | WES |
| Enard et al. (2010) | GWSS | Four primates | Novel version of HKA test | SNP |
| Westram et al. (2014) | ESS |
Littorina saxatilis (marine snail) |
FST outlier | WES |
| Rhesus macaque Genome Sequencing and Analysis Consortium (2007) | GWSS | Macaca mulatta | dN/dS likelihood ratio test | WGS |
| George et al. (2011) | ESS | Numerous primates |
dN/dS for each orthologous set of genes |
WES |
| Branca et al. (2011) | GWSS |
Medicago truncatula (a legume, Barrel clover) |
Extreme 100 kb windows for π, recombination and LD |
WGS |
| Yoder et al. (2014) | GWSS-GLMM |
Medicago truncatula (a legume, Barrel clover) |
GLMM (climatic variables) | SNP |
| Srivastava et al. (2012) | ess |
Melospiza melodia (song sparrow) |
Comparative genomics | SNP |
| Puzey & Vallejo-Marin (2014) | GWSS |
Mimulus guttatus (monkey flower) |
SFS | WGS |
| Ihle et al. (2006) | gwss | Mus musculus (house mouse) | lnRV and lnRH | STR |
| Teschke et al. (2008) | gwss |
Mus musculus domesticus and Mus musculus musculus (house mouse) |
lnRH | STR |
| Limborg et al. (2014) | gwss |
Oncorhynchus gorbuscha (pink salmon) |
FST outlier | SNP |
| Lv et al. (2014) | gwss-glmm | Ovis aries (sheep) | GLMM | SNP |
| Eckert et al. (2010) | ess | Pinus taeda (Loblolly pine) | GLMM (heterozygosity of SNPs) | SNP |
| Frichot et al. (2013) | ess-glmm | Pinus taeda (loblolly pine) | Novel GLMM approach: latent factor mixed models |
SNP |
| Ochola et al. (2010) | ESS | Plasmodium falciparum | HKA test; SFS | WGS |
| Amambua-Nqwa et al. (2012) | GWSS | Plasmodium falciparum | SFS | WES |
| Park et al. (2012) | GWSS | Plasmodium falciparum | XP-EHH on isolates resistant to >1 of 12 antimalarial drugs |
SNP |
| Nygaard et al. (2010) | GWSS |
Plasmodium spp. (seven species) |
Modified McDonald–Kreitman test; SFS |
WGS |
| Fraser et al. (2015) | GWSS | Poecilia reticulata (guppy) | FST outlier | WGS |
| Evans et al. (2014) | GWSS |
Populus trichocarpa (black cottonwood) |
FST outlier; iHS | SNP |
| Cai et al. (2013) | GWSS |
Pseudopodoces humilis (Ground tit) |
Comparative genomics | SNP |
| Vincent et al. (2013) | gwss-glmm | Salmo salar (Atlantic salmon) | GLMM (49 environmental variables) | SNP |
| Zueva et al. (2014) | gwss-glmm | Salmo salar (Atlantic salmon) |
FST outlier and LFMM (Frichot et al. 2013) |
SNP |
| Casa et al. (2005) | gwss | Sorghum bicolor | Ewens–Watterson test; lnRH; FST outlier |
STR |
| Thomas et al. (2012) | GWSS |
Staphylococcus aureus (bacterium) |
SFS (balancing sel.): Tajima’s D > 2.03; π/K > 0.12 |
WGS |
| Dong et al. (2014) | GWSS | Sus scrofa (pig) | FST outlier | SNP |
| Cavagnagh et al. (2013) | ess | Triticum aestivum (wheat) | FST outlier; pairwise haplotype sharing | SNP |
| Sun et al. (2013) | ESS |
Tursiops truncates (common bottlenose dolphin) |
dN/dS | WES |
| Vigouroux et al. (2002) | ess | Zea mays (maize) | Ewens–Watterson test | STR |
Regarding type of scan: ESS, exonic scan for selection; GLMM, use of generalized linear mixed model methodology; lowercase indicates a relatively small number of markers used. Regarding methodology: CLRT, composite likelihood ratio test; iHS, integrated haplotype statistics; EHH, extended haplotype homozygosity; XP-EHH, cross-population EHH; LRH, long-range haplotype test; WGRLH, whole-genome LRH; SFS, site frequency spectrum statistic(s); PBS, population branch statistic; XP-CLR, cross-population composite likelihood ratio; HKA, Hudson–Kreitman–Aguade test. Regarding marker type: STR, microsatellite (short tandem repeat); CNV, copy number variant; WGS, whole-genome sequence; WES, whole-exome sequence.
The majority of GWSS in Table 1 rely on intraspecific data and methods that compare genetic variation between populations to identify targets of natural selection. Of these methods, the most common are (i) simple FST-outlier approaches, in which SNPs with extreme FST among pairs of populations are associated with linked selection, and (ii) the cross-population extended haplotype homozygosity test (XP-EHH; Sabeti et al. 2007). Given that it is easier to obtain data from a single population, this trend suggests that biologists prefer to apply analyses that rely on multipopulation comparative data. One reason for this may be that statistics of the site frequency spectrum require a relatively large number of genetic markers to estimate. On the other hand, an FST comparison can be made for every marker included regardless of the total number of markers. Importantly, the scope of comparative approaches has expanded with the advent of recently developed methods that use generalized linear mixed models (GLMMs; Hancock et al. 2008, 2010; Frichot et al. 2013), which seek correlations between environmental parameters (potential selective pressures) and genetic variants across multiple populations exposed to different values of these parameters. In these studies, samples are often drawn from individuals spanning wide geographic distances.
Our survey of the GWSS literature also reveals a strong methodological and biological bias towards attempting to detect positive, directional natural selection. Very few of the studies included in our survey address, or attempt to analyse, other forms of natural selection. Exceptions include a small number of scans that intentionally sought signatures of balancing selection. Bubb et al. (2006) identified 16 regions of high SNP density outside of the human leucocyte antigen (HLA) system and loci for ABO blood antigens that provided suggestive evidence for balancing selection within human populations. Intriguingly, Andres et al. (2009) performed an ESS in which signatures of long-term balancing selection in humans were discovered in loci related to cellular structure, including keratins. Leffler et al. (2013) discovered 125 regions in addition to loci of the HLA system in which humans and chimpanzee (Pan troglodytes) shared haplotypes, suggesting long-term balancing selection. Parasites and infectious organisms are relatively overrepresented for scans focused on balancing selection, presumably because loci with greater-than-average genetic variation are critical to the successful infection of host organisms. In various species of Plasmodium, the causative parasite of malaria, two separate scans identified loci subject to balancing selection based on summaries of the site frequency spectrum, including loci involved in host–parasite interaction (Nygaard et al. 2010; Ochola et al. 2010). Thomas et al. (2012) scanned 16 strains of the bacterium Staphylococcus aureus, which can become methicillin resistant (MRSA) and generate serious threats to health care (David & Daum 2010); based on summaries of the site frequency spectrum, the authors discovered 186 windows in 99 genes putatively affected by balancing selection.
Early GWSS focused on the human genome used relatively small numbers of markers (Huttley et al. 1999; Akey et al. 2002; Payseur et al. 2002; Kayser et al. 2003; Shriver et al. 2004; Storz et al. 2004); before 2005, the largest number of markers applied in a human GWSS was 26 530 SNPs (Akey et al. 2002). Preferences for marker number and type changed dramatically in 2005 with the advent of new technologies and large publically available data sets. With one exception (Mattiangeli et al. 2006), all GWSS focused on human with a publication date of 2005 or later used SNPs or whole-genome sequences; in cases where SNPs were used, >50 000 SNPs were genotyped and the majority of studies used ~1 million SNPs. In other species, where comparative data are lacking, it is difficult to assess the strength of this trend, but certainly, other species are now genotyped or sequenced at high coverage with some frequency: 8.3 million SNPs in honeybee, Apis mellifera (Wallberg et al. 2014), and whole-genome sequences for >100 guppies of the species Poecilia reticulata (Fraser et al. 2015).
Interpreting GWSS results: the case of hypoxia as a selective pressure
Humans have adapted to hypoxic conditions in three distinct high-altitude environments: the Tibetan Plateau, the Ethiopian highlands and the Andean Altiplano (Bigham et al. 2010). We compared the results of GWSS in human populations living in these regions as well as several recent studies focused on yak, cattle, pig and ground tit in these same geographic regions (Tables 2 and S2, Supporting information). The GWSS included in Table 1 are too disparate to serve as the basis of a meaningful meta-analysis. However, focusing on this relatively small number of studies in which animals have adapted to the same, unequivocally strong selective pressure revealed valuable insights regarding the interpretation of GWSS results. First, even when a strong selective pressure exists to aid interpretation of results, discrepancies still arise among studies. Second, and more positively, this example shows it is possible to delimit different evolutionary genetic responses to a common selective pressure.
Table 2.
Genes of the hypoxia-inducible factors (HIF) pathway producing signatures of positive selection in various high-altitude adapted populations
| References Species Region |
Wuren et al. (2014) Human Tibet |
Eichstaedt et al. (2014) Human Andes |
Scheinfeldt et al. (2012) Human Ethiopia |
Simonson et al. (2010) Human Tibet |
Beall et al. (2010) Human Tibet |
Bigham et al. (2010) Human Tibet |
Bigham et al. (2010) Human Andes |
Xu et al. (2010) Human Tibet |
Yi et al. (2010) Human Tibet |
Qiu et al.(2012) Yak Tibet |
Edea et al.(2014) Cattle Ethiopia |
Dong et al.(2014) Pig Tibet |
Cai et al.(2013) Ground Tit Tibet |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EGLN1 | X | X | X | X | X | X | |||||||
| ELGN3 | X | ||||||||||||
| PTEN | X | ||||||||||||
| HIF1A | X | X | |||||||||||
| EPAS1 | X | X | X | X | X | X | |||||||
| PPARA | X | X | |||||||||||
| EDNRA | X | ||||||||||||
| ANGPTL4 | X | ||||||||||||
| VEGFB | X | ||||||||||||
| VEGFC | X | ||||||||||||
| MMP3 | X | ||||||||||||
| PRKAA1 | X | ||||||||||||
| NOS2A | X |
Genes in bold face are members of the HIF pathway. Underlined genes are downstream targets of HIF pathway genes.
See Table S2 (Supporting information) for extended findings of these studies.
Humans from low-altitude regions of the world acclimate to hypoxic conditions via erythropoiesis and thereby increased haemoglobin concentrations (Storz 2010). Key to this acclimation (rather than adaptation) response is a regulatory pathway whose central transcription factors are known as hypoxia-inducible factors (HIFs). However, the quick physiological fix of increased erythropoiesis represents a short-term solution; blood viscosity increases with the greater number of erythrocytes, which ultimately hampers blood flow and limits tissue oxygenation (Villafuerte et al. 2004). Surprisingly, Tibetan natives possess haemoglobin concentrations similar to individuals living at sea level, while Andean natives possess significant increases in haemoglobin concentrations relative to low-altitude populations (Beall et al. 1998). This suggests that the genetic architecture of high-altitude adaptation may be different in Andeans and Tibetans.
Indeed, while EPAS1—which codes for the oxygen-sensitive domain of the transcription factor HIF-2—is a top adaptive hit in all GWSS focused on humans of the Tibetan Plateau (Beall et al. 2010; Bigham et al. 2010; Simonson et al. 2010; Xu et al. 2010; Yi et al. 2010; Wuren et al. 2014), the results of GWSS focused on Andean and Ethiopian high-altitude populations do not identify EPAS1 as a target of positive selection (Bigham et al. 2010; Scheinfeldt et al. 2012; Eichstaedt et al. 2014). EGLN1, which codes for a repressor of EPAS1 production, showed signatures of natural selection in both Andeans and Tibetans, but the adaptive patterns of genetic variation at EGLN1 are clearly distinct between the two populations (Bigham et al. 2010). Note that Eichstaedt et al. (2014) did not uncover EGLN1, which shows that distinct studies using different samples, genetic markers and/or methods can arrive at different results despite the strong selective pressure acting to shape relevant genomic regions. Furthermore, Bigham et al. (2010) identified 14 and 37 1 Mb regions of significance based on multiple, corroborating signatures of selection in Tibetans and Andeans, respectively. None of these regions overlapped with each other. Further evidence of the variable genetic architecture of high-altitude adaptation was provided by a GWSS focused on native populations of the Ethiopian highlands, where no enrichment for HIF pathway genes was discovered (Scheinfeldt et al. 2012)—a result that was distinct from both Andeans and Tibetans. Interestingly, top signatures of selection included genes related to immune function, suggesting that distinct pathogenic exposures at high altitude might represent as great a selective pressure as hypoxia itself (Scheinfeldt et al. 2012).
Several GWSS have also examined the effect of high-altitude environment on the evolution of domesticated animals. A comparison of genetic variation between yaks of the Tibetan Plateau and lowland cattle revealed that HIF1A, a subunit of the HIF-1 transcription factor, was targeted by positive selection in yaks (Qiu et al. 2012); the same gene appears to be targeted by selection in human populations of the Tibetan Plateau (Beall et al. 2010). In addition, downstream targets of HIF pathway regulation such as ARG2 as well as numerous proteins key to the metabolism of polysaccharides, amino acids and fatty acids appear to be targeted by selection in yaks. Similarly, metabolic genes of cattle living in the Ethiopian highlands bear strong signatures of selection (Edea et al. 2014). Pigs living in the Tibetan Plateau also bear signatures of selection for genes involved in angiogenesis, response to hypoxia and nucleic acid metabolism (Dong et al. 2014).
In a telling comparison with these mammalian examples, calculation of dN/dS ratios for the ground tit (Pseudopodoces humilis), a bird living in the Tibetan Plateau, in comparison with numerous avian species of low altitude revealed positive selection on genes associated with cardiac function and hormone behaviour (Cai et al. 2013). Putative targets of selection in the ground tit genome were not enriched for (i) metabolic genes, as in high-altitude domesticated mammals, or (ii) genes of the HIF pathway, as in all mammals.
The results from this focused set of studies provide several important insights regarding the interpretation of GWSS. First, several studies mentioned here relied upon an a priori list of candidate genes to filter the list of genes deemed significant in the GWSS (Simonson et al. 2010; Eichstaedt et al. 2014). At face value, this approach may seem strange, as it counters the unbiased nature of GWSS. However, GWSS provide a list of putative regions targeted by selection. Depending on the number of markers, these regions may be quite large and include numerous genes and regulatory regions. Moreover, the list is likely to contain numerous false positives. Then, what approach should we take to filter the list for the most likely targets of selection? One common approach, found frequently in the human GWSS literature, is to find reassurance in the fact that well-established targets of natural selection such as LCT (lactase) are present in the list of significant hits and then suggest that the rest of the list is sure to include numerous true targets of selection. Even if this inference is correct, this approach does little to further our understanding of human biology and the selective forces that have helped shape human adaptation throughout the history of the human lineage. We have trouble connecting selected genes to the causative selective pressure precisely because the unbiased method of GWSS makes no a priori assumptions regarding what classes of genes might be targeted by selection. Indeed, it is in this very situation that the researcher is tempted to cherry-pick the list of significant hits for genes with interesting functions and construct plausible though likely fanciful stories of adaptation (Barrett & Hoekstra 2011; Pavlidis et al. 2012).
Rather than relying upon potentially spurious a posteriori evaluations of a list of selected genes, it therefore seems prudent to list our a priori assumptions of the genes or pathways we expect to find before performing a GWSS. Furthermore, interpretation of the results of GWSS focused on high-altitude adaptation has the advantage of dealing with a clearly defined, strong selective pressure. In this context, when a gene such as EPAS1 is shown to bear the top signature of selection (Beall et al. 2010; Yi et al. 2010), researchers have little reason to doubt the validity of this result. Knowledge of the primary selective pressures acting on a population also facilitates the further exploration of the correlation between phenotype and putatively selected genotype. Beall et al. (2010), for example, showed that the single-nucleotide variants at high frequency in the sequence of EPAS1 in high-altitude Tibetans were associated with low haemoglobin concentration. This finding is congruent with the counterintuitive fact that Tibetans possess low haemoglobin concentrations (Beall et al. 1998), particularly given that Andeans show high haemoglobin concentration (Storz 2010) and GWSS of Andean genomes revealed no selection on EPAS1 (Bigham et al. 2010; Eichstaedt et al. 2014).
Second, intraspecific and interspecific comparisons of adaptive response to hypoxic conditions make clear the radically different genetic architectures that can result from an identical selective pressure. Hypoxic conditions in the Tibetan Plateau, Andean Altiplano and Ethiopian highlands have apparently all elicited highly different genetic adaptations in response to this selective pressure (Bigham et al. 2010; Scheinfeldt et al. 2012; Eichstaedt et al. 2014). Moreover, consideration of domesticated mammals and a wild bird species widens the adaptive response even further. Finally, GWSS focused on humans of the Tibetan Plateau yielded largely similar sets of significant hits—namely genes central to the HIF pathway (Table 2; Beall et al. 2010; Bigham et al. 2010; Simonson et al. 2010; Wuren et al. 2014; Xu et al. 2010; Yi et al. 2010). Reassuringly, this suggests that GWSS reliably uncover adaptive genes of large effect despite varied methodological approaches. However, we again note that the focus of these studies on a key, unambiguous selective pressure makes the adaptive evolution of HIF pathway genes more convincing.
Recommendations and best practices
The genetic complications outlined above make clear that identification of adaptive alleles in species or populations using GWSS is made difficult by genomic heterogeneity in mutation, recombination, selection and the genetic architecture of adaptive phenotypes. By definition, GWSS cover the entire genome. Therefore, GWSS methodology should better incorporate aspects of genomewide heterogeneity.
We provide two recommendations. First, information about key determinants of genomic diversity can be used to adjust genomewide patterns. For example, local estimates of rates of recombination and deleterious mutation could be used to fit a model of background selection to levels of polymorphism across the genome (Reed et al. 2005). The best-fit model could serve as a new null model for identifying instances of positive selection (Comeron 2014). Because measures of genomic heterogeneity are immediately available for genetic model organisms and therefore can be incorporated into analyses, we believe these species are currently the best targets for GWSS. In other organisms, we recommend using surrogates of genomic heterogeneity to improve the interpretation of results. For example, in many species, recombination rates are highly correlated with the distance from the centromere. Even if no recombination rate estimates are available for a species, researchers conducting GWSS could use distance from the centromere as a rough gauge of relative recombination rate. Our most general recommendation is to simply be aware that a set of GWSS results are shaped by genomic heterogeneity. In addition to the use of empirical measurements of genomic heterogeneity in mutation or recombination, it is also important to consider selective targets other than single-nucleotide variants; researchers should be aware of the differing effects selection can have on these genetic variants and, in some cases, methods that have been developed to aid in their detection (Sebat et al. 2004; Feuk et al. 2006; Haasl & Payseur 2013; Haasl et al. 2014).
A second general recommendation is to measure the consequences of heterogeneity in mutation, recombination, selection and the genetic architecture for genomic patterns of diversity using simulations that sample a range of reasonable parameter values. Because genomic heterogeneity forms patterns along chromosomes, whole-chromosome simulations should be especially useful. In empirical cases in which information about genomic heterogeneity is available, these simulations could be built directly into the GWSS inference procedure, using an approach such as approximate Bayesian computation. Simulation results could establish useful guidelines for interpreting GWSS even in the absence of genomic heterogeneity measures. For example, the level of variation in mutation rate that produces high false-positive rates could be determined for a range of selective sweep scenarios the investigator wishes to detect in the GWSS. In this manner, the plausibility of alternatives to selection could be gauged.
Genomewide scans for natural selection can also be improved by considering knowledge of potential selective agents. In our discussion of GWSS focused on adaptation to high-altitude conditions, we suggested that a major advantage of these studies was the presence of an unequivocal selective pressure affecting the subject populations. This advantage came to bear near the end of these studies during interpretation of the results of each GWSS. The known selective agent facilitated the identification of plausible targets of selection from the list of significant genomic regions. Yet, the Tibetan Plateau, Andean Altiplano and Ethiopian highlands represent some of the most extreme terrestrial environments on the Earth. Is it possible to identify unambiguous selective pressures affecting populations in more pedestrian regions of Earth? The short answer is no.
However, a suite of recently developed methods do not require a priori determination of selective pressures. Instead, these approaches, which employ GLMMs, simply require that the researcher identify a set of environmental parameters that may act as selective pressures (Hancock et al. 2008, 2010; Frichot et al. 2013). These approaches search for correlations between values of these environmental parameters (or synthetic combinations of them) and genetic variation in individuals sampled from across a geographic range that includes substantial variation in these environmental parameters. The results of GWSS-GLMMs simultaneously identify the most likely selective pressures and the genomic regions subject to natural selection as a result of these pressures. Again, the main advantage to this type of approach is that it links the otherwise anonymous list of putative selective targets with ecological and biological information. This combination of information makes it less tempting to tell stories about adaptation (Pavlidis et al. 2012) and to scan the genomic palimpsest for signatures of selection that are biologically relevant.
Supplementary Material
Acknowledgments
We thank the editors and two anonymous reviewers for their constructive criticism, which has helped improve the quality of this review.
Footnotes
R.J.H. performed the literature search for GWSS and assembled figures and tables.
Data accessibility
All data used are listed in Tables 1, 2, S1 and S2 (Supporting information).
Additional supporting information may be found in the online version of this article.
Table S1 Additional details regarding the 132 GWSS included in this paper.
Table S2 Expanded table showing the genes identified by studies of high-altitude adaptation.
References
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Research. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey JM, Ruhe AL, Akey DT, et al. Tracking footprints of artificial selection in the dog genome. Proceedings of the National Academy of Sciences of the USA. 2010;107:1160–1165. doi: 10.1073/pnas.0909918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtsen A, Moltke I, Nielsen R. Natural selection and the distribution of identity-by-descent in the human genome. Genetics. 2010;186:295–308. doi: 10.1534/genetics.110.113977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, Liu X, Pillai EN, et al. Characterizing the genetic differences between two distinct migrant groups from Indo-European and Dravidian speaking populations in India. BMC Genetics. 2014;15:86. doi: 10.1186/1471-2156-15-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amambua-Nqwa A, Tettech KK, Manske M, et al. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genetics. 2012;8:e1002992. doi: 10.1371/journal.pgen.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato R, Pinelli M, Monticelli A, Marino D, Miele G, Cocozza S. Genome-wide scan for signatures of human population differentiation and their relationship with natural selection, functional pathways and diseases. PLoS ONE. 2009;4:e7927. doi: 10.1371/journal.pone.0007927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen KG, Shylakhter I, Tabrizi S, Grossman SR, Happi CT, Sabeti PC. Genome-wide scans provide evidence for positive selection of genes implicated in Lassa fever. Philosophical Transactions of the Royal Society of London B. Biological Sciences. 2012;367:868–877. doi: 10.1098/rstb.2011.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres AM, Hubisz MJ, Indap A, et al. Targets of balancing selection in the human genome. Molecular Biology and Evolution. 2009;26:2755–2764. doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backström N, Forstmeier W, Schielzeth H, et al. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Research. 2010;20:485–495. doi: 10.1101/gr.101410.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RD, Hoekstra HE. Molecular spandrels: test of adaptation at the genetic level. Nature Reviews Genetics. 2011;12:767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- Beall CM, Brittenham GM, Strohl KP, et al. Hemoglobin concentration of high-altitude Tibetans and Bolivian Aymara. American Journal of Physical Anthropology. 1998;106:385–400. doi: 10.1002/(SICI)1096-8644(199807)106:3<385::AID-AJPA10>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Beall CM, Cavalleri GL, Deng L, et al. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proceedings of the National Academy of Sciences of the USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biology. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JJ, Coop G. A population genetic signal of polygenic adaptation. PLoS Genetics. 2014;10:e1004412. doi: 10.1371/journal.pgen.1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia G, Tandon A, Patterson N, et al. Genome-wide scan of 29,141 African Americans finds no evidence of directional selection since admixture. American Journal of Human Genetics. 2014;95:437–444. doi: 10.1016/j.ajhg.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A, Bauchet M, Pinto D, et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genetics. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branca A, Paaper TD, Zhou P, et al. Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proceedings of the National Academy of Sciences of the USA. 2011;108:E864–E870. doi: 10.1073/pnas.1104032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. American Journal of Human Genetics. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks LD, Marks RW. The organization of genetic variation for recombination in Drosophila melanogaster. Genetics. 1986;114:525–547. doi: 10.1093/genetics/114.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb KL, Bovee D, Buckley D, et al. Scan of the human genome reveals no new loci under ancient balancing selection. Genetics. 2006;173:2165–2177. doi: 10.1534/genetics.106.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Caballero A, Quesada H, Rolan-Alvarez E. Impact of amplified fragment length polymorphism size homoplasy on the estimation of population genetic diversity and the detection of selective loci. Genetics. 2008;179:539–554. doi: 10.1534/genetics.107.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Qian X, Lang Y, et al. Genome sequence of ground tit Pseudopodoces humilis and its adaptation to high altitude. Genome Biology. 2013;14:R29. doi: 10.1186/gb-2013-14-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CD, Chong JX, Malig M, et al. Estimating the human mutation rate using autozygosity in a founder population. Nature Genetics. 2012;44:1277–1281. doi: 10.1038/ng.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann HM, De Toma C, Cazes L, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Thomas DJ, Eberle MA, et al. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Research. 2005;15:1553–1565. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casa AM, Mitchell SE, Hamblin MT, et al. Diversity and selection in sorghum: simultaneous analyses using simple sequence repeats. Theoretical and Applied Genetics. 2005;111:23–30. doi: 10.1007/s00122-005-1952-5. [DOI] [PubMed] [Google Scholar]
- Cavagnagh CR, Chao S, Wang S, et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proceedings of the National Academy of Sciences of the USA. 2013;110:8057–8062. doi: 10.1073/pnas.1217133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Pashley CH, Wenzler J, et al. A genomic scan for selection reveals candidates for genes involved in the evolution of cultivated sunflower (Helianthus annuus) Plant Cell. 2008;20:2931–2945. doi: 10.1105/tpc.108.059808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Nordborg M, Charlesworth D. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genetical Research. 1997;70:155–174. doi: 10.1017/s0016672397002954. [DOI] [PubMed] [Google Scholar]
- Chavez-Galarza J, Henriques D, Johnston JS, et al. Signatures of selection in the Iberian honey bee (Apis mellifera iberiensis) revealed by a genome scan analysis of single nucleotide polymorphisms. Molecular Ecology. 2013;22:5890–5907. doi: 10.1111/mec.12537. [DOI] [PubMed] [Google Scholar]
- Chen CH, Chuang TJ, Liao BY, Chen FC. Scanning for signatures of positive selection for human-specific insertions and deletions. Genome Biology and Evolution. 2009;1:415–419. doi: 10.1093/gbe/evp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Research. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;203:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Clemente FJ, Cardona A, Inchley CE, et al. A selective sweep on a deleterious mutation in CPT1A in arctic populations. American Journal of Human Genetics. 2014;95:584–589. doi: 10.1016/j.ajhg.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna V, Ayub Q, Chen Y, et al. Human genomic regions with exceptionally high levels of population differentiation identified in 911 whole-genome sequences. Genome Biology. 2014;15:R88. doi: 10.1186/gb-2014-15-6-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM. Background selection as baseline for nucleotide variation across the Drosophila genome. PLoS Genetics. 2014;10:e1004434. doi: 10.1371/journal.pgen.1004434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM, Ratnappan R, Bailin S. The many landscapes of recombination in Drosophila melanogaster. PLoS Genetics. 2012;8:e1002905. doi: 10.1371/journal.pgen.1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- Corbett-Detig RB, Hartl DL, Sackton TB. Natural selection constrains neutral diversity across a wide range of species. PLOS Biology. 2015;13:e1002112. doi: 10.1371/journal.pbio.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Payseur BA. Genomic signatures of selection at linked sites: unifying the disparity among species. Nature Reviews Genetics. 2013;14:262–274. doi: 10.1038/nrg3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and consequences of an emerging epidemic. Clinical Microbiology Reviews. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwiche CF. The genomic palimpsest: genomics in evolution and ecology. BioScience. 2004;54:991–1001. [Google Scholar]
- Dong K, Yao N, Pu Y, et al. Genomic scan reveals loci under altitude adaptation in Tibetan and Dahe pigs. PLoS ONE. 2014;9:e110520. doi: 10.1371/journal.pone.0110520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert AJ, Bower AD, Gonzalez-Martinez SC, Wegrzyn JL, Coop G, Neale DB. Back to nature: ecological genomics of loblolly pine (Pinus taeda, Pinaceae) Molecular Ecology. 2010;19:3789–3805. doi: 10.1111/j.1365-294X.2010.04698.x. [DOI] [PubMed] [Google Scholar]
- Edea Z, Dadi H, Kim SW, et al. Linkage disequilibrium and genomic scan to detect selective loci in cattle populations adapted to different ecological conditions in Ethiopia. Journal of Animal Breeding and Genetics. 2014;131:358–366. doi: 10.1111/jbg.12083. [DOI] [PubMed] [Google Scholar]
- Eichstaedt CA, Antao T, Pagani L, et al. The Andean adaptive toolkit to counteract high altitude maladaptation: genome-wide and phenotypic analysis of the Collas. PLoS ONE. 2014;9:e93314. doi: 10.1371/journal.pone.0093314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Genome sequencing and population genomics in non-model organisms. Trends in Ecology and Evolution. 2014;29:51–63. doi: 10.1016/j.tree.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Enard D, Depaulis F, Roest Crollius H. Human and nonhuman primate genomes share hotspots of positive selection. PLoS Genetics. 2010;6:e1000840. doi: 10.1371/journal.pgen.1000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard D, Messer PW, Petrov DA. Genome-wide signals of positive selection in human evolution. Genome Research. 2014;234:885–895. doi: 10.1101/gr.164822.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LM, Slavov GT, Rodgers-Melnick E, et al. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nature Genetics. 2014;46:1089–1096. doi: 10.1038/ng.3075. [DOI] [PubMed] [Google Scholar]
- Fangy M, Patin E, Enard D, Barreiro LB, Quintana-Murci L, Laval G. Exploring the occurrence of classic selective sweeps in humans using whole-genome sequencing data sets. Molecular Biology and Evolution. 2014;31:1850–1868. doi: 10.1093/molbev/msu118. [DOI] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nature Reviews Genetics. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Fledel-Alon A, Wilson DJ, Broman K, et al. Broad-scale recombination patterns underlying proper disjunction in humans. PLoS Genetics. 2009;5:e1000658. doi: 10.1371/journal.pgen.1000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser BA, Kunstner A, Reznick DN, Dreyer C, Weigel D. Population genomics of natural and experimental populations of guppies (Poecilia reticulata) Molecular Ecology. 2015;24:389–408. doi: 10.1111/mec.13022. [DOI] [PubMed] [Google Scholar]
- Frichot E, Schoville SD, Bouchard G, Francois O. Testing for associations between loci and environmental gradients using latent factor mixed models. Molecular Biology and Evolution. 2013;30:1687–1699. doi: 10.1093/molbev/mst063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Sironi M, Pozzoli U, Ferrer-Admetlla A, Pattini L, Nielsen R. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genetics. 2011;7:e1002355. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire PA, Normandeau E, Cote C, Moller Hansen M, Bernatchez L. The genetic consequences of spatially varying selection in the panmictic American eel (Anguilla rostrata) Genetics. 2012a;190:725–736. doi: 10.1534/genetics.111.134825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire PA, Normandeau E, Bernatchez L. Comparative genomics reveals adaptive protein evolution and a possible cytonuclear incompatibility between European and American eels. Molecular Biology and Evolution. 2012b;29:2909–2919. doi: 10.1093/molbev/mss076. [DOI] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RD, McVicker G, Diederich R, et al. Trans genomic capture and sequencing of pirmate exomes reveals new targets of positive selection. Genome Research. 2011;21:1686–1694. doi: 10.1101/gr.121327.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granka JM, Henn BM, Gignoux CR, Kidd JM, Bustamante CD, Feldman MW. Limited evidence for classic selective sweeps in African populations. Genetics. 2012;192:1049–1064. doi: 10.1534/genetics.112.144071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Orr N, Park SD, Katz LM, et al. A genome scan for positive selection in thoroughbred horses. PLoS ONE. 2009;4:e5767. doi: 10.1371/journal.pone.0005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasl RJ, Paysuer BA. Microsatellites as targets of natural selection. Molecular Biology and Evolution. 2013;30:285–298. doi: 10.1093/molbev/mss247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasl RJ, Johnson RC, Payseur BA. The effects of microsatellites selection on linked sequence diversity. Genome Biology and Evolution. 2014;6:1843–1861. doi: 10.1093/gbe/evu134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenblad J, Olsson M, Parker HG, Ostrander EA, Ellegren H. Population genomics of the inbred Scandinavian wolf. Molecular Ecology. 2009;18:1341–1351. doi: 10.1111/j.1365-294X.2009.04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Gordon AS, et al. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genetics. 2008;4:e32. doi: 10.1371/journal.pgen.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Ehler E, et al. Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proceedings of the National Academy of Sciences of the USA. 2010;107:8924–8930. doi: 10.1073/pnas.0914625107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Alkorta-Aranburu G, et al. Adaptations to climate-mediated selective pressures in humans. PLoS Genetics. 2011a;7:e1001375. doi: 10.1371/journal.pgen.1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Brachi B, Faure N, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011b;334:83–86. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nature Reviews Genetics. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Haygood R, Fedrigo O, Hanson B, Yokoyama KD, Wray GA. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nature Genetics. 2007;39:1140–1144. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- Hebert FO, Renaut S, Bernatchez L. Targeted sequence capture and resequencing implies a predominant role of regulatory regions in the divergence of a sympatric lake whitefish species pair (Coregonus clupeaformis) Molecular Ecology. 2013;22:4896–4914. doi: 10.1111/mec.12447. [DOI] [PubMed] [Google Scholar]
- Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hider JL, Gittelman RM, Shah T, et al. Exploring signatures of positive selection in pigmentation candidate genes in populations of East Asian ancestry. BMC Evolutionary Biology. 2013;13:150. doi: 10.1186/1471-2148-13-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber CD, Nordborg M, Hermisson J, Hellmann I. Keeping it local: evidence for positive selection in Swedish Arabidopsis thaliana. Molecular Biology and Evolution. 2014;31:3026–3039. doi: 10.1093/molbev/msu247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttley GA, Smith MW, Carrington M, O’Brien SJ. A scan for linkage disequilibrium across the human genome. Genetics. 1999;152:1711–1722. doi: 10.1093/genetics/152.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle S, Ravaoarimanana I, Thomas M, Tautz D. An analysis of selective sweeps in natural populations of the house mouse. Molecular Biology and Evolution. 2006;23:790–797. doi: 10.1093/molbev/msj096. [DOI] [PubMed] [Google Scholar]
- Innan H, Kim Y. Pattern of polymorphism after strong artificial selection in a domestication event. Proceedings of the National Academy of Sciences of the USA. 2004;101:10667–10672. doi: 10.1073/pnas.0401720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Wu H, Smith JD, et al. De novo rates and selection of large copy number variation. Genome Research. 2010;20:1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquiery J, Stoeckel S, Nouhaud P, et al. Genome scans reveal candidate regions involved in the adaptation to host plant in the pea aphid complex. Molecular Ecology. 2012;21:5251–5264. doi: 10.1111/mec.12048. [DOI] [PubMed] [Google Scholar]
- Jarvis JP, Scheinfeldt LB, Soi S, et al. Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies. PLoS Genetics. 2012;8:e1002641. doi: 10.1371/journal.pgen.1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nature Genetics. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Furey TS, Payseur BA, et al. Comparative recombination rates in the rat, mouse, and human genomes. Genome Research. 2004;14:528–538. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirimutu, Wang Z, Ding G, et al. Genome sequences of wild and domestic bactrian camels. Nature Communications. 2012;3:1202. doi: 10.1038/ncomms2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A, Gyllensten U. Identification of local selective sweeps in human populations since the exodus from Africa. Hereditas. 2008;145:126–137. doi: 10.1111/j.0018-0661.2008.02054.x. [DOI] [PubMed] [Google Scholar]
- Kane NC, Rieseberg LH. Selective sweeps reveal candidate genes for adaptation to drought and salt tolerance in common sunflower, Helianthus annuus. Genetics. 2007;175:1823–1834. doi: 10.1534/genetics.106.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH. The “hitchhiking effect” revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur T, Rockman MV. Crossover heterogeneity in the absence of hotspots in Caenorhabditis elegans. Genetics. 2014;196:137–148. doi: 10.1534/genetics.113.158857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Stoneking M. A genome scan to detect candidate regions influenced by local natural selection in human populations. Molecular Biology and Evolution. 2003;20:893–900. doi: 10.1093/molbev/msg092. [DOI] [PubMed] [Google Scholar]
- Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Research. 2006;16:980–989. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper KE, Saxton SJ, Bolormaa S, Hayes BJ, Goddard ME. Selection for complex traits leaves little or no classic signatures of selection. BMC Genomics. 2014;15:246. doi: 10.1186/1471-2164-15-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura R, Fujimoto A, Tokunaga K, Ohashi J. A practical genome scan for population-specific strong selective sweeps that have reached fixation. PLoS ONE. 2007;14:e286. doi: 10.1371/journal.pone.0000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura R, Ohashi J, Matsumura Y, et al. Gene flow and natural selection in oceanic human populations inferred from genome-wide SNP typing. Molecular Biology and Evolution. 2008;24:1750–1761. doi: 10.1093/molbev/msn128. [DOI] [PubMed] [Google Scholar]
- Koehler KE, Cherry JP, Lynn A, Hunt PA, Hassold TJ. Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics. 2002;162:297–306. doi: 10.1093/genetics/162.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, et al. A high-resolution recombination map of the human genome. Nature Genetics. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- Langley CH, Stevens K, Cardeno C, et al. Genomic variation in natural populations of Drosophila melanogaster. Genetics. 2012;192:533–598. doi: 10.1534/genetics.112.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Salmela E, Andersen PM, et al. Genomic landscape of positive natural selection in Northern European populations. European Journal of Human Genetics. 2010;18:471–478. doi: 10.1038/ejhg.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latta RG. Differentiation of allelic frequencies at quantitative trait loci affect locally adaptive traits. The American Naturalist. 1998;151:283–292. doi: 10.1086/286119. [DOI] [PubMed] [Google Scholar]
- Le Corre V, Kremer A. Genetic variability at neutral markers, quantitative trait loci and trait in a subdivided population under selection. Genetics. 2003;164:1205–1219. doi: 10.1093/genetics/164.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Corre V, Kremer A. The genetic differentiation at quantitative trait loci under local adaptation. Molecular Ecology. 2012;21:1548–1566. doi: 10.1111/j.1365-294X.2012.05479.x. [DOI] [PubMed] [Google Scholar]
- Leffler EM, Bullaughey K, Matute DR, et al. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biology. 2012;10:e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler EM, Gao Z, Pfeifer S, et al. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science. 2013;339:1578–1582. doi: 10.1126/science.1234070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limborg MT, Waples RK, Seeb JE, Seeb LW. Temporally isolated lineages of pink salmon reveal unique signatures of selection on distinct pools of standing genetic variation. Journal of Heredity. 2014;105:741–751. doi: 10.1093/jhered/esu063. [DOI] [PubMed] [Google Scholar]
- Liu X, Ong RT, Pillai EN, et al. Detecting and characterizing genomic signatures of positive selection in global populations. American Journal of Human Genetics. 2013;92:866–881. doi: 10.1016/j.ajhg.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobreaux S, Melodelima C. Detection of genomic loci associated with environmental variables using generalized linear mixed models. Genomics. 2015;105:69–75. doi: 10.1016/j.ygeno.2014.12.001. [DOI] [PubMed] [Google Scholar]
- López Herráez D, Bauchet M, Tang K, et al. Genetic variation and recent positive selection in worldwide human populations: evidence from nearly 1 million SNPs. PLoS ONE. 2009;4:e7888. doi: 10.1371/journal.pone.0007888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nature Reviews Genetics. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- Lv FH, Agha S, Kantanen J, et al. Adaptations to climate-mediated selective pressures in sheep. Molecular Biology and Evolution. 2014;31:3324–3343. doi: 10.1093/molbev/msu264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen HS, Cano JM, Merita J. Identifying footprints of directional and balancing selection in marine and freshwater three-spined stickleback (Gasterosteus aculeatus) populations. Molecular Ecology. 2008;17:3565–3582. doi: 10.1111/j.1365-294X.2008.03714.x. [DOI] [PubMed] [Google Scholar]
- Marden JH. Nature’s inordinate fondness for metabolic enzymes: why metabolic enzyme loci are so frequently targets of natural selection. Molecular Ecology. 2013;22:5743–5764. doi: 10.1111/mec.12534. [DOI] [PubMed] [Google Scholar]
- Mattiangeli V, Ryan AW, McManus R, Bradley DG. A genome-wide approach to identify genetic loci with a signature of natural selection in the Irish population. Genome Biology. 2006;7:R74. doi: 10.1186/gb-2006-7-8-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genetical Research. 1974;23:23–35. [PubMed] [Google Scholar]
- McKay JK, Latta RG. Adaptive population divergence: markers, QTL and traits. Trends in Ecology and Evolution. 2002;17:285–291. [Google Scholar]
- Metspalu M, Romero IG, Yunusbayev B, et al. Shared and unique components of population structure and genome-wide signals of positive selection in South Asia. American Journal of Human Genetics. 2011;89:731–744. doi: 10.1016/j.ajhg.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliano AB, Romero IG, Metspalu M, et al. Evolution of the pygmy phenotype: evidence of positive selection from genome-wide scans in African, Asian, and Melanesian populations. Human Biology. 2013;85:251–284. doi: 10.3378/027.085.0313. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Myles S, Tang K, Somel M, Green RE, Kelso J, Stoneking M. Identification and analysis of genomic regions with large between-population differentiation in humans. Annals of Human Genetics. 2008;72:99–110. doi: 10.1111/j.1469-1809.2007.00390.x. [DOI] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biology. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard S, Braunstein A, Malsen G, et al. Long- and short-term selective forces on malaria parasite genomes. PLoS Genetics. 2010;6:e1001099. doi: 10.1371/journal.pgen.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochola LI, Tetteh KK, Stewart LB, Riitho V, Marsh K, Conway DJ. Allele frequency-based and polymorphism-versus-divergence indices of balancing selection in a new filtered set of polymorphic genes in Plasmodium falciparum. Molecular Biology and Evolution. 2010;27:2344–2351. doi: 10.1093/molbev/msq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesyk TK, Zhao K, De La Vega FM, Gilbert DA, O’Brien SJ, Smith MW. Identifying selected regions from heterozygosity and divergence using a light-coverage genomic dataset from two human populations. PLoS ONE. 2008;3:e1712. doi: 10.1371/journal.pone.0001712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly PF, Birney E, Balding DJ. Confounding between recombination and selection, and the Ped/Pop method for detecting selection. Genome Research. 2008;18:1304–1313. doi: 10.1101/gr.067181.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution. 1998;52:935–949. doi: 10.1111/j.1558-5646.1998.tb01823.x. [DOI] [PubMed] [Google Scholar]
- Orr HA. The population genetics of adaptation: the adaptation of DNA sequences. Evolution. 2002;56:1317–1330. doi: 10.1111/j.0014-3820.2002.tb01446.x. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. Recombination is proportional to the number of chromosome arms in mammals. Mammalian Genome. 2001;12:318–322. doi: 10.1007/s003350020005. [DOI] [PubMed] [Google Scholar]
- Park DJ, Lukens AK, Neafsey DE, et al. Sequence-based association and selection scans identify drug resistance loci in the Plasmodium falciparum malaria parasite. Proceedings of the National Academy of Sciences of the USA. 2012;109:13052–13057. doi: 10.1073/pnas.1210585109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Jensen JD, Stephan W, Stamatakis A. A critical assessment of storytelling: gene ontology categories and the importance of validating genomic scans. Molecular Biology and Evolution. 2012;29:3237–3248. doi: 10.1093/molbev/mss136. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Cutter AD, Nachman MW. Searching for evidence of positive selection in the human genome using patterns of microsatellite variability. Molecular Biology and Evolution. 2002;19:1143–1153. doi: 10.1093/oxfordjournals.molbev.a004172. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J. Signals of recent positive selection in a worldwide sample of human populations. Genome Research. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras IS, De Montis A, Calo CM, et al. Genome-wide scan with nearly 700,000 SNPs in two Sardinian sub-populations suggest some regions as candidate targets for positive selection. European Journal of Human Genetics. 2012;20:1155–1161. doi: 10.1038/ejhg.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollinger JP, Bustamante CD, Fledel-Alon A, Schmutz S, Gray MM, Wayne RK. Selective sweep mapping of genes with large phenotypic effects. Genome Research. 2005;15:1809–1819. doi: 10.1101/gr.4374505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Corbett-Detig RB, Sugino RP, et al. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genetics. 2012;8:e1003080. doi: 10.1371/journal.pgen.1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Di Rienzo A. Adaptation—not by sweeps alone. Nature Reviews Genetics. 2010;11:665–667. doi: 10.1038/nrg2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution. 2005;59:2312–2323. [PubMed] [Google Scholar]
- Puzey J, Vallejo-Marin M. Genomics of invasion: diversity and selection in introduced populations of monkeyflowers (Mimulus guttauts) Molecular Ecology. 2014;23:4472–4485. doi: 10.1111/mec.12875. [DOI] [PubMed] [Google Scholar]
- Qiu Q, Zhang G, Ma T, et al. The yak genome and adaptation to life at high altitude. Nature Genetics. 2012;44:946–949. doi: 10.1038/ng.2343. [DOI] [PubMed] [Google Scholar]
- Quilez J, Short AD, Martinez V, et al. A selective sweep of >8 Mb on chromosome 26 in the Boxer genome. BMC Genomics. 2011;12:339. doi: 10.1186/1471-2164-12-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj T, Kuchroo M, Replogle JM, Raychaudhuri S, Stranger BE, De Jager PL. Common risk alleles for inflammatory diseases are targets of recent positive selection. American Journal of Human Genetics. 2013;92:517–529. doi: 10.1016/j.ajhg.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed FA, Akey JM, Aquadro CF. Fitting background-selection predictions to levels of nucleotide variation and divergence along the human autosomes. Genome Research. 2005;15:1211–1221. doi: 10.1101/gr.3413205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt JA, Kolaczkowski B, Jones CD, Begun DJ, Kern AD. Parallel geographic variation in Drosophila melanogaster. Genetics. 2014;197:361–373. doi: 10.1534/genetics.114.161463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Roach JC, Glusman G, Smit AFA, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt LB, Soi S, Thompson S, et al. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biology. 2012;13:R1. doi: 10.1186/gb-2012-13-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M, Jonsson H, Chang D, et al. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E5661–E5669. doi: 10.1073/pnas.1416991111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Toge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PLoS Genetics. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA, Huang W, Zhang C, et al. Adaptive genic evolution in the Drosophila genomes. Proceedings of the National Academy of Sciences. 2007;104:2771–2776. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Bell JT, Copley RR, et al. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. Plos Biology. 2006;4:2227–2237. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver MD, Kennedy GC, Parra EJ, et al. The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Human Genomics. 2004;1:274–286. doi: 10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS, Yang Y, Huff CD, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–74. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- Sjostrand AE, Sjodin P, Jakobsson M. Private haplotypes can reveal local adaptation. BMC Genetics. 2014;15:61. doi: 10.1186/1471-2156-15-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukowski CS, Noor MAF. Recombination rate variation in closely related species. Heredity. 2011;107:496–508. doi: 10.1038/hdy.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Wilson Sayres MA, Jordan G, et al. A scan for human-specific relaxation of negative selection reveals unexpected polymorphism in proteasome genes. Molecular Biology and Evolution. 2013;30:1808–1815. doi: 10.1093/molbev/mst098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Winker K, Shaw TI, Jones KL, Glenn TC. Transcriptome analysis of a North American songbird, Melospiza melodia. DNA Research. 2012;19:325–333. doi: 10.1093/dnares/dss015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B, Nederbragt AJ, Jentoft S, et al. The genome of the Atlantic cod reveals a unique immune system. Nature. 2011;477:207–210. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steane DA, Potts BM, McLean E, et al. Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Molecular Ecology. 2014;23:2500–2513. doi: 10.1111/mec.12751. [DOI] [PubMed] [Google Scholar]
- Stephan W, Wiehe T, Lenz MW. The effect of strongly selected substitutions on neutral polymorphism: analytical results based on diffusion theory. Theoretical Population Biology. 1992;47:237–254. [Google Scholar]
- Storz JF. Genes for high altitudes. Science. 2010;329:40–41. doi: 10.1126/science.1192481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Payseur BA, Nachman MW. Genome scans of DNA variability in humans reveal evidence for selective sweeps outside of Africa. Molecular Biology and Evolution. 2004;21:1800–1811. doi: 10.1093/molbev/msh192. [DOI] [PubMed] [Google Scholar]
- Sun JX, Helgason A, Masson G, et al. A direct characterization of human mutation based on microsatellites. Nature Genetics. 2012;44:1161–1165. doi: 10.1038/ng.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YB, Zhou WP, Liu HQ, Irwin DM, Shen YY, Zhang YP. Genome-wide scans for candidate genes involved in the aquatic adaptation of dolphins. Genome Biology and Evolution. 2013;5:130–139. doi: 10.1093/gbe/evs123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo C, Xu H, Khor CC, et al. Natural positive selection and north-south genetic diversity in East Asia. European Journal of Human Genetics. 2007;20:102–110. doi: 10.1038/ejhg.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Thornton KR, Stoneking M. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biology. 2007;5:e171. doi: 10.1371/journal.pbio.0050171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschke M, Mukabayire O, Wiehe T, Tautz D. Identification of selective sweeps in closely related population of the house mouse based on microsatellite scans. Genetics. 2008;180:1537–1545. doi: 10.1534/genetics.108.090811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima KM, Przeworski M. Directional positive selection on an allele of arbitrary dominance. Genetics. 2006;172:713–718. doi: 10.1534/genetics.105.044065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JC, Godfrey PA, Feldgarden M, Robinson DA. Candidate targets of balancing selection in the genome of Staphylococcus aureus. Molecular Biology and Evolution. 2012;29:1175–1186. doi: 10.1093/molbev/msr286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A. CYP3A variation and the evolution of salt-sensitivity variants. American Journal of Human Genetics. 2004;75:1059–1069. doi: 10.1086/426406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura Y, Uchiyama K, Moriguchi Y, Kimura MK, Ueno S, Ujino-Ihara T. Genetic differentiation and evolutionary adaptation in Cryptomeria japonica. G3. 2014;4:2389–2402. doi: 10.1534/g3.114.013896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B, Stergachis AB, Maurano MT, et al. Personal and population genomics of human regulatory variation. Genome Research. 2012;22:1689–1697. doi: 10.1101/gr.134890.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux Y, McMullen M, Hittinger CT, et al. Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proceedings of the National Academy of Sciences of the USA. 2002;99:9650–9655. doi: 10.1073/pnas.112324299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villafuerte FC, Cardenas R, Monge CC. Optimal hemoglobin concentration and high altitude: a theoretical approach for Andean men at rest. Journal of Applied Physiology. 2004;96:1581–1588. doi: 10.1152/japplphysiol.00328.2003. [DOI] [PubMed] [Google Scholar]
- Vincent B, Dionne M, Kent MP, Lien S, Bernatchez L. Landscape genomics in Atlantic salmon (Salmo salar): searching for gene-environment interactions driving local adaptation. Evolution. 2013;67:3469–3487. doi: 10.1111/evo.12139. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biology. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg A, Han F, Wellhagen G, et al. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nature Genetics. 2014;46:1081–1088. doi: 10.1038/ng.3077. [DOI] [PubMed] [Google Scholar]
- Wang ET, Kodama G, Baldi P, Moyzis RK. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JL, Wong C. Mutation of human short tandem repeats. Human Molecular Genetics. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Research. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss KM, Kawasaki K. Reading the palimpsests of life: some relative bear only the faintest trace of their ancestor. Evolutionary Anthropology. 2006;15:207–211. [Google Scholar]
- Westram AM, Galindo J, Alm Rosenblad M, Grahame JW, Panova M, Butlin RK. Do the same genes underlie parallel phenotypic divergence in different Littorina saxatilis populations. Molecular Ecology. 2014;23:4603–4616. doi: 10.1111/mec.12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Lawniczak MK, Cheng C, et al. Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:244–249. doi: 10.1073/pnas.1013648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genetics. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Ruhe AL, Dumont BL, et al. A comprehensive linkage map of the dog genome. Genetics. 2010;184:595–605. doi: 10.1534/genetics.109.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuren T, Simonson TS, Qin G, et al. Shared and unique signals of high-altitude adaptation in geographically distinct Tibetan populations. PLoS ONE. 2014;9:e88252. doi: 10.1371/journal.pone.0088252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Li S, Yang Y, et al. A genome-wide search for signals of high-altitude adaptation in Tibetans. Molecular Biology and Evolution. 2010;28:1003–1011. doi: 10.1093/molbev/msq277. [DOI] [PubMed] [Google Scholar]
- Yi X, Yu L, Huerta-Sanchez E, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JB, Stanton-Geddes J, Zhou P, Briskine R, Young ND, Tiffin P. Genomic signatures of adaptation to climate in Medicago truncatula. Genetics. 2014;196:1263–1275. doi: 10.1534/genetics.113.159319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Pan S, Wang J, et al. Peregrine and saker falcon genome sequences provide insights in evolution of a predatory lifestyle. Nature Genetics. 2013;45:563–566. doi: 10.1038/ng.2588. [DOI] [PubMed] [Google Scholar]
- Zhang C, Bailey DK, Awad T, et al. A whole genome long-range haplotype (WGLRH) test for detecting imprints of positive selection in human populations. Bioinformatics. 2006;22:2122–2128. doi: 10.1093/bioinformatics/btl365. [DOI] [PubMed] [Google Scholar]
- Zhang YB, Li X, Zhang F, Wang DM, Yu J. A preliminary study of copy number variation in Tibetans. PLoS ONE. 2012;7:e41768. doi: 10.1371/journal.pone.0041768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zueva KJ, Lumme J, Veselov AE, Kent MP, Lien S, Primmer CR. Footprints of directional selection in wild Atlantic salmon populations: evidence for parasite-driven evolution? PLoS ONE. 2014;9:e91672. doi: 10.1371/journal.pone.0091672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.