Abstract
Neurodegenerative diseases are a leading cause of death. No disease-modifying therapies are available, and preclinical animal model data have routinely failed to translate into success for therapeutics. Induced pluripotent stem cell (iPSC) biology holds great promise for human in vitro disease modeling because these cells can give rise to any cell in the human brain and display phenotypes specific to neurodegenerative diseases previously identified in postmortem and clinical samples. Here, we explore the potential and caveats of iPSC technology as a platform for drug development and screening, and the future potential to use large cohorts of disease-bearing iPSCs to perform clinical trials in a dish.
Keywords: clinical trials, human pluripotent stem cells, drug development, neurodegeneration
INTRODUCTION
Neurodegenerative diseases (NDDs) are leading causes of death in the United States (1), yet no disease-modifying therapies exist. Clinical trials to identify new drugs for these diseases have famously failed (2, 3). This failure has led to a lack of therapeutics for most NDDs (4, 5) and has discouraged the pharmaceutical industry from investing in new research in this area. The cost and suffering from these diseases will become even more critical as the aged population most at risk for neurodegeneration increases in the coming years (6).
The human central nervous system (CNS) is perhaps the most complex organ in the body, and is mostly inaccessible to manipulation and study. Because the basic biology of the CNS is still being worked out, NDDs create special challenges. Furthermore, it is unclear how understanding the basic biology of the CNS will inform us about age-related brain diseases. Many NDDs appear after reproductive age and may be relatively unshaped by evolution. They might result from a toxic gain of function of an offending protein that has no relationship to the protein’s normal function. The disease condition may be its own unique state, and insights into the basic biology may be uninformative. Thus, accurate and predictive disease models are essential.
NDDs include Alzheimer’s disease (AD), Huntington’s disease (HD), frontotemporal dementia (FTD), Parkinson’s disease (PD), and motor neuron diseases (MNDs) [e.g., amyotrophic lateral sclerosis (ALS)]. Each disease is characterized by dysfunction and death of a specific subtype of neurons and, at the cellular level, displays pathologies that may include cytoplasmic and nuclear protein aggregation, endoplasmic reticulum stress, neuromuscular junction degradation and synaptic defects, proteasome inhibition, axonal transport defects, mitochondrial dysfunction, neurofilament accumulation, increased oxidative stress, glutamate-mediated hyperexcitability, and microglial and astrocyte activation/toxicity (5, 7). NDD mechanisms are not fully understood, and diagnoses depend on clinical manifestations of the disease well after the cellular pathology has begun. Many NDDs have a handful of known familial or genetic causes; however, most are of unknown or sporadic origin. Biomarkers are lacking at all stages, and patient heterogeneity is high. Individual and population differences in the disease-causing agent could be due to genetic, epigenetic, or environmental insults, and unidentified modifiers of disease that could contribute to susceptibility and pathophysiology (8).
Why have so many clinical trials failed? Two primary reasons stand out, and each is a significant challenge. There has been limited success in fully modeling human NDDs; thus, the current preclinical translational pipeline relies heavily on humanized transgenic animal models of disease, which have poor predictive value in a clinical setting. Additionally, a successful trial may require the patient population to be stratified in ways that consider the pathogenic diversity in humans. Both issues highlight the need for human models of NDD that more accurately reflect the disease phenotype in vivo.
Recent advances in stem cell technologies might help to solve these two challenges. Human induced pluripotent stem cells (iPSCs), discovered by Shinya Yamanaka, are particularly exciting. They avoid the cross-species issues of animal models, obviate most ethical concerns with stem cells, and provide a model with a completely human genome and a potentially unlimited source of human subjects. Although iPSCs are not perfect, they provide researchers a tempting method for modeling disease in vitro. Here, we review the potential and challenges of using human iPSCs as a platform for drug development, from the screening of compounds to the use of large cohorts of iPSC lines to perform clinical trials in a dish.
DO PRECLINICAL TESTS IN ANIMAL MODELS CORRELATE WITH CLINICAL RESULTS?
Animal models have been valuable for increasing our understanding of disease processes, but recently, scientists are looking more critically at their value in preclinical testing. Traditionally, efficacy in animal models has been a gatekeeper for clinical trials (9). However, the failure to translate these findings into clinical trials has created concern about their relevance as a predictive model of human disease.
In fact, humans and animals are quite different, and those differences include CNS complexity, lifespan, metabolic rates (including transmitter function), anatomical complexity, and importantly drug absorption, metabolism, and secretion. Even a single amino acid change in proteins between mouse and human can change pharmacological responsiveness (10, 11). Transgenic animal models often express multiple copies of disease-associated mutant human transgenes, but patients generally express only a single allele (12), and even with elevated levels of the mutant human protein, many animal models recapitulate only a subset of the human pathologies. Some rodent models, such as the SOD1 mouse model of ALS (13) and the R6 HD mouse model (14, 15), have led to the discovery of agents that mitigate disease in the laboratory; however, those test agents have nearly all failed in human trials (16, 17). Therefore, recapitulating cytopathological and behavioral features may not be close enough to the human condition and therefore may not be sufficient to make predictive models of disease. Fundamental species-specific differences may be a crucial factor in the failure to translate results from animal models to the clinic.
Additionally, animal models and humans differ in genetic modifiers of disease. Variants in many genes contribute to risk and presumably to pathophysiology (18, 19). Overall, rodent and human genes are conserved, but they differ at the individual nucleotide level (8). Because we do not fully understand the genetic architecture of any of these diseases, serious questions remain. How similar or dissimilar are the genetic environments of mice and humans? How can we tease apart the influence of specific identified genetic mutations versus genetic variation at multiple susceptibility loci?
It is quite profound that aging is the single greatest risk factor for NDDs, and AD, PD, ALS, and HD are not seen in the wild in any of the species we use in the laboratory. Newly appreciated and significant differences in transcriptional changes associated with aging across species, even between human and subhuman primates, are emerging (16, 17, 20). If something as fundamental as aging has species-specific features, this would also contribute to the predictive value of models for NDDs and other diseases associated with aging. We can conclude that nonhuman animal models cannot fully recapitulate human biology, and that the extent of the differences is not fully understood and has likely been underestimated. Accordingly, modeling human CNS diseases in nonhuman models will necessarily have major limitations, which warrant reconsideration of the gatekeeper role that mouse models often play in preclinical drug development.
PATIENT STRATIFICATION
Patient differences are another potential reason that clinical trials fail. Poor phenotyping and endotyping, often because of a lack of robust biomarkers, can lead to the inclusion of patients that do not belong in a trial (9). Several clinical trials for AD failed while attempting to address the role of β-amyloid protein, and the reason was obscured by including patients with a different disease or who were not expressing the protein at sufficient levels (21, 22). It is not clear how much the lack of patient stratification contributes to trial failure. Most cases of AD, ALS, and PD are sporadic, and the cause or causes of the diseases are not known. A drug candidate might provide significant benefit in one case, but not in others. Clinical trials are powered on the assumption that all the enrolled patients are potentially responsive; therefore, without some stratification, the trial could be doomed to failure even though the test agent might be efficacious in a subset of participants. Clearly, new tools are needed to weed out inappropriate participants.
HUMAN PLURIPOTENT STEM CELLS
In the past 15 years, the advent of human pluripotent stem cells (hPSCs) brought new methodologies to modeling human development and disease in vitro. Human embryonic stem cells (hESCs) isolated from the inner cell mass of a human embryo (23) are a self-renewing population of stem cells that give rise to any cell type of the body when given the correct set of signals and growth conditions. hESCs can be modified genetically by introducing a disease gene of interest, and preimplantation diagnosis identifies embryos carrying deleterious alleles (24, 25). However, this approach is hampered by the limited supply and genetic diversity of embryos in the clinic and ethical objections over the destruction of embryos to derive hESCs.
These issues were alleviated to a great extent by the work of Yamanaka and colleagues on somatic cell reprogramming (26). They showed that human fibroblasts derived from skin samples could be reprogrammed to an embryonic-like state by using viruses to introduce pluripotency-inducing transcription factors. The resulting iPSCs are very similar to hESCs in their self-renewal and pluripotent differentiation potential. Rapid adoption of iPSC technology demonstrated the robust nature of reprogramming, and iPSCs can now be made with many gene combinations and delivery methods, including excisable transposons and adenovirus; importantly, many of these methods do not require genetic integration of the transgenes, such as delivery by RNA, protein, and nonintegrating plasmids (27, 28). As the field streamlines these protocols and large iPSC banks are created, there will be a potentially unlimited and diverse source of patient lines poised for enrollment in clinical trials in a dish.
iPSC DIFFERENTIATION TO BRAIN CELLS
iPSCs can theoretically be differentiated into essentially any cell type, including CNS cells. Thus, this technology provides the ability to model all forms of a given disease. Familial and sporadic patients can be represented and bring along with them a true representation of the diversity of genetic modifiers in a given population that can never be achieved with mice.
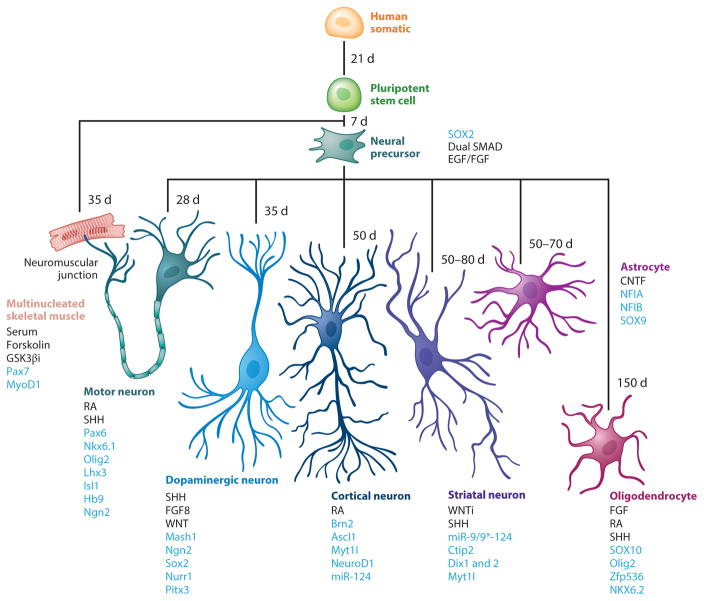
Differentiating iPSCs to CNS cells of interest is a key first step in the development of iPSCs as a tool for in vitro drug development and clinical trials. Protocols have built on lessons from developmental studies that elucidated the combination and timing of instructive queues to drive differentiation of specific lineages. Approaches generally involve growth factors or small molecules to recapitulate the ontogeny of the cell type of interest, for example, neurons (29) and astrocytes (30) (Figure 1). This can also be accomplished by directly reprogramming somatic cells with ectopic expression of genetic drivers toward a given lineage, such as neural precursor cells (31), neurons (32, 33), and astrocytes (34) (Figure 1). Microglia, oligodendrocytes, and Schwann cells have been more difficult to derive from hPSCs, but new reports are emerging (35–39). Specific subtypes of CNS cells, including layer-specific cortical neurons (40), dopaminergic neurons (41, 42), motor neurons (MNs; 43–45), striatal neurons (46), and cortical interneurons (47), can also be derived (Figure 1). As these cells are differentially vulnerable in FTD, PD, MNDs, HD, and AD, they are important for disease modeling and drug development.
Figure 1.
iPSC differentiation flow chart describing differentiation protocols, from somatic cells through iPSC derivation and differentiation to neural, neuronal, and glial lineages. Growth factors driving differentiation are shown in black, and blue indicates exogenously expressed factors used in direct reprogramming protocols (31–33, 38, 40, 44, 49, 59, 64, 75, 93, 94, 135–150). Abbreviations: CNTF, ciliary neurotrophic factor; d, days of growth factor–driven differentiation; EGF, epidermal growth factor; FGF, fibroblast growth factor; FGF8, fibroblast growth factor 8; GSK3βi, glycogen synthase kinase-3β inhibitor; iPSC, induced pluripotent stem cell; RA, retinoic acid; SHH, sonic hedgehog; SMAD, intracellular proteins that transduce extracellular signals from TGFβ signaling; WNT, family of Wnt signaling pathways; WNTi, inhibitors of Wnt signaling pathways.
It is useful to consider MNDs in more depth: Protocols to differentiate iPSCs to MNs are common. They were built on the pioneering work by Wicheterle and Jessell, who derived MNs from mouse ESCs based on embryoid body formation (43, 44, 48). However, monolayer protocols that allow visualization of all stages of development were strongly desired and followed rapidly (49). Generally, MN derivation protocols take from 3 to 10 weeks to obtain the maximum number of postmitotic MNs, with a yield of 10–30% (44, 48), although recent reports indicated 75–80% pure MN derivation in 14–20 days (50, 51). Characterizations by gene and protein expression and electrophysiological properties suggest these protocols are deriving cells whose maturity roughly corresponds to embryonic or fetal cells in vivo. Thus, the requirements for in vitro maturation might not yet have been fully worked out. This important caveat to iPSC-based disease modeling is discussed in more depth below.
Finally, several groups reported derivation of skeletal muscle from iPSCs (52–54). This is not a CNS cell type, but skeletal muscle interacts with MNs at the neuromuscular junction (Figure 1), a point of early vulnerability in several NDDs (55–57); because of this, they are of great interest as a potential model of physiologically relevant coculture systems.
DISEASE-ASSOCIATED PHENOTYPIC END POINTS
The next step in designing a clinical trial in a dish is to determine clinically relevant end points. Current work is focused on determining how well disease-bearing iPSC lines model the cellular phenotypes seen in NDDs in vivo. Most papers reported one or more disease-related phenotypes, such as protein aggregation, defects in neurite growth, axonal transport, protein clearance and electrophysiology, and reduced survival in NDD iPSCs (Table 1). Neural precursor cells or neurons have been described for several diseases, including AD (58), FTD (37), HD (59), and PD (41, 60, 61) (Table 1).
Table 1.
Examples of neurodegenerative iPSC modeling
| Disease | Cell type | Phenotype | References |
|---|---|---|---|
| Alzheimer’s disease | Neurons | Increased deposition of pathological markers amyloid-β and phosphorylated Tau protein Activated glycogen synthase kinase-3β Endosomal abnormalities |
58, 125 |
| Frontotemporal dementia | Motor and forebrain neurons | Impaired axonal transport C9ORF72 repeat region containing RNA foci Altered gene expression Diminished neuronal firing capacity |
37, 72 |
| Huntington’s disease | Neural precursor cells, forebrain and striatal neurons, astrocytes | Reduced survival Altered gene expression Altered cytoskeleton, adhesion, energetics and neurite outgrowth Reduced neuronal firing capacity Increased susceptibility to stressors Cytoplasmic, electron clear vacuoles in astrocytes |
59, 65, 126 |
| Motor neuron diseases: amyotrophic lateral sclerosis Charcot-Marie-Tooth disease giant axonal neuropathy hereditary spastic paraplegia myotonic dystrophy type I spinal-bulbar muscular atrophy spinal muscular atrophy spinal muscular atrophy with respiratory distress type 1 | Motor neurons, astrocytes | Reduced survival Cytoplasmic and nuclear protein aggregation Altered gene expression Neurite degeneration/abnormal growth/decreased complexity Increased oxidative stress and susceptibility to stressors, including PI3K inhibition Altered subcellular axonal transport Activation of endoplasmic reticulum stress and unfolded protein response pathways C9ORF72 repeat region containing RNA foci Intrinsic membrane hyperexicitability and diminished firing capacity Noncell autonomous toxicity from disease-bearing astrocytes |
34, 62, 64, 66, 67, 69, 86, 127–134 |
| Parkinson’s disease | Dopaminergic neurons | Reduced survival Increased susceptibility to stressors Mitochondrial dysfunction Elevated disease-associated α-synuclein protein Reduced glucocerebrosidase enzymatic activity Reduced synthesis and release of dopamine Increased monoamine oxidase B expression Impaired intrinsic network activity |
41, 60, 61, 94, 97 |
In one example, the HD iPSC Consortium used several methods, including gene microarray profiling, to uncover multiple disease-relevant phenotypes that revealed differences in control and disease lines in gene expression as early as the neural precursor cell stage of development. They also described gene expression and survival differences between differentiated iPSCs from patients with early onset of the disease with higher numbers of CAG repeats relative to those from patients with late onset and lower numbers of CAG repeats (59), elucidating potential factors behind the differences in age of onset seen in patients carrying different repeat lengths. Importantly, one study looked at 16 ALS lines (which are representative of ~90% of the patient population) from sporadic patients. Three had intranuclear aggregates that contained a disease-associated marker, phosphorylated TDP-43, in the iPSC-derived MNs that resembled those in postmortem ALS patients (62, 63).
Disease modeling in other types of CNS cells has also been reported. iPSC-derived astrocytes have been described with cell autonomous and nonautonomous toxicity and, in one report, a reduction in iPSC-derived astrocytes from an ALS patient (34, 64, 65). Interestingly, astrocyte-mediated toxicity was found in control MNs cultured with astrocytes made from iPSC lines of sporadic ALS patients and those with SOD1 mutations (34). However, a different ALS-associated mutation in the TDP-43 gene did not appear to cause cell nonautonomous toxicity (64). These findings could be helpful in stratifying patients before clinical trials. However, more models need to be investigated to clarify the contribution of genetics to pathogenesis in vitro.
Early onset childhood diseases, such as spinal muscular atrophy, have very robust survival defects (66–68). Adult-onset diseases, such as AD and ALS, although they show many aspects of cellular dysfunction, are more variable in neuronal loss (69–72). Because a hallmark of NDDs is loss of a specific neuronal subtype, it would seem to be an important readout. Interestingly, the risk of death of MNs and astrocytes bearing a mutation in the disease-related protein TDP-43 was greater than that of controls in a highly sensitive single-cell longitudinal imaging platform that assessed survival (64, 73–76). Thus, in adult-onset diseases, multiple factors may limit our ability to detect disease-related death, such as maturation of the cell at risk; however, these might be overcome with an improvement in technology.
LIMITATIONS OF iPSC TECHNOLOGY
Of course, iPSCs are not without caveats. hPSC science is a young field, and technical issues abound. iPSC-based cell culture is considerably more time and money intensive than the culture of other cell lines and primary cells. There is low efficiency of derivation and high variability in maintenance and differentiation to a mature cell of interest. Derivation methods have evolved rapidly, and they include standardization of media formulations, substrates, and small molecules to promote and maintain pluripotency. The field has moved away from the integration-dependent viral system first reported because aberrant expression from integrated transgenes can alter iPSC growth and differentiation characteristics (77); now, transgenes are mainly introduced by episomal plasmids (27) or noncytopathic viral delivery with viruses, such as Sendai virus (78). Derivation is geared toward creating iPSCs in an integration-free manner, which is essential to ensure that iPSCs are reliable in a clinical setting.
Other iPSC issues include differences in gene expression, self-renewal, differentiation potential, copy number variants, genomic instability, retention of epigenetic signature of the parental somatic cell, and incomplete epigenetic remodeling (79–82). These differences are seen in clones from the same individual and across iPSCs, but close examination of a large number of clones (>10) reveals clones with the best pluripotency and differentiation potential (83). It may be necessary to characterize a large number of clones after derivation to ensure selection of lines with an optimal pluripotency profile. Finally, although many improvements have been made, some problems may be inherent in the starting material or in other stochastic events that are not well understood but are required for reprogramming, and these problems are therefore more difficult to overcome (79). As our understanding of the reprogramming process improves, we hope that such black-box issues will be clarified.
Genomic instability is a major concern with iPSCs and their differentiated progeny, as it could affect results in disease modeling and also diminishes the promise of iPSC-derived cells as a transplantation therapy (84). Instability can be particularly troubling when reprogramming patient samples from diseases, such as HD, spinal-bulbar muscular dystrophy, C9ORF72-associated ALS, and FTD, that are caused by expansion of repeat regions within the disease-causing gene (Table 1). In these lines, the repeat region is often not stable during and after reprogramming, and the instability can continue during differentiation (85, 86). For example, in one study, six iPSC lines with spinal-bulbar muscular atrophy were derived, but only three were used in which the disease associated poly-Q-androgen receptor expansion was stable over multiple passages (86). Chromosomal abnormalities and novel mutations may be unavoidable with long-term culture of iPSCs regardless of the disease being modeled (87); therefore, early passages of iPSCs must be banked immediately after characterization and continually validated while in use.
Control lines are generally derived from patient-matched samples, such as a relative or a healthy individual from the population. They do not have the disease in question, but genome-wide association studies have shown us that everyone carries variants at numerous susceptibility loci (88). We do not fully understand to what extent other loci from the genome contribute to phenotypic differences that arise in disease-bearing iPSCs relative to controls.
One way to address this is to generate isogenic control lines from disease-bearing iPSCs with gene-editing enzymes, such as zinc-finger nucleases (ZFNs), transcriptional activator-like effector nucleases (TALENs), or clustered regulatory interspaced short palindromic repeat (CRISPR)/Cas–based RNA-guided DNA endonucleases (89).
These nucleases use the cellular processes of DNA double-strand breaks and their repair to edit targeted regions of interest, such as NDD-causing genetic mutations. Reversal of the disease phenotype is a proof of principle in itself, and it also provides a control line that is a genetic match to the patient line still bearing the disease-associated mutation. A ZFN-mediated correction of a mutation in the gene coding the Tau protein, which is implicated in several NDDs, rescued Tau-associated abnormalities in neurons with the mutation (90). Furthermore, a ZFN-derived line that was homozygous for the mutation had intensified tauopathies. The technology provided multiple benefits, including a more severe version of the original disease model and genetic rescue of the original lesion while creating a genetically corrected control. However, as mentioned above, there are caveats similar to those with any derivation, as well as the potential for off-target effects to occur (91, 92), and expensive characterization is needed to avoid these pitfalls. Furthermore, isogenic controls may not always be the best option. It may be necessary to ask what will be more informative for a given question: isogenic pairs of lines that eliminate noise in the system or a large number of unique control and disease lines in which you might capture more genetic diversity and a better sense of the robustness of effects.
Other difficulties of iPSCs are protocol efficiency, reproducibility, end-product maturation, and heterogeneity at all stages. As outlined above, there are abundant differentiation protocols. Differences have been described in starting material, media and growth factor cocktails, substrates, and monolayer versus suspension cultures. Additional steps can involve purification of specific lineages during differentiation to ensure maximum yield (93, 94). In new hands, all steps may require optimization, even when the exact protocol and reagents are used (87). A recent comparison of multiple protocols to derive forebrain neurons found that a neurosphere approach promoted highest yield of the neural lineage, but efficient and timely maturation required coculture with astrocytes (95). More reports like this are needed, but for now, each group may need to perform a smaller version of this analysis to assess the best method for their question. Standardized yield and end-point maturity must be achieved across iPSCs if in vitro clinical trials are to be successful.
The large-scale production required for drug screening or in vitro drug trials presents other problems. Batch-to-batch variations in the differentiation protocols could affect results. One strategy is to make enough material initially to freeze down batches of cells at specific critical times in the protocol; this would create enough material to make it through multiple experiments or trials from the same starting material. For this approach to be successful, validation of relevant markers of the lineage must be examined on each batch to ensure post-thaw stability of the population of interest.
Many iPSC lines are based on genetic variants of NDDs; however, in the case of AD, ALS, and PD, the majority of the cases are sporadic. Currently, we have no rational way to model sporadic disease in nonhuman systems; therefore, iPSCs offer hope. However, if the environment/aging has a major role in pathogenesis and that role is to impose epigenetic marks, those contributions might disappear during reprogramming. Initial work is promising, as several reports have described phenotypes in sporadic iPSCs (58, 62).
Finally, most NDDs take decades for disease phenotypes to manifest. Why would anyone think one could model a disease that typically appears in the fifth to seventh decade of life after just a few weeks of reprogrammed stem cell differentiation? Of course, there is tremendous redundancy and plasticity in the CNS. Thus, the age of symptom onset for NDDs is when coping responses that mask symptoms fail, which is much later than when the disease actually begins. In HD, a genetic mutation is expressed from before birth, and evidence suggests that abnormalities can be seen during development (96). Therefore, symptoms in HD could reflect the cumulative effects of the mutation, posing no conceptual obstacle to the idea that one can measure disease-relevant phenotypes in a dish, even in a relatively immature cell. Indeed, this was described in vitro as seen by the aberrant gene expression in neural precursors derived from HD iPSCs (59). These findings are promising, but maturity and aging are still concerns. At a minimum, some phenotypes or functions might only be expressed in mature cells, so an immature cell might not be capable of generating a phenotype of interest. Brain cells produced by current protocols generally display characteristics of embryonic and fetal stage development, perhaps mirroring the human developmental timeline (47), and are difficult to mature. Although strong phenotypes are reported, such as neuronal loss in disease models from childhood-onset diseases [e.g., spinal muscular atrophy (SMA)] (66), these have been harder to detect in iPSC-derived cells from late-onset diseases unless they were captured by a highly sensitive detection platform (59, 64, 75, 76). iPSC derivation involves epigenetic processes, and marks of lineage, aging, or the environment may be lost (79, 97).
These may be problems when modeling a disease that emerges in old age or involves changes to the capacity of homeostatic systems that are reduced with aging. Symptoms or phenotypes may only emerge once the homeostatic capacity is expended. Efforts to age iPSC-derived brain cells have focused on increasing oxidative stress with chemical treatments to mimic aging (58, 60, 98). These did promote some disease phenotypes and cellular loss, but the effect was variable. Another strategy is to use progerin, which induces premature aging in vitro when ectopically expressed (99). Indeed, when progerin was ectopically expressed in iPSC-derived dopaminergic neurons, it induced age-related characteristics, including condensed nuclei and decreased dendrite length. This occurred in both control and PD lines; however, the effect was more pronounced in PD neurons, which suggests that both age and genetic susceptibility to PD contributed to the phenotype (97). It remains to be seen if progerin works in all NDDs or in other neurological diseases with adult onset. It is one of the few tools that promote an aged phenotype, but premature aging in a late-onset disease may have other consequences that have not yet been uncovered. For drug development and in vitro clinical trials, the aged population will likely be of high interest, and modeling iPSCs to resemble the age at onset of clinical phenotype, although perhaps not essential for all disease pathologies, is desirable.
Future goals in the field will be to lower cost and to improve efficiency of deriving, maintaining, and differentiating iPSCs; this includes the ability to recapitulate the relevant biology and disease state of the cell(s) affected in NDDs. One important step in the attempt to fully model defects in brain biology will be to create coculture and spatial models that mimic the complex interactions and dimensionality seen in normal and diseased brains. Coculture of hPSC-derived neurons with astrocytes has been reported (34, 45, 64), and it uncovered important pathological relationships; however, most protocols focused on deriving one type of cell. Structures such as the optic cup self-organize during hPSC differentiation (100), and it would be interesting to see if simpler protocols can move toward greater cellular complexity. Lancaster et al. (101) recently used a three-dimensional cell culture protocol to make a minibrain that mimicked a young cerebral cortex with the neuronal layering seen in cortical development. Minibrains from microcephalic patient iPSCs were smaller than controls, as expected. Hopefully, these and other techniques will continue to evolve and will provide answers to modeling complex cellular interactions seen in NDDs.
Although iPSCs have limitations, the field is advancing quickly to improve derivation and differentiation methods while continuing to build and characterize collections of iPSC lines from many individuals, within and across disease (59, 62, 84). Hundreds of reports detail derivations of iPSCs from many different somatic cell lineages (102), and many examples of disease-relevant phenotypes, have been reported. The number of validated and characterized iPSC lines is growing exponentially, and they are being banked in centers around the world, with support from government, universities, nonprofit institutes, and private companies (e.g., NIMH Stem Cell Center, Kyoto University Center for iPS Cell Research and Application, Harvard Stem Cell Institute, Corriel Institute, American Type Cell Collection, and Wi-Cell). In 2014, 200–250 lines were banked, and numbers are expected to rise into the tens of thousands within 5 years (103). Therefore, within 10 years of the first report of human iPSCs (26), investigators will likely have thousands of iPSC lines to select from, and this will provide the material to recruit patients for in vitro clinical trials.
ASSAY DEVELOPMENT
In vitro drug development and clinical trials will also benefit from well-designed methods of data collection. Protocols have been reported for adapting iPSCs for high-throughput screening (HTS) and high-content screening (HCS) (104). HTS uses automation techniques to quickly interrogate the effects of a large number of compounds, approximately 10,000 or more agents per day, on a relatively simple biological outcome of interest (105). HCS uses cellular imaging to extract quantitative data from complex biological systems and generally measures multiple end points (106). HTS can be difficult in hPSCs owing to the amount of material needed to assay thousands of compounds. Therefore, HCS approaches have been more commonly used when uncovering novel disease pathologies and new disease-modifying therapeutics, including treatments that promote cellular fate in iPSCs (106, 107). However, as systems evolve methods to deal with the heterogeneity of iPSC systems, HTS in iPSCs will become more common as a first pass screening tool to test large libraries of potential therapeutic agents. The focus of iPSC use has been drug discovery, in which a few lines are used to interrogate many potential therapeutics. These assays should translate well as the field shifts to clinical trials that test candidate drugs in many iPSC lines.
Drug discovery has been dominated by two approaches: phenotypic and target-based assays. Phenotypic assays measure the effects of drugs against a cellular or disease-related phenotype without necessarily knowing the molecular target(s) of the perturbagens being tested. Target-based assays measure the effects of compounds on a specific molecular target, often a protein implicated in disease. In recent years, target-based approaches have dominated, but phenotypic screens have been an important player in first-in-kind drug discovery in the past 10 years (108), and are making a strong comeback. Phenotypic assays can identify effects in complex systems, even in a disease setting in which the biology is not fully understood. This may be particularly suitable in iPSC-derived systems that model NDDs, given that much of the etiology in NDDs is unresolved, involves complex interactions, and often has multiple cellular phenotypes (109–112).
Phenotypic assays generally interrogate one to several cellular features of interest. They most commonly use chemical or functional genomics approaches. Chemical genomics screens libraries of well-annotated small molecules for alteration of a biological process or disease phenotype after treatment (112). Functional genomics uses the same approach, but with libraries of nucleic acids, such as small interfering RNAs, that will knockdown genes and that often cover the whole genome (113). Knockdown of each gene gives information on how loss of that gene impacts a cellular phenotype. These two approaches can identify modifiers of cell fate or survival and disease progression and can lead to the discovery of genes that are good drug targets or even promising compounds that can be developed into drugs. The end-point assay often depends on the cell type in question and includes fluorescence-activated cell sorting (114, 115), gene expression with reverse transcription polymerase chain reaction, imaged-based immunofluorescence on cells fixed at discrete times, and longitudinal imaging of live cells that contain a fluorescent reporter(s) that identifies the cell and/or disease process that is being followed.
ANALYSIS
Extracting useful information from cell-based assays can be challenging, particularly when those assays are on a scale as grand as in HTS and HCS. These settings generate vast amounts of data, and computerized data management, analysis, and statistical programs are necessary to gain full value and detect phenotypic differences (116, 117). Careful experimental design starts with consideration of the end-point sensitivity, the effect size, the desired level of confidence, and the rationale behind sample size determination (118). Assay quality can be determined by the Z′ factor, which is a statistical measure that combines assay robustness with the variability of signal. Z′ > 0.5 is optimal, although this applies only to assays with one time point (112). Many platforms track and quantify cells (74, 117). In general, data are collected and organized, and then cells are identified relative to the background, often through a user-defined thresholding and segmentation process (110, 119). Once identified, the cell can be assessed for cellular phenotypes, or live imaging can be used to track the cell’s position and status. Cellular phenotypes can be based on survival, morphology, or changes to a marker of biological significance, such as disease-associated protein expression, mislocalization, or aggregation.
The next step is to apply an algorithm to extract a particular feature and to quantify a phenotype. In some cases, such as when there is a decrease in cell number of a particular cell type, the phenotype is relatively easy for a computer to identify. This is even easier when there are very good positive and negative controls to set the relevant parameters. However, in complex disease settings such as NDDs, it may be necessary for experienced biologists to determine and define novel phenotypes caused by the experimental conditions/treatments (116), and this can be challenging in large data sets. One answer is to use machine learning, in which iterative feedback can be used to hone the system’s ability to accurately identify and cluster novel, rare, or subtle phenotypes (120). The program uses information defined by the researcher’s classification of randomly chosen examples and presents a potential rule for classifying cells. Several rounds of feedback on how rules classify a subset of the data set result in a final set of rules for each phenotype of interest that can be applied to the full data set (120). Tools such as machine learning will provide the analytical support needed to work through large complex data sets generated from in vitro drug testing and clinical trials, and they have the added potential to uncover novel or rare phenotypes undetected by humans.
ROBOTIC MICROSCOPY
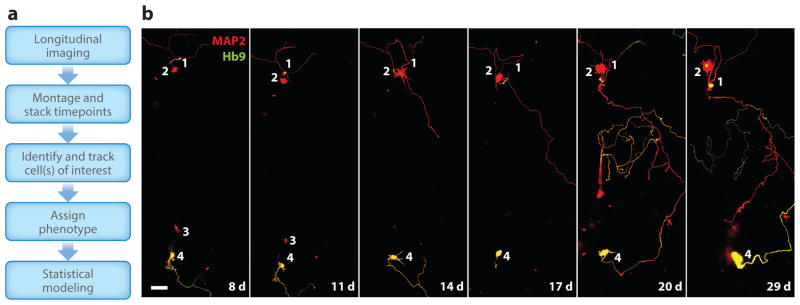
Our group developed an automated longitudinal live-cell imaging and analysis system called robotic microscopy (RM) that incorporates the sensitivity and resolution of single-cell analysis with the speed of HTS/HCS (74). This system allows us to interrogate multiple functions with protein-specific fluorescent markers, creating a live-cell multivariate profile across time that can be related to a cell’s fate (109, 110). Key innovations include an autofocus system to collect many images per well, a fiduciary point that allows the system to find the same cell at different times, and robotics to transfer plates to the microscope from the incubator and back, which allows the system to operate 24/7 without human intervention (110). In-house software packages collect and organize the image files, stitch them into a montaged image, stack the montages based on time, automatically identify cells of interest (which are often neurons expressing a cell-specific morphological marker), and track them over time (Figure 2) (109).
Figure 2.
Acquisition, analysis, and modeling in robotic microscopy (RM). (a) Typical flow of data capture, organization, analysis, binning by phenotype, and statistical modeling with RM. (b) Longitudinal imaging of a control induced pluripotent stem cell (iPSC)-derived motor neuron differentiation 8–29 days (d) posttransfection with the neuronal marker MAP2 (red) and the motor neuron–specific marker Hb9 (green). Note that cell 2 is positive only for MAP2, and cell 4 is positive for both MAP2 and Hb9. Also note that cell 3 is no longer in the analysis after day 11. Scale bar indicates 25 μm.
Longitudinal analysis of single cells can be thought of as similar to evaluation of an individual in a clinical trial, and we use similar analysis tools (i.e., survival analysis and Cox proportional hazards analysis) (121). When used in RM, they enable us to develop statistical models that determine how the biological events measured during the experiment predict a particular outcome (74, 110). We used this approach initially to answer a nagging question in the field of NDD. Are protein aggregations or inclusion bodies in HD protective or pathogenic mechanisms? Indeed, with the increased sensitivity of single-cell longitudinal imaging, we found that diffuse rather than aggregated mutant proteins were toxic to the cell, and the formation of inclusion bodies improved survival (73).
We also discovered that RM is a particularly powerful tool in heterogeneous cell systems, which is one of the more vexing problems with iPSCs. Using a primary rodent system ectopically expressing disease-associated mutant proteins, we discovered that RM is about 100–1,000-fold more sensitive at detecting disease phenotypes and disease-modifying genes or small-molecule therapeutics than other methods (61, 122, 123). In fact, approximately eight cells per well can predict the overall performance of a well with about 90% accuracy in some applications. Indeed, with single-cell analysis, we uncovered survival phenotypes in iPSC-derived neurons that others have not, without the use of stressors (59, 61, 64, 75, 76). This has a significant effect on our ability to conduct HTS/HCS assays in which we interrogate iPSC-derived cultures. As an example, there can be extensive variability in the maturity of differentiated iPSCs within the same well (124). If aspects of the disease emerge as the cell matures, population-level analysis at discrete times may not capture this shift to a disease phenotype, or the heterogeneity of the culture may make it difficult to parse out differences. RM will capture change as it occurs in an individual cell and will stratify the cells into populations on the basis of protein expression, maturation, survival/disease progression, and even responder/nonresponder classification in the case of drug treatments. RM is therefore an example of a highly sensitive new technology that can be exploited to interrogate complex biological systems derived from iPSCs.
CLINICAL TRIALS IN A DISH
It is a long journey from iPSC derivation and differentiation to the cell type of interest, disease modeling, drug development, and finally in vitro clinical trials. The ultimate goal is to harness the power of iPSC technology to allow for multiple levels of interrogation, which are outlined in Figure 3. A primary screen of drug candidates, generally in one iPSC line, can be followed by hit validation, optimization, toxicology, and mechanistic studies in several lines. However, as the numbers of iPSC lines grow, it will be possible to implement clinical trials in a dish and to test the candidate drugs identified in many patient lines. We hope that these studies will bridge the gap between preclinical data and clinical success. By uncovering in vitro efficacy and toxicology across a human population and giving rise to patient stratification schemes, clinical trials in a dish could vastly improve in vivo clinical trial design, which should contribute to better translation of drug candidates in human subjects suffering from NDDs and other devastating diseases.
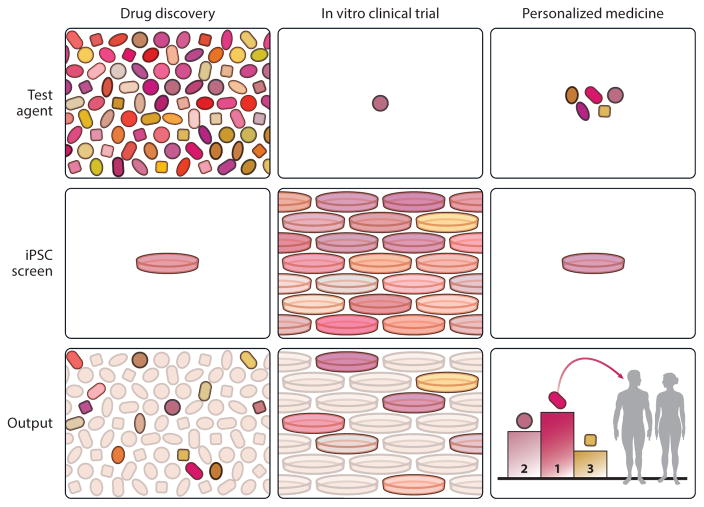
Figure 3.
Clinical trials in a dish. The first column depicts the primary screen, done in a single patient-derived iPSC line from a patient with a particular disease, such as a type of ALS. After additional validation steps (not shown), the agent that emerges as the top candidate moves to the second column, the clinical trial in a dish. The candidate is tested on many iPSCs, each from different people with the same clinical syndrome but possibly with different underlying causes, to identify the subset of responders versus nonresponders. Knowledge of these patients and their response phenotype can be used to create a stratification scheme to enroll patients in the eventual in vivo clinical trials and to improve the chances of demonstrating efficacy in a predefined subset of patients. The third column depicts the hypothetical day when multiple effective medications, resulting from the work done in the first and second columns, can be tailored to a particular patient. In that case, the therapeutic options are tested against the patient’s iPSCs, and the safest, most effective one is chosen. This is the concept of personalized/precision medicine. Abbreviations: ALS, amyotrophic lateral sclerosis; iPSC, induced pluripotent stem cell.
SUMMARY POINTS.
NDDs are leading causes of death in the United States, but no disease-modifying therapies exist.
Clinical trials to identify new drugs for these diseases have failed, possibly because of poor translation of animal model preclinical data to human patients and failure to properly stratify patients in clinical trials.
Human iPSCs provide an in vitro platform to model human development and disease in previously inaccessible cells, such as neurons and glial cells, which make up the brain.
Imaging systems, such as RM, are sensitive enough to overcome the heterogeneity of iPSC differentiations and to detect differences in survival and other disease phenotypes between control and disease-bearing iPSC-derived brain cells.
iPSCs provide a potentially unlimited and genetically diverse source of patients to use for in vitro drug discovery, patient stratification, and clinical trials in a dish. The goal is to eventually utilize iPSC across the drug development process: from drug screening in one line, to candidate testing in many, and finally back to a single patient and the concept of personalized medicine.
FUTURE ISSUES.
Current differentiation protocols used to make iPSCs into brain cells can be expensive and lengthy and produce relatively young cells that may be problematic when modeling diseases of old age. These protocols are rapidly improving, and they include the use of genetic drivers that directly reprogram a somatic cell to the cell type of interest and to factors that age the cells.
Multicellular coculture systems that model complex cellular interactions, such as the neuromuscular junction, which may be a site of early vulnerability in NDDs, are essential. These may require additional dimensionality that will be challenging to high-throughput imaging systems.
Robust numbers of fully characterized and validated clinically sound iPSC lines that exemplify the population of interest must be available. As yet, there is not a coordinated approach to this issue.
NDDs have diverse origins, and genetic, epigenetic, and environmental factors must be considered. Full-genome sequencing, epigenomics, and proteomics on a wide selection of iPSCs will be very helpful.
Acknowledgments
We thank members of the Finkbeiner lab, G. Maki for graphical support, and G. Howard for editorial input. S.F. was supported by NIH grants U01 MH1050135, U54 HG008105, R01 NS083390, 3 R01 NS039074, 2 R01 NS045091, and U24 NS078370; an NSF BRAIN EAGER award; the ALS association; the Michael J. Fox Foundation; the Hellman Family Foundation; and the Taube/Koret Center for Neurodegenerative Disease. K.M.H was supported by grants from the Roddenberry Foundation, and is a postdoctoral fellow of the California Institute of Regenerative Medicine (T2-0003).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review. The findings and conclusions presented in this review are those of the authors and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
LITERATURE CITED
- 1.Kochanek KD, Murphy SL, Xu J, Arias E. Mortality in the United States, 2013. Vol. 178. Natl. Cent. Health Stat; Hyattsville, MD: 2014. Data Brief. [PubMed] [Google Scholar]
- 2.Jucker M. The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat Med. 2010;16(11):1210–14. doi: 10.1038/nm.2224. [DOI] [PubMed] [Google Scholar]
- 3.Mitsumoto H, Brooks BR, Silani V. Clinical trials in amyotrophic lateral sclerosis: why so many negative trials and how can trials be improved? Lancet Neurol. 2014;13(11):1127–38. doi: 10.1016/S1474-4422(14)70129-2. [DOI] [PubMed] [Google Scholar]
- 4.Finkbeiner S. Bridging the Valley of Death of therapeutics for neurodegeneration. Nat Med. 2010;16(11):1227–32. doi: 10.1038/nm.2222. [DOI] [PubMed] [Google Scholar]
- 5.Mason AR, Ziemann A, Finkbeiner S. Targeting the low-hanging fruit of neurodegeneration. Neurology. 2014;83(16):1470–73. doi: 10.1212/WNL.0000000000000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattson MP, Magnus T. Ageing and neuronal vulnerability. Nat Rev Neurosci. 2006;7(4):278–94. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown GC, Neher JJ. Inflammatory neurodegeneration and mechanisms of microglial killing of neurons. Mol Neurobiol. 2010;41(2–3):242–47. doi: 10.1007/s12035-010-8105-9. [DOI] [PubMed] [Google Scholar]
- 8.Gusella JF, Macdonald ME, Lee J-M. Genetic modifiers of Huntington’s disease. Mov Disord. 2014;29(11):1359–65. doi: 10.1002/mds.26001. [DOI] [PubMed] [Google Scholar]
- 9.Pankevich DE, Altevogt BM, Dunlop J, Gage FH, Hyman SE. Improving and accelerating drug development for nervous system disorders. Neuron. 2014;84(3):546–53. doi: 10.1016/j.neuron.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattson MP, Rychlik B, You JS, Sisken JE. Sensitivity of cultured human embryonic cerebral cortical neurons to excitatory amino acid–induced calcium influx and neurotoxicity. Brain Res. 1991;542(1):97–106. doi: 10.1016/0006-8993(91)91003-j. [DOI] [PubMed] [Google Scholar]
- 11.Mertens J, Stüber K, Wunderlich P, Ladewig J, Kesavan JC, et al. APP processing in human pluripotent stem cell–derived neurons is resistant to NSAID-based γ-secretase modulation. Stem Cell Rep. 2013;1(6):491–98. doi: 10.1016/j.stemcr.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matus S, Medinas DB, Hetz C. Common ground: stem cell approaches find shared pathways underlying ALS. Cell Stem Cell. 2014;14(6):697–99. doi: 10.1016/j.stem.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Gurney ME. Transgenic-mouse model of amyotrophic lateral sclerosis. N Engl J Med. 1994;331(25):1721–22. doi: 10.1056/NEJM199412223312516. [DOI] [PubMed] [Google Scholar]
- 14.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87(3):493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 15.Li J-Y, Popovic N, Brundin P. The use of the R6 transgenic mouse models of Huntington’s disease in attempts to develop novel therapeutic strategies. NeuroRx. 2005;2(3):447–64. doi: 10.1602/neurorx.2.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGonigle P. Animal models of CNS disorders. Biochem Pharmacol. 2014;87(1):140–49. doi: 10.1016/j.bcp.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Perrin S. Preclinical research: make mouse studies work. Nature. 2014;507(7493):423–25. doi: 10.1038/507423a. [DOI] [PubMed] [Google Scholar]
- 18.Clabough EBD. Huntington’s disease: the past, present, and future search for disease modifiers. Yale J Biol Med. 2013;86(2):217–33. [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper-Knock J, Kirby J, Ferraiuolo L, Heath PR, Rattray M, Shaw PJ. Gene expression profiling in human neurodegenerative disease. Nat Rev Neurol. 2012;8(9):518–30. doi: 10.1038/nrneurol.2012.156. [DOI] [PubMed] [Google Scholar]
- 20.Winner B, Marchetto MC, Winkler J, Gage FH. Human-induced pluripotent stem cells pave the road for a better understanding of motor neuron disease. Hum Mol Genet. 2014;23(R1):R27–34. doi: 10.1093/hmg/ddu205. [DOI] [PubMed] [Google Scholar]
- 21.Jack CR, Jr, Holtzman DM. Biomarker modeling of Alzheimer’s disease. Neuron. 2013;80(6):1347–58. doi: 10.1016/j.neuron.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vellas B, Carrillo MC, Sampaio C, Brashear HR, Siemers E, et al. Designing drug trials for Alzheimer’s disease: what we have learned from the release of the phase III antibody trials: a report from the EU/US/CTAD Task Force. Alzheimer’s Dement. 2013;9(4):438–44. doi: 10.1016/j.jalz.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–47. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Yosef D, Malcov M, Eiges R. PGD-derived human embryonic stem cell lines as a powerful tool for the study of human genetic disorders. Mol Cell Endocrinol. 2008;282(1–2):153–58. doi: 10.1016/j.mce.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Frumkin T, Malcov M, Telias M, Gold V, Schwartz T, et al. Human embryonic stem cells carrying mutations for severe genetic disorders. In Vitro Cell Dev Biol Anim. 2010;46(3–4):327–36. doi: 10.1007/s11626-010-9275-5. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8(5):409–12. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 28.Coatti GC, Beccari MS, Olávio TR, Mitne-Neto M, Okamoto OK, Zatz M. Stem cells for amyotrophic lateral sclerosis modeling and therapy: myth or fact? Cytometry. 2015;87(3):197–211. doi: 10.1002/cyto.a.22630. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S-C, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19(12):1129–33. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 30.Krencik R, Weick JP, Liu Y, Zhang Z-J, Zhang S-C. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat Biotechnol. 2011;29(6):528–34. doi: 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, et al. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11(1):100–9. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, et al. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9(2):113–18. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer K, Ferraiuolo L, Miranda CJ, Likhite S, McElroy S, et al. Direct conversion of patient fibroblasts demonstrates non-cell autonomous toxicity of astrocytes to motor neurons in familial and sporadic ALS. PNAS. 2014;111(2):829–32. doi: 10.1073/pnas.1314085111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keirstead HS, Nistor G, Bernal G, Totoiu M, Cloutier F, et al. Human embryonic stem cell-derived oligodendrocyte progenitor cell transplants remyelinate and restore locomotion after spinal cord injury. J Neurosci. 2005;25(19):4694–705. doi: 10.1523/JNEUROSCI.0311-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu B-Y, Du Z-W, Zhang S-C. Differentiation of human oligodendrocytes from pluripotent stem cells. Nat Protoc. 2009;4(11):1614–22. doi: 10.1038/nprot.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126(3):385–99. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, Bates J, Li X, Schanz S, Chandler-Militello D, et al. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Stem Cell. 2013;12(2):252–64. doi: 10.1016/j.stem.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thoma EC, Merkl C, Heckel T, Haab R, Knoflach F, et al. Chemical conversion of human fibroblasts into functional Schwann cells. Stem Cell Rep. 2014;3(4):539–47. doi: 10.1016/j.stemcr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y, Kirwan P, Smith J, Robinson HPC, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15(3):477–86. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci Transl Med. 2012;4(141):141ra90. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kriks S, Shim J-W, Piao J, Ganat YM, Wakeman DR, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480(7378):547–51. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110(3):385–97. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 44.Singh Roy N, Nakano T, Xuing L, Kang J, Nedergaard M, Goldman SA. Enhancer-specified GFP-based FACS purification of human spinal motor neurons from embryonic stem cells. Exp Neurol. 2005;196(2):224–34. doi: 10.1016/j.expneurol.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 45.Giorgio FPD, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3(6):637–48. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 46.Aubry L, Bugi A, Lefort N, Rousseau F, Peschanski M, Perrier AL. Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. PNAS. 2008;105(43):16707–12. doi: 10.1073/pnas.0808488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12(5):573–86. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu B-Y, Zhang S-C. Differentiation of spinal motor neurons from pluripotent human stem cells. Nat Protoc. 2009;4(9):1295–304. doi: 10.1038/nprot.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27(3):275–80. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qu Q, Li D, Louis KR, Li X, Yang H, et al. High-efficiency motor neuron differentiation from human pluripotent stem cells and the function of Islet-1. Nat Commun. 2014;5:3449. doi: 10.1038/ncomms4449. [DOI] [PubMed] [Google Scholar]
- 51.Maury Y, Côme J, Piskorowski RA, Salah-Mohellibi N, Chevaleyre V, et al. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat Biotechnol. 2014;33(1):86–93. doi: 10.1038/nbt.3049. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka A, Woltjen K, Miyake K, Hotta A, Ikeya M, et al. Efficient and reproducible myogenic differentiation from human iPS cells: prospects for modeling Miyoshi myopathy in vitro. PLOS ONE. 2013;8(4):e61540. doi: 10.1371/journal.pone.0061540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abujarour R, Bennett M, Valamehr B, Lee TT, Robinson M, et al. Myogenic differentiation of muscular dystrophy-specific induced pluripotent stem cells for use in drug discovery. Stem Cells Transl Med. 2014;3(2):149–60. doi: 10.5966/sctm.2013-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yasuno T, Osafune K, Sakurai H, Asaka I, Tanaka A, et al. Functional analysis of iPSC-derived myocytes from a patient with carnitine palmitoyltransferase II deficiency. Biochem Biophys Res Commun. 2014;448(2):175–81. doi: 10.1016/j.bbrc.2014.04.084. [DOI] [PubMed] [Google Scholar]
- 55.Dupuis L, Loeffler JP. Neuromuscular junction destruction during amyotrophic lateral sclerosis: insights from transgenic models. Curr Opin Pharmacol. 2009;9(3):341–46. doi: 10.1016/j.coph.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Dadon-Nachum M, Ben-Yaacov K, Ben-Zur T, Barhum Y, Yaffe D, et al. Transplanted modified muscle progenitor cells expressing a mixture of neurotrophic factors delay disease onset and enhance survival in the SOD1 mouse model of ALS. J Mol Neurosci. 2014;55(3):788–97. doi: 10.1007/s12031-014-0426-0. [DOI] [PubMed] [Google Scholar]
- 57.Thomson SR, Wishart TM, Patani R, Chandran S, Gillingwater TH. Using induced pluripotent stem cells (iPSC) to model human neuromuscular connectivity: promise or reality? J Anat. 2012;220(2):122–30. doi: 10.1111/j.1469-7580.2011.01459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482(7384):216–20. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.HD iPSC Consortium. Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11(2):264–78. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8(3):267–80. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skibinski G, Nakamura K, Cookson MR, Finkbeiner S. Mutant LRRK2 toxicity in neurons depends on LRRK2 levels and synuclein but not kinase activity or inclusion bodies. J Neurosci. 2014;34(2):418–33. doi: 10.1523/JNEUROSCI.2712-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burkhardt MF, Martinez FJ, Wright S, Ramos C, Volfson D, et al. A cellular model for sporadic ALS using patient-derived induced pluripotent stem cells. Mol Cell Neurosci. 2013;56(C):355–64. doi: 10.1016/j.mcn.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–33. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 64.Serio A, Bilican B, Barmada SJ, Ando DM, Zhao C, et al. Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. PNAS. 2013;110(12):4697–702. doi: 10.1073/pnas.1300398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Juopperi TA, Kim W, Chiang C-H, Yu H, Margolis RL, et al. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol Brain. 2012;5(1):17. doi: 10.1186/1756-6606-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ebert AD, Yu J, Rose FF, Mattis VB, Lorson CL, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457(7227):277–80. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corti S, Nizzardo M, Simone C, Falcone M, Nardini M, et al. Genetic correction of human induced pluripotent stem cells from patients with spinal muscular atrophy. Sci Transl Med. 2012;4(165):165ra162. doi: 10.1126/scitranslmed.3004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sareen D, Ebert AD, Heins BM, McGivern JV, Ornelas L, Svendsen CN. Inhibition of apoptosis blocks human motor neuron cell death in a stem cell model of spinal muscular atrophy. PLOS ONE. 2012;7(6):e39113. doi: 10.1371/journal.pone.0039113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen H, Qian K, Du Z, Cao J, Petersen A, et al. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell. 2014;14(6):796–809. doi: 10.1016/j.stem.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SSW, et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7(1):1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, et al. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med. 2012;4(145):145ra104. doi: 10.1126/scitranslmed.3004052. [DOI] [PubMed] [Google Scholar]
- 72.Alami NH, Smith RB, Carrasco MA, Williams LA, Winborn CS, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81(3):536–43. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–10. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 74.Arrasate M, Finkbeiner S. Automated microscope system for determining factors that predict neuronal fate. PNAS. 2005;102(10):3840–45. doi: 10.1073/pnas.0409777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bilican B, Serio A, Barmada SJ, Nishimura AL, Sullivan GJ, et al. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. PNAS. 2012;109(15):5803–8. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10(8):677–85. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 78.Nishimura K, Sano M, Ohtaka M, Furuta B, Umemura Y, et al. Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming. J Biol Chem. 2011;286(6):4760–71. doi: 10.1074/jbc.M110.183780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10(6):678–84. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 80.Gore A, Li Z, Fung H-L, Young JE, Agarwal S, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471(7336):63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim K, Doi A, Wen B, Ng K, Zhao R, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467(7313):285–90. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2104;470(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144(3):439–52. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Inoue H, Nagata N, Kurokawa H, Yamanaka S. IPS cells: a game changer for future medicine. EMBO J. 2014;33(5):409–17. doi: 10.1002/embj.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sareen D, O’Rourke JG, Meera P, Muhammad AKMG, Grant S, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5(208):208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grunseich C, Zukosky K, Kats IR, Ghosh L, Harmison GG, et al. Stem cell-derived motor neurons from spinal and bulbar muscular atrophy patients. Neurobiol Dis. 2014;70(C):12–20. doi: 10.1016/j.nbd.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Santostefano KE, Hamazaki T, Biel NM, Jin S, Umezawa A, Terada N. A practical guide to induced pluripotent stem cell research using patient samples. Lab Investig. 2014;95(1):4–13. doi: 10.1038/labinvest.2014.104. [DOI] [PubMed] [Google Scholar]
- 88.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fong H, Wang C, Knoferle J, Walker D, Balestra ME, et al. Genetic correction of tauopathy phenotypes in neurons derived from human induced pluripotent stem cells. Stem Cell Rep. 2013;1(3):226–34. doi: 10.1016/j.stemcr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ding Q, Lee Y-K, Schaefer EAK, Peters DT, Veres A, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12(2):238–51. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mandal PK, Ferreira LMR, Collins R, Meissner TB, Boutwell CL, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Stem Cell. 2014;15(5):643–52. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ebert AD, Shelley BC, Hurley AM, Onorati M, Castiglioni V, et al. EZ spheres: a stable and expandable culture system for the generation of pre-rosette multipotent stem cells from human ESCs and iPSCs. Stem Cell Res. 2013;10(3):417–27. doi: 10.1016/j.scr.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Woodard CM, Campos BA, Kuo S-H, Nirenberg MJ, Nestor MW, et al. iPSC-derived dopamine neurons reveal differences between monozygotic twins discordant for Parkinson’s disease. Cell Rep. 2014;9(4):1173–82. doi: 10.1016/j.celrep.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Muratore CR, Srikanth P, Callahan DG, Young-Pearse TL. Comparison and optimization of hiPSC forebrain cortical differentiation protocols. PLOS ONE. 2014;9(8):e105807. doi: 10.1371/journal.pone.0105807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Humbert S. Is Huntington disease a developmental disorder? EMBO Rep. 2010;11(12):899. doi: 10.1038/embor.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13(6):691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell. 2013;12(4):487–96. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 99.Liu G-H, Barkho BZ, Ruiz S, Diep D, Qu J, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472(7342):221–25. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nakano T, Ando S, Takata N, Kawada M, Muguruma K, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10(6):771–85. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 101.Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2014;501(7467):373–79. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singh VK, Kalsan M, Kumar N, Saini A, Chandra R. Induced pluripotent stem cells: applications in regenerative medicine, disease modeling, and drug discovery. Front Cell Dev Biol. 2015;3:2. doi: 10.3389/fcell.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Solomon S, Pitossi F, Rao MS. Banking on iPSC—is it doable and is it worthwhile. Stem Cell Rev Rep. 2015;11(1):1–10. doi: 10.1007/s12015-014-9574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Studer L, Desbordes SC. Adapting human pluripotent stem cells to high-throughput and high-content screening. Nat Protoc. 2012;8(1):111–30. doi: 10.1038/nprot.2012.139. [DOI] [PubMed] [Google Scholar]
- 105.Engle SJ, Puppala D. Integrating human pluripotent stem cells into drug development. Cell Stem Cell. 2013;12(6):669–77. doi: 10.1016/j.stem.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 106.Zanella F, Lorens JB, Link W. High content screening: Seeing is believing. Trends Biotechnol. 2010;28(5):237–45. doi: 10.1016/j.tibtech.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 107.Rubin LL, Haston KM. Stem cell biology and drug discovery. BMC Biol. 2011;9(1):42. doi: 10.1186/1741-7007-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10(7):507–19. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 109.Daub A, Sharma P, Finkbeiner S. High-content screening of primary neurons: ready for prime time. Curr Opin Neurobiol. 2009;19(5):537–43. doi: 10.1016/j.conb.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sharma P, Ando DM, Daub A, Kaye JA, Finkbeiner S. High-throughput screening in primary neurons. Methods Enzymol. 2012;506:331–60. doi: 10.1016/B978-0-12-391856-7.00041-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McGivern JV, Ebert AD. Exploiting pluripotent stem cell technology for drug discovery, screening, safety, and toxicology assessments. Adv Drug Deliv Rev. 2014;69–70(C):170–78. doi: 10.1016/j.addr.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 112.Engle SJ, Vincent F. Small molecule screening in human induced pluripotent stem cell-derived terminal cell types. J Biol Chem. 2014;289(8):4562–70. doi: 10.1074/jbc.R113.529156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moffat J, Sabatini DM. Building mammalian signalling pathways with RNAi screens. Nat Rev Mol Cell Biol. 2006;7(3):177–87. doi: 10.1038/nrm1860. [DOI] [PubMed] [Google Scholar]
- 114.Edwards BS, Oprea T, Prossnitz ER, Sklar LA. Flow cytometry for high-throughput, high-content screening. Curr Opin Chem Biol. 2004;8(4):392–98. doi: 10.1016/j.cbpa.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 115.Sklar L, Carter M, Edwards B. Flow cytometry for drug discovery, receptor pharmacology and high-throughput screening. Curr Opin Pharmacol. 2007;7(5):527–34. doi: 10.1016/j.coph.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li F, Yin Z, Jin G, Zhao H, Wong STC. Chapter 17: bioimage informatics for systems pharmacology. PLOS Comp Biol. 2013;9(4):e1003043. doi: 10.1371/journal.pcbi.1003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 118.Steward O, Balice-Gordon R. Rigor or mortis: best practices for preclinical research in neuroscience. Neuron. 2014;84(3):572–81. doi: 10.1016/j.neuron.2014.10.042. [DOI] [PubMed] [Google Scholar]
- 119.Galimberti D, Scarpini E. Clinical phenotypes and genetic biomarkers of FTLD. J Neural Transm. 2012;119(7):851–60. doi: 10.1007/s00702-012-0804-0. [DOI] [PubMed] [Google Scholar]
- 120.Finkbeiner S, Frumkin M, Kassner PD. Cell-based screening: extracting meaning from complex data 2015. Neuron. 2015;86(1):160–74. doi: 10.1016/j.neuron.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Klein JP, Moeschberger ML. Techniques for censored and truncated data. Lifetime Data Anal. 1995;1(2):195–204. doi: 10.1007/BF00985770. [DOI] [PubMed] [Google Scholar]
- 122.Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA, Finkbeiner S. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. PNAS. 2010;107(39):16982–87. doi: 10.1073/pnas.1004498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbeiner S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30(2):639–49. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Koyanagi-Aoi M, Ohnuki M, Takahashi K, Okita K, Noma H, et al. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. PNAS. 2013;110(51):20569–74. doi: 10.1073/pnas.1319061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515(7526):274–78. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang QC, Yeh T-L, Leyva A, Frank LG, Miller J, et al. A compact model of Huntingtin toxicity. J Biol Chem. 2011;286(10):8188–96. doi: 10.1074/jbc.M110.192013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, et al. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell. 2014;14(6):781–95. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mitne-Neto M, Machado-Costa M, Marchetto MCN, Bengtson MH, Joazeiro CA, et al. Down-regulation of VAPB expression in motor neurons derived from induced pluripotent stem cells of ALS8 patients. Hum Mol Genet. 2011;20(18):3642–52. doi: 10.1093/hmg/ddr284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nihei Y, Ito D, Okada Y, Akamatsu W, Yagi T, et al. Enhanced aggregation of androgen receptor in induced pluripotent stem cell-derived neurons from spinal and Bulbar muscular atrophy. J Biol Chem. 2013;288(12):8043–52. doi: 10.1074/jbc.M112.408211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cortes CJ, Miranda HC, Frankowski H, Batlevi Y, Young JE, et al. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat Neurosci. 2014;17(9):1180–89. doi: 10.1038/nn.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Saporta MA, Dang V, Volfson D, Zou B, Xie XS, et al. Axonal Charcot-Marie-Tooth disease patient-derived motor neurons demonstrate disease-specific phenotypes including abnormal electrophysiological properties. Exp Neurol. 2015;263(C):190–99. doi: 10.1016/j.expneurol.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Havlicek S, Kohl Z, Mishra HK, Prots I, Eberhardt E, et al. Gene dosage-dependent rescue of HSP neurite defects in SPG4 patients’ neurons. Hum Mol Genet. 2014;23(10):2527–41. doi: 10.1093/hmg/ddt644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Denton KR, Lei L, Grenier J, Rodionov V, Blackstone C, Li X-J. Loss of spastin function results in disease-specific axonal defects in human pluripotent stem cell-based models of hereditary spastic paraplegia. Stem Cells. 2014;32(2):414–23. doi: 10.1002/stem.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Johnson-Kerner BL, Ahmad FS, Diaz AG, Greene JP, Gray SJ, et al. Intermediate filament protein accumulation in motor neurons derived from giant axonal neuropathy iPSCs rescued by restoration of gigaxonin. Hum Mol Genet. 2015;24(5):1420–31. doi: 10.1093/hmg/ddu556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xu C, Tabebordbar M, Iovino S, Ciarlo C, Liu J, et al. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell. 2013;155(4):909–21. doi: 10.1016/j.cell.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Borchin B, Chen J, Barberi T. Derivation and FACS-mediated purification of PAX3+/PAX7+ skeletal muscle precursors from human pluripotent stem cells. Stem Cell Rep. 2013;1(6):620–31. doi: 10.1016/j.stemcr.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9(10):2329–40. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Carri AD, Onorati M, Lelos MJ, Castiglioni V, Faedo A, et al. Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development. 2012;140(2):301–12. doi: 10.1242/dev.084608. [DOI] [PubMed] [Google Scholar]
- 139.Kriks S, Shim J-W, Piao J, Ganat YM, Wakeman DR, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480(7378):547–51. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Krencik R, Weick JP, Liu Y, Zhang Z-J, Zhang S-C. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat Biotechnol. 2011;29(6):528–34. doi: 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 142.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 143.Li W, Jiang K, Ding S. Concise review: a chemical approach to control cell fate and function. Stem Cells. 2012;30(1):61–68. doi: 10.1002/stem.768. [DOI] [PubMed] [Google Scholar]
- 144.Liu X, Li F, Stubblefield EA, Blanchard B, Richards TL, et al. Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 2011;22(2):321–32. doi: 10.1038/cr.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Caiazzo M, Dell’anno MT, Dvoretskova E, Lazarevic D, Taverna S, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476(7359):224–27. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 146.Victor MB, Richner M, Hermanstyne TO, Ransdell JL, Sobieski C, et al. Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron. 2014;84(2):311–23. doi: 10.1016/j.neuron.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9(3):205–18. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Caiazzo M, Giannelli S, Valente P, Lignani G, Carissimo A, et al. Direct conversion of fibroblasts into functional astrocytes by defined transcription factors. Stem Cell Rep. 2015;4(1):25–36. doi: 10.1016/j.stemcr.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang N, Zuchero JB, Ahlenius H, Marro S, Ng YH, et al. Generation of oligodendroglial cells by direct lineage conversion. Nat Biotechnol. 2013;31(5):434–39. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Najm FJ, Zaremba A, Caprariello AV, Nayak S, Freundt EC, et al. Rapid and robust generation of functional oligodendrocyte progenitor cells from epiblast stem cells. Nat Methods. 2011;8(11):957–62. doi: 10.1038/nmeth.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]