Abstract
Smoking is a risk factor in pancreatic disease, however, the biochemical mechanisms correlating smoking with pancreatic dysfunction remain poorly understood. Strategies using multiplexed isobaric tag-based mass spectrometry facilitate the study of drug-induced perturbations on biological systems. Here, we present the first large scale analysis of the proteomic and phosphoproteomic alterations in pancreatic stellate cells following treatment with two nicotinic acetylcholine receptor (nAChR) ligands: nicotine and α-bungarotoxin. We treated cells with nicotine or α-bungarotoxin for 12hr in triplicate and compared alterations in protein expression and phosphorylation levels to mock treated cells using a tandem mass tag (TMT9plex)-based approach. Over 8,100 proteins were quantified across all nine samples of which 46 were altered in abundance upon treatment with nicotine. Proteins with increased abundance included those associated with neurons, defense mechanisms, indicators of pancreatic disease and lysosomal proteins. In addition, we measured differences for ∼16,000 phosphorylation sites across all nine samples using a titanium dioxide-based strategy, of which 132 sites were altered with nicotine and 451 with α-bungarotoxin treatment. Many altered phosphorylation sites were involved in nuclear function and transcriptional events. This study supports the development of future targeted investigations to establish a better understanding for the role of nicotine and associated receptors in pancreatic disease.
Keywords: Tandem mass tags, pancreatitis, pancreatic cancer, multiplexing, phosphopeptide enrichment, Fusion, synchronous precursor selection, SPS
Introduction
Cigarette smoking is a prominent risk factor that is correlated with the development of pancreatic cancer (1, 2). Coincidently, smoking promotes the development of pancreatic disease by increasing the severity of calcifications which can further damage the pancreas (3). Notwithstanding, investigations on the association of smoking and conditions such as chronic pancreatitis and pancreatic cancer are scarce. Understanding the underlying mechanisms that dictate the course of pancreatic disease is vital to developing counteracting intervention.
Cigarette smoke consists of a mixture of greater than 4,000 compounds, for which over 50 are suspected carcinogens (4). The toxic compounds in tobacco smoke are absorbed into the bloodstream, and several have been detected in pancreatic fluid (5). Nicotine, a major component of tobacco, is readily absorbed by the lungs, as well as distal organs via systemic circulation (6). Various cancers and chronic diseases have been linked to nicotine, yet the mechanisms of such remain poorly understood (7, 8). Concerning pancreatic disease, nicotine may induce fibrosis and metastasis, hallmarks of chronic pancreatitis and pancreatic adenocarcinoma, respectively (9).
Physiologically, nicotine binds specifically to nicotinic acetylcholine receptors (nAChR) with varying affinities and downstream effectors that are involved in signal transduction resulting in intracellular phosphorylation cascades. nAChR can bind distinct ligands and evidence suggests that each subtype has a unique set of interacting proteins, a wide array of ligands, and varying affinities to nicotine (10). Nicotine binding to these cell surface receptors transduces extracellular signals to the intracellular space, resulting in ion transport and/or initiation of phosphorylation cascades and other signaling pathways (11-13). Moreover, these receptors, particularly the α7 subtype, have been found also in non-neuronal cells and are thought to be important regulators of cellular function (14-16).
nAChR can bind dozens of small molecule ligands, such as nicotine, acetylcholine and derivatives thereof, as well as small proteins, such as snail and snake-venom derived toxins. α-Bungarotoxin is a neurotoxin isolated from the venom of the Taiwanese banded krait, Bungarus multicinctus (17). This protein binds irreversibly and competitively to select subtypes of nAChR, including those with α1, α7, and α9 subunits. Although traditionally considered to bind specifically to these nicotinic receptors, recent evidence suggests that α-bungarotoxin may act as an inhibitor of gamma-aminobutyric type-A receptors (GABAAR) (18).
Pancreatic stellate cells (PaSC) are myofibroblast-like cells that reside in exocrine areas of the pancreas and participate in tissue repair activities (19). PaSC may play a role in the pathogenesis of pancreatitis and pancreatic cancer (20-23). In fact, PaSC, including the human cell line (RLT-PSC) (24) used herein, have been shown previously as being a model for pancreatic cancer (25) and pancreatic fibrosis that is associated with chronic pancreatitis (26). In addition, previous study has shown that hepatic stellate cells (HSC) and PaSC are approximately 99% similar at the mRNA level (27, 28). Moreover, nAChR have been shown to express in HSC (29), but hitherto nAChR have not been identified in PaSC. As a nAChR antagonist, α-bungarotoxin can only alter cellular functions in the presence of agonist that binds to the receptor. Interestingly, PaSC have been shown previously to secrete acetylcholine at a rate to ∼120 pM/million cells (30). Acetylcholine production by PaSC allows for nAChR-mediated responses and thereby inhibition due to α-bungarotoxin.
In the present study, we used quantitative mass spectrometry-based techniques to investigate the effects of a 12-hr treatment of nicotine and α-bungarotoxin on the proteome and phosphoproteome of PaSC. We analyzed three groups of samples - control, nicotine-, and α-bungarotoxin-treated PaSC - in triplicate. In total, we quantified over 8,100 proteins across all nine samples. In addition, we quantified over 16,000 phosphorylation sites using a titanium dioxide-based enrichment strategy. We conclude that a 12 hr treatment with nicotine alters both the proteome and phosphoproteome of PaSC, while α-bungarotoxin has a more pronounced effect on the phosphoproteome than does nicotine. Further studies focusing on the targets identified may reveal insights into the mechanisms relating smoking and pancreatic disease.
Experimental Section
Materials
Tandem mass tag (TMT) isobaric reagents were from ThermoFisher Scientific (Waltham, MA). Water and organic solvents were from J.T. Baker (Center Valley, PA). Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) were from LifeTechnologies (Waltham, MA). Unless otherwise noted, all other chemicals were from Sigma (St. Louis, MO). The antibodies used in this experiment were purchased from SantaCruz Biotechnology (Dallas, Texas): α1 (sc-65829), α2 (sc-365251), α3 (sc-365479), α4 (sc-1772), α5 (sc-376979), α6 (sc-376966), α7 (sc-5544), α9 (sc-13806) or Cell Signaling Technology (Beverley, MA): actin (4968). The human PaSC cell line used in this experiment was RLT-PSC (24).
Cell growth and harvesting of pancreatic stellate cells
Our experimental strategy is outlined in Figure 1. Cells were harvested following 12 hr serum starvation and subsequent 12 hr treatment with either nicotine or α-bungarotoxin. Cells were lysed and methanol-chloroform extracted proteins were digested and labeled with TMT for mass spectrometry analysis.
Figure 1. Experimental overview.
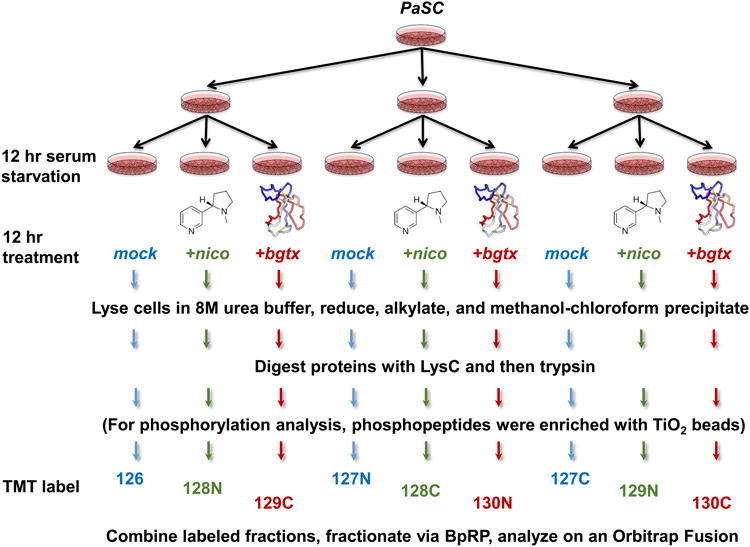
Samples for TMT analysis were prepared as illustrated above. PaSC were harvested following 12 hr serum starvation and subsequent 12 hr treatment with either nicotine or α-bungarotoxin. Cells were harvested and lysed so that proteins could be extracted and prepared for TMT-based mass spectrometry analysis. The additional step of TiO2 enrichment was performed for phosphopeptide analysis. Control samples were labeled with 126, 127N, and 127C; nicotine-treated samples were labeled with 128N, 128C, and 129N, while α-bungarotoxin-treated samples were labeled with 129C, 130N, and 130C.
Methods of cell growth and propagation followed previously utilized techniques (31, 32). In brief, cells were propagated in Dulbecco's modified Eagle's-F12 medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Upon achieving 80% confluency, the growth media was aspirated and the cells were washed 3 times with ice-cold phosphate-buffered saline (PBS). Cells were incubated for 12 hr in serum-free media. The media was then exchanged to complete media containing 10% FBS. Designated cell culture dishes were supplemented with 1 μM nicotine or 1 μg/mL α-bungarotoxin, while control cultures were mock treated with an equal volume of sterile deionized water. Twelve hours after the addition of fresh media, the cells were dislodged with a non-enzymatic reagent, harvested by trituration following the addition of 10 mL PBS, pelleted by centrifugation at 2,000 × g for 5 min at 4°C, and the supernatant was removed. One milliliter of TBSp (50 mM HEPES, 150 mM NaCl, pH 7.4 supplemented with 1X Roche Complete protease inhibitors and PhosphoStop phosphatase inhibitors), 2% SDS were added per each 10 cm cell culture dish.
Cell lysis and protein digestion
Cells were homogenized by 20 passes through a 27 gauge (1.25 inches long) needle. The homogenate was sedimented by centrifugation at 20,000 × g for 5 min at 4°C. Protein concentrations were determined using the bicinchoninic acid (BCA) assay (ThermoFisher Scientific). Proteins were subjected to disulfide bond reduction with 5 mM dithiotreitol (37°C, 25 min) and alkylation with 10 mM iodoacetamide (room temperature, 30 min in the dark). Excess iodoacetamide was quenched with 15 mM dithiotreitol (room temperature, 15 min in the dark).
Methanol-chloroform precipitation was performed prior to protease digestion. In brief, 4 parts neat methanol was added to each sample and vortexed, 1 part chloroform was added to the sample and vortexed, and 3 parts water was added to the sample and vortexed. The samples were centrifuged at 10,000 RPM for 15 min at room temperature and subsequently washed twice with 100% methanol, prior to air-drying. Samples were resuspended in 50 mM HEPES, pH 8.5 and digested at room temperature for 16 hrs with LysC protease at a 100:1 protein-to-protease ratio. Trypsin was then added at a 100:1 protein-to-protease ratio and the reaction was incubated 6 hr at 37°C.
Phosphopeptide enrichment
Phosphopeptides were enriched following digestion and prior to TMT labeling using a method based on that of Kettenbach and Gerber (33) and described elsewhere (34, 35). In brief, Titanosphere TiO2 5 μm beads (GL Biosciences, Tokyo, Japan) were washed three times with 2 M lactic acid/50% acetonitrile. Peptides were resuspended in 2.5 mL of 2 M lactic acid / 50 % acetonitrile. For ∼5 mg of peptide digest, 40 mg beads were added and incubated with gentle rotation for 1 hr at room temperature. Beads were washed twice with 2.5 mL of 2 M lactic acid/50% acetonitrile, then twice with 2.5 mL of 50% acetonitrile/0.1% TFA, and finally twice with 2.5 mL of 25% acetonitrile/0.1% TFA. Enriched phosphopeptides were eluted twice with 500 μL of 50 mM K2HPO4 pH 10 and vacuum centrifuged to dryness.
Isobaric labeling with tandem mass tags (TMT)
TMT reagents (0.8 mg) were dissolved in anhydrous acetonitrile (40 μL) of which 10 μL was added to the peptides along with 20 μL of acetonitrile to achieve a final acetonitrile concentration of approximately 30% (v/v). Control samples were labeled with 126, 127N, and 127C, nicotine-treated samples were labeled with 128N, 128C, and 129N, while α-bungarotoxin treated samples were labeled with 129C, 130N, and 130C. Following incubation at room temperature for 1 hr, the reaction was quenched with hydroxylamine to a final concentration of 0.3% (v/v). The TMT-labeled samples were pooled at a 1:1 ratio across all samples. The sample was vacuum centrifuged to near dryness and subjected to C18 solid-phase extraction (SPE) (Sep-Pak, Waters).
Off-line basic pH reversed-phase (BPRP) fractionation
We fractionated the pooled TMT-labeled peptide sample via BPRP HPLC. We used an Agilent 1100 pump equipped with a degasser and a photodiode array (PDA) detector (set at 220 and 280 nm wavelength) from ThermoFisher Scientific (Waltham, MA). Peptides were subjected to a 50 min linear gradient from 5% to 35% acetonitrile in 10 mM ammonium bicarbonate pH 8 at a flow rate of 0.8 mL/min over an Agilent 300Extend C18 column (5 μm particles, 4.6 mm ID and 22 cm long). The peptide mixture was fractionated into a total of 96 fractions, which were consolidated to 12. These fractions were subsequently acidified with 1% formic acid and vacuum centrifuged to near dryness. Each fraction was also desalted via StageTip, dried via vacuum centrifugation, and reconstituted in 5% acetonitrile, 5% formic acid for LC-MS/MS processing.
Liquid chromatography and tandem mass spectrometry
Our mass spectrometry data were collected using an Orbitrap Fusion mass spectrometer (ThermoFisher Scientific, San Jose, CA) coupled to a Proxeon EASY-nLC 1000 liquid chromatography (LC) pump (ThermoFisher Scientific). Peptides were fractionated on a 75 μm inner diameter microcapillary column packed with ∼0.5 cm of Magic C4 resin (5 μm, 100 Å, Michrom Bioresources) followed by ∼35 cm of GP-18 resin (1.8 μm, 200 Å, Sepax, Newark, DE). For each analysis, we loaded ∼1 μg onto the column.
Peptides were separated using a 3 hr gradient of 6 to 26% acetonitrile in 0.125% formic acid at a flow rate of ∼350 nL/min. Each analysis used the multi-notch MS3-based TMT method (36) on an Orbitrap Fusion mass spectrometer. The scan sequence began with an MS1 spectrum (Orbitrap analysis; resolution 120,000; mass range 400−1400 m/z; automatic gain control (AGC) target 2 × 105; maximum injection time 100 ms). Precursors for MS2/MS3 analysis were selected using a TopSpeed of 2 sec. MS2 analysis consisted of collision-induced dissociation (quadrupole ion trap analysis; AGC 4 × 103; normalized collision energy (NCE) 35; maximum injection time 150 ms). The isolation window for MS/MS was 0.7 Da. Following acquisition of each MS2 spectrum, we collected an MS3 spectrum using our recently developed method in which multiple MS2 fragment ions were captured in the MS3 precursor population using isolation waveforms with multiple frequency notches (36). The isolation window for MS3 was 2.5 Da. MS3 precursors were fragmented by high energy collision-induced dissociation (HCD) and analyzed using the Orbitrap (NCE 55; AGC 5 × 104; maximum injection time 150 ms, resolution was 60,000 at 400 Th).
Proteomic and phosphoproteomic data analysis
Mass spectra were processed using a Sequest-based in-house software pipeline (37). Spectra were converted to mzXML using a modified version of ReAdW.exe. Database searching included all entries from the human uniprot database (March 11, 2014). This database was concatenated with one composed of all protein sequences in the reversed order. Searches were performed using a 50 ppm precursor ion tolerance for total protein level analysis. The product ion tolerance was set to 0.9 Da. These wide mass tolerance windows were chosen to maximize sensitivity in conjunction with Sequest searches and linear discriminant analysis (37, 38). TMT tags on lysine residues and peptide N termini (+229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as a variable modification. For phosphoprotein analysis, +79.966 Da was set as a variable modification on serine, threonine, and tyrosine residues.
Peptide-spectrum matches (PSMs) were adjusted to a 1% false discovery rate (FDR) (39, 40). PSM filtering was performed using a linear discriminant analysis, as described previously (37), while considering the following parameters: XCorr, ΔCn, missed cleavages, peptide length, charge state, and precursor mass accuracy. For TMT-based reporter ion quantitation, we extracted the signal-to-noise (S/N) ratio for each TMT channel and found the closest matching centroid to the expected mass of the TMT reporter ion. For protein-level comparisons, PSMs were identified, quantified, and collapsed to a 1% peptide false discovery rate (FDR) and then collapsed further to a final protein-level FDR of 1%. Moreover, protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for all observed peptides.
Proteins were quantified by summing reporter ion counts across all matching PSMs using in-house software, as described previously (37). Briefly, a 0.003 Th window around the theoretical m/z of each reporter ion (126: 126.127 Th, 127N: 127.124 Th, 127C: 127.131 Th, 128N: 128.128 Th, 128C: 128.134 Th, 129N: 129.131 Th, 129C: 129.138 Th, 130N: 130.135 Th, 130C: 130.141 Th) was scanned for ions, and the maximum intensity nearest the theoretical m/z was used. PSMs with poor quality, MS3 spectra with more than six TMT reporter ion channels missing, MS3 spectra with TMT reporter summed signal-to-noise ratio that is less than 100, or no MS3 spectra were excluded from quantitation (41). Protein quantitation values were exported for further analysis in Excel, JMP, or MatLab. Each reporter ion channel was summed across all quantified proteins and normalized assuming equal protein loading of all 9 samples. One-way ANOVA was then used to identify proteins that were differentially expressed across the two treatments and control. Student t-tests were used to determine statistical significance between each treatment and controls. In both cases, a p-value <0.01 was considered statistically significant. A second threshold based on a log2 fold change of greater than 1.5-fold or less than -1.5 fold was chosen so as to focus the data analysis on a small set of proteins and phosphopeptides with the largest alterations in abundance.
Phosphopeptide motif analysis
For the phosphorylation dataset, site localization was evaluated via AScore (38). Sites with an Ascore >13 were analyzed further using the PhosphoSitePlus (42). Sequences were centered at the phosphorylated residue and extended six amino acids on each side, thereby resulting in a total of 13 amino acids for each phosphorylation site sequence.
Data access
Supplemental Tables 1 and 2 list the proteins and peptides, respectively, as well as normalized TMT reporter ion intensities used for quantitative analysis. RAW files are available upon request.
Results and Discussion
Western blotting revealed that nicotine-binding nAChR subunits were expressed in PaSC
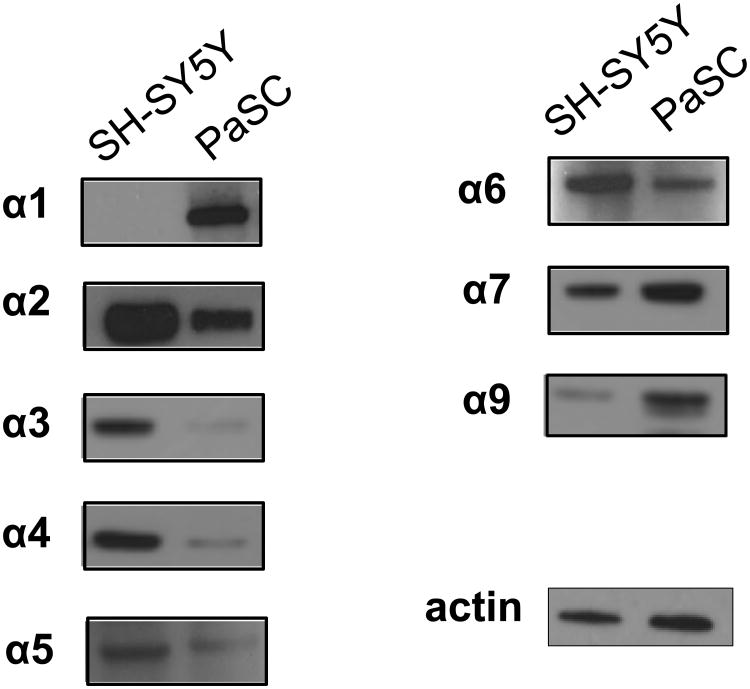
We performed western blotting to access the expression of nAChR subunits in PaSC (Figure 2). For comparison, we used the neuroblastoma cell line SH-SY5Y, which express various nAChR and are regularly used in the study of such receptors. The amount of protein separated by SDS-PAGE was equal for both cell lines, as determined by a bicinchoninic acid (BCA) assay, allowing for relative comparison between the two cell lines. These two cell lines were of different origins and therefore were expected to express diverse populations of nAChR subtypes. One striking difference was the expression of the muscle type subunit α1 in PaSC and not the SH-SY5Y cell line (43). Western blotting showed that PaSC had relatively lower expression of α2, α3, α4, and α5, but higher expression of the homopentameric α7 and α9 nAChR relative to SH-SY5Y cells. An approximately equal amount of α6 and of the loading control actin was detected in both cell lines.
Figure 2. Western blotting analysis revealed nAChR protein expression in PaSC.
The expression levels of the nicotine-binding α-subunits of nAChR in PaSC were assessed via western blotting. The neuroblastoma-like SH-SY5Y cell line was used for comparison.
In addition to binding nicotine, only nAChR subtypes that included the α1, α7, or α9 subunits can bind the neurotoxin α-bungarotoxin. As mentioned above, we determined these subunits as being expressed higher in PaSC than in SH-SY5Y cells. Our TMT multiplexing strategy allowed up to 3 groups to be analyzed in triplicate in a single experiment. Although our main goal was to compare the effects of acute nicotine-induced alterations in PaSC, we included α-bungarotoxin as a second treatment to explore the effects of this alternative ligand on the proteome and phosphoproteome of PaSC.
Minimal protein alterations were observed after 12 hours of nicotine and α-bungarotoxin treatment
We compared untreated, nicotine-treated, and α-bungarotoxin-treated PaSC in biological triplicate with mass spectrometry-based protein quantitation using TMT isobaric tagging (Methods and Figure 1). In total, we identified a total of 140,850 sequenced peptides, of which 79,119 were unique and corresponded to 8120 proteins that were quantified across all nine samples (Table 1).
Table 1. Protein expression level analysis after the 12 hr treatment.
| Category | |
|---|---|
| Quantified proteins across all channels | 8120 |
| Unique peptides | 79,119 |
| Total peptides | 140,850 |
| Significantly altered proteinsa | |
| nicotine vs. control | |
| up | 43 |
| down | 3 |
| α-bungarotoxin vs. control | |
| up | 2 |
| down | 0 |
p-value <0.01, fold change > ±1.5
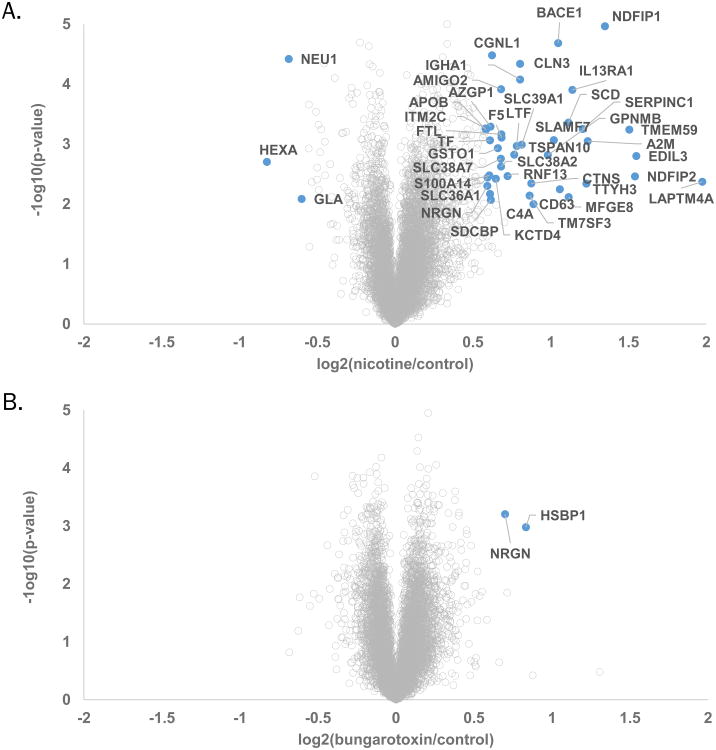
We set our statistical significance level at p-value<0.01 with a minimal fold change of ±1.5. As such, we determined 43 proteins to be of higher abundance when PaSC were treated with nicotine, while only 3 proteins were of lower abundance. Graphically, we illustrated these altered proteins as volcano plots. These plots display -log10(p-value) versus log2 (ratio of average nicotine to average untreated signal-to-noise). In addition, we annotated those proteins with the highest fold-changes (Figure 3A). Moreover, our TMT analysis allowed us to observe α-bungarotoxin-induced protein alterations compared to wildtype. Using again the same significance thresholds (p-value<0.01 and ±1.5-fold change), we determined only two proteins to be of higher abundance - NRGN and HSBP1 - when PaSC were treated with α-bungarotoxin, while no proteins were of lower abundance within our significance thresholds. Again, we displayed proteins in volcano plots and annotated those proteins with altered expression values that fulfilled our significance criteria (Figure 3B).
Figure 3. Volcano plots illustrate statistically significant protein abundance differences in cells treated with nicotine or α-bungarotoxin.
These volcano plots display the –log10 p-value versus the log2 of the relative protein abundance of A) average nicotine or B) average α-bungarotoxin to average control. Blue circles represent proteins with changes in abundance of greater than 1.5 fold and p-value >0.01.
Proteins altered in abundance as a result of nicotine treatment were classified into several groups with similar functions. The three proteins that were down-regulated with nicotine treatment-NEU1 (Sialidase-1), HEXA (β-hexosaminidase subunit alpha), and GLA (α-galactosidase A)-all function in carbohydrate / lipid catabolism. In contrast, the 43 proteins that were up-regulated with nicotine treatment were segregated into various categories. Several proteins that were up-regulated following nicotine-treatment were common to neurons, including amphoterin-induced protein 2 (AMIGO2), β-secretase 1 (BACE1), and neurogranin (NRGN). Defense response proteins were also prevalent, such as Complement C4-A (C4A), Ig α1 chain C region (IGHA1) and Interleukin-13 receptor subunit α1 (IL13RA1). Some proteins, specifically apolipoprotein B (APOB) and α-2-macroglobulin (A2M), have been identified previously as potential biomarkers of chronic pancreatitis (44).
As a consequence of the low number of proteins with altered expression following treatment with α-bungarotoxin, we were unable to segregate these proteins into distinct categories. While no proteins were significantly down-regulated upon the addition of α-bungarotoxin, two proteins - heat shock factor-binding protein 1 (HSBP1) and neurogranin (NRGN) – demonstrated significantly higher protein expression levels. HSBP1 is a negative regulator of the heat shock response transcription from RNA polymerase II promoter (45); but, its role in calcium signaling, pancreas, or nicotine-related functions is currently unknown. However, NRGN is known to bind calmodulin in the absence of calcium (46). Correspondingly, α-bungarotoxin inhibits the α7 nAChR, a calcium channel, thus decreases the calcium influx into the cell, thereby providing evidence for a potential link between α-bungarotoxin binding and NRGN function.
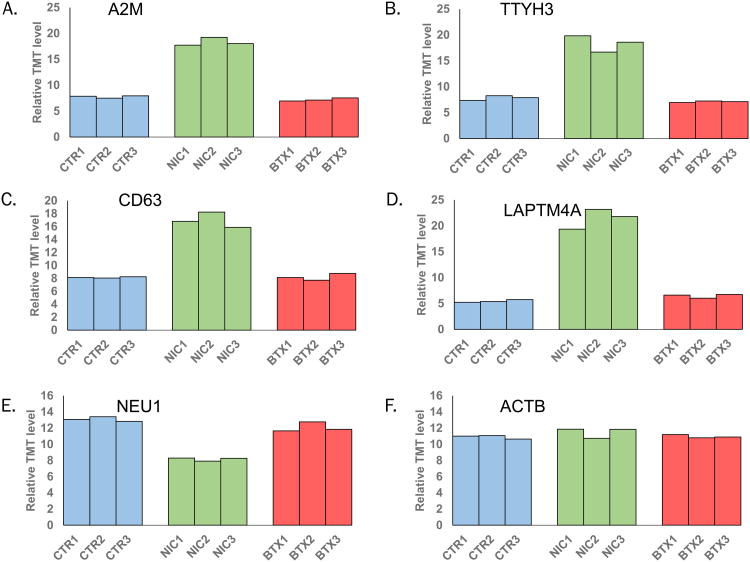
In Figure 4, we illustrated several proteins which showed significant alterations in abundance, including alpha-2-macroglobulin (A2M), tweedy homolog 3 (TTYH3), tetraspanin-30 (CD63), and lysosomal protein transmembrane 4 (LAPTM4A). A2M (Figure 4A) is an antiprotease that can inactivate a variety of serine-, cysteine-, aspartic- and metalloproteinases. This protein functions as an inhibitor of fibrinolysis (47). Alterations in extracellular matrix remodeling can enhance disease progression, including that of the pancreas (48). TTYH3 (Figure 4B) may have a function in Ca2+ signal transduction, as it is a large-conductance Ca2+-activated chloride channel (49). As with NRGN, modulation of calcium influx due to ligand-interaction with the α7 nAChR may be associated with the differential expression of this protein. Similarly, CD63 (Figure 4C) is a member of the tetraspanin family and is a mediator of signal transduction events regulating cell growth and motility (50). CD63 functions as a cell surface receptor for extracellular matrix reorganizing tissue inhibitor of metalloproteinases, such as TIMP1. Several members of the transmembrane 4 superfamily (TM4SF) have been reported as regulating tumor progression and metastasis (50). LAPTM4A (Figure 4D) also has four predicted transmembrane domains, but is of lysosomal origin. Autophagy has been shown independently to contribute to pancreatic disease (51) and to be affected by nicotine (52), but the function of LAPTM4A has not yet been determined in nicotine-associated pancreatic disease. While 43 proteins displayed a significant increase in expression with nicotine treatment, only three showed a decrease, of which we highlight lysosomal sialidase (NEU1). Lysosome-localized NEU1 (Figure 4E) catalyzes the hydrolysis of terminal sialic acid residues of glycoproteins (53). This protein is a tumor suppressor gene (54), and it is noteworthy that we observed a decrease upon nicotine treatment. In Figure 4F, we included actin as control which was not expected to change upon nicotine treatment (55).
Figure 4. Representative proteins displaying altered expression due to nicotine treatment.
Plotted are relative TMT signal-to-noise levels for the selected proteins across the three controls, three nicotine-treated, and three α-bungarotoxin-treated cell cultures. The proteins highlighted include: A) alpha-2-macroglobulin (A2M), B) tweety homolog 3 (TTYH3), C) CD63, D), lysosomal protein transmembrane 4 alpha (LAPTM4A), E) lysosomal sialidase (NEU1), and G) actin (ACTB). Ctr, control; nic, nicotine; bxt, α-bungarotoxin.
Alterations in peptide phosphorylation levels were observed after 12 hours of nicotine and α-bungarotoxin treatment
We used a strategy analogous to that of the protein-level analysis described above to compare alterations in phosphorylation levels of peptides from untreated, nicotine-treated, and α-bungarotoxin-treated PaSC in biological triplicate. Sample preparation preceded in a manner similar to that of the protein analysis with the main differences being that we used 5 mg of starting material (opposed to 50 μg) and enriched phosphopeptides with titanium dioxide prior to TMT labeling. Our phosphopeptide enrichment strategy resulted in greater than 86% of identified peptides being phosphorylated.
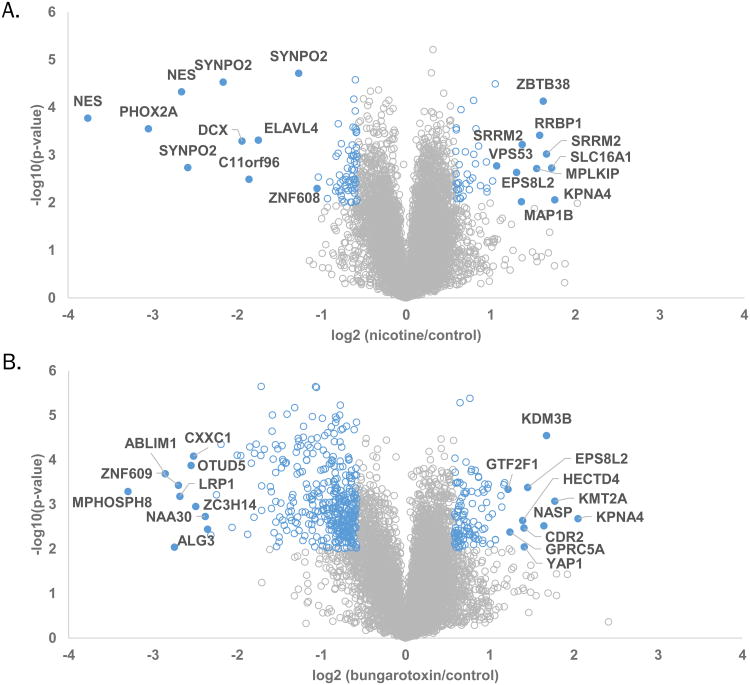
In total, we identified 21,556 phosphorylation sites, of which 16,622 were quantified across all nine samples, as summarized in Table 2. Tallying the phosphorylated residues revealed 14,251 phosphorylation sites for phosphorylated serine (pSer), 2063 for phosphorylated serine (pThr), and 308 for phosphorylated tyrosine (pTyr). Of the 16,622 phosphorylation sites that were quantified across all nine samples, 11,999 of these sites were singly phosphorylated and 4623 contained more than one phosphorylated residue. As with the protein-level analysis, for our phosphorylation site analysis, we set our statistical significance level at p-value<0.01 and a fold change threshold of ±1.5. As such, we determined that the phosphorylation level differences of 59 peptides were higher when treated with nicotine, while that of 73 peptides were lower. Similarly, we determined the phosphorylation levels of 314 peptides to be higher when PaSC were treated with α-bungarotoxin, while that of 137 peptides were lower. Analogous to the protein level analysis, we displayed the phosphorylation data as volcano plots and annotated proteins having phosphorylated peptides with the highest fold-changes following treatment with nicotine (Figure 5A) or α-bungarotoxin (Figure 5B).
Table 2. Phosphorylation level analysis after the 12 hr treatment.
| Category | |
|---|---|
| Phosphopeptide enrichment | >86% |
| Quantified phosphorylation sites across all channels | 16,622 |
| Percentage of pS : pT : pY | 86% : 12% : 2% |
| Singly phosphorylated peptides | 11,999 |
| Multiply phosphorylated peptides | 4,623 |
| Significantly altered phosphorylation levelsa | |
| nicotine vs. control | |
| up | 59 |
| down | 73 |
| α-bungarotoxin vs. control | |
| up | 314 |
| down | 137 |
p-value <0.01, fold change > ±1.5
Figure 5.
Volcano plots illustrate statistically significant phosphopeptide level differences in cells treated with nicotine or α-bungarotoxin. These volcano plots display the –log10 p-value versus the log2 of the relative peptide phosphorylation level differences of A) average nicotine or B) average α-bungarotoxin to average control. Blue circle represent proteins with phosphorylated peptides which show changes in phosphorylation levels of greater than 1.5 fold and p-value >0.01. Closed blue circles indicate those with the highest fold changes.
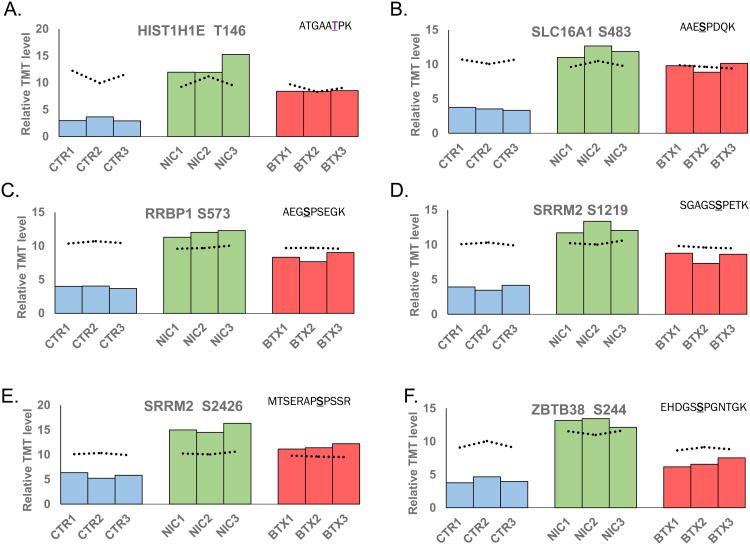
As done for protein expression, we highlighted several phosphorylated peptides that exhibited statistically significant differences in phosphorylation levels (Figure 6). We plotted the percent of total TMT signal-to-noise for these phosphorylated peptides across all nine samples. For comparison, we plotted a dashed line representing the relative protein concentration of the associated protein for each sample. We highlighted six phosphopeptides, which represent five different proteins: Histone H1.4 (HIST1H1E), monocarboxylate transporter 1 (SLC16A1), ribosome binding protein 1 (RRBP1), serine/arginine repetitive matrix 2 (SRRM2), and zinc finger and BTB domain containing protein 38 (ZBTB38). HIST1H1E (Figure 6A) is a histone that regulates gene transcription through DNA methylation, chromatin remodeling, and nucleosome spacing. Although this protein may have a regulatory effect, its role in pancreatic diseases or nicotine-induced alterations has not been defined (56). SLC16A1 (Figure 6B) is a membrane transporter for lactate, pyruvate, and other monocarboxylates, thereby having a role in cellular metabolism (57). In regard to the pancreas, SLC16A1 has been implicated with congenital hyperinsulinism (58, 59). RRBP1 (Figure 6C) is a ribosome-binding protein associated with lung (60), colorectal (61), and breast (62) cancer. SRRM2 (Figure 6D and E) is involved in pre-mRNA splicing, likely at the catalytic center of the spliceosome (63). In addition, this protein has been linked with nicotine receptor-related Parkinson's disease (64). ZBTB38 (Figure 6F) is a transcriptional activator that binds methylated DNA. This protein may be involved in the differentiation and/or survival of late postmitotic neurons (65). Although not significant, the protein expression level of ZBTB38 appeared elevated in the nicotine-treated sample relative to control and α-bungarotoxin-treated samples. Although there was some evidence correlating the function of these proteins with pancreatic disease and/or nicotine/neuronal-related mechanisms, studies targeting the roles of these proteins must be performed to understand better these associations.
Figure 6. Representative peptides displaying altered peptide phosphorylation levels due to nicotine or α-bungarotoxin treatment.
Plotted are the resulting TMT signal-to-noise level for the select phosphorylated peptides across the three controls, three nicotine-treated and three α-bungarotoxin-treated cell cultures. The phosphorylated peptides include: A) T146 from HIT1H1E, B) S483 from SLC16A1, C) S573 from RRBP1, D) S1219 from SRRM2, E) S2426 from SRRM2, and F) S244 from ZBTB38. In addition, the dotted line indicates the protein expression level corresponding to the graphed peptide. The underlined amino acid represents the phosphorylation site. CTR, control; NIC, nicotine; BTX, α-bungarotoxin.
Several proteins exhibit multiple phosphorylation sites with high fold-changes, however for some of these sites no associated protein level quantitation was determined. Although the protein level data revealed that only a small fraction of proteins demonstrated altered abundances, we were unable to confidently access that altered phosphorylation levels for peptides without corresponding protein level quantitation were indeed related to signaling changes and not protein expression levels. Examples included phosphorylated peptides from NES (nestin), Phox2A (Paired mesoderm homeobox protein 2A), and SYNPO2 (Synaptopodin-2), all of which demonstrate a decrease in phosphorylation level upon nicotine treatment. NES has a role in regulation of the assembly and disassembly of intermediate filaments, and participates in cellular remodeling. Of particular interest, NES is expressed in metastatic lesions of pancreatic cancer patients (66) and may suppress pancreatic cell migration, invasion and metastasis (67), thus its down-regulation was expected. Phox2A has a central role in development of the autonomic nervous system (68). In regard to nicotine, Phox2A regulates transcription of the α3 nAChR gene (69), but its role in the pancreas has not yet been elucidated. SYNPO2 has actin-binding activity and shuttles between the nucleus and the cytoplasm in a stress-induced manner (70). This protein has been studied in neurons, but no evidence is available supporting its function in the pancreas. Like many enrichment strategies, the use of TiO2 may allow for the identification of specific proteins or peptides that are not typically observed due to low abundance or poor ionization. Further investigation may be necessary to confirm that these altered phosphorylation events were due to only signaling and not also protein expression.
Phosphorylation motif analysis revealed enriched local consensus sequences
We subjected our significantly-altered phosphorylation sites to PhosphoSitePlus analysis to determine consensus sites displaying altered phosphorylation (42). Examining sites showing nicotine-induced increases in phosphorylation events revealed enrichment in SP motifs, which are typically regulated by proline-directed kinases (Supplemental Figure 1A). In addition, the RxxS motif, corresponding to arginine-directed kinases (e.g., cAMP-dependent protein kinase (PKA), cGMP-dependent protein kinase (PKG), protein kinase C, Akt, and RSK), also were enriched. Similarly, nicotine-induced decreases in phosphorylation events were dominated by SP sites, which can be phosphorylated by a variety of proline-directed kinases, but also showed a modest enrichment of the RxxS motif (Supplemental Figure 1B). Such a result is not necessarily contradictory, as different kinases may be active only under certain conditions. Additionally, the glycogen synthase kinase 3 (GSK3) motif, SxxxS, showed a decrease in phosphorylation resulting from nicotine treatment.
In addition, we examined the consensus phosphorylation motifs altered by α-bungarotoxin. Similar to the nicotine-induced increases in phosphorylation, SP and RxxS motifs were enriched in phosphorylation events by α-bungarotoxin treatment (Supplemental Figure 1C). In addition, the RRxS motif, corresponding to PKA phosphorylation, also exhibited increased phosphorylation due to α-bungarotoxin treatment. In contrast to the other three scenarios described above (i.e., higher phosphorylation levels with nicotine, lower phosphorylation levels with nicotine, higher phosphorylation levels with nicotine), SP-containing motifs did not dominate the phosphorylation sites which were decreased by α-bungarotoxin treatment (Supplemental Figure 1D). Here serine phosphorylation predominated in regions highly populated with basic residues, including the SxxE motif which is regulated by casein kinase II (CK2).
Conclusions
We examined the effects of nicotine and α-bungarotoxin on cultured pancreatic cells. We have shown that several nicotine-binding subunits of nAChR are expressed in PaSC, including the α-bungarotoxin-binding α1, α7, and α9 subunits. Moreover, we use isobaric tag-based multiplexing, TMT, to investigate the effects of nicotine and α-bungarotoxin on the global proteome and phosphoproteome of PaSC. In total, we quantified over 8,100 proteins and over 16,000 phosphorylation sites across all nine samples. In addition, 46 proteins were altered in abundance upon treatment with nicotine, while 132 phosphorylation sites were altered with nicotine treatment and 451 with α-bungarotoxin. Many of the proteins demonstrating a high abundance and/or peptide phosphorylation level alterations have been associated previously with pancreatic disease. Here we have assembled a collection of proteins and phosphorylation sites that can be validated and investigated further using targeted methods, such as western blotting, immunofluorescence or mass spectrometry-based targeted MS2, specifically parallel reaction monitoring (PRM), and used as a starting point for a more in-depth studies of the associated pathway and mechanisms (69).
Our multiplexing strategy can be considered a template from which to design other experiments. For example, dozens of alternative ligands, aside from nicotine and α-bungarotoxin, can be used in analogous experiments. Likewise, cells can be treated with the ligands at different time points and temporal alterations maybe observed. Longer time points would reveal proteins and/or phosphorylation changes reflecting a state of chronic exposure to selected ligands. Whereas shorter time points would be more relevant for quantifying the changes regulating early events in signaling cascades. Using a shorter time frame also limits protein turnover, and as such allows us to isolate better the changes in phosphorylated peptides as being a consequence of signaling, rather than protein expression level alterations. Moreover, different cell lines can be assayed to assess whether the observed changes in the proteome and phosphoproteome are global or specific to the cell line under investigation.
In summary, we have identified nAChR in PaSC and investigated the global proteomic and phosphoproteomic effects of two nicotinic receptor ligands, nicotine and α-bungarotoxin. Among the nAChR detected in PaSC included all three α-bungarotoxin-binding subunits α1, α7, and α9. Using a TMT-based strategy, we determined proteins that were significantly altered in expression following nicotine or α-bungarotoxin treatment. In addition, TiO2 enrichment followed by TMT-labeling, allows for the analysis of alterations in phosphorylated peptide levels due to treatment with either ligand. Many of the differentially-expressed proteins function in pathways relevant to pancreatic disease and/or via nicotine-related mechanisms, while others are novel and may merit further investigation. Moreover, we provide potential protein and phosphorylation targets for which hypotheses can be derived for future investigations. Furthermore, the TMT-based mass spectrometry techniques outlined here offer a valuable tool to address questions concerning the effects of multiple drugs on biological models in a single experiment.
Supplementary Material
Supplemental Figure 1: Phosphorylation motif analysis reveal enrichment of certain residues surrounding phosphorylated sites that correspond to targets of specific kinases. For enrichment analysis, we subjected motifs - consisting of the phosphorylated residue and seven flanking residues – of phosphorylated sites with A) higher abundance upon nicotine treatment, B) lower abundance upon nicotine treatment, C) higher abundance upon α-bungarotoxin treatment, and D) lower abundance upon α-bungarotoxin treatment.
Supplemental Table 1: Proteins identified in this dataset. Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), protein description/name (Description), number of peptides identified per protein (peptides), the normalized summed signal-to-noise for each of the 9 channels (126_sn_sum to 130C_sn_sum), log2 ratios for each comparison pair, and p-values of t-test for each comparison pair. Proteins with statistically significant alterations upon nicotine treatment have protein name and gene symbol cells shaded in green, while those altered with α-bungarotoxin have red-colored font.
Supplemental Table 2: Phosphopeptides identified in this dataset. Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), protein description (Description), single or multiple phosphorylation site (type), sequence position of phosphorylation site (site position), sequence of phosphorylated motif (motif), the phosphorylated residue (PhosRes), ASCORE (ASCORE), peptide sequence (peptide sequence), number of quantified peptides (num_quant), the normalized summed signal to noise for each of the 10 channels (126_sn_norm to 130C_sn_norm) log2 ratios for each comparison pair, and p-values of t-test for each comparison pair. Phosphopeptides with statistically significant alterations upon nicotine treatment have the phosphopeptide sequence cells shaded in green, while those altered with α-bungarotoxin have red-colored font.
Acknowledgments
We would also like to thank members of the Gygi Lab at Harvard Medical School. This work was funded in part by an NIH/NIDDK grant K01 DK098285 (J.A.P.). The PaSC cell line (RLT-PSC) was a gift from Dr. Ralf Jesnowski (German Cancer Research Center).
Footnotes
Conflicts Of Interest: The authors acknowledge no conflict of interest.
References
- 1.Yadav D, Hawes RH, Brand RE, Anderson MA, Money ME, Banks PA, Bishop MD, Baillie J, Sherman S, DiSario J, Burton FR, Gardner TB, Amann ST, Gelrud A, Lawrence C, Elinoff B, Greer JB, O'Connell M, Barmada MM, Slivka A, Whitcomb DC. Alcohol consumption, cigarette smoking, and the risk of recurrent acute and chronic pancreatitis. Arch Intern Med. 2009;169(11):1035–45. doi: 10.1001/archinternmed.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Law R, Parsi M, Lopez R, Zuccaro G, Stevens T. Cigarette smoking is independently associated with chronic pancreatitis. Pancreatology. 2010;10(1):54–9. doi: 10.1159/000225927. [DOI] [PubMed] [Google Scholar]
- 3.Lin Y, Tamakoshi A, Hayakawa T, Ogawa M, Ohno Y. Cigarette smoking as a risk factor for chronic pancreatitis: a case-control study in Japan. Research Committee on Intractable Pancreatic Diseases. Pancreas. 2000;21(2):109–14. doi: 10.1097/00006676-200008000-00001. [DOI] [PubMed] [Google Scholar]
- 4.National Toxicology P. Tobacco-related exposures: tobacco smoking. Report on carcinogens : carcinogen profiles / US Dept of Health and Human Services, Public Health Service, National Toxicology Program. 2011;12:408–410. [PubMed] [Google Scholar]
- 5.Prokopczyk B, Hoffmann D, Bologna M, Cunningham AJ, Trushin N, Akerkar S, Boyiri T, Amin S, Desai D, Colosimo S, Pittman B, Leder G, Ramadani M, Henne-Bruns D, Beger HG, El-Bayoumy K. Identification of tobacco-derived compounds in human pancreatic juice. Chem Res Toxicol. 2002;15(5):677–85. doi: 10.1021/tx0101088. [DOI] [PubMed] [Google Scholar]
- 6.Chowdhury P, Rayford PL. Smoking and pancreatic disorders. Eur J Gastroenterol Hepatol. 2000;12(8):869–77. doi: 10.1097/00042737-200012080-00006. [DOI] [PubMed] [Google Scholar]
- 7.Momi N, Ponnusamy MP, Kaur S, Rachagani S, Kunigal SS, Chellappan S, Ouellette MM, Batra SK. Nicotine/cigarette smoke promotes metastasis of pancreatic cancer through alpha7nAChR-mediated MUC4 upregulation. Oncogene. 2013;32(11):1384–95. doi: 10.1038/onc.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu KM, Cho CH, Shin VY. Nicotine and gastrointestinal disorders: its role in ulceration and cancer development. Curr Pharm Des. 2013;19(1):5–10. doi: 10.2174/13816128130103. [DOI] [PubMed] [Google Scholar]
- 9.Apte M, Pirola R, Wilson J. New insights into alcoholic pancreatitis and pancreatic cancer. J Gastroenterol Hepatol. 2009;24(3):S51–6. doi: 10.1111/j.1440-1746.2009.06071.x. [DOI] [PubMed] [Google Scholar]
- 10.Changeux JP. Nicotine addiction and nicotinic receptors: lessons from genetically modified mice. Nat Rev Neurosci. 2010;11(6):389–401. doi: 10.1038/nrn2849. [DOI] [PubMed] [Google Scholar]
- 11.Al-Wadei MH, Al-Wadei HA, Schuller HM. Pancreatic Cancer Cells and Normal Pancreatic Duct Epithelial Cells Express an Autocrine Catecholamine Loop that Is Activated by Nicotinic Acetylcholine Receptors alpha3, alpha5, and alpha7. Mol Cancer Res. 2012;10(2):239–49. doi: 10.1158/1541-7786.MCR-11-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Browne CJ, Sharma N, Waters KA, Machaalani R. The effects of nicotine on the alpha-7 and beta-2 nicotinic acetycholine receptor subunits in the developing piglet brainstem. Int J Dev Neurosci. 2010;28(1):1–7. doi: 10.1016/j.ijdevneu.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Paulo JA, Brucker WJ, Hawrot E. Proteomic analysis of an alpha7 nicotinic acetylcholine receptor interactome. J Proteome Res. 2009;8(4):1849–58. doi: 10.1021/pr800731z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heeschen C, Weis M, Aicher A, Dimmeler S, Cooke JP. A novel angiogenic pathway mediated by non-neuronal nicotinic acetylcholine receptors. J Clin Invest. 2002;110(4):527–36. doi: 10.1172/JCI14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macklin KD, Maus AD, Pereira EF, Albuquerque EX, Conti-Fine BM. Human vascular endothelial cells express functional nicotinic acetylcholine receptors. J Pharmacol Exp Ther. 1998;287(1):435–9. [PubMed] [Google Scholar]
- 16.Sharma G, Vijayaraghavan S. Nicotinic receptor signaling in nonexcitable cells. J Neurobiol. 2002;53(4):524–34. doi: 10.1002/neu.10114. [DOI] [PubMed] [Google Scholar]
- 17.Moise L, Piserchio A, Basus VJ, Hawrot E. NMR structural analysis of alpha-bungarotoxin and its complex with the principal alpha-neurotoxin-binding sequence on the alpha 7 subunit of a neuronal nicotinic acetylcholine receptor. J Biol Chem. 2002;277(14):12406–17. doi: 10.1074/jbc.M110320200. [DOI] [PubMed] [Google Scholar]
- 18.Hannan S, Mortensen M, Smart TG. Snake neurotoxin alpha-bungarotoxin is an antagonist at native GABA receptors. Neuropharmacology. 2015 doi: 10.1016/j.neuropharm.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson JS, Pirola RC, Apte MV. Stars and stripes in pancreatic cancer: role of stellate cells and stroma in cancer progression. Front Physiol. 2014;5:52. doi: 10.3389/fphys.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu K. Mechanisms of pancreatic fibrosis and applications to the treatment of chronic pancreatitis. J Gastroenterol. 2008;43(11):823–32. doi: 10.1007/s00535-008-2249-7. [DOI] [PubMed] [Google Scholar]
- 21.Patel M, Fine DR. Fibrogenesis in the pancreas after acinar cell injury. Scand J Surg. 2005;94(2):108–11. doi: 10.1177/145749690509400205. [DOI] [PubMed] [Google Scholar]
- 22.Ellenrieder V, Schneiderhan W, Bachem M, Adler G. Fibrogenesis in the pancreas. Rocz Akad Med Bialymst. 2004;49:40–6. [PubMed] [Google Scholar]
- 23.Apte MV, Wilson JS. Mechanisms of pancreatic fibrosis. Dig Dis. 2004;22(3):273–9. doi: 10.1159/000082799. [DOI] [PubMed] [Google Scholar]
- 24.Jesnowski R, Furst D, Ringel J, Chen Y, Schrodel A, Kleeff J, Kolb A, Schareck WD, Lohr M. Immortalization of pancreatic stellate cells as an in vitro model of pancreatic fibrosis: deactivation is induced by matrigel and N-acetylcysteine. Lab Invest. 2005;85(10):1276–91. doi: 10.1038/labinvest.3700329. [DOI] [PubMed] [Google Scholar]
- 25.Haqq J, Howells LM, Garcea G, Metcalfe MS, Steward WP, Dennison AR. Pancreatic stellate cells and pancreas cancer: current perspectives and future strategies. Eur J Cancer. 2014;50(15):2570–82. doi: 10.1016/j.ejca.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 26.Phillips P. Pancreatic stellate cells and fibrosis. In: Grippo PJ, Munshi HG, editors. Pancreatic Cancer and Tumor Microenvironment. Trivandrum (India): 2012. [PubMed] [Google Scholar]
- 27.Mathison A, Liebl A, Bharucha J, Mukhopadhyay D, Lomberk G, Shah V, Urrutia R. Pancreatic stellate cell models for transcriptional studies of desmoplasia-associated genes. Pancreatology. 2010;10(4):505–16. doi: 10.1159/000320540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omary MB, Lugea A, Lowe AW, Pandol SJ. The pancreatic stellate cell: a star on the rise in pancreatic diseases. J Clin Invest. 2007;117(1):50–9. doi: 10.1172/JCI30082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soeda J, Morgan M, McKee C, Mouralidarane A, Lin C, Roskams T, Oben JA. Nicotine induces fibrogenic changes in human liver via nicotinic acetylcholine receptors expressed on hepatic stellate cells. Biochem Biophys Res Commun. 2012;417(1):17–22. doi: 10.1016/j.bbrc.2011.10.151. [DOI] [PubMed] [Google Scholar]
- 30.Phillips PA, Yang L, Shulkes A, Vonlaufen A, Poljak A, Bustamante S, Warren A, Xu Z, Guilhaus M, Pirola R, Apte MV, Wilson JS. Pancreatic stellate cells produce acetylcholine and may play a role in pancreatic exocrine secretion. Proc Natl Acad Sci U S A. 2010;107(40):17397–402. doi: 10.1073/pnas.1000359107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulo JA, Urrutia R, Banks PA, Conwell DL, Steen H. Proteomic analysis of a rat pancreatic stellate cell line using liquid chromatography tandem mass spectrometry (LC-MS/MS) J Proteomics. 2011;75(2):708–17. doi: 10.1016/j.jprot.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulo JA, Urrutia R, Banks PA, Conwell DL, Steen H. Proteomic analysis of an immortalized mouse pancreatic stellate cell line identifies differentially-expressed proteins in activated vs nonproliferating cell states. J Proteome Res. 2011;10(10):4835–44. doi: 10.1021/pr2006318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kettenbach A, Gerber S. Rapid and reproducible single-stage phosphopeptide enrichment of complex peptide mixtures: application to general and phosphotyrosine-specific phosphoproteomics experiments. Analytical chemistry. 2011;83(20):7635–7644. doi: 10.1021/ac201894j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulo JA, Gygi SP. A comprehensive proteomic and phosphoproteomic analysis of yeast deletion mutants of 14-3-3 orthologs and associated effects of rapamycin. Proteomics. 2015;15(2-3):474–86. doi: 10.1002/pmic.201400155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulo JA, McAllister FE, Everley RA, Beausoleil SA, Banks AS, Gygi SP. Effects of MEK inhibitors GSK1120212 and PD0325901 in vivo using 10-plex quantitative proteomics and phosphoproteomics. Proteomics. 2015;15(2-3):462–73. doi: 10.1002/pmic.201400154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. MultiNotch MS3 Enables Accurate, Sensitive, and Multiplexed Detection of Differential Expression across Cancer Cell Line Proteomes. Anal Chem. 2014;86(14):7150–8. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–89. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–92. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 39.Elias JE, Gygi SP. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol Biol. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 41.McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, Gygi SP. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal Chem. 2012;84(17):7469–78. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40(Database issue):D261–70. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoepfer R, Luther M, Lindstrom J. The human medulloblastoma cell line TE671 expresses a muscle-like acetylcholine receptor. Cloning of the alpha-subunit cDNA. FEBS Lett. 1988;226(2):235–40. doi: 10.1016/0014-5793(88)81430-3. [DOI] [PubMed] [Google Scholar]
- 44.Paulo JA, Kadiyala V, Lee LS, Banks PA, Conwell DL, Steen H. Proteomic analysis (GeLC-MS/MS) of ePFT-collected pancreatic fluid in chronic pancreatitis. J Proteome Res. 2012;11(3):1897–912. doi: 10.1021/pr2011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Satyal SH, Chen D, Fox SG, Kramer JM, Morimoto RI. Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev. 1998;12(13):1962–74. doi: 10.1101/gad.12.13.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baudier J, Deloulme JC, Van Dorsselaer A, Black D, Matthes HW. Purification and characterization of a brain-specific protein kinase C substrate, neurogranin (p17). Identification of a consensus amino acid sequence between neurogranin and neuromodulin (GAP43) that corresponds to the protein kinase C phosphorylation site and the calmodulin-binding domain. J Biol Chem. 1991;266(1):229–37. [PubMed] [Google Scholar]
- 47.de Boer JP, Creasey AA, Chang A, Abbink JJ, Roem D, Eerenberg AJ, Hack CE, Taylor FB., Jr Alpha-2-macroglobulin functions as an inhibitor of fibrinolytic, clotting, and neutrophilic proteinases in sepsis: studies using a baboon model. Infect Immun. 1993;61(12):5035–43. doi: 10.1128/iai.61.12.5035-5043.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol. 2011;3(12) doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki M, Mizuno A. A novel human Cl(-) channel family related to Drosophila flightless locus. J Biol Chem. 2004;279(21):22461–8. doi: 10.1074/jbc.M313813200. [DOI] [PubMed] [Google Scholar]
- 50.Sho M, Adachi M, Taki T, Hashida H, Konishi T, Huang CL, Ikeda N, Nakajima Y, Kanehiro H, Hisanaga M, Nakano H, Miyake M. Transmembrane 4 superfamily as a prognostic factor in pancreatic cancer. Int J Cancer. 1998;79(5):509–16. doi: 10.1002/(sici)1097-0215(19981023)79:5<509::aid-ijc11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 51.Gukovskaya AS, Gukovsky I. Autophagy and pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2012;303(9):G993–G1003. doi: 10.1152/ajpgi.00122.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hung SY, Huang WP, Liou HC, Fu WM. Autophagy protects neuron from Abeta-induced cytotoxicity. Autophagy. 2009;5(4):502–10. doi: 10.4161/auto.5.4.8096. [DOI] [PubMed] [Google Scholar]
- 53.Pshezhetsky AV, Richard C, Michaud L, Igdoura S, Wang S, Elsliger MA, Qu J, Leclerc D, Gravel R, Dallaire L, Potier M. Cloning, expression and chromosomal mapping of human lysosomal sialidase and characterization of mutations in sialidosis. Nat Genet. 1997;15(3):316–20. doi: 10.1038/ng0397-316. [DOI] [PubMed] [Google Scholar]
- 54.Bera A, VenkataSubbaRao K, Manoharan MS, Hill P, Freeman JW. A miRNA signature of chemoresistant mesenchymal phenotype identifies novel molecular targets associated with advanced pancreatic cancer. PLoS One. 2014;9(9):e106343. doi: 10.1371/journal.pone.0106343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paulo JA. Nicotine alters the proteome of two human pancreatic duct cell lines. JOP. 2014;15(5):465–74. doi: 10.6092/1590-8577/2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Albig W, Kardalinou E, Drabent B, Zimmer A, Doenecke D. Isolation and characterization of two human H1 histone genes within clusters of core histone genes. Genomics. 1991;10(4):940–8. doi: 10.1016/0888-7543(91)90183-f. [DOI] [PubMed] [Google Scholar]
- 57.Garcia CK, Goldstein JL, Pathak RK, Anderson RG, Brown MS. Molecular characterization of a membrane transporter for lactate, pyruvate, and other monocarboxylates: implications for the Cori cycle. Cell. 1994;76(5):865–73. doi: 10.1016/0092-8674(94)90361-1. [DOI] [PubMed] [Google Scholar]
- 58.Yorifuji T. Congenital hyperinsulinism: current status and future perspectives. Ann Pediatr Endocrinol Metab. 2014;19(2):57–68. doi: 10.6065/apem.2014.19.2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Halestrap AP. The SLC 16 gene family - structure, role and regulation in health and disease. Mol Aspects Med. 2013;34(2-3):337–49. doi: 10.1016/j.mam.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 60.Tsai HY, Yang YF, Wu AT, Yang CJ, Liu YP, Jan YH, Lee CH, Hsiao YW, Yeh CT, Shen CN, Lu PJ, Huang MS, Hsiao M. Endoplasmic reticulum ribosome-binding protein 1 (RRBP1) overexpression is frequently found in lung cancer patients and alleviates intracellular stress-induced apoptosis through the enhancement of GRP78. Oncogene. 2013;32(41):4921–31. doi: 10.1038/onc.2012.514. [DOI] [PubMed] [Google Scholar]
- 61.Krasnov GS, Oparina N, Khankin SL, Mashkova TD, Ershov AN, Zatsepina OG, Karpov VL, Beresten SF. Colorectal cancer 2D-proteomics: identification of altered protein expression. Mol Biol (Mosk) 2009;43(2):348–56. doi: 10.1134/s0026893309020186. [DOI] [PubMed] [Google Scholar]
- 62.Telikicherla D, Marimuthu A, Kashyap MK, Ramachandra YL, Mohan S, Roa JC, Maharudraiah J, Pandey A. Overexpression of ribosome binding protein 1 (RRBP1) in breast cancer. Clin Proteomics. 2012;9(1):7. doi: 10.1186/1559-0275-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blencowe BJ, Issner R, Nickerson JA, Sharp PA. A coactivator of pre-mRNA splicing. Genes Dev. 1998;12(7):996–1009. doi: 10.1101/gad.12.7.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shehadeh LA, Yu K, Wang L, Guevara A, Singer C, Vance J, Papapetropoulos S. SRRM2, a potential blood biomarker revealing high alternative splicing in Parkinson's disease. PLoS One. 2010;5(2):e9104. doi: 10.1371/journal.pone.0009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26(1):169–81. doi: 10.1128/MCB.26.1.169-181.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsuda Y, Yoshimura H, Ueda J, Naito Z, Korc M, Ishiwata T. Nestin delineates pancreatic cancer stem cells in metastatic foci of NOD/Shi-scid IL2Rgamma(null) (NOG) mice. Am J Pathol. 2014;184(3):674–85. doi: 10.1016/j.ajpath.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsuda Y, Naito Z, Kawahara K, Nakazawa N, Korc M, Ishiwata T. Nestin is a novel target for suppressing pancreatic cancer cell migration, invasion and metastasis. Cancer Biol Ther. 2011;11(5):512–23. doi: 10.4161/cbt.11.5.14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hirsch MR, Tiveron MC, Guillemot F, Brunet JF, Goridis C. Control of noradrenergic differentiation and Phox2a expression by MASH1 in the central and peripheral nervous system. Development. 1998;125(4):599–608. doi: 10.1242/dev.125.4.599. [DOI] [PubMed] [Google Scholar]
- 69.Benfante R, Flora A, Di Lascio S, Cargnin F, Longhi R, Colombo S, Clementi F, Fornasari D. Transcription factor PHOX2A regulates the human alpha3 nicotinic receptor subunit gene promoter. J Biol Chem. 2007;282(18):13290–302. doi: 10.1074/jbc.M608616200. [DOI] [PubMed] [Google Scholar]
- 70.Schroeter MM, Beall B, Heid HW, Chalovich JM. In vitro characterization of native mammalian smooth-muscle protein synaptopodin 2. Biosci Rep. 2008;28(4):195–203. doi: 10.1042/BSR20080079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Phosphorylation motif analysis reveal enrichment of certain residues surrounding phosphorylated sites that correspond to targets of specific kinases. For enrichment analysis, we subjected motifs - consisting of the phosphorylated residue and seven flanking residues – of phosphorylated sites with A) higher abundance upon nicotine treatment, B) lower abundance upon nicotine treatment, C) higher abundance upon α-bungarotoxin treatment, and D) lower abundance upon α-bungarotoxin treatment.
Supplemental Table 1: Proteins identified in this dataset. Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), protein description/name (Description), number of peptides identified per protein (peptides), the normalized summed signal-to-noise for each of the 9 channels (126_sn_sum to 130C_sn_sum), log2 ratios for each comparison pair, and p-values of t-test for each comparison pair. Proteins with statistically significant alterations upon nicotine treatment have protein name and gene symbol cells shaded in green, while those altered with α-bungarotoxin have red-colored font.
Supplemental Table 2: Phosphopeptides identified in this dataset. Columns include: Uniprot protein identification number (proteinID), gene symbol (Gene Symbol), protein description (Description), single or multiple phosphorylation site (type), sequence position of phosphorylation site (site position), sequence of phosphorylated motif (motif), the phosphorylated residue (PhosRes), ASCORE (ASCORE), peptide sequence (peptide sequence), number of quantified peptides (num_quant), the normalized summed signal to noise for each of the 10 channels (126_sn_norm to 130C_sn_norm) log2 ratios for each comparison pair, and p-values of t-test for each comparison pair. Phosphopeptides with statistically significant alterations upon nicotine treatment have the phosphopeptide sequence cells shaded in green, while those altered with α-bungarotoxin have red-colored font.