Abstract
Little attention has been paid to the evolutionary consequences of the colonizing dynamics and succession processes following glacier retreat. Here we studied hybrid populations that have recently formed and established on glacier forefields of the European Alps owing to secondary contact of a lowland colonizer with a subalpine species. We analyzed the composition of two hybrid populations between Salix purpurea and Salix helvetica with nine microsatellite markers by using Bayesian methods (structure and NewHybrids), and simulations. We also studied niche differentiation between the hybrids and the parental species based on indicator values, soil pH and water retention potential measurements. Allelic structure of hybrids confirms the assumed parentage and in situ origin of the crosses on two independent sites within the last decades. Both hybrid populations comprised F1 and later generation hybrids (F2 and backcrosses), confirming hybrid fertility. The parental species showed significant differences in niche characteristics for temperature, soil pH, nutrients and moisture. Remarkably, the hybrids exhibited a higher tolerance to cold temperatures, nutrient-poor and acidic soils than either parent. Our results show that willow hybrids originated after glacier retreat and have established persistent populations within a few decades. One factor contributing to hybrid establishment in sympatry with their parents is their ability to occupy more extreme niches than either parental species within a mosaic-like pattern of microhabitats on the forefield. Introgression and/or transgressive segregation may have resulted in novel genotypes that are able to expand the ecological spectrum of either parent.
Introduction
Though natural hybridization is a quite common phenomenon in plants (Arnold, 1997; Mallet, 2005), the evolutionary role of hybrid formation is, however, still controversial and in details poorly explored (Mallet, 2005). Hybridization is often regarded as a more or less accidental breakdown of crossing barriers that is maladaptive owing to low hybrid fertility (Mallet, 2005). Other authors, by contrast, emphasize the potential creative role of hybridization in generating ‘evolutionary novelty' (Arnold, 1997; Rieseberg and Willis, 2007; Soltis and Soltis, 2009). When the genomes of divergent species merge during hybridization, a broad range of recombinant genotypes is produced, which is then subject to selection (Burke and Arnold, 2001). Although polyploid hybrids are reproductively isolated from their parents by postzygotic crossing barriers (Mallet, 2007), homoploid hybrids, on the other hand, require ecological or chromosomal crossing barriers against their parents to persist in the long term (Rieseberg and Willis, 2007). Segregating hybrid populations can, for example, occupy extreme habitats relative to their parents. This requires that hybrids reach the F2 or later generations so that transgressive segregation of genes can generate individuals with novel characteristics enabling coexistence of the hybrids in distinct ecological niches (Rieseberg and Willis, 2007; Soltis and Soltis, 2009). If crossing barriers against the parents are permeable, homoploid hybrids can also backcross with one or both parental species. This facilitates the introgression of genes from one species into the other, probably leading to the transfer of adaptive genes between species (Arnold and Bennett, 1993; Barton, 2001).
Hybridization is often triggered by habitat disturbance, which facilitates secondary contact of species and creates new niches for establishment (Nolte and Tautz, 2010). In the European Alps, global warming has led to a severe retreat of glaciers, which is exceptionally rapid since the 1980s (Paul et al., 2004). The ice-free glacier forefields provide unoccupied territory for colonization by various species that previously might not have been in contact with each other. Another well-documented impact of global warming on mountain regions is the upward shift of tree lines, vegetation zones and species' distribution ranges (Parmesan and Yohe, 2003; Kelly and Goulden, 2008; Lenoir et al., 2008; Harsch et al., 2009). The speed of migration differs among species, and it has been observed that species from lower elevations advance more rapidly than those of higher altitudes are retreating, which leads to alterations of the community composition in higher elevations (Root et al., 2003) and a (transient) increase in species richness (Walther et al., 2002; Pauli et al., 2012). Species richness increases only temporally because seed establishment may be limited in high altitudes that are above the current distribution range of species (Sedlacek et al., 2014), or because cold-adapted species will eventually retreat from lower elevations (Gottfried et al., 2012).
Many studies have investigated establishment and succession on glacier forefields and identified important factors for colonization and primary succession in those environments (Moreau et al., 2005; Raffl et al., 2006) or observed gradual changes in species composition and richness (Cannone et al., 2008; Erschbamer et al., 2008). However, though the conditions on glacier forefields are particularly suited to enhance formation and establishment of hybrids, strikingly little research has been done on evolutionary processes taking place in these pioneer habitats. So far, detailed population genetic studies of such sites are missing, and hardly any study exists that explores evolution of the first hybrid generations after their origin.
Willows represent good models for such studies. Many willow species are fast-growing shrubs and appear as pioneers on disturbed sites such as quarries, embankments, alluvial plains or screes (Karrenberg et al., 2003; Hörandl et al., 2012). They also produce large numbers of lightweight, wind-dispersed seeds, which have been observed to arrive in large amounts on glacier forefields (Stöcklin and Bäumler, 1996). Hence, willows are among the first woody colonizers found on glacier forefields (Burga, 1999). Unsurprisingly, glacier forefields have been reported to be willow diversity hotspots with up to 20 different species occurring in close spatial proximity (Hörandl et al., 2012). Crossing barriers in Salix are in general mostly due to ecological and geographical isolation (Argus, 1973, 1974) and break down if otherwise isolated willow species occur in sympatry on disturbed glacier forefields. Homoploid hybridization, even among distantly related willow species, has been reported frequently in the genus (Skvortsov, 1999; Hörandl et al., 2012; Oberprieler et al., 2013).
In this study, we want to focus on the climate change-mediated hybridization of two Salix species, S. helvetica and S. purpurea, for which hybrid zones on glacier forefields in the Swiss Alps have been detected. The two species are morphologically distinct and belong to two different, unrelated sections within the genus (Skvortsov, 1999). Despite a long and intense research history on alpine willows (for example, Buser, 1940), the hybrid between S. purpurea and S. helvetica yet represents a novel combination to the literature. We therefore assume in situ origin of the hybrid as a consequence of recent re-colonization processes. Salix helvetica, a subalpine to alpine shrub, occurs on glacier forefields within its normal altitudinal range, whereas the widespread pioneer species S. purpurea is invading from lower altitudes. Under the particular conditions on the glacier forefield, populations of both species meet and interbreed. Here, we study two populations of homoploid hybrids in detail using a combination of genetic and ecological methods to address the following questions: (i) does the genetic structure of the hybrid population support the hypothesis of recent in situ origin? (ii) do the hybrid populations consist of a stable, recurrently originating F1 generation, or do the hybrids establish a population of later generations, potentially also backcrossing with the parents? (iii) do the hybrids occupy different ecological niches in comparison with the parents?
Materials and methods
Sampling and identification
We collected leaves in silica gel from a total of 343 individuals at four sampling sites in the European Alps (Table 1). The two sites containing mixed populations of S. purpurea, S. helvetica and their hybrid were located at the forefields of the Rhône Glacier and the Morteratsch Glacier. These locations were chosen because other glacier forefields we visited were in earlier stages of succession so that S. helvetica occurred only in low numbers and S. purpurea was absent. Numerous individuals of S. purpurea and S. helvetica were present at both sampling sites. However, although we sampled 51 individuals with a hybrid phenotype at the Rhône Glacier, we found only six of them at the Morteratsch Glacier. The sampling also included pure stands of S. purpurea and S. helvetica as reference populations representing the genetic diversity of large, natural, outcrossing populations. A pure stand of S. purpurea was sampled at low elevation (1100 m ASL), where S. helvetica does not occur, along a natural area of the Inn River near San Niclá, Switzerland. On this site it could be assumed that the population actually emerged from natural seed dispersal, and not from a planting of cuttings, for which S. purpurea is often used (Hörandl et al., 2012). A population of S. helvetica was sampled at higher elevation (2100 m ASL) at the Gepatschferner in Austria, an area completely devoid of S. purpurea. In the field, species were classified as S. purpurea or S. helvetica based on their morphology (Hörandl et al., 2012). Individuals were classified as hybrids if they showed an intermediate phenotype between both parental species (Supplementary Table S1). In addition, we performed a detailed morphometric analysis to confirm the field classification. For this purpose, 17 quantitative characters were measured on voucher specimens (Supplementary Table S2, Supplementary Figure S1). Remarkably, we observed well-developed fruits and seeds in female hybrid individuals, indicating hybrid fertility. A detailed study of the fertility of the natural hybrids on the glacier forefield will be presented elsewhere (Gramlich et al. in prep.). Herbarium vouchers of each specimen were deposited in the herbarium of the University of Vienna (WU) and the University of Göttingen (GOET).
Table 1. Names, geographical coordinates, mean altitude, species and number of sampled individuals for the four sampling locations.
| Sampling location | Country | Coordinates | Altitude | Species and no. of individuals sampled |
|---|---|---|---|---|
| Rhône Glacier | Switzerland | 46° 34' 03.3'' N 08° 22' 12.3'' E | 1770 m | S. purpurea, n=65; S. helvetica, n=66; Hybrids, n=51 |
| Morteratsch Glacier | Switzerland | 46° 26' 14.1'' N 09° 56' 01.8'' E | 2000 m | S. purpurea, n=39; S. helvetica, n=37; Hybrids, n=6 |
| Inn near San Niclà | Switzerland | 46° 51' 28.1'' N 10° 25' 28.4'' E | 1070 m | S. purpurea, n=52 |
| Gepatschferner | Austria | 46° 52' 45.9'' N 10° 44' 30.7'' E | 2065 m | S. helvetica, n=27 |
DNA extraction and microsatellite analysis
DNA was extracted using the Invisorb Spin Plant Mini Kit (Stratec, Berlin, Germany) or following a slightly modified CTAB protocol after Doyle and Doyle (1987). We screened a set of 150 microsatellite primers, which were originally developed for diverse Salix or Populus species, for their applicability in S. purpurea and S. helvetica. Out of these, we chose nine primer pairs that amplified polymorphic loci in both species: GCPM 1255, GCPM 1413-2, GCPM 1812, GCPM 2041-1, ORPM 301 and ORPM 446 developed for P. trichocarpa (http://www.ornl.gov/sci/ipgc/ssr_resource.htm), SB 199 and SB 233 developed for S. burjatica (Barker et al., 2003), and gSIMCT024 developed for S. lanata (Stamati et al., 2003). A three-primer system was used to label the PCR products fluorescently. This system consisted of forward primers with a CAG-tag added to the 5′ end, CAG-primers labeled with fluorescent dyes (6-FAM, NED, PET, VIC), and unlabeled reverse primers. Microsatellite loci were amplified in 25 μl polymerase chain reactions containing 10 × NH4 reaction buffer (Bioline, Luckenwalde, Germany), 3 mM MgCl2 (Bioline), 0.2 mM of each dNTP (Carl Roth, Karlsruhe, Germany), 1.25 U BIOTAQ DNA Polymerase (Bioline), 0.08 μM forward primer (Eurofins, Ebersberg, Germany), 0.4 μM of the reverse (Eurofins) and CAG-primer (Applied Biosystems, Woolston, Cheshire, UK), respectively. Microsatellite fragments were amplified on a BIO-RAD thermocycler (Bio Rad, München, Germany) with a touchdown PCR program consisting of 95 °C for 5 min; 5 cycles at 95 °C for 30 s, primer and species-specific initial annealing temperature for 30 s, 72 °C for 30 s; 15 cycles (touchdown) at 95 °C for 30 s, primer and species-specific TA-1 °C/cycle for 30 s, 72 °C for 30 s; 15 cycles at 95 °C for 30 s, 50 °C for 30 s, 72 °C for 30 s; 72 °C for 5 min. PCR products were multiplexed and run on an ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster city, CA, USA), using GeneScan 500 LIZ Size Standard (Applied Biosystems). An overview of the applied microsatellite primers and detailed reaction conditions is included in Supplementary Table S3. Fragment lengths were determined using GeneMarker v2.6.0 (Softgenetics, State College, PA, USA). All parental and hybrid individuals exhibited a maximum of two alleles per locus, as expected for diploid plants. As both parental species were always reported as diploid (2n=38±1; Tropicos, http://www.tropicos.org) all statistical analyses and conclusions were based on a case of homoploid (diploid) hybridization.
Statistical analyses
Basic frequency statistics like the number of alleles, the effective number of alleles, the number of private alleles, and also the Principal Coordinate Analysis based on pairwise genetic distances between individuals were calculated with GenAlEx 6.5 (Peakall and Smouse, 2006, 2012). Owing to the unequal sample sizes we also calculated allelic richness with ADZE 1.0 (Szpiech et al., 2008), which corrects the number of alleles by the rarefaction method. The hybrid population from the Morteratsch Glacier was excluded from this analysis owing to small sample size. Further, ARLEQUIN 3.5 (Excoffier and Lischer, 2010) was used to calculate Ho, He and FST values, and to conduct the analysis of molecular variance. We used GENEPOP 4.3 (Raymond and Rousset, 1995) to calculate the inbreeding coefficient (FIS), to test for deviations from the Hardy–Weinberg equilibrium at each locus in each population, and to perform a global test for deviation from the Hardy–Weinberg equilibrium across loci in each population with the specific alternative hypothesis of heterozygote deficiency. The observed heterozygosity between S. purpurea, S. helvetica and the hybrids was compared with one-way analysis of variance (ANOVA) using SPSS Statistics 22 (IBM Corp., Armonk, NY, USA). The effective number of alleles was compared between S. purpurea, S. helvetica and the hybrid individuals using a linear mixed-effect model with species as fixed effect and sampling site as random effect. The linear mixed-effect model was calculated in R (R Core Team, 2015) using the R package lme4 (Bates et al., 2015). To check for effects of spatial distances on population structure within the sites, Mantel tests were performed to test for a correlation between genetic and geographic distance between individuals of S. purpurea and S. helvetica on the two glacier forefields. Geographic distances were calculated based on the geographic coordinates of the sampled individuals using the Geographic Distance Matrix Generator v 1.2.3 (Ersts, 2009). Pairwise individual genetic distances and Mantel tests were calculated in GenAlEx 6.5. The Bayesian clustering methods implemented in the program STRUCTURE v 2.3.4 (Pritchard et al., 2000) were applied to estimate the number of gene clusters (K) in data sets containing admixed or purebred populations, and to distinguish purebred and admixed individuals on the basis of their multilocus genotypes. All analyses were run under the admixture model without prior population information, a burn-in period of 20 000 and 100 000 iterations. We assumed independent allele frequencies except for the data set containing only S. helvetica from all three sampling locations. To estimate the number of populations (K) in a data set, STRUCTURE was run with K ranging from 1 to 5 with three iterations per K-value. Altogether, four different data sets were analyzed for population structure. Two data sets contained the admixed populations of the Rhône Glacier and the Morteratsch Glacier, respectively. For these data sets, the most likely value of K was determined using the method of Evanno et al. (2005). To test for cryptic population structure within the purebred species, two data sets were created that contained only individuals of S. purpurea or S. helvetica, respectively, from the three sampling locations. For those data sets, the method described in the STRUCTURE manual was used to determine the most likely value of K because ΔK cannot be calculated for K=1 following the method of Evanno et al. (2005). In order to distinguish purebred and hybrid individuals in the admixed populations of the Morteratsch and Rhône Glacier, STRUCTURE was run with the most likely K-value (K=2) in five replicates. To precisely determine the hybrid categories of the individuals identified as hybrids in STRUCTURE, the methods implemented in the program NewHybrids (Anderson and Thompson, 2002) were used. NewHybrids calculates for each individual the posterior probabilities of belonging to each of up to six predefined categories. The chosen categories were the purebred parental species, F1 hybrids, F2 hybrids and backcrosses to each parent. NewHybrids was run five times for each of the two data sets containing admixed populations. The program was run with Jeffreys-like priors, a burn-in of 10 000, and 500 000 iterations after burn-in. Like in STRUCTURE, no prior population information was included in the data sets.
Simulations
In order to determine the resolution that can be expected in STRUCTURE and NewHybrids with our genetic data, and to decide where to set the threshold values for a reliable identification of purebred species and hybrids, STRUCTURE and NewHybrids were run with data sets containing simulated hybrid individuals. From the population of the Rhône Glacier, we selected 65 genotypes of S. purpurea and 61 genotypes of S. helvetica that had been unequivocally assigned to the purebred category both in runs with STRUCTURE (q>0.95) and NewHybrids (P>0.95). These individuals were used for the simulation of hybrid individuals in the program HYBRIDLAB 1.1 (Nielsen et al., 2006). We simulated 50 F1 individuals, 30 F2 individuals and 30 backcrosses to each parental species. For the runs in STRUCTURE and NewHybrids 25 F1 individuals, 5 F2 individuals and 5 backcrosses each were selected by chance. Only few F2 hybrids and backcrosses were simulated and selected because we did not expect many later generation hybrids in the real data set owing to low age of the total population. The total size of the data set for the simulation runs was 166 individuals with 24% hybrids, which resembled our real data set of 188 individuals with 27% hybrids. Simulations were performed 10 times independently, so that we created ten data sets, which were run in structure and NewHybrids. We then calculated accuracy, efficiency and overall performance for diverse threshold values, according to Vähä and Primmer (2006). The term ‘efficiency' reflects the strength of the algorithm to correctly detect individuals of a certain group in the sample, and ‘accuracy' denotes the percentage of individuals correctly assigned to a certain category (Vähä and Primmer, 2006).
Ecological data
To analyze the potential ecological divergence among the hybrids and their parental species, we established a set of 58 quadratic plots of 64 m2 (that is, 8 m × 8 m) on the forefield of the Rhône Glacier (Supplementary Figure S2). Forty-eight of these plots contained at least one hybrid individual and one individual of either the one or the other parental species. The remaining 10 plots were selected such that five of them were dominated by one, and the other five by the other parental species, respectively. For each plot, the abundance of parental species and hybrids (classified as sensu lato, without discriminating further classes) was estimated as the percentage area covered by all the individuals of the respective taxon. Moreover, we collected a complete list of all vascular plant species occurring in the plots apart from the willows and took samples from the uppermost soil layer (c. 10 cm) directly below the parental and the hybrid individuals.
The soil samples were used to measure water retention capacity (WRC) as well as the actual and potential pH. For the assessment of WRC, 5 g of fresh soil were put into sealed syringes and soaked with distilled water for 2 h. After draining of excess water, samples were weighted. For measurement of soil dry weight, 1 g of fresh soil of each plot was dried at 80 °C for 24 h. The difference between water-saturated and dry masses of the soil samples was used to calculate WRC in percent. For the measurement of actual pH, an Eppendorf tube of soil was mixed with 1 ml of distilled water. For the assessment of potential pH, 1 ml 0.01 m aqueous CaCl2 was added to the same amount of soil. After 24 h, both were measured with a pH-meter (Sentron Europe BV, VD Leek, The Netherlands).
For an additional or alternative characterization of the site conditions of the sampling plots, respectively, we used the collected plant species lists to calculate so-called Ellenberg indicator values (Ellenberg et al., 1992). Ellenberg indicator values represent a nine-level ordinal classification of plants according to the position of their realized ecological niche along different environmental gradients. Such indicator values are mainly based on expert judgement but have been proven to be reliable and useful indicators of local-scale environmental conditions in many studies (Koerner et al., 1997; Diekmann, 2003; Duprè et al., 2010; Lenoir et al., 2013). Here, we calculated indicator values for temperature, moisture, soil pH and soil nitrogen content (T, F, R, N) for each sampling plot as an average of the indicator values of all plant species recorded in the respective plot.
To characterize the niches of the parental species and the hybrids along these abiotic gradients (WRC, pH, T, F, R, N), we used a kernel smoothing procedure adapted from Broennimann et al. (2012) by replacing occurrence frequencies in bins of environmental gradients by species' cover values on our plots. The approach uses the sampled data and a (Gaussian) kernel density function to (i) compute species densities along each abiotic gradient and (ii) to compute the densities of particular gradient values. Species density is then divided by gradient density to scale species distribution along the gradient by the availability of particular conditions (Broennimann et al., 2012). Based on these density functions, we then tested for niche overlap between each pair of taxa by means of the D-metric (Schöner's D, Warren et al., 2008), and subsequently used a permutation test to evaluate the significance of niche differences, that is, we randomly re-shuffled the cover values of both species in the pair across the plots 100 times and recalculated the D-metric. In doing so, we hence assessed if the two species of the pair are more different from each other than the two species distributed at random across the sampling plots. We accepted this to be the case if the observed D-value was in the lower 5%-quantile of the permuted D-values.
Finally, to analyze the niches of the parental species and the hybrids in a broader context, we did the same analyses for a larger data set that not only comprised the 58 plots from the Rhône Glacier but also additional 46 reference plots for each parental species (that is, 150 plots in total). The reference plots were randomly selected from all those containing either species in the Austrian Vegetation Database (Willner et al., 2012) and can hence be assumed to represent the current ecological distribution of each parental species. However, analyses for this larger sample could only be conducted for the gradients represented by Ellenberg indicator values as we had no WRC and pH-data available for the 92 additional plots. The tests for niche overlap were done with two different data sets. In one data set, all hybrids were pooled, in the second data set, the hybrids were divided in F1 hybrids and later generation hybrids, according to the results of the genetic analysis. Splitting of the data set reduced the sample size weakening the reliability of the results. Thus, we focused on the results of the data set with the pooled hybrids and added the results of the other data set to the supplementary information (Supplementary Figure S3 and S4, Supplementary Table S4 and S5).
Results
Genetic diversity and differentiation
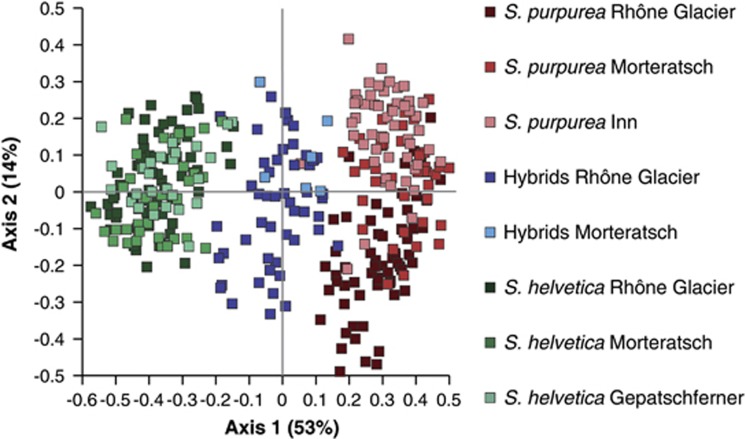
The population genetic analyses showed marked differences in the genetic diversity between the alpine species S. helvetica and the lowland species S. purpurea (Table 2). Overall, the effective number of alleles was significantly lower in S. purpurea than in S. helvetica (Tukey test after ANOVA, P<0.001) or the hybrids (Tukey test after ANOVA, P<0.01), whereas there was no difference in the effective number of alleles between S. helvetica and the hybrids. Moreover, the number of private alleles was lower in S. purpurea than in S. helvetica. S. helvetica had 34 private alleles in total so that each locus possessed at least one private allele. S. purpurea, in contrast, had only five private alleles that were located at five different loci. There were no unique alleles in the hybrids. Further, the observed heterozygosity (Ho) was smallest in S. purpurea, whereas it was highest for the hybrid populations. The Ho was significantly smaller in S. purpurea than in S. helvetica (Games–Howell test after ANOVA, P<0.01) or the hybrids (Games–Howell test after ANOVA, P<0.001). Nearly all purebred populations of both species showed significant deviations from Hardy–Weinberg equilibrium due to heterozygote deficiency (Table 2). Accordingly, FIS values were slightly positive in all purebred populations, whereas those in the hybrid populations were slightly negative. The FST values revealed a certain degree of genetic differentiation between the two parental species (mean=0.30). Accordingly, S. purpurea and S. helvetica form two well-separated clusters in the Principal Coordinate Analysis with hybrid individuals situated between the parental clusters (Figure 1). The intermediate position of the hybrid individuals is also the genetic confirmation that the hybrids are indeed crosses between S. purpurea and S. helvetica. The genetic differentiation between the hybrids and S. purpurea (mean FST=0.13) was quite similar to that between the hybrids and S. helvetica (mean FST=0.11). Although there was no differentiation among the populations of S. helvetica (mean FST=0.02) and among the hybrid populations (mean FST=0.03), there was a slight differentiation among the populations of S. purpurea (mean FST=0.11, Table 3). In contrast to S. helvetica, S. purpurea also showed a higher level of variation among populations (11.6%) in the analysis of molecular variance (Table 4). These differences also become apparent from the clustering of individuals in the Principal Coordinate Analysis. Although the S. helvetica individuals form a quite dense cluster, the S. purpurea individuals are scattered over a much wider range. Further, within the populations of S. purpurea, the populations of the Morteratsch and the Inn overlap to some extent, whereas the population of the Rhône Glacier seems to be relatively separated. The Mantel tests showed that at both glacier forefields there was no significant correlation between genetic and geographic distance of individuals of the purebred species (Rhône Glacier, r=−0.001, P>0.05; Morteratsch Glacier, r=0.029, P>0.05).
Table 2. Population genetic diversity measures of the willow populations based on nine microsatellite loci, including the mean number of alleles per locus (NA), allelic richness, the number of effective alleles per locus (Ne), observed heterozygosity (H o), expected heterozygosity (H e), the number of loci deviating from HWE in each population, the test for heterozygote deficiency in each population and inbreeding coefficient (F IS).
| Sampling location | Species | NA mean | Allelic richness | Ne mean | Ho | He | Loci deviating from HWE | Test het. deficit | FIS |
|---|---|---|---|---|---|---|---|---|---|
| Rhône Glacier | Salix purpurea | 4.1 | 3.3 | 2.3 | 0.488 | 0.515 | 2 | P<0.001 | 0.052 |
| Salix helvetica | 8.8 | 6.0 | 3.4 | 0.616 | 0.689 | 2 | P<0.001 | 0.104 | |
| Hybrid | 8.3 | 6.1 | 3.9 | 0.732 | 0.712 | 5 | P<0.05 | −0.028 | |
| Morteratsch Glacier | Salix purpurea | 3.2 | 2.9 | 1.9 | 0.402 | 0.441 | 1 | n.s. | 0.090 |
| Salix helvetica | 8.8 | 6.4 | 3.9 | 0.612 | 0.707 | 3 | P<0.001 | 0.134 | |
| Hybrid | 4.3 | n/a | 3.1 | 0.685 | 0.677 | 1 | n.s. | −0.014 | |
| Inn River | Salix purpurea | 4.8 | 3.8 | 2.4 | 0.479 | 0.518 | 2 | P<0.01 | 0.077 |
| Gepatschferner | Salix helvetica | 8.0 | 6.3 | 3.5 | 0.629 | 0.684 | 2 | P<0.01 | 0.074 |
Abbreviations: n/a, not available; n.s., non-significant.
Figure 1.
PCoA plot of purebred populations of S. purpurea (Inn) and S. helvetica (Gepatschferner), and two admixed populations containing hybrid individuals (Rhône Glacier, Morteratsch). Percentages of variance are given in brackets.
Table 3. Genetic differentiation among and between species collected at different sampling locations, given by pairwise FST values.
| Species | Sampling location |
Salix purpurea |
Salix helvetica |
Hybrid |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Rhône Gl. | Morteratsch | Inn River | Rhône Gl. | Morteratsch | Gepatschferner | Rhône Gl. | Morteratsch | ||
| Salix purpurea | Rhône Gl. | 0 | |||||||
| Morteratsch | 0.116 | 0 | |||||||
| Inn River | 0.136 | 0.077 | 0 | ||||||
| Salix helvetica | Rhône Gl. | 0.318 | 0.348 | 0.318 | 0 | ||||
| Morteratsch | 0.316 | 0.36 | 0.326 | 0.013 | 0 | ||||
| Gepatschferner | 0.325 | 0.364 | 0.318 | 0.022 | 0.021 | 0 | |||
| Hybrid | Rhône Gl. | 0.095 | 0.159 | 0.139 | 0.099 | 0.092 | 0.094 | 0 | |
| Morteratsch | 0.146 | 0.135 | 0.109 | 0.125 | 0.121 | 0.125 | 0.027 | 0 | |
Table 4. Analysis of molecular variance among and within populations of S. purpurea and S. helvetica from three sampling locations.
| Species | Source of variation | Sum of squares | Variance components | % variation |
|---|---|---|---|---|
| S. purpurea | Among populations | 56.797 | 0.262 | 11.635 |
| Within populations | 611.130 | 1.989 | 88.365 | |
| S. helvetica | Among populations | 15.399 | 0.058 | 1.821 |
| Within populations | 784.618 | 3.121 | 98.179 |
Simulations
In structure, all purebred individuals of S. purpurea were assigned to one group with q>0.97, whereas all purebred S. helvetica individuals were assigned to a second group with q>0.95. All simulated F1 and F2 hybrids were recognized as hybrids, whereas some of the simulated backcrosses were assigned to the parental categories. Consequently, a threshold value of 0.95 seems to ensure that no F1 and F2 individuals are assigned to the parental categories, but there may be some undetected backcrosses in the sample. Based on a threshold value of q>0.95 for the assignment to the parental categories, the mean accuracy for S. purpurea and S. helvetica was 0.99. Consequently, in the runs with the real data, there will be ~1% incorrectly assigned backcrosses among all individuals assigned to one of the parental categories. The overall performance of STRUCTURE for the simulated data set was 0.99 for S. purpurea and S. helvetica, and 0.97 for admixed individuals.
In NewHybrids, with a threshold value of 0.95 for the unequivocal assignment to one of the six categories, simulated F1 hybrids were detected in the data set with a mean efficiency of 0.65, and F2 hybrids with an efficiency of 0.30, whereas backcrosses were never correctly identified. Nevertheless, the mean accuracy was quite high with 0.97 for F1 hybrids and 1.00 for F2 hybrids. In many cases, F1 hybrids could not be assigned to this category with P>0.95, but yet P-values for the F1 category were higher (0.71<P<0.94) than for any other category in nearly all cases. Some of the backcrosses were assigned to the purebred categories, but in the most cases backcrosses and F2 hybrids could not be assigned to any category with high probability.
Population assignment in STRUCTURE
As expected owing to the presence of two species in the data set, the most likely number of populations was K=2 in each of the admixed populations of the Rhône Glacier (Figure 2a) and Morteratsch Glacier (Figure 3a). In the analysis containing only the purebred individuals of one species, the most likely number of populations was one in S. helvetica from the three different sampling locations, but two in S. purpurea from the three sampling locations. In the STRUCTURE analysis of the mixed populations, we used a threshold value of ⩾0.95 for the assignment of an individual to one of the purebred species, so that individuals with q<0.95 were regarded as admixed. In the admixed population of the Rhône Glacier, all 51 individuals that had been classified as hybrids based on their phenotype were also classified as hybrids in the STRUCTURE analysis. Of the 66 individuals that had been classified as S. helvetica, only 60 were clearly assigned to this group, whereas six had q-values ranging from 0.830 to 0.945. Finally, all 65 individuals classified as S. purpurea were assigned to this group in STRUCTURE. Of the 82 individuals sampled at the Morteratsch Glacier, 39 individuals could be clearly assigned to purebred S. purpurea (range of q 0.989–0.997), 37 individuals could be assigned to purebred S. helvetica (range of q 0.959–0.997) and 6 individuals had q-values below the threshold and were classified as hybrids. One individual that had been classified as S. purpurea in the field was reclassified as a hybrid in the genetic analysis. STRUCTURE was also run with a data set containing the purebred reference stands of S. purpurea from the Inn and of S. helvetica from the Gepatschferner. The two populations were clearly separated with minimum q-values of 0.973 for S. purpurea and 0.976 for S. helvetica.
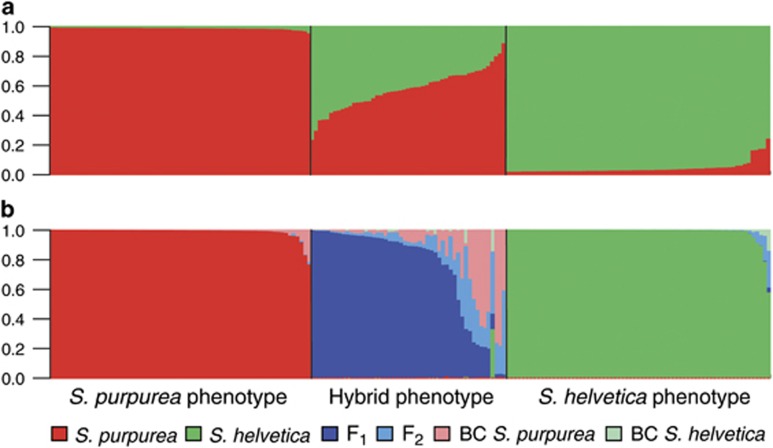
Figure 2.
Group assignment of the individuals sampled at the Rhône Glacier (N=188) in STRUCTURE (a) and NewHybrids (b). Each bar denotes one individual and the height of the color corresponds to the admixture proportion (qi) (a) or the posterior probability of the respective genotype frequency class (b). Individuals are sorted by descending q-values, so that the order is different in both charts.
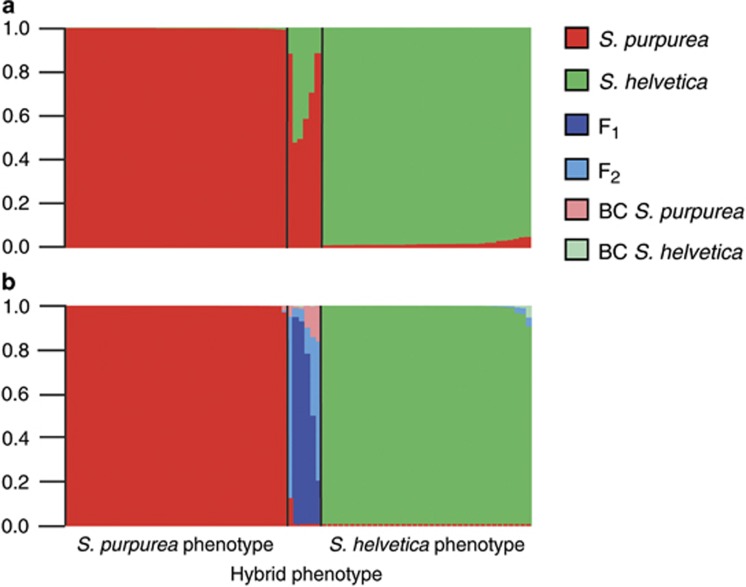
Figure 3.
Group assignment of the individuals sampled at the Morteratsch Glacier (N=82) in STRUCTURE (a) and NewHybrids (b). Each bar denotes one individual and the height of the color corresponds to the admixture proportion (qi) (a) or the posterior probability of the respective genotype frequency class (b). Individuals are sorted by descending q-values, so that the order is different in both charts.
Classification of individuals using NewHybrids and STRUCTURE
The results of STRUCTURE and NewHybrids for the population of the Rhône Glacier were almost congruent (Figure 2). The S. purpurea individuals were generally assigned to the purebred category with very high posterior probabilities (mean 0.99). Nevertheless, there were three outliers with a comparatively low probability of 0.764–0.912 of belonging to purebred S. purpurea. By rechecking the herbarium vouchers of these individuals, we indeed detected sparse hair at the ventral side on the leaves of one individual. This observation leads to the conclusion that this individual may be a backcross to S. purpurea, which was not detected as such by NewHybrids as predicted by the simulations. Nearly all S. helvetica individuals were also assigned to the purebred category with high probability (mean 0.99). Compared with the STRUCTURE results, there were only five individuals with a lower probability of being pure S. helvetica (range of p 0.574–0.945). The clear assignment of two of those individuals seems to be impeded by the fact that their genotypes comprise many alleles that are present in both species. In the genotypes of the remaining three individuals, there are one or two alleles that have a low frequency in S. helvetica but a high frequency in S. purpurea, so that it is not clear if the genotypes just contain rare alleles or if these individuals are indeed backcrosses. Of the 51 hybrids, 17 could be classified as F1 hybrids on the threshold of 0.95. The remaining 34 individuals could not be assigned to any of the six categories with high probability. At least, they had zero probability of belonging to one of the purebred categories, so that they can still be regarded as some kind of admixed individuals, probably F2 individuals or backcrosses. For the population at the Morteratsch, the group assignments of STRUCTURE were confirmed in nearly all cases in NewHybrids (Figure 3). Only one individual of S. helvetica was assigned to this group with a lower probability of 0.90. Of the six hybrid individuals, one could be assigned to the F1 category with high probability, another one could be assigned to this category on a relaxed level (P=0.93). Another individual had a quite high probability of being an F2 hybrid (P=0.83). The three remaining individuals could not be assigned to any of the six categories with high probability.
Ecology
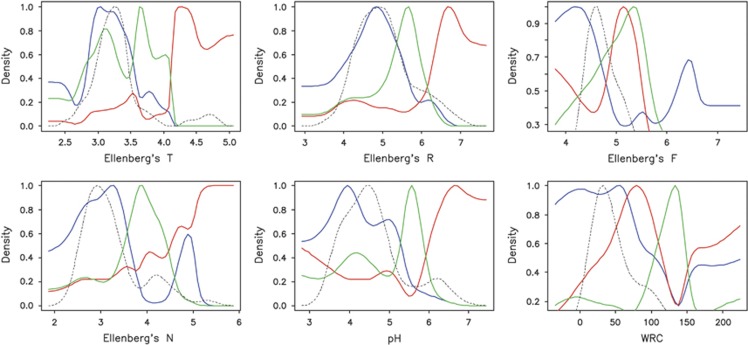
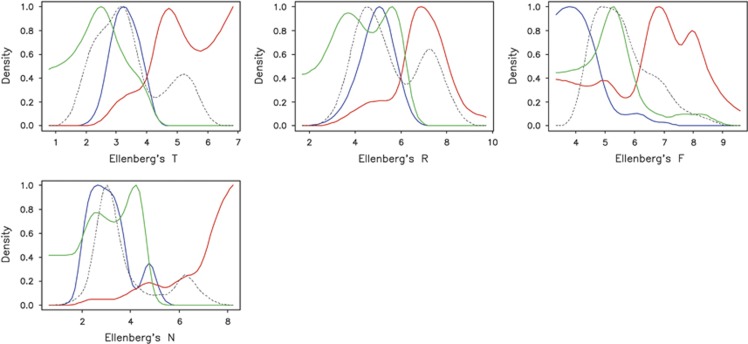
The abundance density plots along Ellenberg indicator value gradients showed that S. purpurea individuals occupied the warmest, most basic and most nutrient-rich sites, whereas S. helvetica individuals were occurring on sites with cool to intermediate temperature, intermediate nutrient supply and soil reaction values (Figure 4). Strikingly, along these gradients, the hybrid preferred the most extreme habitats on the Rhône Glacier forefield, which were scattered over the whole area in a mosaic-like pattern (Supplementary Figure S2). The situation was a bit different along Ellenberg's F value (moisture) where the parental species' niches were very similar and the hybrid seemed to have a bimodal distribution with the more pronounced peak at the dry margin of the gradient. The results along the measured pH and WRC gradients were very similar to those at the Ellenberg R and F gradients, demonstrating the validity of the indicator value approach. Interestingly, the density curves of the hybrid were closest to the densities of most environmental factors on the glacier forefield.
Figure 4.
Smoothed density of species abundances along six environmental gradients as calculated from data of 58 sampling plots at the Rhône Glacier forefield. Ellenberg's T, R, F and N values are indicator values of temperature, pH, soil moisture and nutrient availability as computed from the accompanying vegetation. The green, red and blue curves represent densities of S. helvetica, S. purpurea and the hybrid (pooled), respectively. The dashed curve represents the density of the respective environmental variable.
Permutation tests demonstrated that the apparent niche differences among the parental species were larger than random for T, R, N and pH. The hybrid was generally closer to S. helvetica, but still differed significantly from it along the nutrient availability (N) and WRC gradients. Conversely, the hybrid was significantly distinct from S. purpurea along all measured gradients except F and WRC (Supplementary Table S6).
In the analysis covering the current ecological distribution of the parental species, as represented by the larger sample, the results changed only slightly. Although the hybrid still occupied extreme conditions with respect to moisture (F) and nutrient availability (N), its position along the temperature (T) gradient now appeared intermediate between the parental species, and in respect to soil reaction (R) the hybrid was close to S. helvetica (Figure 5). Permutation tests again underpinned the significant difference between the parental species with respect to all tested niche dimensions. The hybrids were distinct from both parental species along the T gradient, whereas they are more similar to S. helvetica than to S. purpurea regarding all other environmental factors (R, F, N) (Supplementary Table S7).
Figure 5.
Smoothed density of species abundances along four environmental gradients as calculated from 58 sampling plots taken at the Rhône Glacier forefield plus 92 additional samples (46 containing S. helvetica and 46 containing S. purpurea) randomly selected from the Austrian vegetation database. Ellenberg's T, R, F and N values are indicator values of temperature, pH, soil moisture and nutrient availability as computed from the accompanying vegetation. The green, red and blue curves represent densities of S. helvetica, S. purpurea and the hybrid (pooled), respectively. The dashed curve represents the density of the respective environmental variable.
Discussion
Origin of the hybrid
Our genetic data confirm the hybrid nature of individuals with phenotypes intermediate between the subalpine species S. helvetica and the lowland species S. purpurea. Remarkably, such intermediate phenotypes have neither been mentioned by Buser (1940) for the Swiss Alps, nor by Lautenschlager-Fleury and Lautenschlager (1985) in their extensive survey of willow populations on the Rhône Glacier forefield. Further, a vegetation survey conducted between 1979 and 1982 at the Rhône Glacier forefield reported that the moraines were at that time still dominated by a herbaceous pioneer vegetation typical for alluvials (Epilobietum fleischeri community; Schubiger-Bossard, 1988). During this period, S. purpurea occurred only sporadically, and S. helvetica was rather restricted to older moraines and the flanking slopes. It can therefore be assumed that hybridization between these willow species is a recent development that has taken place on glacier forefields only over the last three decades, which were also characterized by a pronounced warming trend (Casty et al., 2005). Further, the genotypes of the hybrids just combined alleles of the parents and did not contain any private alleles. This pattern does neither support a scenario of mutational change over many generations, nor does it support the possibility of an earlier origin of the hybrid elsewhere and a secondary colonization of the forefields. We thus conclude that the two hybrid populations are probably not more than 20–30 years old, that they originated in situ, and that origin and establishment of those hybrids were facilitated by global warming and glacier retreat. This study therefore investigates genetic patterns and ecology of the first hybrid generations after their origin.
Composition of the hybrid zones
Despite their recent emergence, the hybrids have already established stable populations. The genetic analyses of two willow hybrid populations formed by crosses of S. purpurea and S. helvetica revealed similar patterns in both populations. Besides identifying F1 hybrids, we could also show the presence of later generation hybrids in the samples. We could not ascertain with high probability whether these later generation hybrids represent F2 hybrids or backcrosses owing to the low number of loci. However, test runs with simulated data sets revealed that individuals that cannot be assigned to a single category can at least be regarded as admixed individuals including F2 hybrids as well as backcrosses to each parental species. It can therefore be assumed that unassigned individuals hint at the presence of further later generation hybrids in a sample. Considering the short timespan since the onset of hybridization, it seems unlikely that many following generation hybrids (F3, F4 and so on) have been generated. Thus, the willow hybrid populations apparently comprise a mixture of F1 hybrids, F2 hybrids and backcrosses. Hybrid populations with a similar structure have also been discovered in Populus (Hersch-Green et al., 2014) and Ulmus (Brunet et al., 2013), among other tree species. By contrast, Milne et al. (2003) genotyped a Rhododendron hybrid population and found exclusively F1 hybrids that excluded other genotypes owing to habitat-mediated fitness advantage. With regard to the willow population, we conclude that on the glacier forefield no habitat-mediated selection acts against the establishment of later generation hybrids. In contrast, differentiation of microhabitats on the glacier forefield could facilitate establishment of different hybrid classes (see detailed discussion on ecology below). Lo (2010) found only F1 hybrids in a mangrove hybrid population but assumed strong postzygotic crossing barriers as the cause. Between the willow species, postzygotic barriers are apparently weak, as hybrids are fertile and do form later generations. The prezygotic crossing barriers readily broke down after secondary contact. In Salix, morphologically and ecologically differentiated species show only a shallow phylogenetic divergence (for example, Wu et al., 2015), which may facilitate rapid hybrid formation upon secondary contact. Indeed, S. purpurea and S. helvetica are genetically close though being placed in different sections of the genus (Skvortsov, 1999; Hörandl et al., 2012). Further, both parental species are diploid and share the same chromosome number (2n=38). Hybridization between willow species with different ploidy levels has of course been observed in artificial (Serapiglia et al., 2014) and natural hybrids (Hörandl et al., 2012). However, an equal ploidy level of the parents facilitates successful hybrid establishment in Salix (Hörandl et al., 2012). The strength of the reproductive barrier between the parental species determines the strength of the reproductive barrier between the hybrids and their parents (reviewed in Rieseberg, 1997). Thus, we expect the reproductive barrier between the willow hybrids and their parents to be weak, facilitating the frequent production of backcrosses. Buerkle et al. (2000) modeled the mechanisms underlying homoploid hybrid speciation and found that hybridization leads more often to introgression than to speciation. In his review, Rieseberg (1997) concluded that recombinational speciation was even possible with very low hybrid fertility but that outcrossing would delay the process of speciation. The willow hybrids seem to be fertile (Gramlich et al. in prep.) so that a further evolution of the hybrids could generally be possible. But owing to the obvious lack of crossing barriers and the spatial proximity to the parental species, introgression seems to be more likely than speciation. At the moment it is impossible to predict the real trajectory of the hybridization process because of the young age of the willow hybrid population and also because the population occurs in a constantly changing environment.
Yet, we have already discovered two hybrid populations that also showed similar patterns in the genetic analyses. A similar pattern in two populations is more likely caused by similar forces than by random processes (Hersch-Green et al., 2014). These findings imply that hybridization between S. purpurea and S. helvetica is a repeated process that could also occur at other locations in the Alps with the same outcome. The hybrid population at the Morteratsch Glacier was much smaller than that at the Rhône Glacier, which can be related to the different altitudes of the glacier forefields. The forefield of the Morteratsch Glacier is situated ~300 m higher than that of the Rhône Glacier, so that the processes of colonization and first hybrid formation are probably delayed owing to the higher altitude. S. purpurea occurred only sporadically at the Rhône Glacier in the early successional stages (Schubiger-Bossard, 1988), but today it can be found in high numbers. This suggests that in the future, with progressing global warming and succession, large numbers of hybrids between S. purpurea and S. helvetica could also be formed on other glacier forefields. In such hybrid zones, backcrossing could affect the gene pools of both species throughout the Alps.
Besides the hybrids, S. purpurea is also a relatively new colonizer found on glacier forefields. Our results show that the populations of S. purpurea on the glacier forefields exhibit a reduced number of alleles compared with the lowland population due to the founder effect. Heterozygosity, however, appears to be less affected, probably owing to enforced outcrossing in a dioecious species. Generally, the gene pool of S. purpurea on glacier forefields could be affected by two processes simultaneously: the reduction of genetic diversity due to colonization and the probable introgression of alleles owing to backcrossing with the hybrids. Introgression of alleles could restore the genetic diversity of S. purpurea on glacier forefields faster than genetic exchange with the source population or novel mutations. Hersch-Green et al. (2014) studied a Populus hybrid zone and found evidence for bidirectional gene flow between the parental species and hypothesized the occurrence of adaptive introgression. On the other hand, introgression can also have detrimental effects. In a study on hybridization between native and introduced elm species in Italy, Brunet et al. (2013) discovered that introgression altered the genetic structure of the native species. Whereas S. purpurea, which is a widespread lowland species, could benefit from introgression in its populations on glacier forefields, the gene pool of the alpine species S. helvetica could suffer irreversible changes by introgression of alien alleles. Intensive introgressive hybridization has been observed in a comparable study on hybridizing Senecio species along an elevational transect with the complete introgression of one parental species into the other one (Oberprieler et al., 2015).
The sampled S. purpurea populations also showed a slight genetic structuring, which is probably due to colonization from different source populations and a limitation of gene flow by the mountain chains. Actually, the Morteratsch Glacier valley belongs to the catchment area of the Inn River, and the connection of the valleys probably allowed gene flow between these willow populations. The Rhône Glacier, however, is connected to the valley of the river Rhône, which is separated from the Inn River by several mountain ridges. Thus, impeded gene flow between the populations at the two river systems led to a detectable genetic structuring within S. purpurea.
Ecology and evolution of hybrids
Despite its recent emergence, the hybrid population on the Rhône Glacier forefield already occupies ecologically distinct sites. The hybrid sites are spatially not concentrated, but appear scattered over the whole area of the alluvial plains (Supplementary Figure S2), in a mosaic-like spatial pattern. Our Mantel tests confirmed the lack of geographical structure on the forefield. The willow hybrids and their parental species occur evenly dispersed over the whole study area in a pattern of small-scale habitat differentiation and thus may constitute a mosaic hybrid zone, as described by Harrison (1986). This pattern is probably facilitated by the local availability of microhabitats, which is typical for alpine environments (Körner, 2003). Indeed, Little et al. (2015) found that nutrient availability varied greatly and unpredictably among alpine microhabitats. S. purpurea and S. helvetica tend to flower at the same time (pers. obs. of S. Gramlich and E. Hörandl) so that marked effects of microhabitat on phenology (Cortés et al., 2014) do not seem to occur. In the analysis of ecological data from this sampling area, the hybrids show different and partly transgressive ecological requirements compared with both parents concerning most of the assessed factors. With respect to soil pH, nutrients and temperature the hybrids occur under more extreme environmental conditions than the parental species. The ability to grow under more extreme conditions could be explained by formation of novel genotypes, either by transgressive segregation in later generation hybrids (Rieseberg and Willis, 2007) or by backcrossing that could already have produced some genotypes that can tolerate extreme conditions (Soltis and Soltis, 2009). The occupation of harsh sites could result from exclusion by the parents (especially S. helvetica) on the more favorable sites. The bimodal distribution of hybrids along the moisture and nutrient availability gradients is particularly suggestive in this respect as the hybrid obviously has a broad ecological tolerance but is nevertheless rare in intermediate sites where the parental species concentrate. Probably individuals of the parental species just arrived earlier on the glacier forefield and have already occupied the favorable spots before the first hybrid generation originated. Direct competition between individuals of the same age may have played a subordinate role in hybrid establishment on the glacier forefield because the open area still offers plenty of opportunities for settlement. Accordingly, most of the mixed populations were found on the alluvial sites, where still plenty of open ground is available, whereas on the adjacent slopes, which are already covered by other shrubs and tall-herbs, the willow stands were dominated by S. helvetica, whereas individuals of S. purpurea and the hybrid were absent. The exclusion of S. purpurea and hybrids from the slopes could be explained by the course of re-colonization (see Schubiger-Bossard, 1988) and does not seem to be due to facilitation of S. helvetica by the adjacent vegetation (Wheeler et al., 2015). The alpine species S. helvetica arrived earlier than the lowland species S. purpurea and could become established on the slopes when these were only scarcely vegetated. In general, the hybrid population seems to be well adapted to the conditions prevailing on the Rhône Glacier forefield as its density is closest to the predominant site conditions along most of the abiotic gradients.
Integrating the current ecological distribution of the parental species in the analysis, the optima of the parents differ considerably, especially with respect to temperature. This result is hardly surprising because S. purpurea occurs most frequently in lower altitudes, whereas S. helvetica is restricted to subalpine and alpine environments. In this broader perspective, the hybrid therefore takes an intermediate position along the temperature gradient, whereas on the Rhône Glacier forefield, where the realized temperature gradient is much narrower, it prefers the cooler sites, similar to S. helvetica. Temperature differences among the plots on the forefield of the Rhône Glacier could be due to local exposure of sites to freezing conditions, as differences or gradients of altitude within the mostly plain area of the forefield are negligible. It has been shown that freezing conditions vary greatly among alpine microhabitats and that they affect the performance of alpine shrub communities (Wheeler et al., 2014; Little et al., 2015). In the soil conditions, we found a differentiation of parents and hybrids in a patchy mosaic of microhabitats. In a study on two hybridizing Rhododendron species, Milne and Abbott (2008) found that the parents preferred acidic or alkaline soil, respectively, and that hybrid zones occurred on soil with intermediate pH. However, the authors also discovered that the soil in hybrid zones consisted of a mosaic of patches with varying pH values enabling the co-occurrence of both parental species and the hybrids similar as in our study.
Overall, the hybrid population on the Rhône Glacier forefield appears to be established and is able to coexist with the parental species via habitat differentiation. The formation of later generation hybrids, either via introgression or segregation, may have resulted in novel genotypes with allelic combinations that are favorable in the novel environment. Thus, our study seems to support the idea of ‘evolutionary novelty' by Arnold (1997). Considering continued glacier retreat and upwards moving of vegetation zones in the Alps, it can be envisioned that further sites at higher altitudes with ecological opportunities for the hybrids will become available in the near future, enabling them to expand their range and establish a stable lineage.
Conclusion
Our results suggest a recent origin of the hybrids between S. purpurea and S. helvetica in situ, which is due to secondary contact and the breakdown of ecological crossing barriers on glacier forefields. Despite the young age of the hybrid population, hybridization has already proceeded beyond the F1 generation. The formation of later generation hybrids was facilitated by weak pre- or postzygotic crossing barriers and the occurrence of parental species and hybrids in sympatry. Further, the glacier forefield, together with the fine-scale environmental variation typical for Alpine ecosystems, offers many unoccupied niches for hybrid establishment. The hybrids seem to have a broad ecological amplitude and are able to grow under more extreme conditions (regarding soil pH, moisture and nutrient supply) than either parental species enabling the coexistence of both parental species and the hybrids in a patchy habitat. Future research should use more genetic markers to achieve a better resolution of the post F1 hybrids. When the actual hybrid class is known, population genomic markers could be applied to identify segregation patterns, loci under selection and signatures of adaptation.
Data archiving
Microsatellite data set available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.dn53j.
Acknowledgments
We thank Anne-Caroline Cosendai, Ruth Flatscher and Jennifer Krüger for technical help, Gert Bachmann for assisting with pH and WRC measurements, and three anonymous reviewers whose comments improved the quality of the manuscript. The study was supported by a grant of the Austrian Academy of Sciences (ÖAW, Global Change Program), and by the Ursula Hofmann Foundation at the University of Göttingen (both to EH).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Anderson EC, Thompson EA. (2002). A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160: 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argus GW. (1973). The genus Salix in Alaska and the Yukon. National Museums of Canada, Publications in Botany 2.
- Argus GW. (1974). An experimental study of hybridization and pollination in Salix (willow). Can J Bot 52: 1613–1619. [Google Scholar]
- Arnold ML. (1997) Natural Hybridization and Evolution. Oxford University Press: Oxford, England. [Google Scholar]
- Arnold ML, Bennett BD. (1993) Natural hybridization in Louisiana irises: genetic variation and ecological determinants. In: Harrison RG (ed). Hybrid Zones and the Evolutionary Process. Oxford University Press: Oxford, England, pp 115–139. [Google Scholar]
- Barker J, Pahlich A, Trybush S, Edwards K, Karp A. (2003). Microsatellite markers for diverse Salix species. Mol Ecol Notes 3: 4–6. [Google Scholar]
- Barton NH. (2001). The role of hybridization in evolution. Mol Ecol 10: 551–568. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. (2015). Fitting linear mixed-effects models using lme4. J Stat Softw 67: 1–48. [Google Scholar]
- Broennimann O, Fitzpatrick MC, Pearman PB, Petitpierre B, Pellissier L, Yoccoz NG et al. (2012). Measuring ecological niche overlap from occurrence and spatial environmental data. Glob Ecol Biogeogr 21: 481–497. [Google Scholar]
- Brunet J, Zalapa JE, Pecori F, Santini A. (2013). Hybridization and introgression between the exotic Siberian elm, Ulmus pumila, and the native Field elm, U. minor, in Italy. Biol Invasions 15: 2717–2730. [Google Scholar]
- Buerkle CA, Morris RJ, Asmussen MA, Rieseberg LH. (2000). The likelihood of homoploid hybrid speciation. Heredity 84: 441–451. [DOI] [PubMed] [Google Scholar]
- Burga C. (1999). Vegetation development on the glacier forefield Morteratsch (Switzerland). Appl Veg Sci 2: 17–24. [Google Scholar]
- Burke J, Arnold M. (2001). Genetics and the fitness of hybrids. Annu Rev Genet 35: 31–52. [DOI] [PubMed] [Google Scholar]
- Buser R. (1940). Kritische Beiträge zur Kenntnis der schweizerischen Weiden (W Koch, Ed). Büchler.
- Cannone N, Diolaiuti G, Guglielmin M, Smiraglia C. (2008). Accelerating climate change impacts on alpine glacier forefield ecosystems in the European Alps. Ecol Appl 18: 637–648. [DOI] [PubMed] [Google Scholar]
- Casty C, Wanner H, Luterbacher J, Esper J, Böhm R. (2005). Temperature and precipitation variability in the European Alps since 1500. Int J Climatol 25: 1855–1880. [Google Scholar]
- Cortés AJ, Waeber S, Lexer C, Sedlacek J, Wheeler JA, van Kleunen M et al. (2014). Small-scale patterns in snowmelt timing affect gene flow and the distribution of genetic diversity in the alpine dwarf shrub Salix herbacea. Heredity 113: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann M. (2003). Species indicator values as an important tool in applied plant ecology - a review. Basic Appl Ecol 4: 493–506. [Google Scholar]
- Doyle J, Doyle JL. (1987). Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem Bull 19: 11–15. [Google Scholar]
- Duprè C, Stevens CJ, Ranke T, Bleeker A, Peppler-Lisbach C, Gowing DJG et al. (2010). Changes in species richness and composition in European acidic grasslands over the past 70 years: the contribution of cumulative atmospheric nitrogen deposition. Glob Change Biol 16: 344–357. [Google Scholar]
- Ellenberg H, Weber H, Düll R, Wirth V, Werner W, Paulissen D. (1992). Zeigerwerte von Pflanzen in Mitteleuropa. Scr Geobot 18: 1–248. [Google Scholar]
- Erschbamer B, Niederfriniger Schlag R, Winkler E. (2008). Colonization processes on a central Alpine glacier foreland. J Veg Sci 19: 855–862. [Google Scholar]
- Ersts PJ. (2009). Geographic Distance Matrix Generator (version 1.2.3). American Museum of Natural History, Center for Biodiversity and Conservation. Available from http://biodiversityinformatics.amnh.org/open_source/gdmg/. Accessed on 15 December 2015.
- Evanno G, Regnaut S, Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Gottfried M, Pauli H, Futschik A, Akhalkatsi M, Barančok P, Benito Alonso JL et al. (2012). Continent-wide response of mountain vegetation to climate change. Nat Clim Change 2: 111–115. [Google Scholar]
- Harrison RG. (1986). Pattern and process in a narrow hybrid zone. Heredity 56: 337–349. [Google Scholar]
- Harsch MA, Hulme PE, McGlone MS, Duncan RP. (2009). Are treelines advancing? A global meta-analysis of treeline response to climate warming. Ecol Lett 12: 1040–1049. [DOI] [PubMed] [Google Scholar]
- Hersch-Green EI, Allan GJ, Whitham TG. (2014). Genetic analysis of admixture and patterns of introgression in foundation cottonwood trees (Salicaceae) in southwestern Colorado, USA. Tree Genet Genomes 10: 527–539. [Google Scholar]
- Hörandl E, Florineth F, Hadacek F. (2012) Weiden in Österreich und angrenzenden Gebieten. 2nd edn, Arbeitsbereich Ingenieurbiologie und Landschaftsbau, Inst. für Landschaftsplanung und Ingenieurbiologie, Univ. für Bodenkultur: Wien, Austria. [Google Scholar]
- Karrenberg S, Kollmann J, Edwards PJ, Gurnell AM, Petts GE. (2003). Patterns in woody vegetation along the active zone of a near-natural Alpine river. Basic Appl Ecol 4: 157–166. [Google Scholar]
- Kelly AE, Goulden ML. (2008). Rapid shifts in plant distribution with recent climate change. Proc Natl Acad Sci USA 105: 11823–11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner W, Dupouey JL, Dambrine E, Benoît M. (1997). Influence of past land use on the vegetation and soils of present day forest in the Vosges mountains, France. J Ecol 85: 351–358. [Google Scholar]
- Körner C. (2003) Alpine plant life: functional plant ecology of high mountain ecosystems, 2nd edn, Springer: Berlin, Germany. [Google Scholar]
- Lautenschlager-Fleury D, Lautenschlager E. (1985). Der Gletschboden, ein Weidenparadies. Bauhinia 8: 89–98. [Google Scholar]
- Lenoir J, Graae BJ, Aarrestad PA, Alsos IG, Armbruster WS, Austrheim G et al. (2013). Local temperatures inferred from plant communities suggest strong spatial buffering of climate warming across Northern Europe. Glob Change Biol 19: 1470–1481. [DOI] [PubMed] [Google Scholar]
- Lenoir J, Gégout JC, Marquet PA, de Ruffray P, Brisse H. (2008). A significant upward shift in plant species optimum elevation during the 20th century. Science 320: 1768–1771. [DOI] [PubMed] [Google Scholar]
- Little CJ, Wheeler JA, Sedlacek J, Cortés AJ, Rixen C. (2015). Small-scale drivers: the importance of nutrient availability and snowmelt timing on performance of the alpine shrub Salix herbacea. Oecologia,. (2015) e-pub ahead of print 4 August 2015. [DOI] [PubMed]
- Lo EYY. (2010). Testing hybridization hypotheses and evaluating the evolutionary potential of hybrids in mangrove plant species. J Evol Biol 23: 2249–2261. [DOI] [PubMed] [Google Scholar]
- Mallet J. (2005). Hybridization as an invasion of the genome. Trends Ecol Evol 20: 229–237. [DOI] [PubMed] [Google Scholar]
- Mallet J. (2007). Hybrid speciation. Nature 446: 279–283. [DOI] [PubMed] [Google Scholar]
- Milne RI, Abbott RJ. (2008). Reproductive isolation among two interfertile Rhododendron species: low frequency of post-F1 hybrid genotypes in alpine hybrid zones. Mol Ecol 17: 1108–1121. [DOI] [PubMed] [Google Scholar]
- Milne RI, Terzioglu S, Abbott RJ. (2003). A hybrid zone dominated by fertile F1s: maintenance of species barriers in Rhododendron. Mol Ecol 12: 2719–2729. [DOI] [PubMed] [Google Scholar]
- Moreau M, Laffly D, Joly D, Brossard T. (2005). Analysis of plant colonization on an arctic moraine since the end of the Little Ice Age using remotely sensed data and a Bayesian approach. Remote Sens Environ 99: 244–253. [Google Scholar]
- Nielsen EE, Bach LA, Kotlicki P. (2006). Hybridlab (Version 1.0): a program for generating simulated hybrids from population samples. Mol Ecol Notes 6: 971–973. [Google Scholar]
- Nolte AW, Tautz D. (2010). Understanding the onset of hybrid speciation. Trends Genet 26: 54–58. [DOI] [PubMed] [Google Scholar]
- Oberprieler C, Dietz L, Harlander C, Heilmann J. (2013). Molecular and phytochemical evidence for the taxonomic integrity of Salix alba, S. fragilis, and their hybrid S. × rubens (Salicaceae) in mixed stands in SE Germany. Plant Syst Evol 299: 1107–1118. [Google Scholar]
- Oberprieler C, Heine G, Bässler C. (2015). Can divergent selection save the rare Senecio hercynicus from genetic swamping by its spreading congener S. ovatus (Compositae, Senecioneae)? Flora - Morphol Distrib Funct Ecol Plants 210: 47–59. [Google Scholar]
- Parmesan C, Yohe G. (2003). A globally coherent fingerprint of climate change impacts across natural systems. Nature 421: 37–42. [DOI] [PubMed] [Google Scholar]
- Paul F, Kääb A, Maisch M, Kellenberger T, Haeberli W. (2004). Rapid disintegration of Alpine glaciers observed with satellite data. Geophys Res Lett 31: 12–15. [Google Scholar]
- Pauli H, Gottfried M, Dullinger S, Abdaladze O, Akhalkatsi M, Benito Alonso JL et al. (2012). Recent plant diversity changes on Europe's mountain summits. Science 336: 353–355. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. (2006). GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. (2012). GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Raffl C, Mallaun M, Mayer R, Erschbamer B. (2006). Vegetation succession pattern and diversity changes in a glacier valley, Central Alps, Austria. Arctic, Antarct Alp Res 38: 421–428. [Google Scholar]
- Raymond M, Rousset F. (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249. [Google Scholar]
- Rieseberg LH. (1997). Hybrid origins of plant species. Annu Rev Ecol Syst 28: 359–389. [Google Scholar]
- Rieseberg LH, Willis JH. (2007). Plant speciation. Science 317: 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root TL, Price JT, Hall KR, Schneider SH, Rosenzweig C, Pounds JA. (2003). Fingerprints of global warming on wild animals and plants. Nature 421: 57–60. [DOI] [PubMed] [Google Scholar]
- Schubiger-Bossard CM. (1988). Die Vegetation des Rhonegletschervorfeldes, ihre Sukzession und naturräumliche Gliederung. Flück-Wirth.
- Sedlacek JF, Bossdorf O, Cortés AJ, Wheeler JA, van Kleunen M. (2014). What role do plant-soil interactions play in the habitat suitability and potential range expansion of the alpine dwarf shrub Salix herbacea? Basic Appl Ecol 15: 305–315. [Google Scholar]
- Serapiglia MJ, Gouker FE, Smart LB. (2014). Early selection of novel triploid hybrids of shrub willow with improved biomass yield relative to diploids. BMC Plant Biol 14: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvortsov AK. (1999) Willows of Russia and adjacent countries. University of Joensuu: Joensuu, Finland. [Google Scholar]
- Soltis PS, Soltis DE. (2009). The role of hybridization in plant speciation. Annu Rev Plant Biol 60: 561–588. [DOI] [PubMed] [Google Scholar]
- Stamati K, Blackie S, Brown JWS, Russell J. (2003). A set of polymorphic SSR loci for subarctic willow (Salix lanata, S. lapponum and S. herbacea). Mol Ecol Notes 3: 280–282. [Google Scholar]
- Stöcklin J, Bäumler E. (1996). Seed rain, seedling establishment and clonal growth strategies on a glacier foreland. J Veg Sci 7: 45–56. [Google Scholar]
- Szpiech ZA, Jakobsson M, Rosenberg NA. (2008). ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vähä J-P, Primmer CR. (2006). Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol Ecol 15: 63–72. [DOI] [PubMed] [Google Scholar]
- Walther G-R, Post E, Convey P, Menzel A, Parmesan C, Beebee TJC et al. (2002). Ecological responses to recent climate change. Nature 416: 389–395. [DOI] [PubMed] [Google Scholar]
- Warren DL, Glor RE, Turelli M. (2008). Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution 62: 2868–2883. [DOI] [PubMed] [Google Scholar]
- Wheeler JA, Hoch G, Cortés AJ, Sedlacek J, Wipf S, Rixen C. (2014). Increased spring freezing vulnerability for alpine shrubs under early snowmelt. Oecologia 175: 219–229. [DOI] [PubMed] [Google Scholar]
- Wheeler JA, Schnider F, Sedlacek J, Cortés AJ, Wipf S, Hoch G et al. (2015). With a little help from my friends: community facilitation increases performance in the dwarf shrub Salix herbacea. Basic Appl Ecol 16: 202–209. [Google Scholar]
- Willner W, Berg C, Heiselmayer P. (2012). Austrian vegetation database. Biodivers Ecol 4: 333. [Google Scholar]
- Wu J, Nyman T, Wang D-C, Argus GW, Yang Y-P, Chen J-H. (2015). Phylogeny of Salix subgenus Salix s.l. (Salicaceae): delimitation, biogeography, and reticulate evolution. BMC Evol Biol 15: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.