Abstract
The cyclic AMP receptor protein family of transcription factors regulates various metabolic pathways in bacteria, and also play roles in response to environmental changes. Here, we identify four homologs of the CRP family in Deinococcus radiodurans, one of which tolerates extremely high levels of oxidative stress and DNA-damaging reagents. Transcriptional levels of CRP were increased under hydrogen peroxide (H2O2) treatment during the stationary growth phase, indicating that CRPs function in response to oxidative stress. By constructing all CRP single knockout mutants, we found that the dr0997 mutant showed the lowest tolerance toward H2O2, ultraviolet radiation, ionizing radiation, and mitomycin C, while the phenotypes of the dr2362, dr0834, and dr1646 mutants showed slight or no significant differences from those of the wild-type strain. Taking advantage of the conservation of the CRP-binding site in many bacteria, we found that transcription of 18 genes, including genes encoding chromosome-partitioning protein (dr0998), Lon proteases (dr0349 and dr1974), NADH-quinone oxidoreductase (dr1506), thiosulfate sulfurtransferase (dr2531), the DNA repair protein UvsE (dr1819), PprA (dra0346), and RecN (dr1447), are directly regulated by DR0997. Quantitative real-time polymerase chain reaction (qRT-PCR) analyses showed that certain genes involved in anti-oxidative responses, DNA repair, and various cellular pathways are transcriptionally attenuated in the dr0997 mutant. Interestingly, DR0997 also regulate the transcriptional levels of all CRP genes in this bacterium. These data suggest that DR0997 contributes to the extreme stress resistance of D. radiodurans via its regulatory role in multiple cellular pathways, such as anti-oxidation and DNA repair pathways.
Introduction
Cyclic AMP (cAMP) receptor proteins (CRPs) are global transcriptional regulators that are widely distributed in bacteria[1, 2]. They play important roles in the regulation of many biological processes, including adaptation to starvation and extreme temperatures, energy metabolism, cell division, and toxin production[3–7]. As a transcription factor, the CRP/FNR family has diverse functions in different bacteria[8–11].
In Escherichia coli, CRP directly or indirectly controls the transcription of more than 400 genes, and it has been studied extensively[12, 13]. It regulates transcription through binding to the effector cAMP, which is synthesized by a membrane-bound adenylate cyclase (CyaA) in the absence of glucose[14]. Crystallographic studies of E. coli CRP have been performed to determine the structure of a CRP-cAMP complex, its target DNA, and the carboxyl-terminal domain of the RNA polymerase (RNAP) α-subunit[15–19]. It is shown that CRP contains a helix-turn-helix DNA-binding motif in its carboxyl-terminal domain and a cAMP-binding site in its amino-terminal domain[20–22]. Binding of cAMP causes a conformational change in CRP that leads to the formation of the CRP-cAMP complex[23–25], which can interact with an ~22-bp DNA-binding site with the consensus sequence 5′–AAATGTGATCTAGATCACATTT–3′[26]. However, the consensus DNA-binding sequences of CRP homologs from Corynebacterium glutamicum GlxR[27], Mycobacterium tuberculosis RV3676[28], and Haemophilus influenza CRP[29] differ slightly from that of E. coli CRP. Biochemical analyses reveal that CRP interacts with the carboxyl-terminal domain of the RNAP α-subunit[30]. This interaction facilitates binding of RNAP to the promoter, resulting in the initiation of transcription.
Deinococcus radiodurans, which belongs to the phylum Deinococcus-Thermus, is characterized by its extreme resistance to ionizing radiation, ultraviolet (UV) irradiation, desiccation, hydrogen peroxide (H2O2), and other DNA-damaging agents[31, 32]. Because of its efficient DNA repair ability and extreme stress resistance, D. radiodurans is generally considered to be an ideal model organism for studying bacterial resistance mechanisms under various stress conditions. There exists many predicted open reading frames (ORFs) encoding transcriptional factors in the D. radiodurans genome [33], whereas their functions, activities, and binding sites have rarely been elucidated.
D. radiodurans is predicted to contain four CRP family proteins, including DR0997, DR1646, DR2362, and DR0834. Interestingly, the transcriptional level of dr0997, also referred to as ddrI (DNA damage response gene I), increases 38-fold after ionizing radiation treatment[34], indicating that it might be involved in post-ionizing radiation recovery.
In this study, we constructed all the single mutants of CRPs and examined their roles in anti-oxidation, DNA repair, and other important cellular pathways. It was demonstrated that one of the CRP homologs, DR0997, acts as an important transcriptional activator that is involved in diverse cellular pathways, including cell growth, oxidative stress response, and DNA damage repair.
Materials and Methods
Bacterial strains, plasmids, media, and growth conditions
The D. radiodurans wild-type R1 (American Type Culture Collection 13939, Rockville, MD, USA), E. coli DH5α, and E. coli BL21 (DE3)pLysS strains were available in our laboratory. All D. radiodurans strains were grown at 30°C in tryptone-glucose-yeast extract (TGY) broth (0.5% tryptone, 0.1% glucose, 0.3% yeast extract) with aeration, or on TGY medium solidified with 1.5% w/v agar. When necessary, antibiotics were added to D. radiodurans cultures as follows: 30 μg mL–1 kanamycin, 10 μg mL–1 streptomycin, and 3.4 μg mL–1 chloramphenicol. All E. coli strains were grown at 37°C in Luria–Bertani medium (1% tryptone, 0.5% yeast extract, 1% NaCl) and supplemented with 30 μg mL–1 chloramphenicol, 50 μg mL–1 kanamycin, or 50 μg mL–1 ampicillin where appropriate. All strains and plasmids used in this study are listed in Table 1.
Table 1. Strains and plasmids used in this study.
| Strains and plasmids | Description | Source |
|---|---|---|
| D. radiodurans | ||
| R1 | ATCC13939 | [35] |
| dr0997 mutant | D. radiodurans dr0997 gene knockout mutant | This work |
| dr2362 mutant | D. radiodurans dr2362 gene knockout mutant | This work |
| dr1646 mutant | D. radiodurans dr1646 gene knockout mutant | This work |
| dr0834 mutant | D. radiodurans dr0834 gene knockout mutant | This work |
| dr0997 mutant Cwt | dr0997 mutant complement with pRADdr0997 | This work |
| E. coli | ||
| DH5α | Host for cloning vectors | Takara |
| Bl21 (DE3) plysS | Host for expressing proteins | Takara |
| DR0997-HMT BL21 | BL21 containing expression plasmid pET28a-HMTdr0997 | This work |
| DR0615 BL21 | BL21 containing expression plasmid pET28a-dr0615 | This work |
| TEV-pET28a BL21 | BL21 containing expression plasmid TEV-pET28a | [35] |
| Plasmids | ||
| pET28a | Expression vector | Takara |
| pET28a-HMT | Expression vector reformed by pET28a with insertion of maltose affinity protein | [35] |
| pRADK | pRADZ3 derivative in which lacZ is replaced by the kanamycin gene | [36] |
| pRADdr0997 | pRADK derivative in which kanamycin gene is replaced by the dr0997 gene | This work |
| pET28a-dr0615 | pET28a expression plasmid containing BamHI-NdeI fragment of dr0615 | This work |
| pET28a-HMTdr0997 | pET28a-HMT expression plasmid containing BamHI-NdeI fragment of dr0997 | This work |
CRP gene mutation and complementation in D. radiodurans
CRP null mutants, including dr0997, dr2362, dr1646, and dr0834 null mutant, were constructed using the deletion replacement method as described previously[37]. Take the dr0997 null mutant for example. Briefly, ~500-bp DNA fragments immediately upstream and downstream of dr0997 were amplified from the genome of the wild-type R1 strain. After gel extraction, the two fragments were digested with BamHI and HindIII respectively, and ligated with a kanamycin resistance cassette fragment containing the groEL promoter, which was digested with the same enzymes. The kanamycin resistance cassette was obtained from pRADK, a shuttle plasmid derived from pRADZ3. The ligated product was then transformed into competent D. radiodurans cells in the exponential growth phase using CaCl2 as described previously[37]. The mutant strain was confirmed by genomic polymerase chain reaction (PCR) with the primers dr0997upF and dr0997downR (S1 Table). The full-length dr0997 gene was amplified from the genome of the wild-type R1 strain with primers Comdr0997F and Comdr0997R (S1 Table) and digested with NdeI and BamHI. The predigested shuttle vector pRADZ3 was then ligated with the dr0997 fragment and transformed into the dr0997 null mutant to form the dr0997-complemented strain.
Growth curves and survival tests
Growth curves of the CRP null mutant strains were determined by measuring the optical density at 600 nm (OD600). All strains were cultured in 5 ml of liquid TGY medium until the OD600 reached ~2.0, and then they were diluted 1:500 into new flasks containing 100 ml of TGY medium. The cultures were incubated at 30°C with shaking at 220 rpm, and samples were taken every 2 h to measure the OD600.
H2O2, UV, and mitomycin C (MMC) were used to test the mutants’ stress resistances. The survival rates of the CRP mutants after exposure to H2O2 were determined with a previously described method [38]. Briefly, cells were diluted to an appropriate concentration with 10 mM MgSO4. H2O2 (Sigma-Aldrich, St. Louis, MO, USA) was added to the cell suspension to a final concentration of 50 mM, and the cells were incubated at room temperature. Samples were obtained at 0, 5, 10, 20, and 40 min, and catalase (30 mg/ml) (Sigma-Aldrich) was added to inactive the H2O2. The survival fraction (%) was calculated using the following equation: survival fraction (%) = Nsample/Ncontrol × 100%, where Ncontrol is the number of wild-type bacteria and Nsample is the number of mutant bacteria. For the UV sensitivity test, cultures with an OD600 of ~1.0 were diluted with 10 mM MgSO4. After plating onto TGY agar plates, the plates were exposed to different dosages of UV radiation by calculating the exposure time as described previously[38]. For the MMC sensitivity assay, cells were diluted with 10 mM MgSO4 and treated with a final concentration of 5 μg/ml MMC (Sigma-Aldrich), and then plated onto TGY agar plates every 10 min. All the plates were incubated at 30°C for 3 d. All data shown are the means ± SDs of three independent experiments.
Measurement of catalase and total antioxidant capacity
Cells were cultured at 30°C in 100 ml of TGY medium for at least 12 h until the OD600 reached ~1.0. Half of the culture was treated with 30 mM H2O2 for 30 min, harvested, washed twice with phosphate buffer (20 mM, pH 7.4), and suspended in 2 ml of phosphate buffer.
Catalase activity was determined as described previously[39]. The collected cells were disrupted on ice with an ultrasonic cell disruptor at an output of 450 W for 15 min. Debris was removed by centrifugation, and the concentration of soluble protein was determined using the Bradford method. The soluble supernatant was diluted with an appropriate chromogenic reagent mixture (Beyotime, China). The quantity of H2O2 remaining in the mixture is determined by the oxidative production of N-(4-antipyryl)-3-chloro-5-sulfonate-p-benzoquinonemonoimine in the presence of H2O2 when catalyzed by horseradish peroxidase in the chromogenic reagent mixture. After a 15-min incubation, the resultant product was quantified at 520 nm.
The total antioxidant capacity was assayed as reported previously[40]. Briefly, 2, 2′-azino-bis (3-ethylbenzthiazoline-6-sulfonic acid (ABTS)) was used as a dye solution. An appropriately diluted protein solution was mixed with the oxidant regent for 6 min, and the generation of ABTS.+ was measured at 734 nm. The total antioxidant capacity was presented by comparing it with the antioxidant activity of Trolox (Beyotime).
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR) assay
Total RNA extraction and data processing were conducted as described previously[41]. Briefly, the wild-type strain and dr0997 mutant strain were cultured until the OD600 reached 0.4, and then harvested. Total RNA was extracted using the Whole RNA Extraction kit (Promega, Madison, WI USA). RNA quality and quantity were evaluated by measuring the A260/A280 ratio with a NanoDrop-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA) and Denaturing Agarose Gel Electrophoresis.
The qRT-PCR assay utilized RNA samples that obtained under different conditions. The wild-type and mutant strains were grown in TGY until the OD600 reached 0.5. Each culture was divided in two halves: one half of the culture was treated with H2O2 at a final concentration of 50 mM, while the other half was used as the non-treated control. Total RNA was extracted as mentioned previously. First-strand cDNA synthesis was conducted in 20-μl reactions containing 1 μg of purified RNA and 3 mg of random hexamers. The SYBR Green PCR kit (Tiangen, Beijing, China) was used for PCR amplification according the manufacturer’s instructions, and all assays were performed using the Mx3005PTM Real-time Detection System (Stratagene, La Jolla, CA, USA).
Protein expression and purification
Proteins were induced and purified as described previously[42, 43]. Briefly, the dr0997 fragment was obtained by digesting pRAD-dr0997 with NdeI and BamHI, and it was ligated with pET28a-HMT. pET28a-HMT is a pET-28a derivative that encodes maltose binding protein (MBP) containing specific tobacco etch virus (TEV) protease recognition sites. E. coli BL21 (DE3) pLysS cells carrying plasmid pET28a-HMTdr0997 were grown to an OD600 of 0.6 at 37°C and induced with 200 μM isopropyl β-D-1-thiogalactopyranoside at 37°C for at least 12 h. Cells were harvested by centrifugation, washed twice with phosphate buffer (20 mM, pH 7.4), and resuspended in binding buffer (500 mM NaCl, 20 mM Tris, 5% glycerol, pH 8.0). Cells were disrupted on ice with an ultrasonic cell disruptor at an output of 350 W for 20 min. After centrifugation at 15,000 × g for 20 min at 4°C, the supernatant was loaded onto a HisTrap HP column (GE Healthcare, Little Chalfont, UK). The purified DR0997-MBP fusion protein was digested with a proper concentration of TEV protease at 4°C overnight, and then loaded onto a maltose-affinity column. The eluted protein was applied to a HiTrap Q HP column to remove undigested protein and imidazole. The purity of the protein sample was determined using 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and only the fraction containing pure DR0997 protein was used for further experiments.
Electrophoretic mobility-shift assay (EMSA)
EMSA was performed with DNA fragments (100 ng) mixed with 2 μM purified DR0997 or DR0615 protein in a total volume of 10 μl. The binding buffer contained 10 mM Tris-HCl (pH7.5), 10 mM KCl, 200 mM NaCl, 5 mM MgCl2, 5 mM MnCl2, 10 μg mL–1 bovine serum albumin, 200 μM cAMP, and 100 μM dithiothreitol[44]. The reaction mixture was incubated at 30°C for 30 min and then loaded onto 12% (w/v) polyacrylamide gels in 1×Tris-borate-ethylenediamine tetraacetic acid buffer. Electrophoresis was performed at 100 V for 6 h at 4°C, and the gel was stained with ethidium bromide and photographed (Typhoon 9500, GE Healthcare). The E. coli CRP-binding site was used to identify predicted CRP-regulated genes in D. radiodurans. The promoter of dr1998 was used as a positive control of DR0615. The dr0167 promoter and coding region of dr0167 (dr0167C) were used as negative controls. All primers are listed in S1 File.
Results
Characterization of D. radiodurans CRPs
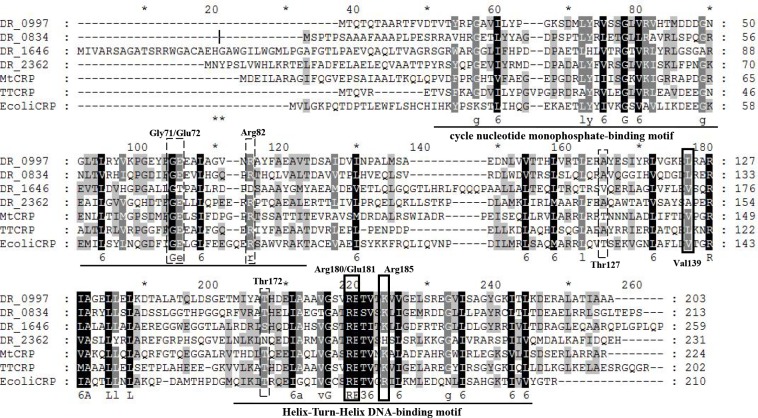
The dr0997 ORF is predicted to encode a 260-amino acid protein (National Center for Biotechnology Information accession no. AAF10573.1). However, based on our sequencing results, a base was missing at the 543-bp site, resulting in the shortening of the dr0997 ORF (S1 Fig). The corrected DR0997, DR2362, DR0834, and DR1646 exhibit nearly 23% identity to E. coli CRP, as indicated by a protein Basic Local Alignment Search Tool (BLASTP) analysis. The conserved helix-turn-helix and the nucleotide monophosphate-binding regions are underlined in Fig 1. Based on the crystal structure of CRP-cAMP, CRP (cAMP)-DNA and CRP-CTD-DNA complex., the residues required for cAMP binding were marked with dotted frame, while the residues required for DNA binding were marked with solid frame It is observed that 4 residues (Gly71, Glu72, Arg82 and Thr172) required for cAMP binding and 2 residues (Arg180 and Glu181) required for DNA binding are conserved in E. coli CRP [45–49]. Unlike most of the CRP/FNR family proteins, there is no cysteine residue in D. radiodurans CRP that could sense oxygen or redox variations [50–52].
Fig 1. Alignment of CRP homologs from different organisms.
ClustalX software was used to align the amino acid sequence of D. radiodurans R1 DR0997, DR0834, DR1646, and DR2362, M. tuberculosis RV3676, T. thermophilus HB8 TTHA1357, and E. coli CRP. The conservative residues Gly71, Glu72, Arg82 and Thr172 (dotted frames) of E. coli CRP are required for cAMP binding, while Arg180 and Glu181 (solid frames) are required for DNA binding, based on the crystal structure of E. coli CRP.
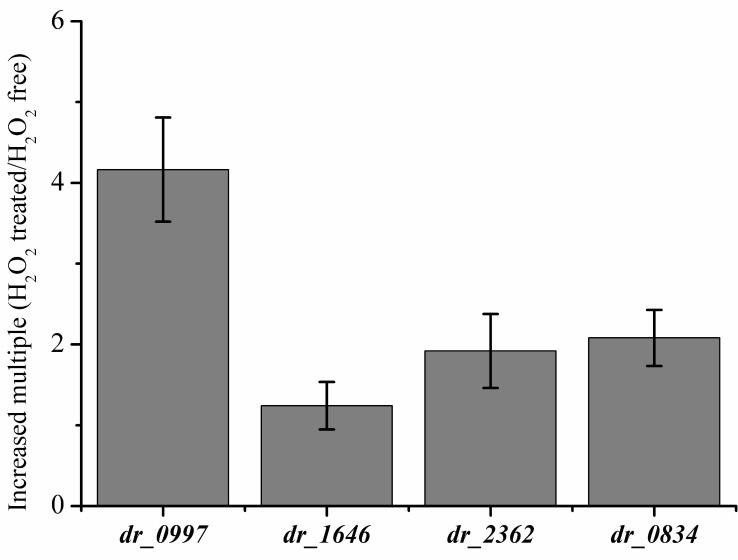
The alteration of expression of CRP genes in D. radiodurans in response to H2O2 treatment were detected compared to the untreated sample, and statistical analyses were applied for data processing (S1 Table). It was demonstrated that the expression levels of CRP genes are increased to different extents when the bacteria were exposed to oxidizing agents. Specifically, the expression of dr0997 is induced 4-fold, indicating that it might be a crucial player in the oxidative stress response (Fig 2). Meanwhile, the expression of dr0997 increases greatly following post-irradiation recovery [34]. These results demonstrate the important roles of CPRs in stress resistance in D. radiodurans.
Fig 2. Increased multiple of transcriptional levels of CRP genes in D. radiodurans R1 after exposure to H2O2 compared with the untreated samples.
Data represent the averages and standard deviations of three independent experiments.
The dr0997 mutant exhibits a growth defect
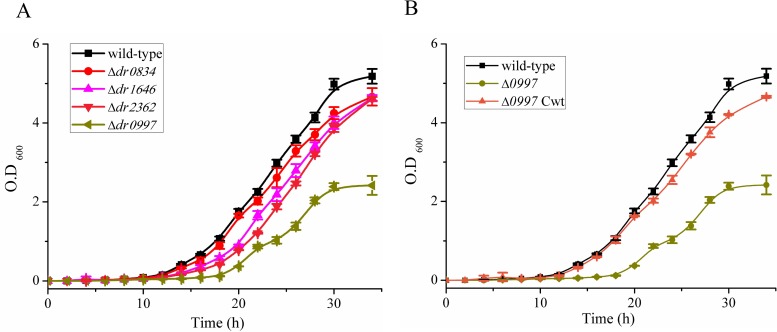
Through a double crossover recombination strategy, four CRP mutants were generated by replacing the CRP-encoding genes with a kanamycin cassette (S2 Fig). After growth in TGY broth without antibiotics, bacterial growth rates were monitored using a spectrophotometer at 2-h internals. As shown in Fig 3, only the dr0997 mutant exhibited an obvious growth defect, and the wild-type growth rate was restored in the dr0997-complemented strain. The dr2362, dr0834, and dr1646 mutants only showed slight growth defects. These data indicated that dr0997 is necessary for normal growth.
Fig 3. Growth curves of D. radiodurans strains.
(A) Wild-type and four CRP mutants were individually grown in TGY medium, and the OD600 was measured every 2 h. (B) Growth rates of the wild-type, dr0997 mutant, and the dr0997 mutant complemented with the dr0997 gene (dr0997 mutant Cwt) were recorded every 2 h.
Deletion of dr0997 results in sensitivity to various stresses
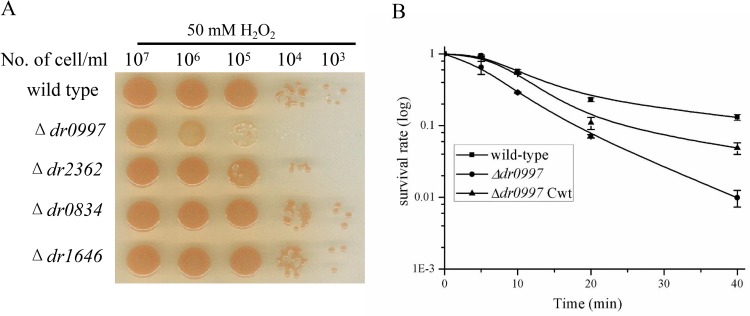
To identify the role of D. radiodurans CRPs in stress resistances, the survival of the CRP mutants were measured and the statistical analyses were applied for data processing (S2 Table). The results showed that the dr0997 mutant was more susceptible to H2O2 than the wild-type strain, as it exhibited a more than 10-fold decrease in survival after treatment with 50 mM H2O2 for 20 min (Fig 4A). The H2O2 resistance of the mutant recovered partly when complemented with the dr0997 gene (pRAD0997) (Fig 4B), and the statistical analysis also indicated the complemented action has statistical significance, suggesting that dr0997 is responsible for the oxidative resistance of the bacterium. However, the dr2362 mutant was only slightly sensitive to oxidative stress (S3 Fig and S3 Table), while the dr0834 and dr1646 mutants were not sensitive to oxidative stress (Data not shown).
Fig 4. Survival curves of D. radiodurans strains exposed to 50 mM H2O2.
(A) Wild-type and CRP mutants exposed to 50 mM H2O2 for 20 min, and then plated on TGY agar plates. (B) Wild-type, dr0997 mutant, and the dr0997 mutant complemented with the dr0997 gene (dr0997 mutant Cwt) were exposed to 50 mM H2O2 for different periods of time. Data represent the averages and standard deviations of three independent experiments.
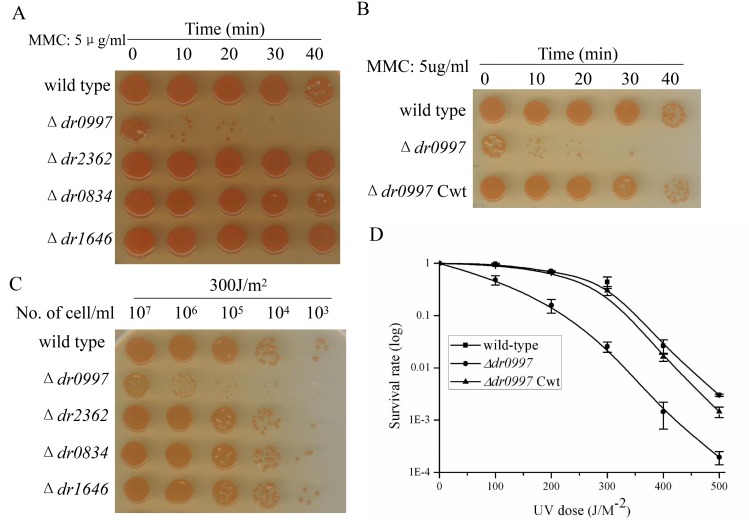
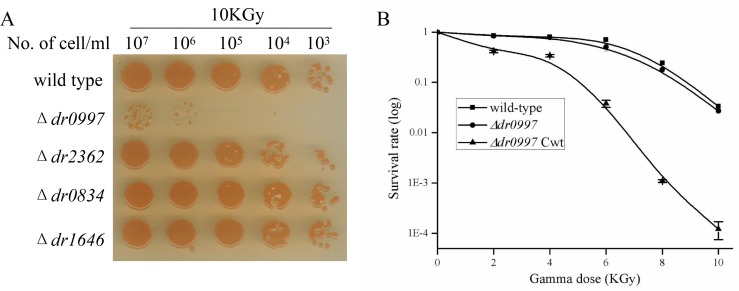
Similar results were obtained in response to other stresses, such as UV radiation, ionizing radiation, and MMC. Disruption of dr0997 resulted in a dramatic increase in sensitivity to UV, gamma radiation, and MMC, which was restored by complementation with the dr0997 gene (Figs 5 and 6). Either 200 J m−2 of UV radiation or 5 kGy of gamma ray radiation were sufficient to kill 95% of the dr0997 mutant cells. Meanwhile, the dr0997 mutant only survived 5 μg/ml MMC for 20 min, while the other CRP mutants did not shown any survival differences at this dosage. These results reveal the key role of dr0997 in stress resistances.
Fig 5. Survival of CRP mutants exposed to MMC and UV radiation.
(A) Survival of CRP mutants and the wild-type strain after exposure to 5 μg/ml MMC for different periods of time. (B) Survival of the dr0997 mutant and the dr0997 mutant complemented with the dr0997 gene (dr0997 mutant Cwt) after MMC exposure. (C) Survival of the CRP mutants and the wild-type strain after exposure to UV radiation. (D) Survival curves of the wild-type, dr0997 mutant, and dr0997 mutant Cwt strains after exposure to UV radiation (0 to 500 Jm−2). Data represent the averages and standard deviations of three independent experiments.
Fig 6. Survival of CRP mutants after exposure to gamma radiation.
(A) Survival of CRP mutants and the wild-type strain after exposure to 10 kGy of gamma radiation. (B) Survival curves of the dr0997 mutant and the dr0997 mutant complemented with the dr0997 gene (dr0997 mutant Cwt) after exposure gamma radiation. Data represent the averages and standard deviations of three independent experiments.
Disruption of dr0997 lowers anti-oxidation activity
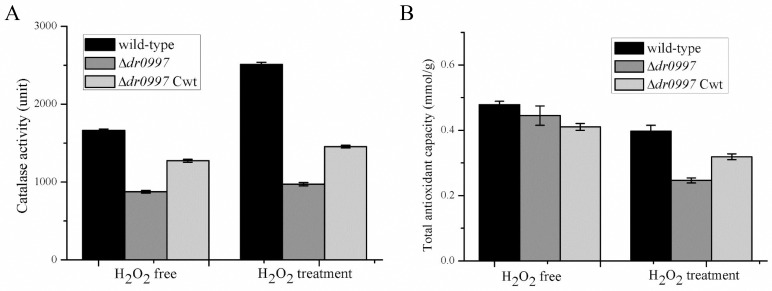
The dr0997 mutant of D. radiodurans displayed decreased H2O2 resistance, suggesting that DR0997 may be involved in the cellular anti-oxidative response. Under normal conditions, as shown in Fig 7A, the dr0997 mutant did not show any significant decrease in total anti-oxidant capacity. However, when treated with 30 mM H2O2 for 30 min, the anti-oxidative activity of the dr0997 mutant decreased by nearly 50%, compared with a 20% decrease in the wild-type and dr0997-complemented strains. These data are consistent with the H2O2 survival phenotype of the dr0997 mutant.
Fig 7. Effect of dr0997 disruption on the anti-oxidant ability of D. radiodurans.
(A) Total antioxidant capacity of the wild-type R1, dr0997 mutant, and dr0997 mutant complemented with the dr0997 gene (dr0997 mutant Cwt) strains after exposure to 30 mM H2O2 treatment. (B) Catalase activities after 30 mM H2O2 treatment. Data represent the average and standard deviations of three independent experiments.
Catalase activity was measured under normal growth conditions or in response to H2O2 treatment. The results showed that the dr0997 mutant exhibited less catalase activity than the wild-type strain under normal conditions (Fig 7B). When treated with 30 mM H2O2 for 30 min, the wild-type strain showed a significant increase (approximately 40%) in catalase activity. However, catalase activity was barely induced in the dr0997 mutant, whereas the catalase activity in the dr0997-complemented strain was nearly equal to that of the wild-type strain, indicating that cells lacking the dr0997 gene lose the ability to cope with oxidative stress. The data were subjected to statistical analysis, and the differences were statistically significant (S4 Table).
DR0997 regulates numerous genes via a specific CRP-binding site
CRP has been shown to be a global transcriptional regulator that is involved in many cellular pathways, such as carbon utilization, lycopene synthesis, antibiotic production, virulence factor regulation, and anti-oxidative stress responses[3, 53–55]. Based on the phenotype of the dr0997 mutant in response to oxidative stress, DNA damage, and other stresses, we focused on the characterization of dr0997 in anti-oxidative and DNA damage response pathways.
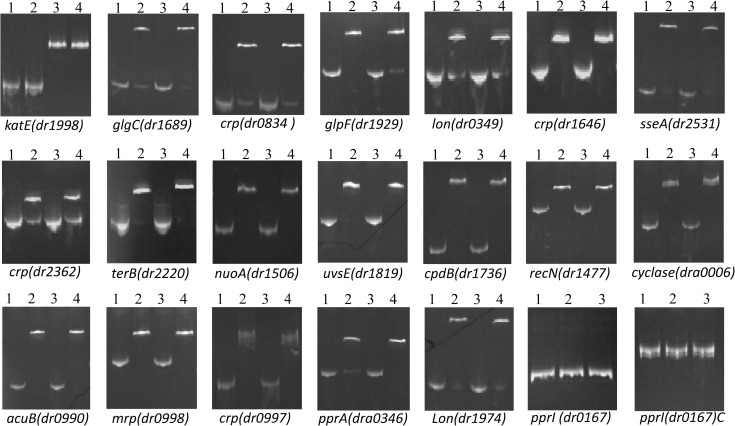
By virtue of the consensus CRP-binding site in E. coli, promoters of numerous anti-oxidation or DNA repair related genes were checked to see whether they contain similar CRP binding sites. Meawhile, the homologues which has been already proved to be regulated by CRP in other bacteria were also analysed[3, 52, 56, 57]. Finally, 18 genes containing CRP-binding sites in their promoters were shown to be directly regulated by DR0997 via EMSAs (Fig 8). Interestingly, most of these promoters contain at least two similar CRP-binding sites. To verify the precise binding site, the regions containing two or more predicted CRP binding sites were segmented into two or more DNA frgments with their own primers respectively, and re-tested by EMSA. It was demonstrated that in most of promoters including dr1506, dr1477, dr0998, dra0006, dr0990, and dr1689, both segments could be bound by DR0997. However, in other promoters including dr1819, dr1974, dr1929, dr1736, only one segment would interact with DR0997. The confirmed CRP-binding sites (S4 Fig) are listed in S2 File. Based on the confirmed binding sites, the CRP-binding consensus sequence in D. radiodurans was predicted in Fig 9. Classification of the genes that are directly regulated by DR0997 revealed that nine genes are involved in stress resistance, and eight genes are involved in metabolic pathways.
Fig 8. Electrophoretic mobility shift assays of DR0997 binding to the upstream regions of different genes with predicted CRP binding sites.
Lane 1, 100 ng of the indicated DNA fragment; lane 2, 100 ng of the indicated DNA fragment with the DR0997 protein. Lane 3, 100 ng of the indicated DNA fragment with the DR0615 protein. Lane 4, 100 ng of the indicated DNA fragment with the DR0997 and DR0615 proteins. The promoter of dr1998 was used as a positive control of DR0615. The dr0167 promoter and coding region of dr0167 (dr0167C) were used as negative controls.
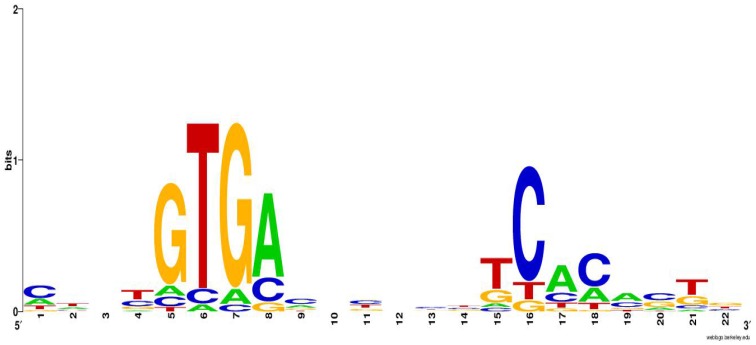
Fig 9. Sequence logo of the predicted CRP-binding sites in D. radiodurans.
The height of each stack of letters represents the degree of sequence conservation measured in bits. The height of each letter within a stack is proportional to its frequency at that position in the binding site. The letters are sorted with the most frequent on top. This sequence logo was generated using the online WebLogo (Univ. of California at Berkeley, Berkeley, CA, USA) program (http://weblogo.berkeley.edu/logo.cgi).
Induction of anti-oxidation-related genes is attenuated in the dr0997 mutant
As a predicted global transcription regulator, DR0997 may control a series of genes that is directly or indirectly involved in oxidative resistance. Using reverse transcription-PCR method, the transcriptional levels of a series of anti-oxidative enzymes, including katE (dr1998 and dra0259), katA (dra0146), sodC (dr1546 and dra0202), sodA (dr1279), terB (dr2220), and NADH dehydrogenase (dr1506) were measured. As shown in Table 2, prior to H2O2 treatment, the transcriptional levels of katE (dr1998 and dra0259), katA (dra0146), and sodC (dra0202) were 2-fold lower in the mutant than in the wild-type strain. After treatment with 50 mM H2O2, the transcriptional levels in the mutant were still lower than those of the wild-type strain, which is in accordance with the aforementioned catalase activity.
Table 2. qRT PCR of anti-oxidation-related genes in the dr0997 mutant relative to wild-type R1 with or without H2O2 treatment.
| Locus | Annotation | Fold change (±SD) | |
|---|---|---|---|
| H2O2 free | H2O2 treatment | ||
| Δ0997(-H)/R1(-H) | Δ0997(+H)/R1(+H) | ||
| DR1998 | Catalase (KatE) | -5.47 (±1.20) | -2.30 (±0.30) |
| DRA0259 | Catalase (KatE) | -2.04 (±0.37) | -1.09 (±0.11) |
| DRA0146 | Catalase (CatA) | -2.00 (±0.24) | -1.33 (±0.20) |
| DR1546 | Cu/Zn family superoxide dismutase (SodC) | 0.86 (±0.02) | 0.81 (±0.03) |
| DRA0202 | Cu/Zn family superoxide dismutase (SodC) | -3.41 (±0.65) | -1.26 (±0.07) |
| DR1279 | Mn family superoxide dismutase (SodA) | -1.27 (±0.03) | -1.34 (±0.09) |
| DR2220 | Tellurite resistance protein TerB | 0.74 (±0.22) | -3.52 (±0.65) |
| DR1506 | NADH-quinone oxidoreductase subunit A | 0.76 (±0.14) | -1.38 (±0.24) |
Functional annotation is based on KEGG (http://www.genome.jp/kegg/).
Specifically, the dr1506 and dr2220 genes were directly regulated by DR0997. DR1506 is an NADH-quinone oxidoreductase that is involved in redox maintenance. The DR2220 (TerB) knockout strain exhibited reduced H2O2 resistance compared with the wild-type strain (S5 Fig). Combined with the aforementioned survival results, we propose that certain antioxidant enzymes, such as catalase or anti-oxidation-related proteins like TerB and NADH dehydrogenase, are dependent on the expression of DR0997, suggesting that DR9997 might act as a positive regulator under oxidative stress.
Induction of DNA repair-related genes is attenuated in the dr0997 mutant
To reveal how the dr0997 deletion affects the bacterial survival rate in response to DNA-damaging agents, such as UV radiation, gamma radiation, and MMC, the transcriptional levels of an array of DNA damage response and DNA repair genes were measured by qRT-PCR under normal conditions and in response to gamma radiation treatment. Twenty-five genes were selected for the qRT-PCR analysis, including recA, recN, recG, recF, recJ, recO, uvrB, uvrC, uvsE, ddrO, priA, drRRA, ddrB, ddrC, and ddrD (Table 3).
Table 3. qRT PCR of DNA repair related genes in the dr0997 mutant relative to wild-type R1 with or without gamma treatment.
| Locus | Annotation | Fold change (±SD) | |
|---|---|---|---|
| γ free | γ treatment | ||
| Δ0997(-γ)/R1(-γ) | Δ0997(+γ)/R1(+γ) | ||
| DR1477 | DNA repair protein RecN | -3.46 (±0.32) | -2.38 (±0.03) |
| DRA0346 | DNA damage repair protein PprA | -2.79 (±0.29) | -1.77 (±0.19) |
| DR1819 | putative UV damage endonuclease UvsE | -5.19 (±0.45) | -1.36 (±0.17) |
| DR1771 | excinuclease ABC subunit A UvrA | -1.11 (±0.30) | 0.81 (±0.06) |
| DR2275 | excinuclease ABC subunit B UvrB | -3.21 (±0.39) | -1.87 (±0.21) |
| DR1354 | excinuclease ABC subunit C UvrC | -5.95 (±0.53) | -2.99 (±0.31) |
| DR1274 | Holliday junction DNA helicase RuvA | -1.13 (±0.02) | -0.93 (±0.13) |
| DR0596 | Holliday junction DNA helicase RuvB | -1.09 (±0.06) | 0.98 (±0.09) |
| DR0440 | crossover junction endodeoxyribonuclease RuvC | -7.54 (±0.98) | -4.57 (±0.83) |
| DR0400 | cell division protein FtsK | -1.36 (±0.12) | -1.35 (±0.09) |
| DR1916 | ATP-dependent DNA helicase RecG | -2.68 (±0.16) | -2.78 (±0.09) |
| DR2340 | recombination protein RecA | -15.56 (±0.75) | -8.27 (±0.39) |
| DR2606 | primosome assembly protein PriA | -2.58 (±0.38) | -2.57 (±0.11) |
| DR2418 | DNA-binding response regulator DrRRA | -1.61 (±0.04) | -1.81(±0.02) |
| DR0167 | Radiation response protein PprI | -1.50 (±0.63) | -1.05 (±0.02) |
| DR0003 | tellurite resistance protein TerB | -14.59 (±1.32) | -11.08 (±1.24) |
| DR0070 | radiation induced single-stranded DdrB | -6.35 (±0.27) | -2.06 (±0.14) |
| DR1440 | cation-transporting ATPase | 0.86 (±0.07) | -5.68 (±0.35) |
| DR2574 | XRE family transcriptional regulator DdrO | -3.82 (±0.25) | -1.00 (±0.03) |
| DR0326 | radiation-induced protein | -7.13 (±0.65) | -2.19 (±0.14) |
| DR1126 | single-stranded-DNA-specific exonuclease | -2.02 (±0.16) | -3.25 (±0.56) |
| DR1089 | DNA replication and repair protein RecF | -4.81 (±0.15) | -2.11 (±0.03) |
| DR0819 | DNA replication and repair protein RecO | -2.64 (±0.05) | 0.94 (±0.01) |
| DR0198 | DNA replication and repair protein RecR | 0.78 (±0.06) | 0.76 (±0.02) |
Functional annotation is based on KEGG (http://www.genome.jp/kegg/).
The expression of 17 of the 25 genes was significantly down- or up-regulated in the dr0097 mutant, suggesting that these genes are transcriptionally affected by DR0997. Under normal conditions, the transcriptional levels of recA, priA, and recG were 23-, 10-, and 4-fold lower, respectively, in the dr0997 mutant than those of the wild type strain. RuvABC is required for the formation of Holliday junctions, as well as for recombination[58]. UvrABC has been reported to be involved in DNA excision repair[59, 60]. The transcriptional levels of these genes were all lower in the dr0997 mutant. The expression of a newly identified repressor of DNA damage response gene, ddrO[61], was 3.8-fold lower in the dr0997 mutant. DdrO and PprI mediate a novel DNA damage response pathway that differs from the classic LexA-mediated SOS response system in the radiation-sensitive bacterium E. coli [41]. The expression of several other DNA damage response proteins, including ddrB, ddrC and ddrD, was also 2.5-fold lower in the dr0997 mutant. Although their precise functions remain to be illustrated, their important roles in the DNA damage response have already been confirmed [62].
Additionally, the transcriptional level of drRRA was nearly 2.9-fold lower in the dr0997 mutant than in the wild-type strain, indicating that DR0997 might be involved in the feedback regulation of drRRA expression [38]. The transcriptional levels of most of the above genes were also lower in the dr0997 mutant strain under ionizing radiation treatment (Table 3).
Among the detected genes, uvsE (dr1819), pprA (dra0346), and recN (dr1477) were shown to be directly regulated by DR0997 (Fig 8). RecN mainly functions in response to double-stranded DNA breaks, and it acts as a cohesion-like protein that stimulates intermolecular DNA interactions [63, 64]. PprA is regarded as a RecA-independent, DNA repair-related protein in D. radiodurans[62, 65, 66]. UvsE encodes a UV damage endonuclease that is involved in nucleotide excision repair[67]. From the results of the qRT-PCR analysis, the transcript levels of these genes were lower in the dr0997 mutant than in the wild-type strain under normal conditions and in response to ionizing radiation treatment, indicating that they play crucial roles in DNA repair.
Genes in other cellular pathways are also affected in the dr0997 mutant
As previously mentioned, CRP is a global transcriptional regulator that is involved in many cellular pathways [3, 53–55]. Based on the EMSA results, the genes that are directly regulated by DR0997 can be classified into diverse cellular pathways, including oxidative response and DNA repair pathways. The transcriptional levels of these genes were also measured under normal conditions or ionizing radiation treatment (Table 4).
Table 4. qRT PCR of directly regulated genes in the dr0997 mutant relative to wild-type R1.
| Locus | Annotation | Fold change (±SD) | |
|---|---|---|---|
| γ free | γ treatment | ||
| Δ0997(-γ)/R1(-γ) | Δ0997(+γ)/R1(+γ) | ||
| DR0997 | CRP/FNR family transcriptional regulator | 0.12 (±0.01) | 0.17 (±0.01) |
| DR0834 | CRP/FNR family transcriptional regulator | -1.03 (±0.08) | -1.21 (±0.07) |
| DR1646 | CRP/FNR family transcriptional regulator | -1.56 (±0.11) | -2.35 (±0.12) |
| DR2362 | CRP/FNR family transcriptional regulator | -2.78 (±0.09) | -1.95 (±0.12) |
| DR0990 | acetoin utilization protein AcuB | 0.77 (±0.09) | -2.00 (±0.29) |
| DR0998 | ATP-binding protein involved in chromosome partitioning(mrp) | -3.10 (±0.11) | -2.72 (±0.04) |
| DR1736 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase 3'-nucleotidase CpdB | -1.13 (±0.15) | -1.47 (±0.19) |
| DR1974 | ATP-dependent protease LA | -2.71 (±0.22) | -5.48 (±1.06) |
| DRA0006 | cyclase/dehydrase | -3.42 (±0.13) | -1.40 (±0.15) |
| DR1929 | glycerol uptake facilitator protein GlpF | -1.14 (±0.07) | -4.07 (±0.13) |
| DR1689 | glucose-1-phosphate adenylyltransferase GlgC | -22.02 (±1.04) | -24.25 (±0.20) |
| DR2531 | thiosulfate sulfurtransferase | -2.02 (±0.19) | -2.92 (±0.02) |
| DR0349 | ATP-dependent protease LA | -3.78 (±0.13) | -5.54 (±0.93) |
Functional annotation is based on KEGG (http://www.genome.jp/kegg/).
Of the identified genes, glpF (dr1929) and glgC (dr1689) are involved in glycometabolism. Dr0349 and dr1974 encode proteins that belong to the Lon protease family, which can degrade and recycle damaged proteins, suggesting that DR0997 participates in protein degradation. CpdB (dr1736) encodes a cAMP phosphodiesterase that catalyzes the conversion of cAMP to AMP, while dra0006 encodes a CyaA-like cyclase, indicating that DR0997 might regulate cAMP metabolism. Dr0998 encodes an ATP-binding protein that is involved in chromosome partitioning, which implies that it functions in chromosome partitioning during cell division. Meanwhile, the dr2531 gene encodes a thiosulfate sulfurtransferase that was also regulated by DR0997. Because the oxidation of thiols would result in non-native disulfide bond formation in proteins[52] and cause protein damage, it is suggested that DR0997 may have a role in regulating sulfur transfers and the prevention of thiol oxidation.
Furthermore, it is interesting that DR0997 could regulate not only its own expression, but also that of the other CRP homologs in D. radiodurans, indicating that DR0997 can regulate its own expression to cope with changes in the environment.
Discussion
The regulation of gene expression is critical for a cell’s response and survival in various environments. As global transcriptional factors, CRPs have been investigated in diverse bacteria, and they are considered to be important players in the responses to various environmental changes. In this study, an analysis of survival of CRP knockout strains, as well as the transcriptional responses of CRP-regulated genes, revealed that one of the CRPs (DR0997) plays vital roles in D. radiodurans.
The first CRP regulation model was in E. col several decades ago[68]. Just like E. coli CRP, D. radiodurans CRPs contain a cyclic nucleotide-binding domain in their amino-terminus, as well as a helix-turn-helix domain that bindings DNA and recruits RNAP to promoters to activate transcription. The crystal structure of an E. coli CRP-DNA complex showed that the CRP dimer binds to the TGTGA and TCACA blocks of the 22-bp consensus CRP binding site. The consensus CRP-binding sites in other CRP homologs, such as C. glutamicum GlxR, M. tuberculosis Rv3676, and Thermus thermophilus HB8 TTHA1437, are highly similar. Using WebLogo, we obtained the sequence logo of DR0997-binding sites (TGTGA-N6-TCACA) based on the aforementioned EMSA data (Fig 9), which is somewhat similar to the consensus binding site of E. coli CRP (AAATGTGA-N6-TCACATTT) and other CRP family proteins[52, 69]. The specific binding site should be defined by further biochemical experiments. In E. coli CRP, cAMP acts as an essential signaling molecule, and it helps to form the CRP-DNA complex. FNR uses the redox states of bound metals as signals to regulate the expression of other genes[70] The M. tuberculosis genome encodes 15 adenylate cyclases, showing the importance of cAMP[71]. However, D. radiodurans does not contain a classical adenylate cyclase that catalyzes the synthesis of cAMP. Nevertheless, an alternation of the cAMP concentration after radiation has been reported[72], indicating that some uncharacterized proteins encode adenylate cyclases. DR0997 was also observed to regulate the expression of dra0006, which contains domains that are similar to those of adenylate cyclases, and dr1736 (cpdB), which catalyzes the conversion of cAMP to AMP, implying a connection between DR0997-mediated gene regulation and cAMP metabolism.
E. coli CRP has been demonstrated to play its anti-oxidation role by regulating the expression of rpoS, which activates the general stress response[73]. Using an error-prone PCR technique, CRP was engineered to improve oxidative stress resistance[55]. The T. thermophilus HB8 CRP/FNR family protein SdrP is regarded as an oxidative stress-responsive transcriptional activator[74], and it was shown to control the promoters of sodA and a thioredoxin reductase-encoding gene, which are known to participate in redox control and oxidative resistance. However, there are no rpoS homologs in D. radiodurans, and the CRP family protein DR0997 does not bind the promoters of sodA (dr1279) and katE (data not shown). Nevertheless, it was demonstrated that DR0997 binds the promoters of dr1506, which encodes NADH-quinone oxidoreductase subunit A, and terB (dr2220). DR1506 is involved in redox control, while DR2220 was reported to respond to oxidative stress[75]. These data suggest that DR0997 plays a role in transcriptional regulation during anti-oxidant processes.
For decades, CRPs have been regarded to be involved in metabolism and anti-oxidation pathways. However, our investigations revealed that DR0997 also directly regulates the expression of DNA repair-related genes, including recN, pprA, and uvsE. Many DNA damage response and repair genes were indirectly regulated by DR0997. It will be interesting to further clarify the function of DR0997 in DNA damage responses and DNA repair.
Beyond the impact of DR0997 on cells via its regulatory role in oxidation and DNA repair responses, its ability to regulate other cellular pathways could also affect cell survival. The regulation of expression of Lon protease and thiosulfate sulfurtransferase would directly affect cell viability in response to damage, while regulating glycometabolism would affect energy production. The regulation of the cAMP pathway also affects many cell activities. Further studies of DR0997 will contribute to a detailed understanding of its functions on the regulatory network during stress responses.
Supporting Information
(TIF)
(A) All of the mutants were identified using their upF and downR (S1 File) primers, respectively. Lane 1, the wild-type strain; lane 2, the indicated mutant respectively. (B) The complemented strain and the mutant strain were identified using com0997F and com0997R primers, respectively. Lane 1, the dr0997 mutant complemented with dr0997 gene; Lane 2, the Δdr0997 strain.
(TIF)
Wild-type and dr2362 mutant were exposed to 50 mM H2O2 for different periods of time. Data represent the averages and standard deviations of three independent experiments.
(TIF)
Lane 1, 100 ng of the indicated DNA fragment; lanes 2 to 6, 100 ng of the indicated DNA fragment with increasing concentrations of DR0997 (0.1, 0.25, 0.5, 1, and 10 μM, respectively).
(TIF)
(TIF)
Lane 1, protein ladder; lane 2, DR0997 protein.
(TIF)
Lane 1, protein ladder; lane 2, DR0615 protein.
(TIF)
Lane 1, 100 ng of the indicated DNA fragment; lanes 2 to 6, 100 ng of the indicated DNA fragment with increasing concentrations of DR0997 (0.1, 0.25, 0.5, 1, and 10 μM, respectively).
(TIF)
(DOC)
(DOC)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to thank Professor Bingkui Wang and his colleagues (Institute of Crops and Nuclear Technology, Academy of Agricultural Sciences, Zhejiang, China) for their assistance in radiation treatment.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
YH is supported by National Basic Research Program of China (2015CB910600) and a grant from Special Fund for Agro-scientific Research in the Public Interest from the Ministry of Agriculture of China (201103007); YH, LW, and BT are supported by the National Science Foundation (31210103904, 31370102, 31370119, 31570058); LW is supported by the Natural Science Foundation of Zhejiang Province (LY13C010001) and the Public Project of Zhejiang Province (2014C33024).
References
- 1.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–95. [DOI] [PubMed] [Google Scholar]
- 2.Tutar Y. Neglected role of cAMP receptor protein monomer. Mol Biol Rep. 2012;39(4):4261–5. 10.1007/s11033-011-1212-5 [DOI] [PubMed] [Google Scholar]
- 3.Shimada T, Fujita N, Yamamoto K, Ishihama A. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One. 2011;6(6):e20081 10.1371/journal.pone.0020081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abe F, Maeda Y. Precise expression of the cAMP receptor gene, CAR1, during transition from growth to differentiation in Dictyostelium discoideum. FEBS Lett. 1994;342(3):239–41. [DOI] [PubMed] [Google Scholar]
- 5.Kumar S, Prakash N, Sharma VK. Control of minicell producing cell division by cAMP-receptor protein complex in Escherichia coli. Mol Gen Genet. 1979;176(3):449–50. [DOI] [PubMed] [Google Scholar]
- 6.Kim YR, Lee SE, Kim B, Choy HE, Rhee JH. A dual regulatory role of cAMP receptor protein in various virulence traits of Vibrio vulnificus. Microbiol Immunol. 2013. [DOI] [PubMed] [Google Scholar]
- 7.Britos L, Abeliuk E, Taverner T, Lipton M, McAdams H, Shapiro L. Regulatory response to carbon starvation in Caulobacter crescentus. PLoS One. 2011;6(4):e18179 10.1371/journal.pone.0018179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kahramanoglou C, Cortes T, Matange N, Hunt DM, Visweswariah SS, Young DB, et al. Genomic mapping of cAMP receptor protein (CRP Mt) in Mycobacterium tuberculosis: relation to transcriptional start sites and the role of CRPMt as a transcription factor. Nucleic Acids Res. 2014;42(13):8320–9. 10.1093/nar/gku548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mesa S, Hennecke H, Fischer HM. A multitude of CRP/FNR-like transcription proteins in Bradyrhizobium japonicum. Biochem Soc Trans. 2006;34(Pt 1):156–9. [DOI] [PubMed] [Google Scholar]
- 10.Derouaux A, Dehareng D, Lecocq E, Halici S, Nothaft H, Giannotta F, et al. Crp of Streptomyces coelicolor is the third transcription factor of the large CRP-FNR superfamily able to bind cAMP. Biochem Biophys Res Commun. 2004;325(3):983–90. [DOI] [PubMed] [Google Scholar]
- 11.Tagami H, Aiba H. Role of CRP in transcription activation at Escherichia coli lac promoter: CRP is dispensable after the formation of open complex. Nucleic Acids Res. 1995;23(4):599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuang SE, Daniels DL, Blattner FR. Global regulation of gene expression in Escherichia coli. J Bacteriol. 1993;175(7):2026–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Umemoto A, Morita M, Nakazono N, Sugino Y. The effect of the crp genotypes on the transformation efficiency in Escherichia coli. DNA Res. 1996;3(2):93–4. [DOI] [PubMed] [Google Scholar]
- 14.Huynh TT, McDougald D, Klebensberger J, Al Qarni B, Barraud N, Rice SA, et al. Glucose starvation-induced dispersal of Pseudomonas aeruginosa biofilms is cAMP and energy dependent. PLoS One. 2012;7(8):e42874 10.1371/journal.pone.0042874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimoto N, Toyama A, Takeuchi H. Binding modes of cyclic AMP and environments of tryptophan residues in 1:1 and 1:2 complexes of cyclic AMP receptor protein and cyclic AMP. Biopolymers. 2002;67(3):186–96. [DOI] [PubMed] [Google Scholar]
- 16.Tagami H, Aiba H. A common role of CRP in transcription activation: CRP acts transiently to stimulate events leading to open complex formation at a diverse set of promoters. EMBO J. 1998;17(6):1759–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savery NJ, Rhodius VA, Wing HJ, Busby SJ. Transcription activation at Escherichia coli promoters dependent on the cyclic AMP receptor protein: effects of binding sequences for the RNA polymerase alpha-subunit. Biochem J. 1995;309 (Pt 1):77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heyduk E, Heyduk T. Physical studies on interaction of transcription activator and RNA-polymerase: fluorescent derivatives of CRP and RNA polymerase. Cell Mol Biol Res. 1993;39(4):401–7. [PubMed] [Google Scholar]
- 19.Bonarek P, Kedracka-Krok S, Kepys B, Wasylewski Z. Quantitative analysis of the ternary complex of RNA polymerase, cyclic AMP receptor protein and DNA by fluorescence anisotropy measurements. Acta Biochim Pol. 2008;55(3):537–47. [PubMed] [Google Scholar]
- 20.Meiklejohn AL, Gralla JD. Activation of the lac promoter and its variants. Synergistic effects of catabolite activator protein and supercoiling in vitro. J Mol Biol. 1989;207(4):661–73. [DOI] [PubMed] [Google Scholar]
- 21.Gallagher DT, Smith N, Kim SK, Robinson H, Reddy PT. Profound asymmetry in the structure of the cAMP-free cAMP Receptor Protein (CRP) from Mycobacterium tuberculosis. J Biol Chem. 2009;284(13):8228–32. 10.1074/jbc.C800215200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH. Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol. 2004;14(1):10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blazy B, Culard F, Maurizot JC. Interaction between the cyclic AMP receptor protein and DNA. Conformational studies. J Mol Biol. 1987;195(1):175–83. [DOI] [PubMed] [Google Scholar]
- 24.Park SH, Lee TW, Lee BJ. Conformational change of cyclic AMP receptor protein by the binding of cyclic nucleotide. Biochem Mol Biol Int. 1996;40(1):93–100. [DOI] [PubMed] [Google Scholar]
- 25.Arce-Rodriguez A, Durante-Rodriguez G, Platero R, Krell T, Calles B, de Lorenzo V. The Crp regulator of Pseudomonas putida: evidence of an unusually high affinity for its physiological effector, cAMP. Environ Microbiol. 2012;14(3):702–13. 10.1111/j.1462-2920.2011.02622.x [DOI] [PubMed] [Google Scholar]
- 26.Ebright RH, Ebright YW, Gunasekera A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucleic Acids Res. 1989;17(24):10295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letek M, Valbuena N, Ramos A, Ordonez E, Gil JA, Mateos LM. Characterization and use of catabolite-repressed promoters from gluconate genes in Corynebacterium glutamicum. J Bacteriol. 2006;188(2):409–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai G, McCue LA, McDonough KA. Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J Bacteriol. 2005;187(22):7795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cameron AD, Redfield RJ. Non-canonical CRP sites control competence regulons in Escherichia coli and many other gamma-proteobacteria. Nucleic Acids Res. 2006;34(20):6001–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kristensen HH, Valentin-Hansen P, Sogaard-Andersen L. Design of CytR regulated, cAMP-CRP dependent class II promoters in Escherichia coli: RNA polymerase-promoter interactions modulate the efficiency of CytR repression. J Mol Biol. 1997;266(5):866–76. [DOI] [PubMed] [Google Scholar]
- 31.Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, et al. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev. 2001;65(1):44–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkateswaran A, McFarlan SC, Ghosal D, Minton KW, Vasilenko A, Makarova K, et al. Physiologic determinants of radiation resistance in Deinococcus radiodurans. Appl Environ Microbiol. 2000;66(6):2620–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka M, Earl AM, Howell HA, Park MJ, Eisen JA, Peterson SN, et al. Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics. 2004;168(1):21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Zhou J, Omelchenko MV, Beliaev AS, Venkateswaran A, Stair J, et al. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc Natl Acad Sci U S A. 2003;100(7):4191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polayes DA, Parks TD, Johnston SA, Dougherty WG. Application of TEV Protease in Protein Production. Methods in molecular medicine. 1998;13:169–83. 10.1385/0-89603-485-2:169 [DOI] [PubMed] [Google Scholar]
- 36.Huang LF, Hua XT, Lu HM, Gao GJ, Tian B, Shen BH, et al. Functional analysis of helicase and three tandem HRDC domains of RecQ in Deinococcus radiodurans. J Zhejiang Univ Sci B. 2006;7(5):373–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Hu J, Liu M, Yang S, Zhao Y, Cheng K, Xu G, Li M, Tian B, Hua Y. Int J Radiat Biol. Proteomic insights into the functional basis for the response regulator DrRRA of Deinococcus radiodurans. 2016. March 7:1–8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Xu G, Chen H, Zhao Y, Xu N, Tian B, et al. DrRRA: a novel response regulator essential for the extreme radioresistance of Deinococcus radiodurans. Mol Microbiol. 2008;67(6):1211–22. 10.1111/j.1365-2958.2008.06113.x [DOI] [PubMed] [Google Scholar]
- 39.Cao L, Dai C, Li Z, Fan Z, Song Y, Wu Y, et al. Antibacterial activity and mechanism of a scorpion venom peptide derivative in vitro and in vivo. PLoS One. 2012;7(7):e40135 10.1371/journal.pone.0040135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang DY, Luo M, Wang W, Zhao CJ, Gu CB, Zu YG, et al. Variation of active constituents and antioxidant activity in pyrola [P. incarnata Fisch.] from different sites in Northeast China. Food Chem. 2013;141(3):2213–9. 10.1016/j.foodchem.2013.05.045 [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Xu Q, Lu H, Lin L, Wang L, Xu H, et al. Protease activity of PprI facilitates DNA damage response: Mn2+-dependence and substrate sequence-specificity of the proteolytic reaction. PLoS One. 2015;10(3):e0122071 10.1371/journal.pone.0122071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaines PJ, Tang L, Wisnewski N. Insect allantoinase: cDNA cloning, purification, and characterization of the native protein from the cat flea, Ctenocephalides felis. Insect Biochem Mol Biol. 2004;34(3):203–14. [DOI] [PubMed] [Google Scholar]
- 43.Chen H, Xu G, Zhao Y, Tian B, Lu H, Yu X, et al. A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. PLoS One. 2008;3(2):e1602 10.1371/journal.pone.0001602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc. 2007;2(8):1849–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee EJ, Glasgow J, Leu SF, Belduz AO, Harman JG. Mutagenesis of the cyclic AMP receptor protein of Escherichia coli: targeting positions 83, 127 and 128 of the cyclic nucleotide binding pocket. Nucleic Acids Res. 1994;22(15):2894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber IT, Steitz TA. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 A resolution. J Mol Biol. 1987;198(2):311–26. [DOI] [PubMed] [Google Scholar]
- 47.Passner JM, Schultz SC, Steitz TA. Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 A resolution. J Mol Biol. 2000;304(5):847–59. [DOI] [PubMed] [Google Scholar]
- 48.Parkinson G, Wilson C, Gunasekera A, Ebright YW, Ebright RH, Berman HM. Structure of the CAP-DNA complex at 2.5 angstroms resolution: a complete picture of the protein-DNA interface. J Mol Biol. 1996;260(3):395–408. [DOI] [PubMed] [Google Scholar]
- 49.Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991;253(5023):1001–7. [DOI] [PubMed] [Google Scholar]
- 50.Green J, Scott C, Guest JR. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv Microb Physiol. 2001;44:1–34. [DOI] [PubMed] [Google Scholar]
- 51.Kiley PJ, Beinert H. The role of Fe-S proteins in sensing and regulation in bacteria. Curr Opin Microbiol. 2003;6(2):181–5. [DOI] [PubMed] [Google Scholar]
- 52.Agari Y, Kashihara A, Yokoyama S, Kuramitsu S, Shinkai A. Global gene expression mediated by Thermus thermophilus SdrP, a CRP/FNR family transcriptional regulator. Mol Microbiol. 2008;70(1):60–75. 10.1111/j.1365-2958.2008.06388.x [DOI] [PubMed] [Google Scholar]
- 53.Huang L, Pu Y, Yang X, Zhu X, Cai J, Xu Z. Engineering of global regulator cAMP receptor protein (CRP) in Escherichia coli for improved lycopene production. J Biotechnol. 2015;199:55–61. 10.1016/j.jbiotec.2015.02.006 [DOI] [PubMed] [Google Scholar]
- 54.Donovan GT, Norton JP, Bower JM, Mulvey MA. Adenylate cyclase and the cyclic AMP receptor protein modulate stress resistance and virulence capacity of uropathogenic Escherichia coli. Infect Immun. 2013;81(1):249–58. 10.1128/IAI.00796-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basak S, Jiang R. Enhancing E. coli tolerance towards oxidative stress via engineering its global regulator cAMP receptor protein (CRP). PLoS One. 2012;7(12):e51179 10.1371/journal.pone.0051179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J, Beacham IR. Transcription and regulation of the cpdB gene in Escherichia coli K12 and Salmonella typhimurium LT2: evidence for modulation of constitutive promoters by cyclic AMP-CRP complex. Mol Gen Genet. 1990;222(1):161–5. [DOI] [PubMed] [Google Scholar]
- 57.Hollands K, Busby SJ, Lloyd GS. New targets for the cyclic AMP receptor protein in the Escherichia coli K-12 genome. FEMS Microbiol Lett. 2007;274(1):89–94. [DOI] [PubMed] [Google Scholar]
- 58.Shinagawa H. [SOS responses to the mechanism of Holliday junction resolution by RuvABC proteins]. Tanpakushitsu kakusan koso Protein, nucleic acid, enzyme. 2003;48(6):727–32. [PubMed] [Google Scholar]
- 59.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chemical reviews. 2006;106(2):233–52. [DOI] [PubMed] [Google Scholar]
- 60.Weng MW, Zheng Y, Jasti VP, Champeil E, Tomasz M, Wang Y, et al. Repair of mitomycin C mono- and interstrand cross-linked DNA adducts by UvrABC: a new model. Nucleic Acids Res. 2010;38(20):6976–84. 10.1093/nar/gkq576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hua Y, Narumi I, Gao G, Tian B, Satoh K, Kitayama S, et al. PprI: a general switch responsible for extreme radioresistance of Deinococcus radiodurans. Biochem Biophys Res Commun. 2003;306(2):354–60. [DOI] [PubMed] [Google Scholar]
- 62.Selvam K, Duncan JR, Tanaka M, Battista JR. DdrA, DdrD, and PprA: components of UV and mitomycin C resistance in Deinococcus radiodurans R1. PLoS One. 2013;8(7):e69007 10.1371/journal.pone.0069007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pellegrino S, Radzimanowski J, de Sanctis D, Boeri Erba E, McSweeney S, Timmins J. Structural and functional characterization of an SMC-like protein RecN: new insights into double-strand break repair. Structure. 2012;20(12):2076–89. 10.1016/j.str.2012.09.010 [DOI] [PubMed] [Google Scholar]
- 64.Reyes ED, Patidar PL, Uranga LA, Bortoletto AS, Lusetti SL. RecN is a cohesin-like protein that stimulates intermolecular DNA interactions in vitro. J Biol Chem. 2010;285(22):16521–9. 10.1074/jbc.M110.119164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narumi I, Satoh K, Cui S, Funayama T, Kitayama S, Watanabe H. PprA: a novel protein from Deinococcus radiodurans that stimulates DNA ligation. Mol Microbiol. 2004;54(1):278–85. [DOI] [PubMed] [Google Scholar]
- 66.Dulermo R, Onodera T, Coste G, Passot F, Dutertre M, Porteron M, et al. Identification of new genes contributing to the extreme radioresistance of Deinococcus radiodurans using a Tn5-based transposon mutant library. PLoS One. 2015;10(4):e0124358 10.1371/journal.pone.0124358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Earl AM, Rankin SK, Kim KP, Lamendola ON, Battista JR. Genetic evidence that the uvsE gene product of Deinococcus radiodurans R1 is a UV damage endonuclease. J Bacteriol. 2002;184(4):1003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lisenkov AF, Smirnov Iu V. [Study of the regulation of crp gene expression in Escherichia coli K12]. Mol Gen Mikrobiol Virusol. 1990;(3):18–21. [PubMed] [Google Scholar]
- 69.Lavigne M, Kolb A, Buc H. Transcription activation by cAMP receptor protein (CRP) at the Escherichia coli gal P1 promoter. Crucial role for the spacing between the CRP binding site and the -10 region. Biochemistry. 1992;31(40):9647–56. [DOI] [PubMed] [Google Scholar]
- 70.Akhter Y, Yellaboina S, Farhana A, Ranjan A, Ahmed N, Hasnain SE. Genome scale portrait of cAMP-receptor protein (CRP) regulons in mycobacteria points to their role in pathogenesis. Gene. 2008;407(1–2):148–58. [DOI] [PubMed] [Google Scholar]
- 71.Akhter Y, Tundup S, Hasnain SE. Novel biochemical properties of a CRP/FNR family transcription factor from Mycobacterium tuberculosis. Int J Med Microbiol. 2007;297(6):451–7. [DOI] [PubMed] [Google Scholar]
- 72.Kamble VA, Rajpurohit YS, Srivastava AK, Misra HS. Increased synthesis of signaling molecules coincides with reversible inhibition of nucleolytic activity during postirradiation recovery of Deinococcus radiodurans. FEMS Microbiol Lett. 2010;303(1):18–25. 10.1111/j.1574-6968.2009.01855.x [DOI] [PubMed] [Google Scholar]
- 73.Barth E, Gora KV, Gebendorfer KM, Settele F, Jakob U, Winter J. Interplay of cellular cAMP levels, {sigma}S activity and oxidative stress resistance in Escherichia coli. Microbiology. 2009;155(Pt 5):1680–9. 10.1099/mic.0.026021-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agari Y, Kuramitsu S, Shinkai A. Identification of novel genes regulated by the oxidative stress-responsive transcriptional activator SdrP in Thermus thermophilus HB8. FEMS Microbiol Lett. 2010;313(2):127–34. 10.1111/j.1574-6968.2010.02133.x [DOI] [PubMed] [Google Scholar]
- 75.Anaganti N, Basu B, Gupta A, Joseph D, Apte SK. Depletion of reduction potential and key energy generation metabolic enzymes underlies tellurite toxicity in Deinococcus radiodurans. Proteomics. 2015;15(1):89–97. 10.1002/pmic.201400113 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(A) All of the mutants were identified using their upF and downR (S1 File) primers, respectively. Lane 1, the wild-type strain; lane 2, the indicated mutant respectively. (B) The complemented strain and the mutant strain were identified using com0997F and com0997R primers, respectively. Lane 1, the dr0997 mutant complemented with dr0997 gene; Lane 2, the Δdr0997 strain.
(TIF)
Wild-type and dr2362 mutant were exposed to 50 mM H2O2 for different periods of time. Data represent the averages and standard deviations of three independent experiments.
(TIF)
Lane 1, 100 ng of the indicated DNA fragment; lanes 2 to 6, 100 ng of the indicated DNA fragment with increasing concentrations of DR0997 (0.1, 0.25, 0.5, 1, and 10 μM, respectively).
(TIF)
(TIF)
Lane 1, protein ladder; lane 2, DR0997 protein.
(TIF)
Lane 1, protein ladder; lane 2, DR0615 protein.
(TIF)
Lane 1, 100 ng of the indicated DNA fragment; lanes 2 to 6, 100 ng of the indicated DNA fragment with increasing concentrations of DR0997 (0.1, 0.25, 0.5, 1, and 10 μM, respectively).
(TIF)
(DOC)
(DOC)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.