Abstract
Y chromosomes display population variation within and between species. Co-evolution within populations is expected to produce adaptive interactions between Y chromosomes and the rest of the genome. One consequence is that Y chromosomes from disparate populations could disrupt harmonious interactions between co-evolved genetic elements and result in reduced male fertility, sterility or inviability. Here we address the contribution of ‘heterospecific Y chromosomes' to fertility in hybrid males carrying a homozygous region of Drosophila mauritiana introgressed in the Drosophila simulans background. In order to detect Y chromosome–autosome interactions, which may go unnoticed in a single-species background of autosomes, we constructed hybrid genotypes involving three sister species: Drosophila simulans, D. mauritiana, and D. sechellia. These engineered strains varied due to: (i) species origin of the Y chromosome (D. simulans or D. sechellia); (ii) location of the introgressed D. mauritiana segment on the D. simulans third chromosome, and (iii) grandparental genomic background (three genotypes of D. simulans). We find complex interactions between the species origin of the Y chromosome, the identity of the D. mauritiana segment and the grandparental genetic background donating the chromosomes. Unexpectedly, the interaction of the Y chromosome and one segment of D. mauritiana drastically reduced fertility in the presence of Ysim, whereas the fertility is partially rescued by the Y chromosome of D. sechellia when it descends from a specific grandparental genotype. The restoration of fertility occurs in spite of an autosomal and X-linked genome that is mostly of D. simulans origin. These results illustrate the multifactorial basis of genetic interactions involving the Y chromosome. Our study supports the hypothesis that the Y chromosome can contribute significantly to the evolution of reproductive isolation and highlights the conditional manifestation of infertility in specific genotypic combinations.
Introduction
The Y chromosome of Drosophila simulans and other closely related species harbor a high density of transposable elements and megabase long segments of repetitive sequences (Carvalho and Clark, 2005; Smith et al., 2007), with few protein-coding genes and nearly absent protein-coding polymorphism. The Y chromosome is required for male fertility in most Drosophila species (Ashburner et al., 2005) even though it does not contain a male-determining gene. Instead, most if not all of the ~14 protein-coding genes present on the Y chromosomes of Drosophila melanogaster, D. simulans and other closely related species have male-specific functions and are exclusively expressed in the testis (Gepner and Hays, 1993; Carvalho et al., 2000; Carvalho et al., 2001; Vibranovski et al., 2008). Nevertheless, a suite of studies have shown that genetic variation present on the Y of Drosophila underlie phenotypic variation in male fitness (Chippindale and Rice, 2001; Yee et al., 2015), sex ratio distortion (Carvalho et al., 1997; Montchamp-Moreau et al., 2001; Branco et al., 2013a, b), tolerance to temperature extremes (Rohmer et al., 2004; David et al., 2005), behavior (Stoltenberg and Hirsch, 1997; Huttunen and Aspi, 2003), gene expression (Lemos et al., 2008; Sackton et al., 2011; Branco et al., 2013a, b) and chromatin states in somatic tissues (Lemos et al., 2010).
Y-linked regulatory variation (YRV) is a source of gene expression diversity in Drosophila (Ashburner et al., 2005; Lemos et al., 2008). Moreover, analyses of Y chromosome variation in wild-type backgrounds and in genotypes with loss-of-function mutations revealed that the manifestation of YRV is exquisitely sensitive to the genomic background (Jiang et al., 2010; Branco et al., 2013a, b). Moreover, these studies support the expectation that Y-linked variation preferentially affects genes with male-biased expression and modifies male fertility. Interestingly, YRV affects genes with greater expression divergence between D. melanogaster and D. simulans and higher level of expression polymorphism within species (Lemos et al., 2008; Sackton et al., 2011). It is reasonable to expect that YRV will modulate the evolution of fast-evolving genes and contribute to speciation and hybrid incompatibility in Drosophila. Indeed, the Y chromosome of Drosophila has been shown to be involved in hybrid genome incompatibilities. For example, protein binding to the Y chromosome in hybrids of D. simulans and D. mauritiana is linked to local chromatin condensation and hybrid sterility (Bayes and Malik, 2009). Albeit in a different hybrid species system, similar mechanisms might underlie earlier observations that Drosophila mojavensis males with a Y chromosome from Drosophila arizonae are sterile (Vigneault and Zouros, 1986; Pantazidis et al., 1993). Collectively, these results are in agreement with the notion that epistatic interactions between the Y chromosome and the genetic background are prevalent for components of male fitness (Johnson et al., 1992, 1993; Chippindale and Rice, 2001; Sackton et al., 2011; Yee et al., 2015). Molecularly, a variety of mechanisms—involving both RNA mediators and non-coding DNA sequences—have been suggested for Y chromosome modulation of autosomal and X-linked gene expression that is, in turn, expected to modulate male fertility (reviewed in Francisco and Lemos, 2014).
Here we report a quantitative assay of the contribution of a heterospecific Y chromosome to male fertility in hybrid genotypes involving three sister species: D. simulans, D. mauritiana, and D. sechellia. Crosses between any two of these species produce an F1 of fertile females and sterile males (Lachaise et al., 1986; Ashburner et al., 2005). The construction of mixed-genotype lines in the laboratory provides an ideal system to test whether hybrid male fertility deficiency results from specific interactions among defined genomic regions. Intraspecific variation in the Y chromosome can be responsible for modulating hybrid phenotypes (Johnson et al., 1992, 1993; Sackton et al., 2011; Cutter, 2012). However, subtle effects on fertility may go unnoticed and are particularly difficult to ascertain when single-species systems are investigated. The three-species hybrid system has the potential of inciting less subtle effects, which may reveal novel aspects of the Y chromosome contribution to fertility.
Introgression lines of D. mauritiana into the D. simulans background were constructed by multiple generations of backcrossing hybrid females with D. simulans males. Aided by molecular markers, these introgressed segments of D. mauritiana are well delimited (see Tao et al., 2001, 2003). We crossed seven pairs of introgression lines, each carrying a different segment of D. mauritiana's chromosome 3, in order to produce homozygotes of certain portions of these segments. Along the crossing scheme, we also incorporated an exogenous Y chromosome (from D. simulans or D. sechellia) to the introgression genotypes and produced focal males from three sets of grandfathers. Males with different grandfathers, different combinations of homozygous introgressions and the Y chromosome of either D. simulans or D. sechellia were individually tested for fertility.
We found complex interactions between the Y chromosome and D. mauritiana segments that are manifested in male fertility. Notably, one segment of D. mauritiana is more detrimental in the presence of the Ysim (Y of D. simulans) than in the presence of the Ysech (Y of D. sechellia), in spite of most of the autosomal and X-linked genome being from D. simulans. The segment reduces fertility in the Y-simulans background but the infertility is partially rescued by the Y chromosome of D. sechellia. Moreover, the genotype of the grandparental line donating the Y chromosome displayed a significant contribution to male fertility. The complex interactions revealed in our study support the hypothesis that the heterochromatic Y chromosome participates in co-adapted associations within populations and may contribute significantly to the evolution of reproductive isolation as well as to the conditional manifestation of infertility in specific genotypic combinations.
Material and methods
Drosophila stocks
D. simulans stocks w; e (kindly provided by J Coyne) and simB (w; nt; III) were described in Tao et al., (2003). In the simB stock, the third chromosome is isogenic to the third chromosome of the highly inbred stock 13w 1 X 1 JJ (constructed by sib-pair mating for 20 generations from 13w—Liu et al., 1996). This chromosome does not carry a phenotypic marker, while the first and second chromosomes carry markers white (w) and net (nt), respectively (Tao et al., 2003). Drosophila mauritiana stocks (w; P [w+]) contain independent P-element inserts on the third chromosome, which carry a functional copy of the gene white (True et al., 1996). Numerical identifiers of each of these stocks are according to Tao et al. (2003). These stocks were used elsewhere for the construction of introgression lines that allowed the fine mapping of sex-ratio distorter and suppressor genes (Dox and Nmy—Tao et al., 2007a, b), as well as one candidate gene for hybrid male sterility (agt—Araripe et al., 2010). This is a reliable and well-studied system in which the manifestation of incompatibility phenotypes can be tracked to study the influence of Y chromosome variation. All flies were reared on cornmeal–molasses–agar medium sprinkled with yeast grains at room temperature (21±1 °C).
Introgression lines
Segments of the third chromosome of D. mauritiana were introgressed in the genomic background of D. simulans stock simB by repeated backcrosses (see Tao et al., 2003 for details). Aided by molecular markers, these introgressed segments of D. mauritiana had their lengths well delimited in previous work (see Tao et al., 2001, 2003). All introgression lines used here have the genotype of simB, w; nt; P/ III (Nmy), and each has a different segment of D. mauritiana's third chromosome with the semidominant P-element transgene P [w+] inserted (True et al., 1996) in positions that were previously mapped by Araripe et al. (2006). If the introgression overlaps with the suppressor of the Winters sex-ratio distortion, Nmy (Tao et al., 2007a, b), the line will be carrying the lack of function mutant, nmy.
Stocks used as sources of Y chromosomes
The Y chromosome of D. sechellia (Ysech) (stock 3588, Dermitzakis et al., 2000) was introgressed into stocks A14, SR12-2-7 and G23 of D. simulans (Tao et al., 2007a, b). D. simulans stocks A14, SR12-2-7 and G23 were chosen because they carry different combinations of the Winters sex-ratio distortion gene (Dox) and the suppressor of sex-ratio distortion (Nmy), as shown in Table 1 and Figure 1.
Table 1. Grandparental genotypes and distortion phenotypes used as sources for the construction of hybrid and triple hybrid males.
| Line | Genotype | Sex-ratio distortion | Detailed genotype |
|---|---|---|---|
| A14/Ysim and A14/Ysech | dox/nmy | No | w dox/ Y; nt; nmy 1427 |
| SR12-2-7/Ysim and SR12-2-7/Ysech | Dox/nmy | Yes | Dox/ Y; nt; nmy 1427 |
| G23/Ysim and G23/Ysech | Dox/Nmy | No | w/ C(1)yw; nt; Nmy |
| simB/Ysim | Dox/Nmy | No | Dox/ Y; nt; Nmy |
Grandparental genotype is A14, SR12-2-27 and G23. Y chromosome is from D. simulans (Ysim) or D. sechellia (Ysech).
Figure 1.
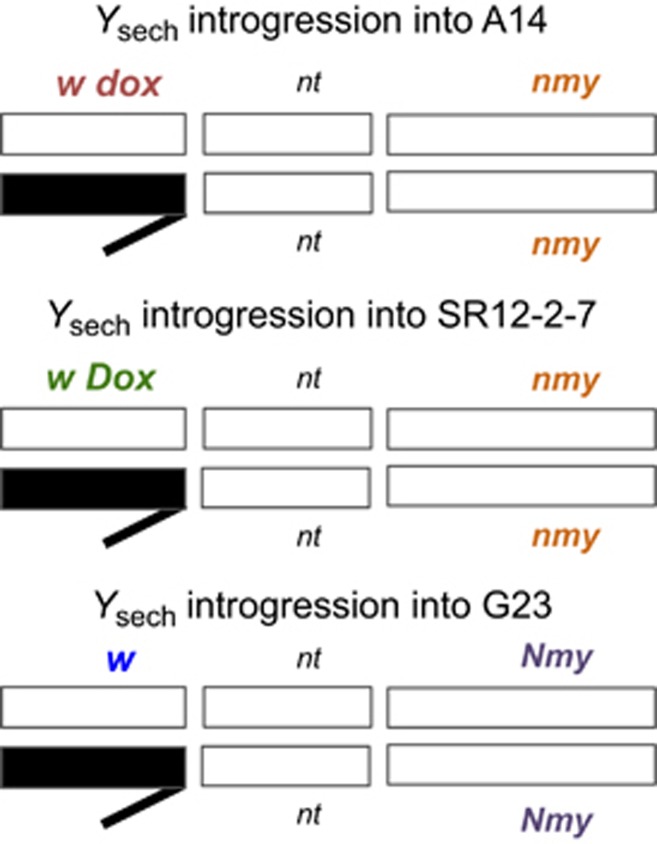
Grandparental genotypes of males with the Y chromosome of D. sechellia introgressed (A14/Ysech, SR12-2-7/Ysech and G23/Ysech). Grandparental strains with Ysim have the same genotype in the X chromosome/autosome background but are not shown here. Grandparental females of the G23 stock carry the attached X chromosome [C(1)RM]. Y chromosomes switch between males and females each generation along the G23 lineage. White bars represent chromosomes of D. simulans, black bars represent the Y chromosome of D. sechellia.
Construction of hybrid and triple-hybrid males
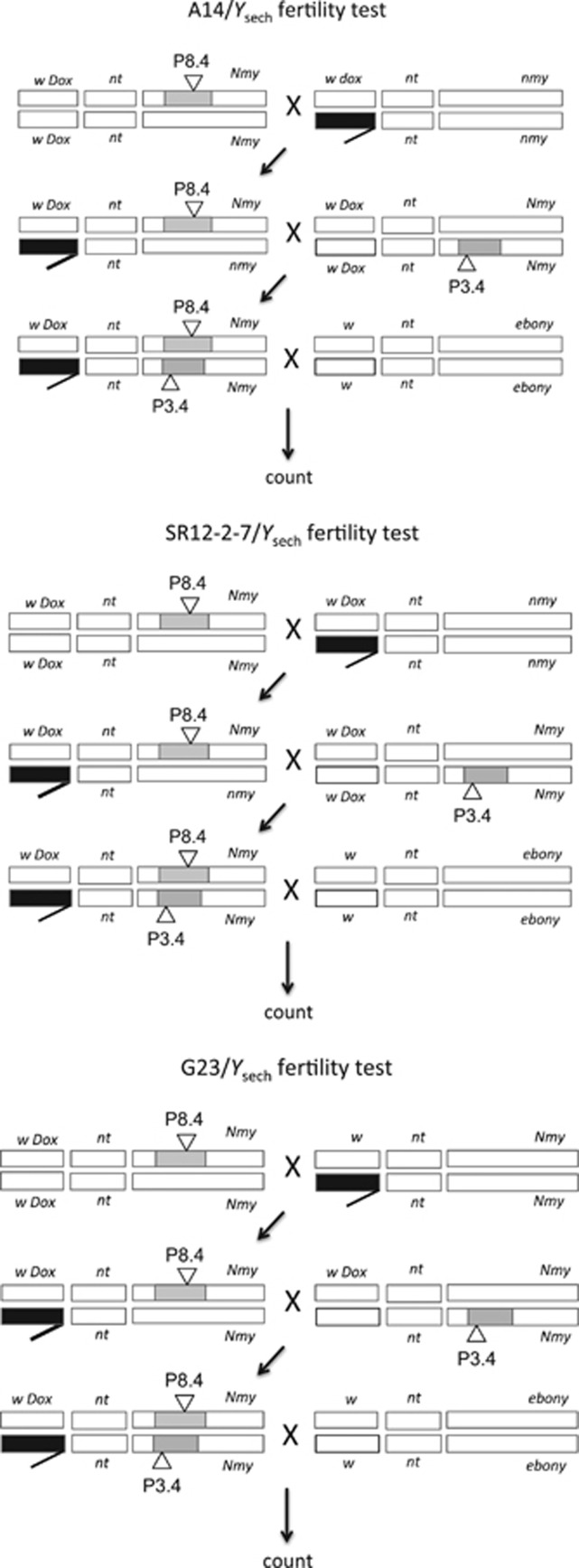
Seven pairs of the heterozygote introgression lines (3.4 and 8.4; 21.12 and 26.14; 38.9 and 38.6; 32.2 and 27.2; 32.2 and 33.3; 27.2 and 26.14; 33.5 and 42.2) were studied. To exemplify the procedure, we describe the steps to construct three of those lines that were examined in greater detail. Males of stocks A14, SR12-2-7 and G23, carrying the exogenous Y chromosome (Ysech or Ysim), were crossed to females of each one of the three heterozygous introgression lines. For this first step, we arbitrarily chose lines 8.4, 21.12 and 38.6 (Figure 2). The F1 males from these crosses carry the X chromosome of simB (from the female with D. mauritiana introgression), the Y chromosome provided by one of the three Ysim grandparental genotypes or one of the three Ysech grandparental genotypes described above, and the third chromosome heterozygote with one copy of 8.4, 21.12 or 38.6 P-elements and one copy of the third chromosome of simB (Figure 3). F1 males with colored eyes (P[w+]) were then crossed to females of each one of the three other heterozygous introgression lines: 3.4, 26.14, and 38.9. The focal males were F2 males with either Ysech or Ysim and homozygote introgressions paired as 3.4/8.4, 21.12/26.14 and 38.6/38.9, adding to a total of 18 genomic combinations to be tested (Figure 3). Focal males were selected by the dark red eyes given by 2P [w+].
Figure 2.
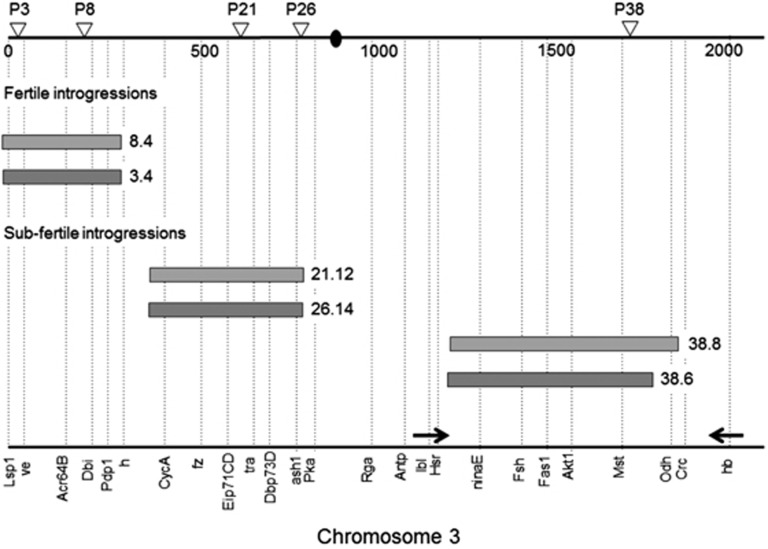
Positions of the D. mauritiana chromosomal segments introgressed into the D. simulans background, on the third chromosome (modified from Tao et al., 2003). Each introgression is tagged by a P [w+] insert (open triangles on the top), followed by an individual number. The introgression ends are defined with the aid of ASO markers (names at the bottom and dotted lines). The ASO markers and P [w+] inserts are positioned according to polytene bands. Cytological positions are based on the standard map of D. melanogaster (Lefevre, 1976), but D. simulans differs from D. melanogaster by an inverted segment (93F6-7 to 84F9, the two arrows facing each other) (Horton, 1939). Filled oval: the centromere.
Figure 3.
Schematics of the construction of focal males for fertility tests for one pair of D. mauritiana introgressions (8.4/3.4) with the Ysech. Identical crosses and male fertility tests with the Ysim chromosome were performed simultaneously (not shown here). Shown here is A14/Ysech in the background genome of simB with homozygous D. mauritiana introgressions 8.4/3.4. Males from lines SR12-2-7/Ysech and G23/Ysech also provided Ysech chromosomes for assay with 8.4/3.4, 21.2/26.14 and 38.6/38.8. Males from lines A14/Ysim, SR12-2-7/Ysim and G23/Ysim provided Ysim chromosomes for the same introgression combinations cited above. Crosses and fertility assays were performed simultaneously for all treatments. Note that the focal male is identical across all cases. Male fertility was tested by crossing one male with three D. simulans w; e females for 7 days and subsequently counting the progeny. White bars represent chromosomes of D. simulans, black bars represent chromosomes of D. sechellia and gray bars represent segments of D. mauritiana third chromosome.
Fertility tests
We focused on quantitative decrease or increase in male fertility, rather than on complete male sterility, as the phenotype resulting from hybridization. Fertility tests were carried out by mating one male with three virgin w; e females for 7 days before being discarded. The progeny was counted up to the twentieth day after cross setup, when all adults had emerged. Fertility was defined as the number of progeny produced (Tao et al., 2003). Fertility tests were conducted with ~15 single males per genotype. To qualitatively validate our observations, crosses made to create the described genotypes were independently conducted a second time for each genotype and essayed for fertility in an experiment with fewer males tested. All fertility tests were performed at room temperature (21±1 °C).
Results
The construction of lines with various combinations of Y chromosome and incompatibility genes could reveal new genomic interactions involving the Y chromosome. These genotypes could also shed light on the contribution of Y chromosome–autosomal epistasis to the process of reproductive isolation and speciation. Here we hypothesized that Y chromosomes of D. simulans or D. sechellia origin might exhibit differential responses to incompatibility elements present in the genomic background. To address the issue, seven pairs of the heterozygote introgression lines (3.4 and 8.4; 21.12 and 26.14; 38.9 and 38.6; 32.2 and 27.2; 32.2 and 33.3; 27.2 and 26.14; 33.5 and 42.2) were crossed in order to make specific regions of the third chromosome homozygote for D. mauritiana. Although the introgression of the whole third chromosome of D. mauritiana into D. simulans causes males to be fully sterile, the seven D. mauritiana homozygote segments described above were shown to generate subfertile-to-fertile males (Tao et al., 2003). This is key for our study, as it allows for quantitative effects of Y chromosome substitution on the hybrid fertility phenotype to be noticed in both directions, that is, either as an increase or a decrease in fertility.
We initially performed a screen for fertility variation emerging from the interaction of each of the seven pairs of D. mauritiana introgressions and Ysim and Ysech chromosomes originating from the A14 grandparental background (Figures 1 and 3). This initial screen with all seven pairs revealed that four of them did not exhibit fertility differences between Ysim and Ysech (mean and s.e.m. of progeny number in 32.2/27.2: Ysim=235.1±13.7 offspring vs Ysech=210.6±25.6 offspring; 32.2/33.3: Ysim=222.0±38.9 vs Ysech=189.6±20.3; 27.2/26.14: Ysim=176.3±35.3 vs Ysech=208.2±20.7; 33.5/42.2: Ysim=193.0±17.5 vs Ysech=218.4±22.3; P>0.05 for all cases, Student's t-test). On the other hand, three pairs of introgressions displayed significant differences between Ysim and Ysech (8.4/3.4: Ysim=246.1±15.6 vs Ysech=72.0±14.1; 21.12/26.14: Ysim=6.5±2.1 vs Ysech=69.0±11.3; 38.9/38.6: Ysim=23.0±7.9 vs Ysech=0.0±0.0; P<0.05 for all cases, Student's t-test). Together these observations highlight the relevance of the genetic background in the emergence of fertility differences mediated by the Y chromosome.
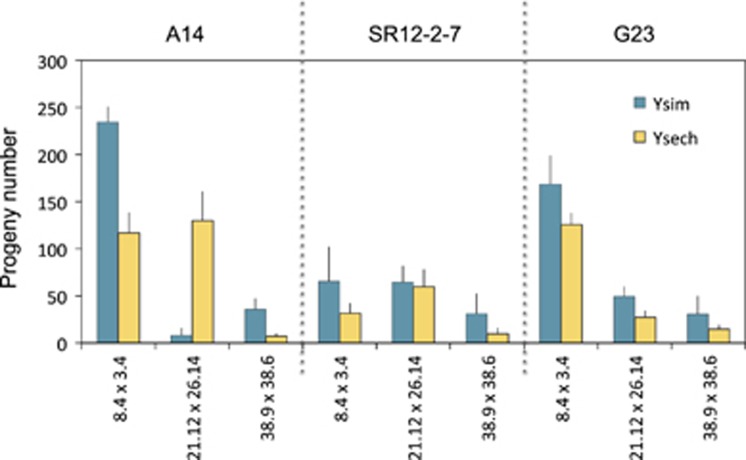
In view of these observations, we chose the three pairs of introgression segments displaying Y chromosome variation in fertility for further analysis. Specifically, these introgressions were further studied in greater detail in 18 focal males that were classified according to: (1) the species origin of the Y chromosome (D. simulans or D. sechellia) and (2) the grandparental genomic background of origin (D. simulans of A14, SR12-2-7 or G23; Figure 1; and the introgressed D. mauritiana segment on D. simulans third chromosome (3.4/8.4, 21.12/26.14 or 38.9/38.6; Figure 2). As expected from the initial screen, differences in progeny number are significant between males carrying Y chromosome from the line A14/Ysim and Y chromosome from the line A14/Ysech for all three hybrid backgrounds re-tested: 8.4/3.4 (t=4.351, P<0.001), 21.12/26.14 (t=−3.815, P<0.001), and 38.9/38.6 (t=2.458, P<0.05) (Figure 4, Table 2). Similarly, as expected from the initial screen, the A14/Ysech exhibited greater fertility than A14/Ysim in the introgression 21.12/26.14. Surprisingly, however, we observed that while the species origin of the Y chromosome had a clear influence on male fertility when originating from some grandparental backgrounds it displayed a milder effect when originating from other backgrounds. For instance, progeny number of SR12-2-7/Ysim males is not significantly different from the progeny number of SR12-2-7/Ysech males (Figure 4). The variation illustrates the multifactorial nature of genomic interactions involving the Y chromosome and autosomes and is further detailed below.
Figure 4.
Progeny number for males carrying either Ysim or Ysech and three pairs of introgressions from D. mauritiana. (for example, 8.4 × 3.4 refers to the final D. mauritiana genotype in the focal male). A14, SR12-2-7 and G23 refer to the grandparental genotype donating the chromosomes to the focal male in which the assays were performed. (Left panel) A14/Ysim vs A14/Ysech, (Center panel) SR12-2-7/Ysim vs SR12-2-7/Ysech and (Right panel) G23/Ysim vs G23/Ysech. Error bars: 1.96 × s.e.m.
Table 2. Average progeny number sired by hybrid males with every arrangement of three factors: Grandparental Genotype (GG), Introgression Combination (IC), and Y chromosome Origin (YO).
| Grandparental Genotype (GG) | Introgression Combination (IC) |
Y chromosome Origin (YO) |
|||
|---|---|---|---|---|---|
| Ysim | Ysech | df | t | ||
| A14 | 8.4 × 3.4 | 234.4 | 116.7 | 17 | 4.353*** |
| 21.12 × 26.14 | 8.0 | 129.9 | 18 | −3.815** | |
| 38.9 × 38.6 | 35.6 | 6.9 | 17 | 2.458* | |
| 8.4 × 3.4 | 65.3 | 31.5 | 7 | 0.166NS | |
| SR12-2-7 | 21.12 × 26.14 | 64.9 | 59.8 | 18 | 0.204NS |
| 38.9 × 38.6 | 31.2 | 9.7 | 18 | 0.965NS | |
| 8.4 × 3.4 | 168.4 | 125.6 | 17 | 1.235NS | |
| G23 | 21.12 × 26.14 | 49.5 | 27.4 | 18 | 1.73NS |
| 38.9 × 38.6 | 30.5 | 14.5 | 18 | 0.804NS | |
| ANOVA | df | F | |||
| YO | 1 | 5.896* | |||
| GG | 2 | 10.461*** | |||
| IC | 2 | 67.208*** | |||
| YO × GG | 2 | 0.179NS | |||
| YO × IC | 2 | 15.801*** | |||
| GG × IC | 4 | 5.745*** | |||
| YO × GG × IC | 4 | 4.669** | |||
| Within | 148 | ||||
| Total | 165 | ||||
Abbreviations: ANOVA, analysis of variance; NS, not significant. Averages were compared between Ysim and Ysech males using a Student's t-test. Three-way ANOVA reveals significant interactions among factors (*P<0.05; **P<0.01; ***P<0.001).
The observation that Ysech chromosome exhibits greater fertility than Ysim in the presence of introgression pair 21.12/26.14 is particularly unexpected. Triple-hybrid males, which carry Ysech and homozygous D. mauritiana introgressions in a D. simulans background, are expected to be less fertile than bi-hybrid males, which carry Ysim instead of Ysech. This is because the former may show three-way interspecific incompatibilities among all three species. Indeed, this was the case for most of the genotype combinations in which the two Y chromosomes were contrasted (Figure 4, Table 2). For instance, the A14/Ysim; 8.4/3.4 bi-hybrid males produced on average 234.4 offspring while the A14/Ysech; 8.4/3.4 tri-hybrid males produced 116.7 offspring (t=4.353, P<0.001). Surprisingly, however, we observed that the tri-hybrid A14/Ysech; 21.12/26.14 produced on average much more offspring per male (129.9) than the bi-hybrid A14/Ysim; 21.12/26.14 (8.0, t=−3.815, P<0.01). This result suggests that the Ysech is more compatible than the Ysim to the 21.12/26.14 portion of D. mauritiana genome and to the background genome of line A14. Evidently, Y chromosomes from D. simulans and D. sechellia must differ in some causative Y-linked sequence that mediates the ability of the D. sechellia Y chromosome to rescue the infertility phenotype.
The D. simulans grandparental background also has a profound effect on fertility of the hybrid and tri-hybrid males. For example, the males constructed with Y chromosomes deriving from the line SR12-2-7 showed low fertility overall, irrespective of the species origin of the Y chromosome and the introgression background (on average 53.8 flies produced by the Ysim and 33.7 produced by the Ysech). These data suggest that the genotype Dox/nmy (Dox distorter/nmy lack-of-function suppressor) has a predominant effect on fertility regardless of the Y chromosome. Interestingly, Branco et al. (2013a) have shown that the D. simulans Y chromosome is polymorphic for its capacity to modulate sex ratio distortion in genotype SR12-2-7. These authors used the same stock SR12-2-7 as a background for the introgression of 78 Y chromosomes from several D. simulans populations and found that there is significant diversity in Y chromosome resistance to sex-ratio distortion: some Y chromosomes carry YRV that overrides the effects of the distorter Dox, while other Y chromosomes are even more sensitive to Dox than the original Y chromosome present in SR12-2-7.
Finally, males with the homozygous introgressions 38.9/38.6 show the lowest fertility (on average 34.4 offspring flies produced by males carrying Ysim and 10.4 produced by males carrying Ysech), irrespective of the Y chromosome species and the genomic background. The 38.9/38.6 introgressions have the hybrid male sterility factors and sex-ratio distortion suppressor too much ying (tmy) (Tao et al., 2001, 2003). When homozygous, tmymau males (the tmy region from D. mauritiana in an otherwise D. simulans background) have very low fertility and offspring with female-biased sex ratio (~75% female). Interestingly, Ysech did not show a ‘rescuing' effect for male sterility caused by tmymau. Similarly, Tao et al. (2007a, b) observed that Ysech and Ysim show the same sensitivity to the sex-ratio distortion by Dox; the same sex ratio is seen in Dox; nmy males regardless of the origin of the Y chromosome.
Discussion
Genetic incompatibilities manifesting within and between populations could provide essential variation leading to speciation. YRV affects rapidly evolving testis-specific genes and is expected to be involved in adaptive processes, with Y chromosome-mediated incompatibility possibly emerging from the disruption of co-adapted networks of genes within populations. These networks are built by interactions between autosomal and sex-linked genes; the contribution of X–Y interactions to the emergence of disrupted hybrid phenotypes has been especially considered (Haldane, 1932; Coyne, 1985; Johnson et al., 1992, 1993; Zeng and Singh, 1993). The identification of specific loci on the X chromosome and autosomes that are incompatible with specific Y chromosome variants could illuminate processes mediating variable fertility. In turn, the extent of Y chromosome-mediated fertility breakdown in hybrid genotypes may reveal mechanisms responsible for a broader spectrum of phenotypic variation.
Genome-wide gene expression in genotypes differing only in Y chromosome origin revealed cryptic Y chromosome diversity (Lemos et al., 2008; Lemos et al., 2010; Jiang et al., 2010; Branco et al., 2013a). These include changes in the expression level of genes involved with functions as varied as metabolism, cell division, immune response and chromatin structure. In D. simulans, Branco et al. (2013a) observed a range of sex ratios in Y chromosome substitution lines of the Winter's sex-ratio system. The data indicate that polymorphic variation residing in the Y chromosome causes resistance to an X-linked distorter. These observations recapitulate previous observations by Montchamp-Moreau et al. (2001) with the Paris system. In addition to sex-ratio distortion, other phenotypes associated with reproductive isolation and hybrid incompatibilities may be modulated by interactions with genic and regulatory sequences of the Y chromosome. These phenotypes, including fertility and YRV, may emerge from non-genic elements on the Y chromosome heterochromatin and may be under selection (Lemos et al., 2010; Francisco and Lemos, 2014). The piRNA pathway is a candidate to mediate Y chromosome effects in hybrids (Castillo et al., 2011; Kelleher et al., 2012) and could reconcile the low polymorphism in Y-linked protein-coding sequences (Zurovcova and Eanes, 1999; Larracuente and Clark, 2013) with YRV. Differences in the extensive blocks of heterochromatin are given by the kinds and quantities of satellite DNA and transposable elements. These elements are predicted to be key players in the phenomenon of YRV, and the mechanisms for their manifestation are likely to be varied (Francisco and Lemos, 2014).
The three sister species in the D. simulans clade are able to intercross, but postzygotic incompatibilities such as hybrid male sterility are evident. One gene involved in the reproductive isolation of D. simulans and D. mauritiana is the fast-evolving X chromosome gene Odysseus site homeobox, first described by Ting et al., (1998). Further investigation found that the protein OdsH binds to Y chromosome heterochromatic sites in hybrids, affecting local chromatin condensation and leading to hybrid sterility (Bayes and Malik, 2009). In the D. simulans sister-species clade, the binding sites of OdsH differ for hybrids of different pairs of species. For instance, the Y chromosomes of D. sechellia and D. simulans were enriched for the protein OdsH of D. mauritiana, whereas the protein of D. simulans did not bind to any of the three Y chromosomes. Given that the divergence of the D. simulans species clade is recent (~250 000 years, Kliman et al., 2000), these results are consistent with the hypothesis that the rise of genomic incompatibilities may be partly driven by the rapid evolution of heterochromatic DNA (Brideau et al., 2006).
Here we studied nine genomic combinations and showed that one segment of the D. mauritiana genome (21.12/26.14) interacts with the Y chromosome of D. simulans and D. sechellia in a unique way, resulting in unexpected reversal of the direction of variation. Moreover, the reversal was conditional on the grandparental genotype. As shown in Figure 2, introgression fragments 21.12 and 26.14 are located on chromosome 3L, between genes hairy (h) and Pka-R1. These introgression fragments overlap through a common segment between 7.8 and 12.2 Mb in length (Tao et al., 2003). At least three hypotheses may guide identification of the genetic elements in the segment that might differentially interact with the Y chromosome of D. simulans and D. sechellia. First, regulatory factors in the introgressed D. mauritiana segment may differentially modulate the expression of Y-linked coding elements (genes, transposable elements, piRNAs and so on). Second, satellite sequences may serve as a differential sink for the binding of protein-coding genes/small RNAs in these chromosomes and may become unavailable in other sites of the genome, including the introgressed segment. Third, interspecific divergence in blocks of repeated sequences embedded in heterochromatin may affect fertility by abnormally affecting chromosome segregation. This appears to be the cause of hybrid female lethality from crosses between D. simulans females and D. melanogaster males, where Zhr gene is actually a 359-bp stretch of satellite sequences on the heterochromatic region of the X chromosome of D. melanogaster that is not present in D. simulans (Ferree and Barbash, 2009).
Our results show that not only the species origin of the Y chromosome generates subfertility in hybrid males but also the magnitude and direction of the effect depends on the genomic background of the grandparent donor line. The influence of the grandparental genome on the modulating capacity of the Y chromosome is implied when we compare the progeny number of males A14/Ysech; 21.12/26.14 with the progeny number of males SR12-2-7/Ysech; 21.12/26.14. Both the Ysech and the autosomic background are the same among these lines, whereas the Y chromosome donor line (A14 vs SR12-2-7) is different. Noteworthy, the second chromosomes from all lines came from the same D. simulans donor and all carry the phenotypic marker net. However, the origin is not strictly immediate and we cannot rule out that new mutations in it might have contributed to the difference in progeny number across grandparental backgrounds. On the other hand, sex chromosomes are sensitive to parental origin in Drosophila (Golic et al., 1998; Maggert and Golic, 2002), including substantial consequences to testis-specific gene expression and epigenetic states elsewhere in the genome (Greil and Ahmad, 2012; Zhou et al., 2012; Lemos et al., 2014). Collectively, our observations raise the prospect that genetic or epigenetic variation acquired during the making of the focal males is partially responsible for modulating hybrid male sterility.
Finally, our results are in concordance with the expectation that the extent of reproductive isolation between a pair of species and hybrid male fertility attributes may be the by-product of a multifactor genomic environment. Relevant components include rapidly evolving repetitive sequences as well as rapidly evolving pathways implicated in the maintenance of heterochromatic chromosomes (Ferree and Barbash, 2009; Bayes and Malik 2009; Castillo et al., 2011). The observation that the heterochromatic Y chromosome from D. sechellia can rescue the fertility loss of a D. mauritiana segment in a D. simulans genome background is unexpected and displays the evident complexity of epistatic interactions with the Y chromosome. We conclude that investigating the diversity of phenotypic outcomes in hybrid genotypes is an important step to uncover the full spectrum of Y chromosome modulation of endogenous regulatory processes.
Data Archiving
Data are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.n2v20.
Acknowledgments
We thank Alan Branco and members of the Lemos laboratory for critical readings of the manuscript, Daniel L Hartl for generously providing facilities and a stimulating environment for the conduct of pilot studies for these experiments, and two anonymous reviewers for valuable comments. LOA is partly supported by the Conselho Nacional de Pesquisa Cientifica (CNPq). BL is partly supported by an Ellison Foundation New Scholar in Aging Award, and a Smith Family Award for Biomedical Research.
The authors declare no conflict of interest.
References
- Araripe LO, Eckstrand ND, Hartl DL, Tao Y. (2006). Flanking regions of P-elements inserted in the 3rd chromosome of Drosophila mauritiana. Dros Inf Ser 89: 54. [Google Scholar]
- Araripe LO, Montenegro H, Lemos B, Hartl DL. (2010). Fine-scale genetic mapping of a hybrid sterility factor between Drosophila simulans and D. mauritiana: the varied and elusive functions of ‘speciation genes'. BMC Evolutionary Biology 10: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Golic KG, Hawley RS (2005). Drosophila: A Laboratory Handbook, 2nd edn. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, p 1408.
- Bayes JJ, Malik HS. (2009). Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science 326: 1538–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco AT, Tao Y, Hartl DL, Lemos B. (2013. a). Natural variation of the Y chromosome suppresses sex ratio distortion and modulates testis-specific gene expression in Drosophila simulans. Heredity 111: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco AT, Hartl DL, Lemos B. (2013. b). Chromatin-associated proteins HP1 and Mod (mdg4) modify Y-linked regulatory variation in the Drosophila testis. Genetics 194: 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA. (2006). Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295. [DOI] [PubMed] [Google Scholar]
- Carvalho AB, Clark AG. (2005). Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science 307: 108–110. [DOI] [PubMed] [Google Scholar]
- Carvalho AB, Vaz SC, Klaczko LB. (1997). Polymorphism for Y-linked suppressors of sex-ratio in two natural populations of Drosophila mediopuntacta. Genetics 146: 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Lazzaro BP, Clark AG. (2000). Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc Natl Acad Sci USA 97: 13239–13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Dobo BA, Vibranovski MD, Clark AG. (2001). Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci USA 98: 13225–13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo DM, Mell JC, Box KS, Blumenstiel JP. (2011). Molecular evolution under increasing transposable element burden in Drosophila: a speed limit on the evolutionary arms race. BMC Evol Biol 11: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippindale AK, Rice WR. (2001). Y chromosome polymorphism is a strong determinant of male fitness in Drosophila melanogaster. Proc Natl Acad Sci USA 98: 5677–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA. (1985). The genetic basis of Haldane's rule. Nature 314: 736–738. [DOI] [PubMed] [Google Scholar]
- Cutter AD. (2012). The polymorphic prelude to Bateson-Dobzhansky-Muller incompatibilities. TREE 27: 209–218. [DOI] [PubMed] [Google Scholar]
- David JR, Araripe LO, Chakir M, Legout H, Lemos B, Pétavy G et al. (2005). Male sterility at extreme temperatures: a significant but neglected phenomenon for understanding Drosophila climatic adaptations. J Evol Biol 18: 838. [DOI] [PubMed] [Google Scholar]
- Dermitzakis ET, Masly JP, Waldrip HM, Clark AG. (2000). Non-Mendelian segregation of sex chromosomes in heterospecific Drosophila males. Genetics 154: 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree PM, Barbash DA. (2009). Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol 7: e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco FO, Lemos B. (2014). How do Y chromosomes modulate genome-wide epigenetic states: genome folding, chromatin sinks, and gene expression. J Genomics 2: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepner J, Hays TS. (1993). A fertility region on the Y chromosome of Drosophila melanogaster encodes a dynein microtubule motor. Proc Natl Acad Sci USA 90: 11132–11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic KG, Golic MM, Pimpinelli S. (1998). Imprinted control of gene activity in Drosophila. Curr Biol 8: 1273–1276. [DOI] [PubMed] [Google Scholar]
- Greil G, Ahmad K. (2012). Nucleolar dominance of the Y chromosome in Drosophila melanogaster. Genetics 191: 1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. (1932) The Causes of Evolution. Longmans, Green & Co.: London, UK. [Google Scholar]
- Horton IH. (1939). A comparison of the salivary gland chromosomes of Drosophila melanogaster and D. simulans. Genetics 24: 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttunen S, Aspi J. (2003). Complex genetic architecture of male courtship song characters in Drosophila virilis. Behav Genet 33: 17. [DOI] [PubMed] [Google Scholar]
- Jiang PP, Hartl DL, Lemos B. (2010). Y not a dead end: epistatic interactions between Y-linked regulatory polymorphisms and genetic background affect global gene expression in Drosophila melanogaster. Genetics 186: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NA, Perez DE, Cabot EL, Hollocher H, Wu C-I. (1992). A test of reciprocal X-Y interactions as a cause of hybrid sterility in Drosophila. Nature 358: 751–753. [DOI] [PubMed] [Google Scholar]
- Johnson NA, Hollocher H, Noonburg E, Wu C-I. (1993). The effects of interspecific Y chromosome replacements on hybrid sterility within the Drosophila simulans clade. Genetics 135: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Edelman NB, Barbash DA. (2012). Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol 10: e1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman RM, Andolfatto P, Coyne JA, Depaulis F, Kreitman M, Berry AJ et al. (2000). The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156: 1913–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D, David JR, Lemeunier F, Tsacas L, Ashburner M. (1986). The reproductive relationships of Drosophila sechellia with Drosophila mauritiana Drosophila simulans and Drosophila melanogaster from the Afrotropical region. Evolution 40: 262–271. [DOI] [PubMed] [Google Scholar]
- Larracuente AM, Clark AG. (2013). Surprising differences in the variability of Y chromosomes in African and cosmopolitan populations of Drosophila melanogaster. Genetics 193: 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre G (1976). A photographic representation and interpretation of the polytene chromosomes of Drosophila melanogaster salivary glands. In: Ashburner M and Novitski E (eds). The Genetics and Biology of Drosophila. Academic Press: New York, NY, USA, pp 31–66.
- Lemos B, Araripe LO, Hartl DL. (2008). Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science 319: 91. [DOI] [PubMed] [Google Scholar]
- Lemos B, Branco AT, Hartl DL. (2010). Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci USA 107: 15826–15831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Branco AT, Jiang PP, Hartl DL, Meiklejohn CD. (2014). Genome-wide gene expression effects of sex chromosome imprinting in Drosophila. G3 (Bethesda) 4: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Mercer JM, Stam LF, Gibson GC, Zeng Z-B, Laurie CC. (1996). Genetic analysis of a morphological shape difference in the male genitalia of Drosophila simulans and D. mauritiana. Genetics 142: 1129–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert KA, Golic KG. (2002). The Y chromosome of Drosophila melanogaster exhibits chromosome-wide imprinting. Genetics 162: 1245–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montchamp-Moreau C, Ginhoux V, Atlan A. (2001). The Y chromosomes of Drosophila simulans are highly polymorphic for their ability to suppress sex- ratio drive. Evolution 55: 728–737. [DOI] [PubMed] [Google Scholar]
- Pantazidis AC, Galanopoulos VK, Zouros E. (1993). An autosomal factor from Drosophila arizonae restores normal spermatogenesis in Drosophila mojavensis males carrying the D. arizonae Y chromosome. Genetics 134: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer C, David JR, Moreteau B, Joly D. (2004). Heat induced male sterility in Drosophila melanogaster: adaptive genetic variations among geographic populations and role of the Y chromosome. J Exp Biol 207: 2735. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Montenegro H, Hartl DL, Lemos B. (2011). Interspecific Y chromosome introgressions disrupt testis-specific gene expression and male reproductive phenotypes in Drosophila. Proc Natl Acad Sci USA 108: 17046–17051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, Shu S, Mungall CJ, Karpen GH. (2007). The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science 316: 1586–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltenberg SF, Hirsch J. (1997). Y chromosome effects on Drosophila geotaxis interact with genetic or cytoplasmic background. Anim Behav 53: 853–864. [DOI] [PubMed] [Google Scholar]
- Tao Y, Araripe L, Kingan SB, Ke Y, Xiao H, Hartl DL. (2007. a). A sex-ratio meiotic drive system in Drosophila simulans. II: An X-linked distorter. PLoS Biol 5: e293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Hartl DL, Laurie CC. (2001). Sex-ratio meiotic drive segregation distortion associated with reproductive isolation in Drosophila. Proc Natl Acad Sci USA 98: 13183–13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Chen S, Hartl DL, Laurie CC. (2003). Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics 164: 1383–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Masly JP, Araripe L, Ke Y, Hartl DL. (2007. b). A sex-ratio meiotic drive system in Drosophila simulans. I: an autosomal suppressor. PLoS Biol 5: e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting C-T, Tsaur S-C, Wu M-L, Wu C-I. (1998). A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282: 1501–1504. [DOI] [PubMed] [Google Scholar]
- True JR, Weir BS, Laurie CC. (1996). A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics 144: 819–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Koerich LB, Carvalho AB. (2008). Two new Y-linked genes in Drosophila melanogaster. Genetics 179: 2325–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneault G, Zouros E. (1986). The genetics of asymmetrical male sterility in Drosophila mojavensis and Drosophila arizonensis hybrids: interaction between the Y chromosome and autosomes. Evolution 40: 1160–1170. [DOI] [PubMed] [Google Scholar]
- Yee WKW, Rogell B, Lemos B, Dowling DK. (2015). Intergenomic interactions between mitochondrial and Y-linked genes shape male mating patterns and fertility in Drosophila melanogaster. Evolution 69: 2876–2890. [DOI] [PubMed] [Google Scholar]
- Zeng LW, Singh RS. (1993). The genetic basis of Haldane's rule and the nature of asymmetric hybrid male sterility among Drosophila simulans Drosophila mauritiana, and Drosophila sechellia. Genetics 134: 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Sackton TB, Martinsen L, Lemos B, Eickbush TH, Hartl DL. (2012). Y chromosome mediates ribosomal DNA silencing and modulates the chromatin state in Drosophila. Proc Natl Acad Sci USA 109: 9941–9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurovcova M, Eanes WF. (1999). Lack of nucleotide polymorphism in the Y-linked sperm flagellar dynein gene Dhc2Yh3 of Drosophila melanogaster and D. simulans. Genetics 153: 1709–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]