Abstract
Climatic differences across a taxon's range may be associated with specific bioenergetic demands and may result in genetics-based metabolic adaptation, particularly in aquatic ectothermic organisms that rely on heat exchange with the environment to regulate key physiological processes. Extending down the east coast of Australia, the Great Dividing Range (GDR) has a strong influence on climate and the evolutionary history of freshwater fish species. Despite the GDR acting as a strong contemporary barrier to fish movement, many species, and species with shared ancestries, are found on both sides of the GDR, indicative of historical dispersal events. We sequenced complete mitogenomes from the four extant species of the freshwater cod genus Maccullochella, two of which occur on the semi-arid, inland side of the GDR, and two on the mesic coastal side. We constructed a dated phylogeny and explored the relative influences of purifying and positive selection in the evolution of mitogenome divergence among species. Results supported mid- to late-Pleistocene divergence of Maccullochella across the GDR (220–710 thousand years ago), bringing forward previously reported dates. Against a background of pervasive purifying selection, we detected potentially functionally relevant fixed amino acid differences across the GDR. Although many amino acid differences between inland and coastal species may have become fixed under relaxed purifying selection in coastal environments rather than positive selection, there was evidence of episodic positive selection acting on specific codons in the Mary River coastal lineage, which has consistently experienced the warmest and least extreme climate in the genus.
Introduction
Evolution by natural selection has a fundamental role in shaping the geographic distribution of lineages (Avise, 2000). Understanding the distribution of genetic diversity across landscapes and patterns of local adaptation among and within lineages is an important component of effective conservation management, informing captive breeding programmes, translocations, ‘genetic rescue' and targeted application of conservation resources (Harrisson et al., 2014). Taxa that span heterogeneous landscapes and/or strong environmental gradients are commonly associated with strong patterns of genetic and/or phenotypic variation and are therefore good candidates for exploring local adaptation (Schoville et al., 2012).
An obvious potential driver of local adaptation is climate, because climatic differences across a taxon's range may be associated with specific bioenergetic demands and involve metabolic adaptations (Franks and Hoffmann, 2012). Because ectothermic organisms in aquatic systems rely on heat exchange with the environment to regulate many key physiological, developmental and behavioural processes (including metabolism), fish are highly sensitive to climatic and thermal conditions (Narum et al., 2013). Thermal regime is therefore considered a key determinant of fish species' distributions and acts as a strong selective agent (Caissie, 2006).
Extending down the east coast of Australia, the Great Dividing Range (GDR) is Australia's most significant mountain range (~3500 km in length, up to ~300 km in width and up to ~2228 m in height) and has had a key role in freshwater fish speciation in eastern Australia (Unmack, 2001) (Figure 1). The GDR has a strong influence on the eastern Australian climate, with topographic relief leading to a typically arid/semi-arid climate inland and more mesic conditions (mean annual precipitation: evaporation ratio of >0.4) along the coast (Byrne et al., 2011). Despite the GDR acting as a strong barrier to fish movement, many species, and species with recent shared ancestries, are found on both sides of the GDR (Unmack, 2001; Faulks et al., 2015). Because the GDR predates most freshwater fish radiations in Australia, faunal similarity across the range is thought to have occurred by dispersal across the range (for example, by river-capture events for species that diverged tens of millions of years ago, or by extreme floods in areas of low elevation for more recently diverged species) (Unmack, 2013; Faulks et al., 2015).
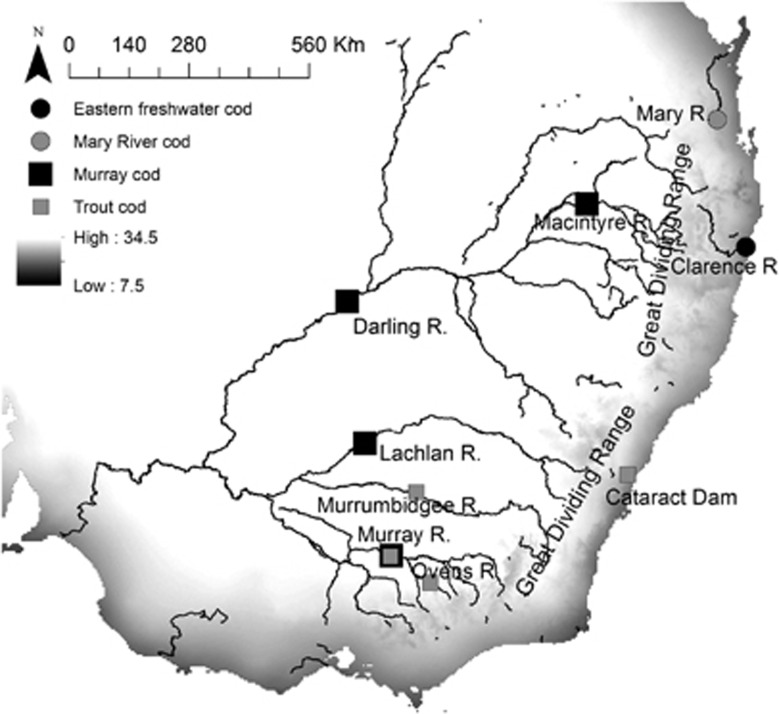
Figure 1.
Sampling locations for sequenced Maccullochella individuals across South Eastern Australia. Major rivers and the GDR are shown. Grey shading reflects the mean annual temperature range (7.5–34.5 °C).
Maccullochella is represented by four extant species of large, predatory freshwater cod in the Percichthyidae family: the Murray cod M. peelii, the trout cod M. macquariensis, the Mary River cod M. mariensis and the eastern freshwater cod M. ikei. Species of Maccullochella occur on both sides of the GDR. The Murray cod and trout cod are endemic to the Murray-Darling Basin on the inland side of the GDR and the eastern freshwater cod and Mary River cod persist in isolated coastal catchments to the east of the GDR (Figure 1) (Lintermans et al., 2005). All Maccullochella cod species have suffered severe declines in population size though the twentieth century because of a range of anthropogenic pressures including habitat loss and degradation, altered flow and temperature patterns, in-stream sedimentation, pollution, introduced fishes, over-fishing and population fragmentation due to in-stream barriers (Lintermans et al., 2005). The two coastal species, the Mary River and eastern freshwater cod, are each now restricted to single isolated drainage systems: the Mary River system in South Eastern Queensland and the Clarence River system in North Eastern New South Wales, respectively (Figure 1). Up until the early twentieth century, Maccullochella also occurred in intermediate drainages between the Clarence and Mary Rivers (predominantly in the Richmond and Brisbane Rivers) (Nock et al., 2010). The inland Murray cod is widespread, but the trout cod is restricted to a handful of locations in the southern parts of the Murray-Darling Basin (Lintermans et al., 2005). A stocked population of trout cod exists outside of its native range in Cataract Dam, east of the GDR (Figure 1).
Previous phylogenetic analyses suggest that the Mary River and eastern freshwater cod are sister taxa, and that the Murray cod is more closely related to the two coastal species than to the other inland congeneric, the trout cod (Nock et al., 2010; Lavoué et al., 2014). On the basis of data from two mitochondrial genes (ATPase 6 and ATPase 8), the two coastal species (eastern freshwater cod and Mary River cod) are thought to have diverged from each other around 300 (120–510) thousand years ago, following a crossing of the GDR by Maccullochella during the early- to mid-Pleistocene, around 1.09 (0.62–1.62) million years ago (Nock et al., 2010). The number and directionality of historical crossings of the GDR by Maccullochella are not clear, but a single crossing in the direction of west to east has previously been hypothesised on the basis of the fossil record (Rowland, 1993; Nock et al., 2010).
The mitochondrial genome, encoding 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs) and 13 structural proteins in vertebrates, has proved a tractable system for exploring adaptive evolution, particularly evolutionary adaptations related to metabolic function (da Fonseca et al., 2008). In coordination with the nuclear genome, the mitochondrial genome contributes substantially to metabolic performance, having a key role in energy and heat production through oxidative phosphorylation. There are five major protein complexes that contribute to the oxidative phosphorylation (OXPHOS) pathway in animals: complexes I, III, IV and V contain nuclear and mitochondrial encoded proteins, whereas complex II contains only nuclear encoded ones. Mitochondrial genes involved in the OXPHOS pathway can have important adaptive roles (for example, related to thermal adaptation and aerobic capacity) and be shaped by natural selection (da Fonseca et al., 2008; Garvin et al., 2014).
Because of its key role in energy production, the mitochondrial genome is under strong functional constraint (Meiklejohn et al., 2007). However, despite a background of strong purifying selection, particular sites in the mitochondrial genome can experience positive selection in response to environmental pressures. Many studies have reported positive selection acting on mitochondrial protein-coding genes in vertebrates in response to environmental factors, including in mammals (da Fonseca et al., 2008), birds (Stager et al., 2014; Morales et al., 2015) and fish (Silva et al., 2014). Many of these studies find that candidate sites for positive selection are disproportionately concentrated in the NADH complex genes (OXPHOS complex I), which produces ~40% of the proton flux required for ATP synthesis (Garvin et al., 2014).
In this study, we sequenced complete mitogenomes from the four extant species of Maccullochella and constructed a dated phylogeny to improve on previously reported divergence estimates based on two mitochondrial genes (Nock et al., 2010). We also explored the relative influences of purifying and positive selection in the evolutionary history of mitogenome divergence among species. Different climatic conditions exist east and west of the GDR, with the inland environment characterised by a highly variable arid or semi-arid climate and the coastal environment characterised by more mesic conditions (Figure 1; Table 1). We hypothesised that divergence of the two coastal Maccullochella species would be associated with evolutionary change in response to a different climate. In particular, we explored whether there was evidence of positive selection acting on mitogenomes in the two coastal species, potentially as a result of adaptation to warmer and more consistent climate and thermal regimes east of the GDR.
Table 1. Geographic coordinates for sampling locations and associated WORLDCLIM (Hijmans et al., 2005) climate data: maximum temperature of warmest month (°C), minimum temperature of coldest month (°C), mean annual precipitation (mm) and mean annual temperature range (°C).
| Region | Location | Latitude | Longitude | Max temp warmest month (°C) | Min temp coldest month (°C) | Precipitation (mm) | Annual temp range (°C) |
|---|---|---|---|---|---|---|---|
| Coastal | Clarence River | −29.49632a | 153.29391a | 27.2 | 8.3 | 36 | 18.9 |
| Coastal | Mary River | −26.83406a | 152.69057a | 27.7 | 5.6 | 45 | 22.0 |
| Coastal | Cataract Dam | −34.26566 | 150.81000 | 25.5 | 3.9 | 28 | 21.7 |
| Inland | Murrumbidgee River | −34.64000 | 146.37400 | 32.3 | 3.4 | 13 | 28.9 |
| Inland | Darling River | −30.64600 | 144.92600 | 35.2 | 5.0 | 27 | 30.2 |
| Inland | Lachlan River | −33.61600 | 145.30000 | 32.7 | 3.6 | 15 | 29.1 |
| Inland | Murray River | −35.96628 | 145.85256 | 31.2 | 3.4 | 17 | 27.8 |
| Inland | Macintyre River | −28.60233 | 149.96233 | 33.8 | 4.5 | 33 | 29.3 |
| Inland | Ovens River | −36.53400 | 146.66933 | 28.7 | 2.0 | 29 | 26.7 |
| Inland | Murray River | −35.98070 | 145.82780 | 31.2 | 3.4 | 16 | 27.8 |
Climate variables are derived from monthly temperature and rainfall data.
Geographic coordinates for the Clarence River and Mary River samples are approximate sampling coordinates as exact GIS locations were unavailable.
Methods
Sampling
Murray cod and trout cod samples were sourced from four and three different inland locations across the Murray-Darling Basin, respectively (Figure 1). Trout cod sampled from the Murrumbidgee and Ovens Rivers represent translocated fish originally of Murray River origin. Three additional trout cod samples were sourced from Cataract Dam, a coastal population outside the trout cod's natural range consisting of stocked fish (Figure 1). The translocations occurred too recently on evolutionary timescales (early twentieth century) for novel adaptive mutations to be likely to have occurred in Cataract Dam, and thus samples from Cataract Dam are classified as inland samples for the purposes of analyses. The specific interest of sampling these trout cod is that the dam was stocked in around 1916 during the ‘Nature's Waste' programme that sought to salvage fish impacted by severe drought: trout cod were translocated from the Murrumbidgee Catchment, where the species is now naturally extinct (Trueman, 2011). The two coastal Mary River cod samples were sourced from the Mary River and the three coastal eastern freshwater cod samples came from the Clarence River (Figure 1). Additional collection details for samples are provided in Supplementary material (Supplementary Table S1). The complete mitogenome sequence of the nightfish Bostockia porosa (sister group to Maccullochella; Lavoué et al., 2014) (Genbank accession AP014529) was used as an outgroup in phylogenetic analyses.
Climatic differences across the GDR
Different climatic conditions exist east and west of the GDR. The Mary River and Clarence River systems are classified as humid subtropical (Peel et al., 2007). In contrast, the majority of the Murray-Darling Basin is considered arid or semi-arid and is characterised by a highly variable climate (Peel et al., 2007). Climate data were downloaded from WorldClim (Hijmans et al., 2005) and visualised in ArcGIS 10.2. Relative to inland locations, coastal locations were associated with narrower annual temperature ranges, lower maximum temperatures in the hottest month, higher minimum temperatures in the coldest month and higher annual precipitation (Figure 1; Table 1).
Mitogenome sequencing and annotations
Genomic DNA was extracted from fin clips using the Qiagen Blood & Tissue Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Purified DNA was physically fragmented to 800 bp using a Covaris220 (Covaris, Woburn, MA, USA) and prepped using the NEXTflex DNA-seq library preparation kit (Bioo Scientific, Houston, TX, USA). The libraries were quantified, normalised, pooled and sequenced on the MiSeq Desktop Sequencer (2 × 250 bp read-length configuration) located at the Monash University Malaysia Genomics Facility. The raw paired-end reads were interleaved and assembled using the MITObim V 1.6 wrapper script (Hahn et al., 2013), which relies on the MIRA 3.4.1.1.assembler for mitochondrial baiting and iterative mapping (Chevreux et al., 1999). The consensus sequence for each assembled mitogenome was defined using the standard MIRA algorithm (with default settings), which accepts or refuses reads on the basis of quality score, number of reads, presence of forward and reverse reads, and any potential inexplicable errors introduced into the consensus (see Supplementary Data 1 in Wilson et al., 2012 for further details). The assembled mitogenomes were subsequently annotated using MITOannotator (http://mitofish.aori.u-tokyo.ac.jp/) (Iwasaki et al., 2013). As crude quality control for potential presence of nuclear pseudogene in the assembly, each mitogenome annotated by MITOannotator was inspected for missing genes and abnormal gene length (because of the presence of early stop codons). Mitogenomes were aligned and manually checked using GENEIOUS 6.1.3 (http://www.geneious.com/). Annotated genomes were submitted to Genbank (accession numbers: KT337324-KT337338).
Genetic diversity and divergence
The number of haplotypes (H), haplotype diversity (Hd) and nucleotide diversity (π) were calculated in DnaSP v.5 (Librado and Rozas, 2009) for each of the 13 protein-coding genes, tRNAs and rRNAs in each species. For each of the protein-coding genes, the number of fixed synonymous and non-synonymous mutations between the species was calculated in DnaSP. Between-lineage p-distances were calculated in MEGA 6.06 (Tamura et al., 2011). Consistent with Nock et al. (2010), we were unable to align the control region (CR) of trout cod with the other three species and so the CR was removed from all analyses.
Phylogenetic analysis
Bayesian phylogenetic analysis (MrBayes) was performed on the concatenated alignment of the 13 protein-coding genes to generate the phylogeny required for input into subsequent codon-based tests for selection. Gaps, intergenic spacers and stop codons were removed from the alignment and overlapping regions between ATPase 8 and ATPase 6 and between ND4L and ND4 were resolved by repeating the overlapping bases to ensure each gene was fully represented and to prevent disruption to the reading frame. ND6 is encoded on the light strand, so for consistency, the reverse-complemented sequence was used in phylogenetic reconstruction. We identified the optimal partitioning scheme, and the optimal models of molecular evolution for each subset partition, using the Bayesian Information Criterion (BIC) as implemented in PartitionFinder (Lanfear et al., 2012) for both the protein-coding dataset and complete mitogenomes (excluding control region). Bayesian phylogenetic reconstruction analysis was then conducted in MrBayes 3.2 (Ronquist et al., 2012). The analysis was run for 10 million generations sampled every 1000 generations with four independent runs to assess convergence. Run convergence was checked using TRACER 1.6.0 (Rambaut and Drummond, 2007). Trees were summarised using the 50% majority rule method after discarding the first 25% of the sample as burnin and visualised using FIGTREE 1.4.2 (Rambaut, 2007). The MrBayes consensus tree with the outgroup removed was used in codon-based selection analyses described below (results were robust to inclusion/exclusion of outgroup). We also inferred amino acid sequences at the ancestral node preceding the split between trout cod and the other three species using the method implemented in CODEML, under the phylogeny obtained from the analysis described above.
Divergence time estimates
We estimated divergence times between Maccullochella cod species using BEAST 1.8.2 (Drummond et al., 2012). Analyses were run using complete mitogenomes (excluding the control region) and the nine partitions determined using PartitionFinder (Supplementary Table S3), with unlinked substitution and clock models but linked tree topologies and applying the Yule process (appropriate for species divergence estimates) starting with a random tree. Because the Yule process assumes each operational taxonomic unit is a different species, inclusion of multiple individuals per species may affect estimates of divergence dates. We therefore ran the BEAST analysis including only one individual per species. Because inclusion of codons under positive selection may bias divergence estimates, we also removed putatively selected codons from the analysed dataset. Clock rates were chosen to encompass a range of mitochondrial time-dependent rates for comparably recent (<3 million years) divergences of galaxiids and other fishes, calibrated on the basis of geologically dated changes in river drainages and isolation events (0.003–0.13 substitutions/site/million years; Burridge et al., 2008). To account for uncertainty in clock rates, we sampled clock rates from a distribution rather than setting fixed rates. We assigned partitions either a ‘slow' or ‘fast' clock rate distribution on the basis of typical patterns of rate variation among positions. The clock rate was sampled from a log-normal distribution with mean (in real time)=0.01 substitutions/site/million years, log standard deviation=0.8 and starting value=0.01 for partitions 1–4, 6 and 8 (‘slow' clock; 1st and 2nd codon positions, tRNAs and rRNAs; Supplementary Table S3), and mean=0.04, log sd=0.8 and starting value=0.04 for partitions 5 and 7 (‘fast' clock; 3rd codon positions; Supplementary Table S3). The prior distribution was truncated to a maximum of 0.5 substitutions/site/million years (that is, the pedigree rate of control region in humans; Santos et al., 2005). The ‘slow' and ‘fast' clock priors corresponded to 95% of molecular clock rates being sampled between 0.002 and 0.04 substitutions/site/million years and between 0.006 and 0.14 substitutions/site/million years, respectively. To check for possible rate heterogeneity among lineages, we initially ran the analysis using an uncorrelated log-normal relaxed clock model. The initial analysis was run for 100 million generations and sampled every 10 000 steps. Because the ucld.stdev parameter estimates for all partitions included zero and/or were not greater than one, rate heterogeneity among branches was rejected and the final analysis was run using a strict clock model (Supplementary Table S4). For the final analysis, five replicate runs were checked for convergence, combined and analysed in Tracer 1.6.0 after discarding the first 10% of samples in each run as burnin.
Codon-based approaches to detecting positive selection associated with divergence across the GDR
Estimating codon-based ω ratios
The ratio of non-synonymous to synonymous substitutions (ω=dN/dS) is a measure widely used to detect selection, with ω>1 indicative of positive selection, ω<1 indicative of purifying selection and ω=1 indicative of neutrality (Yang, 2007). Because strong purifying selection acts to preserve mitochondrial function and can mask positive selection acting on individual sites, we used phylogeny-based models implemented as described below, to estimate ω=dN/dS for each codon rather than for entire genes/genomes (Meiklejohn et al., 2007; da Fonseca et al., 2008).
We used CODEML implemented in PAML4 (Yang, 2007) to run a series of maximum likelihood models and subsequently made model comparisons designed to test for signatures of positive selection associated with particular codons. First, we ran the recommended six M-series site models (M0, M1a, M2a, M7, M8; Table 2) to examine whether there was selection acting on particular codons independent of phylogenetic branch. We compared M0 (one ω ratio) with M1a (nearly neutral; two classes ω ratios: ω0 <1 and ω1=1) to test for evidence of non-neutrality, and compared M1a with M2a (adds a third class of ω ratios to M1a: ω>1) and M7 (beta) with M8 (beta and ω) to test for evidence of positive selection (Yang, 2007) (Table 2). Second, because different lineages may have experienced different selection regimes (for example, inland vs coastal species) we ran the branch model (Model 2). We compared the two-ω branch model (allows different ω for coastal and inland) with M0 as a test for different ω values on the coastal and inland branches. To complement the branch model comparisons, we also used GA-branch model selection implemented in the web-server Datamonkey (Delport et al., 2010). In contrast to CODEML models, GA-branch does not require a priori selection of branches of interest, but instead uses a model selection approach to partition and estimate ω values across the tree. Third, we ran branch-site-model-A-modified in CODEML to test for episodic selection affecting particular sites in particular lineages. In branch-site-model-A-modified, background lineages (defined as inland here) are restricted to ω values⩽1 and the focal foreground branches (defined as coastal here) are allowed ω values>1. A comparison of branch-site-model-A-modified with M1a tests for positive selection in the foreground (coastal) lineages, however, this comparison is unable to distinguish between positive selection and relaxation of purifying selection (Zhang et al., 2005). The branch-site-model-A-modified was thus compared with the null model (in which ω is fixed at 1) as a more stringent test of positive selection as outlined in Zhang et al. (2005). All model comparisons were made using a likelihood ratio test.
Table 2. CODEML model comparisons and associated hypothesis that is being tested are described.
| Model comparison | Log ratio test | Testing for |
|---|---|---|
| Model 0 vs Model 1a | χ2=40.04, P<0.05 | Non-neutrality |
| Model 1a vs Model 2a | χ2=0, NS | Positive selection |
| Model M7 vs Model M8 | χ2=1.43, NS | Positive selection |
| Model 0 vs branch model 2 | χ2=20.55, P<0.001 | Higher ω ratios in pre-specified foreground branches (coastal branches here) |
| MAnull vs branch-sites-model-A-modified | χ2=2327.9, P<0.001 | Positive selection or relaxation of selective constraint affecting certain sites in pre-specified foreground branches (coastal branches here) |
| Model 1a vs branch-sites-model-A-modified | χ2=18.99, P<0.001 | Positive selection (if ω 2a and/or ω2b>1 in foreground), or relaxation of selective constraint (if ω 2a and/or ω2b⩽1 in foreground) affecting certain sites in pre-specified foreground branches (coastal branches here) |
Bold font indicates a significant (P<0.05) log ratio test and support for the stated hypothesis.
In addition to CODEML models, we used the programme HyPhy implemented in the web-server Datamonkey (Delport et al., 2010) to identify codons putatively under selection using Fast Unconstrained Bayesian AppRoximation (FUBAR; Murrell et al., 2013) and Mixed Effects Model of Evolution (MEME; Murrell et al., 2012). FUBAR assigns each codon a posterior probability (PP) of belonging to three classes of ω: ω0 <1, ω1=1 and ω0 >1. Codons with PP>0.9 and ω >1 or ω <1 were inferred to have evolved under positive and purifying selection, respectively. Whilst FUBAR identifies codons putatively under selection across all branches on the phylogeny, MEME allows ω at each codon to vary across branches/lineages and so detects episodic selection (that is, positive selection that varies temporally across branches/lineages). Codons with P<0.05 were considered to have experienced episodic positive selection in certain lineages.
Detecting radical changes in physicochemical properties of amino acids
Because ω ratios >1 may not reflect molecular adaptation per se, we used TREESAAP (Woolley et al., 2003) to look for significant changes in physicochemical properties associated with amino acid replacements in inferred proteins on particular branches in the phylogeny. TREESAAP uses a sliding window approach to compare observed patterns of amino acid replacements and changes in physiochemical properties to those expected under neutrality (Woolley et al., 2003). Amino acid replacements that have strong effects on protein biochemistry are considered candidates for selection (Woolley et al., 2003). We ran the analysis with a sliding window size of 20 codons, increasing in 1-codon increments. To minimise false positives, only amino acid replacements with magnitude categories ⩾6 and z-scores above/below +/−3.09 (P<0.001) were considered as potentially under positive/purifying selection, respectively (McClellan and Ellison, 2010). We only considered amino acid properties with an accuracy of detecting selection >85% (thus, excluding 11 out of the 31 properties tested) (McClellan and Ellison, 2010).
Results
Per-base coverage varied across samples (mean per base coverage=121; range=25–336) and complete mitogenome sequence lengths varied among species: eastern freshwater cod and Mary River cod: 16 437 bp, Murray cod: 16 442 bp and trout cod: 16 479 bp (Supplementary Table S2). Two of the eastern freshwater cod samples (EC002 and EC806) had identical mitogenome sequences (including the CR), so EC806 was removed from analyses. We detected no gene-order rearrangements in any lineage. All Maccullochella cod species had an additional amino acid towards the 3′-end of ND1 and two additional amino acids in ND5, relative to the outgroup sequence B. porosa. Conversely, B. porosa had an additional amino acid at the 3′-end of ND5 and two additional amino acids at the 3′-end of Cytb. The B. porosa outgroup mitogenome sequence was >25% divergent from the Maccullochella cod species.
Genetic diversity and divergence
Within-species nucleotide diversity (π) was low for all the Maccullochella cod species: Murray cod=0.0004, trout cod=0.0003, Mary River cod=0.0003, eastern freshwater cod=0.0001 (Supplementary Figure S1). The majority of within-species diversity was located in protein-coding genes for all species, with no variation at any of the tRNAs or rRNAs in the two coastal species (Supplementary Figure S1).
Mean uncorrected p-distances for the complete mitochondrial genomes (excluding control region) between species were: 12.0% for Murray cod vs trout cod, 1.1% for Murray cod vs Mary River cod, 1.2% for Murray cod vs eastern freshwater cod and 0.04% for Mary River cod vs eastern freshwater cod. There were 13 fixed non-synonymous differences between the coastal (Mary River cod and eastern freshwater cod) and inland (Murray cod and trout cod) lineages (Table 3). In 11 of the 13 instances of non-synonymous fixed differences between inland and coastal species, the same amino acid was maintained in both inland species (Table 3). In addition, there were fixed differences at 12 nucleotide sites in the rRNAs and tRNAs: 12S rRNA (N=1), 16S rRNA (N=7), tRNA-Val (N=2), tRNA Met (N=1) and tRNA-Arg (N=1) (Supplementary Figure S2).
Table 3. Fixed amino acid differences between coastal (eastern freshwater cod and Mary River cod) and inland (Murray cod and trout cod) Maccullochella species.
| OXPHOS complex | Codon | Gene | Ancestral state |
Inland |
Coastal |
TREESAP Significant properties (P<0.001, categories 6–8) | ||
|---|---|---|---|---|---|---|---|---|
| Murray | Trout | Mary River | Eastern freshwater | |||||
| I | 252 | ND1 | S | S | S | L | L | ↑ RF |
| I | 62 | ND2 | V | V | V | I | I | ↓pK' |
| I | 243 | ND2 | L | L | L | F | F | — |
| I | 293 | ND2 | V | V | V | I | I | ↓pK' |
| V | 14 | ATPase 6 | F | F | F | L | L | — |
| V | 48 | ATPase 6 | S | S | S | N | N | — |
| IV | 40 | COIII | M | M | M | T | T | ↓Pα |
| IV | 168 | COIII | V | V | I | A | A | — |
| I | 108 | ND3 | T | T | T | M | M | ↓Pα |
| I | 68 | ND4 | T | T | T | I | I | — |
| I | 357 | ND4 | A | A | A | T | T | ↓Pα |
| I | 395 | ND4 | T | T | T | A | A | ↓Pα |
| I | 448 | ND5 | H | S | H | F | F | — |
Standard amino acid codes are given for each species and the inferred ancestral state (reconstructed using CODEML). Codon number refers to codon number in gene. Bold font indicates properties with evidence of positive selection based on global Z scores. TREESAAP codes for properties are: RF=Chromatographic index, pK'=Equilibrium Constant – ionisation, COOH, Pα=α−helical tendencies. Increase or decrease in amino acid properties from inland to coast is indicated with up and down arrows.
Phylogenetic analysis
PartitionFinder supported 9 partitions for the complete mitochondrial genome sequences (excluding control region), which were used in the BEAST analysis. Eight partitions were supported for the protein-coding dataset, which were subsequently used for phylogeny estimation (the same phylogeny was supported for the complete mitochondrial sequences, but we present only results from the protein-coding dataset because downstream codon-based tests for selection were based on protein-coding sequence) (Supplementary Table S3). All partitions were considered informative for differences between the three closely related cod species (Murray cod, Mary River cod and eastern freshwater cod; Supplementary Table S3). All four cod species were monophyletic and well supported (PP=1.0; Figure 2).
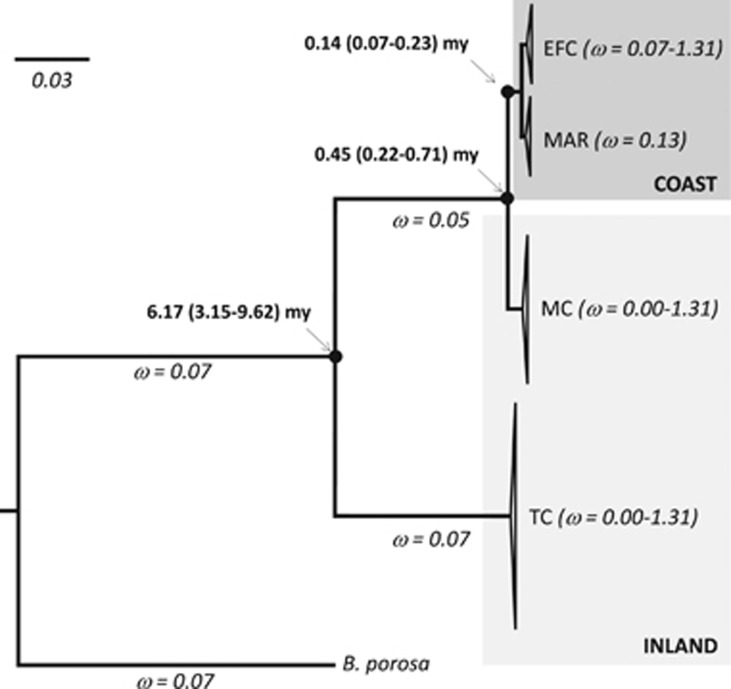
Figure 2.
Rooted phylogeny of the Maccullochella genus based on 13 protein-coding mitochondrial gene sequences, estimated using Bayesian inference in MrBayes. All nodes were well supported (PP=1.0). BEAST 95% HPD estimates for time to most recent common ancestor (in million years) for nodes indicated by black circles are given. Ratios of non-synonymous to synonymous substitutions (ω values) inferred from the branch partitioning scheme chosen through GA-branch model selection are indicated in italics below each branch and in parentheses following species labels for species branches and tips. Very short branches within-species clades have been collapsed for better presentation; triangle height corresponds to number of sequences and triangle width corresponds to diversity within clade. Species codes are: Murray cod (MC, N=4), trout cod (TC, N=6), Mary River cod (MAR, N=2) and eastern freshwater cod (EC, N=2). The outgroup is the nightfish Bostockia porosa.
Divergence time estimates
Posterior estimates of substitution rates ranged from 0.02 (95% HPD 0.01–0.03) substitutions per site per million years for partition 4 (2nd codon positions) to 0.04 (0.02–0.07) substitutions per site per million years for partition 9 (1st codon position for ND6; Supplementary Tables S3 and S4). On the basis of complete mitochondrial genomes excluding control region and the 11 putatively positively selected sites, the mean estimate of time to the most recent common ancestor of all four Maccullochella species was 6.17 (3.15–9.62) million years. The most recent common ancestor of Murray cod and the coastal cod species (Mary River cod and eastern freshwater cod) was 450 (220–710) thousand years and the most recent common ancestor of Mary River cod and eastern freshwater cod was 140 (68–230) thousand years (Figure 2; Supplementary Table S3).
Detecting positive selection associated with divergence across the GDR
Variation in codon-based ω ratios
CODEML site models found support for non-neutrality (model M1a), but no evidence of positive selection (models 2a or 8) (Table 2 and Supplementary Table S6). Under the supported non-neutral model (M1a), 97% of codons (p0) were estimated as being under purifying selection independent of phylogenetic branch (with the remaining 3% behaving neutrally) (Supplementary Table S6). Model selection in GA-Branch supported five different ω values along the tree (Figure 2). Although the majority of inland and coastal branches had ω<1, indicative of purifying selection, the coastal branches were generally associated with higher ω values than the inland branches (Figure 2). Higher ω ratios associated with coastal species compared with inland species was also supported by both CODEML branch and branch-site models. CODEML branch models estimated higher ω ratios in coastal species (ω=0.16) compared with inland species (ω=0.05) (model 2; Table 2 and Supplementary Table S6). The branch-site model found support for variable ω across codons and branches, with higher ω ratios associated with 13.5% of sites in foreground (coastal) branches compared with background (inland) branches (branch-sites-model-A-modified; Table 2 and Supplementary Table S6). Because the estimated values of ω2a and ω2b were <1 in the foreground branches, support for the branch-site-model-A-modified was consistent with relaxation of purifying selection in the foreground (coastal) branches rather than positive selection (Table 2 and Supplementary Table S6).
FUBAR supported strong, pervasive purifying selection acting across all mitochondrial protein-coding genes independent of phylogenetic branch, finding 79.6% of codons showed evidence of purifying selection at posterior probability ⩾0.9 and no codons with evidence of pervasive diversifying selection at posterior probability ⩾0.9 (results not shown).
MEME found evidence of episodic diversifying selection at 11 codons (P<0.05) (Table 4). Seven of these 11 codons were associated with the coastal lineages, 6 of these 7 with divergence of the Mary River cod (Table 4). Four of the seven sites showing evidence of episodic diversifying selection in the coastal lineages were located in OXPHOS complex I and three in OXPHOS complex IV (Table 4).
Table 4. Codons with evidence of episodic positive selection detected by MEME.
| OXPHOS complex | Codon | Gene | Ancestral state |
Inland |
Coastal |
Sample/species (location) affected | MEME P-value | TREESAP Significant properties (P<0.001, categories 6–8) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Murray | Trout | Mary River | Eastern freshwater | |||||||
| I | 261 | ND1 | T | T | T | T | T/Aa | EC002 (Clarence R.) | 0.013 | ↑Pα |
| I | 314 | ND1 | P | P | P | Q | P | MAR (Mary R.) | 0.030 | — |
| IV | 88 | COI | G | G/Va | G | G | G | MCC0105 (Darling R.) | 0.009 | ↓P |
| IV | 366 | COI | V | V | V | I | V | MAR (Mary R.) | 0.046 | ↓pK' |
| IV | 21 | COII | I | I | I | I/La | I | MAR001 (Mary R.) | 0.046 | ↑pK' |
| IV | 107 | COIII | A | A | A | T | A | MAR (Mary R.) | 0.047 | ↓Pα |
| I | 37 | ND5 | A | A/Da | A | A | A | MCC0105 (Darling R.) | 0.008 | ↓Et |
| I | 357 | ND5 | N | N | N/S | N | N | TC_B5 (Ovens R.) | 0.013 | — |
| I | 528 | ND5 | H | H | H | Y | H | MAR (Mary R.) | 0.049 | — |
| III | 80 | Cytb | R | R/Qa | R | R | R | MCC0105 (Darling R.) | 0.013 | — |
| I | 170 | ND6 | L | L | L | I | L | MAR (Mary R.) | 0.026 | ↓pK' |
Abbreviation: MEME, Mixed Effects Model of Evolution. Codon number refers to codon number in gene. OXPHOS complex and gene for each codon, the amino acids present in each of the four cod species and the sample ID/species associated with the amino acid replacement are given. Standard amino acid codes are given for each species and the inferred ancestral state (reconstructed using CODEML). Species codes in sample IDs are: Murray cod (MCC), trout cod (TC), Mary River cod (MAR) and eastern freshwater cod (EC).
MEME P-values are given for codons identified as under episodic positive selection. Significant (P<0.001, categories 6–8) TREESAAP properties associated with the amino acid replacements are given, and the directions of change are indicated. Bold font indicates properties with evidence of positive selection based on global Z scores. TREESAAP codes for properties are: P=Turn tendencies, Et=Total non-bonded energy, pK'=Equilibrium Constant – ionisation, COOH, Pα=α−helical tendencies. Amino acid replacements affecting coastal lineages are shaded.
Indicates only one individual has amino acid replacement.
Radical changes in the physicochemical properties of amino acids
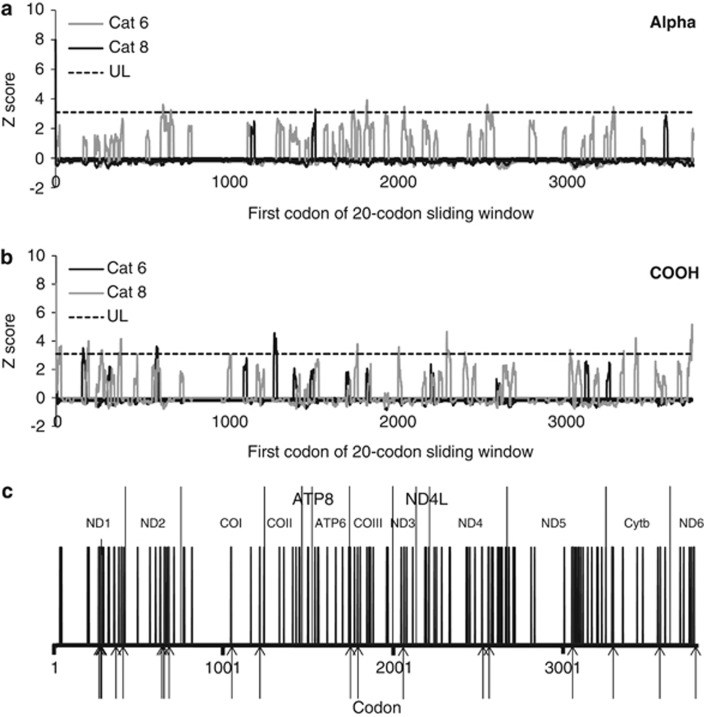
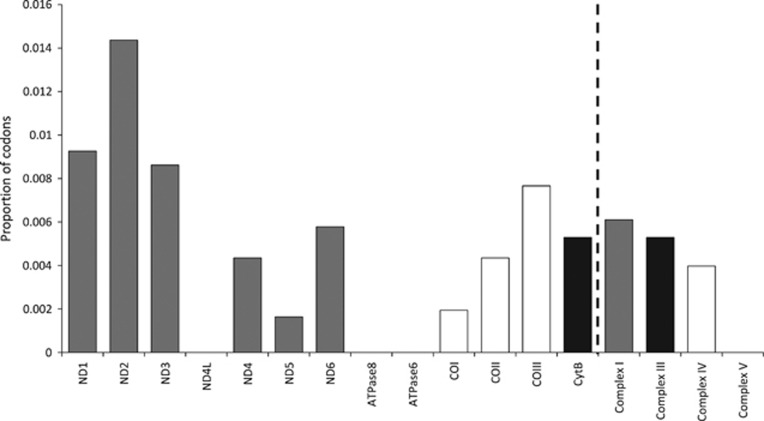
On the basis of global Z scores, nine of the physicochemical properties tested in TREESAAP showed overall patterns of purifying selection across the phylogenetic tree (Supplementary Table S7). Only two of the properties tested showed an overall pattern of positive selection (alpha-helical tendencies and equilibrium constant (ionisation of COOH); Supplementary Table S7 and Figure 3). Specific codons associated with significant (P<0.001) radical (categories 6–8) changes in amino acid properties were detected across all the mitochondrial protein-coding genes (Figure 3). The majority of radical changes detected were associated with divergence of the trout cod (73.7% or 98/133). Divergence between the Murray cod and the ancestor of two coastal species was associated with 14.3% (19/133) of the radical changes detected (Figure 3). Seven of the 13 amino acid differences fixed between the inland and coastal species were associated with significant radical changes in physicochemical properties (Table 3). Seven of the 11 codons detected as showing evidence of episodic positive selection by MEME were also associated with significant radical changes in physicochemical properties in TREESAAP analysis (Table 4). As a proportion of gene length, OXPHOS complex I had a slightly higher proportion of codons showing evidence of significant radical changes in physicochemical properties (Figure 4).
Figure 3.
Evidence of significant changes in physicochemical properties of amino acids detected using TREESAAP analysis. For categories 6 and 8, Z scores greater than 3.09 (above the dotted line) indicate overall significant radical changes in the physicochemical properties: (a) alpha-helical tendencies and (b) equilibrium constant (ionisation of COOH). The x axis for (a) and (b) represents a sliding window (20 codons) increasing in one-codon implements. Codons associated with significant radical amino acid changes on particular branches of the tree are represented by black lines in (c). Arrows below x axis indicate significant radical amino acid changes associated with the coastal branches of the tree. The x axis in (c) represents a specific codon position.
Figure 4.
Proportion of codons relative to gene length in each mitochondrial protein-coding gene/complex (complexes are colour-coded) that show evidence of significant positive disruptive selection in coastal Maccullochella lineages in TREESAAP analysis.
Discussion
Phylogenetic analyses supported mid- to late-Pleistocene divergence of Maccullochella across the GDR (220–710 thousand years ago) and were consistent with the revised taxonomy based on shorter mitochondrial DNA fragments published by Nock et al. (2010), with the two coastal species being sister taxa and, in turn, a sister group to Murray cod (Figure 2). We detected overall strong, pervasive purifying selection acting across the mitogenome in Maccullochella species, as has been observed across many other taxa (Meiklejohn et al., 2007). Although population diversity metrics based on small sample sizes should be interpreted with caution, very strong purifying selection was consistent with very low mitogenome diversity within species, even in the widely distributed Murray cod, for which only six synonymous and four non-synonymous changes were detected across our broadly distributed samples (Figure 1). Although purifying selection appeared to dominate the evolution of the mitochondrial genome in Maccullochella, the strength of selection (ω values) did vary among sites and among branches on the tree and there was evidence of potentially functionally relevant fixed amino acid replacements between the inland and coastal species. Although many of the amino acid differences between inland and coastal species may have been the result of relaxed purifying selection rather than positive selection, we did detect some evidence of episodic positive selection acting on specific codons in specific coastal lineages, particularly associated with divergence of the Mary River cod.
Mid- to late-Pleistocene divergence of Maccullochella across the GDR
Analysis of complete mitochondrial genomes allowed us to generate more precise estimates of divergence times among Maccullochella than those obtained previously by Nock et al. (2010), even while incorporating uncertainty in evolutionary rates. Phylogenetic analysis suggests that divergence between the inland and coastal Maccullochella species occurred 450 (220–710) thousand years ago during the mid- to late-Pleistocene, bringing forward the bounds of previously reported dates based on two mitochondrial genes (0.62–1.62 million years; Nock et al., 2010). Similarly, our data suggested more recent divergence between the two coastal species (~140 (68–230) thousand years ago) than previously estimated (120–510 thousand years; Nock et al., 2010). Comparable estimates of mitogenome divergence across the GDR were obtained for another endemic freshwater fish species, the Macquarie Perch Macquaria australasica, suggesting that the same geological/environmental conditions may have facilitated GDR crossing for multiple species (119–385 thousand years; Pavlova et al., unpublished data). During the Pleistocene, cycles of alternating warm, moist interglacial climate conditions and cool, dry glacial climate conditions shaped fish distributions by influencing the amount of freshwater habitat and connectivity among drainages (Faulks et al., 2015). Movement across the GDR by Maccullochella was most likely facilitated by extreme flooding in low elevation regions of the GDR (Nock et al., 2010; Unmack, 2013). Drainage isolation as a result of increasingly dry conditions through the Pleistocene and/or increased sea levels during interglacial periods may have led to divergence between the Mary River and eastern freshwater cod east of the GDR (Byrne et al., 2011; Faulks et al., 2015).
Although we excluded sites showing evidence of positive selection from the BEAST analysis, strong, pervasive purifying selection acting across the mitogenome detected in our study does violate the molecular clock's assumption of neutrality. However, because pervasive purifying selection is widespread among taxa, it is expected that similar selective processes would have occurred on a comparable timescale in the evolution of the taxa on which we based our clock rate priors (Meiklejohn et al., 2007; Burridge et al., 2008).
Potential relaxation of purifying selection in coastal species may be driven by relaxed metabolic constraints and/or historical population bottlenecks
Relaxed purifying selection in both coastal species was supported by higher ratios of non-synonymous to synonymous changes (ω values) associated with coastal branches in both CODEML and GA-Branch analyses. Reduced selective constraint on OXPHOS mitochondrial genes could be the result of relaxed metabolic constraints in the milder climate east of the GDR and/or reduced effectiveness of selection owing to reductions in effective population size associated with historical population bottlenecks (Hughes, 2007). In addition, lack of strong evidence of positive selection using the CODEML method may reflect low power because of relatively recent divergence of the coastal species and associated low polymorphism (Melo-Ferreira et al., 2014).
Environments and lifestyles with high energy demands (for example, extreme climate, flight ability, migratory behaviour) are typically associated with stronger functional constraint on metabolism (that is, lower ω values associated with codons/genes linked to energy metabolism) (Melo-Ferreira et al., 2014). Given the climatic differences across the GDR, the coastal environment may be associated with a relaxation of selective constraint on genes linked to tolerance of temperature extremes and/or variability. However, historical population bottlenecks (for example, associated with colonisation of coastal environments by Maccullochella) and small effective population size can also lead to reduced efficiency of selection and the accumulation of mildly deleterious changes. Identifying whether relaxed metabolic constraint or reduced efficiency of selection drives signatures of relaxed selection is challenging, especially as the two processes are not mutually exclusive. For example, higher ratios of non-synonymous to synonymous changes in mitogenomes of a number of domesticated (cf. wild) taxa have been attributed to both relaxed selective constraint on metabolic efficiency because of human actions (for example, provision of resources and improved living conditions) and reduced efficiency of selection owing to repeated population bottlenecks during the domestication process (Hughes, 2013). In our case, rigorously distinguishing among metabolic adaptation to coastal environments (positive selection), small effective population in coastal environments (reduced efficiency of selection) and reduced metabolic constraint in coastal environments (relaxed selection) would be impossible without controlled experiments and/or studies of protein biochemistry. For example, better metabolic efficiency of inland than coastal species under more variable temperature regimes would support relaxed selection in the coastal environment as the driving process. Alternatively, greater metabolic efficiency of coastal than inland species under less variable temperature regimes would support positive selection acting on coastal species.
Some evidence of functional differences across the GDR and episodic positive selection in coastal lineages, particularly associated with divergence of the Mary River cod
Although CODEML analyses did not support positive selection acting consistently on mitogenomes across coastal lineages, there was evidence of potentially functionally relevant fixed amino acid differences across the GDR and of possible episodic positive selection acting on specific codons in specific coastal lineages (TREESAAP and MEME analysis), particularly associated with the divergence of the Mary River cod. In the case of the Mary River cod, signatures of potential positive selection acting on the mitogenome have accumulated after a relatively short period of evolutionary time (68–230 thousand years).
Of the 13 non-synonymous fixed differences (that is, amino acids not shared) between inland and coastal species, 11 were represented by the same amino acid in both inland species. Given the strong levels of mitochondrial divergence (~12%) observed between the two inland species (Murray cod and trout cod), conservation of the same amino acid in Murray cod and trout cod for differences that were fixed across the GDR could reflect strong selective pressures for these amino acids in environments west of the GDR that are relaxed or different east of the GDR. Seven of the 13 amino acid differences fixed between the inland and coastal species were associated with significant changes in physicochemical properties (six of these changes relating to properties with evidence of positive selection based on global Z scores) suggesting that functional differences exist between mitogenomes east and west of the GDR that may have consequences for metabolic function (Woolley et al., 2003).
Six of the eleven sites identified by MEME as being putatively under episodic positive selection were associated with the Mary River cod (the more northern of the two coastal species), with five of these being fixed amino acid differences between the Mary River cod and all other Maccullochella species and mostly associated with radical changes in physicochemical properties of amino acids. Amino acid replacements putatively under selection in the Mary River cod were located in complexes I (ND1, ND5 and ND6) and, somewhat atypically, in complex IV (COI, COII and COIII) (Zhang and Broughton, 2013). Whilst amino acid replacements in complex I (ND genes) tend to have minor effects on functional properties of amino acids, amino acid replacements in complex IV (COX genes) tend to have disproportionate effects on fitness, making COX genes typically the most highly conserved in the mitogenome (Zhang and Broughton, 2013). Although sites showing signatures of episodic positive selection detected in the Mary River cod may represent false positives that are in fact deleterious but not (yet) removed by selection because of low effective population size, investigation of candidate sites and adaptive processes in Mary River cod seems warranted (Hughes, 2007).
Concluding remarks
We report mid- to late-Pleistocene divergence of Maccullochella across the GDR (220–710 thousand years ago), followed by relatively recent divergence of the two coastal species (68–230 thousand years ago). We detected potentially functionally relevant fixed amino acid differences between the inland and coastal species of Maccullochella. Although many of the amino acid differences between inland and coastal species may have been the result of relaxed purifying selection rather than positive selection, there was evidence of episodic positive selection acting on specific codons in specific coastal lineages, particularly the Mary River cod, which experiences relatively stable, and the warmest, environments in the genus. Our study establishes a foundation for future experimental investigations into adaptive processes in species of Maccullochella, including the functional effects of candidate amino acid replacements on metabolism.
Data archiving
Sequence data have been submitted to GenBank: accession numbers: KT337324-KT337338.
Acknowledgments
This work was supported by ARC Grant LP110200017 to Monash University, Flinders University of South Australia and the University of Canberra, with Partner Organization University of Montana. Funding and other support was contributed by industry Partner Organizations ACTEW Corporation, Department of Sustainability and Environment (Victoria) and Fisheries Victoria (now within DELWP- Department of Environment, Land, Water & Planning), Melbourne Water. We are grateful to the Holsworth Wildlife Research Endowment - Equity Trustees Charitable Foundation for support to KH. Meaghan Duncan, Joanne Kearns, Dean Gilligan, Stuart Rowland, Cathy Nock, Bronwen Jones, Glenn Wilson, Ian Wooden/Dean Hartwell and Bob Simpson collected and/or provided access to samples. We thank Hernán Morales and A. Gonçalves da Silva for useful discussions. Jian Yen and three anonymous reviewers provided helpful comments on drafts. Funding for MiSeq sequencing was provided in part by the Monash University Malaysia Tropical Medicine and Biology Multidisciplinary Platform.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Avise JC. (2000) Phylogeography: the History and Formation of Species. Harvard University Press: Cambridge, MA, USA.. [Google Scholar]
- Burridge CP, Craw D, Fletcher D, Waters JM. (2008). Geological dates and molecular rates: fish DNA sheds light on time dependency. Mol Biol Evol 25: 624–633. [DOI] [PubMed] [Google Scholar]
- Byrne M, Steane DA, Joseph L, Yeates DK, Jordan GJ, Crayn D et al. (2011). Decline of a biome: evolution, contraction, fragmentation, extinction and invasion of the Australian mesic zone biota. Journal of Biogeography 38: 1635–1656. [Google Scholar]
- Caissie D. (2006). The thermal regime of rivers: a review. Freshwater Biology 51: 1389–1406. [Google Scholar]
- Chevreux B, Wetter T, Suhai S. (1999). Genome sequence assembly using trace signals and additional sequence information. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics 99: 45–56. [Google Scholar]
- da Fonseca RR, Johnson WE, O'Brien SJ, Ramos MJ, Antunes A. (2008). The adaptive evolution of the mammalian mitochondrial genome. Bmc Genomics 9: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delport W, Poon AF, Frost SD, Pond SLK. (2010). Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26: 2455–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulks LK, Gilligan DM, Beheregaray LB. (2015) 'Ragged mountain ranges, droughts and flooding rains': the evolutionary history and conservation of Australian freshwater fishes. In: Stow A, Maclean N, Holwell GI (eds) Austral Ark: The State of Wildlife in Australia and New Zealand. Cambridge University Press: Cambridge. pp 492–511. [Google Scholar]
- Franks SJ, Hoffmann AA. (2012). Genetics of climate change adaptation. Genetics 46: 185–208. [DOI] [PubMed] [Google Scholar]
- Garvin MR, Bielawski JP, Sazanov LA, Gharrett AJ. (2014). Review and meta-analysis of natural selection in mitochondrial complex I in metazoans. Journal of Zoological Systematics and Evolutionary Research 53: 1–17. [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. (2013). Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res 41: e129–e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrisson KA, Pavlova A, Telonis-Scott M, Sunnucks P. (2014). Using genomics to characterize evolutionary potential for conservation of wild populations. Evol Appl 7: 1008–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. (2005). Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25: 1965–1978. [Google Scholar]
- Hughes AL. (2007). Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity 99: 364–373. [DOI] [PubMed] [Google Scholar]
- Hughes AL. (2013). Accumulation of slightly deleterious mutations in the mitochondrial genome: a hallmark of animal domestication. Gene 515: 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP et al. (2013). MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol 30: 2531–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SY, Guindon S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29: 1695–1701. [DOI] [PubMed] [Google Scholar]
- Lavoué S, Nakayama K, Jerry DR, Yamanoue Y, Yagishita N, Suzuki N et al. (2014). Mitogenomic phylogeny of the Percichthyidae and Centrarchiformes (Percomorphaceae): comparison with recent nuclear gene-based studies and simultaneous analysis. Gene 549: 46–57. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lintermans M, Rowland S, Koehn J, Butler G, Simpson B, Wooden I. (2005). Management of Murray Cod in the Murray-Darling Basin: Statement, Recommendations and Supporting Papers. Workshop, 3-4 June, 2004. Murray-Darling Basin Commission Canberra 15–29.
- McClellan DA, Ellison DD. (2010). Assessing and improving the accuracy of detecting protein adaptation with the TreeSAAP analytical software. Int J Bioinform Res Appl 6: 120–133. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Montooth KL, Rand DM. (2007). Positive and negative selection on the mitochondrial genome. Trends Genet 23: 259–263. [DOI] [PubMed] [Google Scholar]
- Melo-Ferreira J, Vilela J, Fonseca MM, da Fonseca RR, Boursot P, Alves PC. (2014). The elusive nature of adaptive mitochondrial DNA evolution of an arctic lineage prone to frequent introgression. Genome Biol Evol 6: 886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales HE, Pavlova A, Joseph L, Sunnucks P. (2015). Positive and purifying selection in mitochondrial genomes of a bird with mitonuclear discordance. Mol Ecol 24: 2820–2837. [DOI] [PubMed] [Google Scholar]
- Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Pond SLK et al. (2013). FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol 30: 1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Pond SLK. (2012). Detecting individual sites subject to episodic diversifying selection. Plos Genet 8: e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narum SR, Campbell NR, Meyer KA, Miller MR, Hardy RW. (2013). Thermal adaptation and acclimation of ectotherms from differing aquatic climates. Mol Ecol 22: 3090–3097. [DOI] [PubMed] [Google Scholar]
- Nock CJ, Elphinstone MS, Rowland SJ, Baverstock PR. (2010). Phylogenetics and revised taxonomy of the Australian freshwater cod genus, Maccullochella (Percichthyidae). Marine and Freshwater Research 61: 980–991. [Google Scholar]
- Peel MC, Finlayson BL, McMahon TA. (2007). Updated world map of the Köppen-Geiger climate classification. Hydrology and Earth System Sciences Discussions 4: 439–473. [Google Scholar]
- Rambaut A. (2007). FigTree v1.3.1. Available at http://tree.bio.ed.ac.uk/software/figtree/.
- Rambaut A, Drummond AJ. (2007). Tracer v1.4 Available at http://beast.bio.ed.ac.uk/Tracer.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland S. (1993). Maccullochella ikei, an endangered species of freshwater cod (Pisces: Percichthyidae) from the Clarence River system, NSW and M. peelii mariensis, a new subspecies from the Mary River system, Qld. Records of the Australian Museum 45: 121–145. [Google Scholar]
- Santos C, Montiel R, Sierra B, Bettencourt C, Fernandez E, Alvarez L et al. (2005). Understanding differences between phylogenetic and pedigree-derived mtDNA mutation rate: a model using families from the Azores Islands (Portugal). Mol Biol Evol 22: 1490–1505. [DOI] [PubMed] [Google Scholar]
- Schoville SD, Bonin A, Francois O, Lobreaux S, Melodelima C, Manel S. (2012). Adaptive genetic variation on the landscape: methods and cases. Annu Rev Ecol Evol S 43: 23–43. [Google Scholar]
- Silva G, Lima FP, Martel P, Castilho R. (2014). Thermal adaptation and clinal mitochondrial DNA variation of European anchovy. Proc Biol Sci 281: 20141093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stager M, Cerasale DJ, Dor R, Winkler DW, Cheviron ZA. (2014). Signatures of natural selection in the mitochondrial genomes of Tachycineta swallows and their implications for latitudinal patterns of the ‘pace of life'. Gene 546: 104–111. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueman WT. (2011). True Tales of the Trout Cod: River Histories of the Murray-Darling Basin. MDBA Publication No. 215/11.
- Unmack PJ. (2001). Biogeography of Australian freshwater fishes. Journal of Biogeography 28: 1053–1089. [Google Scholar]
- Unmack PJ. (2013) Biogeography. In: Humphries P, Walker KF (eds) The Ecology of Australian Freshwater Fishes. CSIRO Publishing: Collingwood. pp 25–48. [Google Scholar]
- Wilson ML, Cai Y, Hanlon R, Taylor S, Chevreux B, Setubal JC et al. (2012). Sequence verification of synthetic DNA by assembly of sequencing reads. Nucleic Acids Res 41: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. (2003). TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics 19: 671–672. [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang F, Broughton RE. (2013). Mitochondrial–nuclear interactions: compensatory evolution or variable functional constraint among vertebrate oxidative phosphorylation genes? Genome Biol Evol 5: 1781–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. (2005). Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 22: 2472–2479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.