Bioluminescence is a versatile imaging platform with applications ranging from metabolite biosensing to whole animal imaging.[1] At the heart of this technology are enzymes (luciferases) that catalyze the oxidation of small molecule substrates (luciferins).[2] During each enzymatic transformation, an electronically excited oxyluciferin is generated that emits a photon of light upon relaxation to the ground state.[3] Since mammalian cells do not produce large numbers of photons in the absence of incident light, bioluminescence can provide an exquisitely sensitive readout on biological processes in these environments.[4] Indeed, luciferase-luciferin pairs have been widely used to report on enzyme activities and gene expression patterns in live cells and tissue lysates.[1] Additionally, since bioluminescence does not require an excitation source, this technology is well suited for noninvasive imaging in whole animals, where delivery of excitation light is often inefficient or impractical.[1a, 5]
The most widely used luciferases for cell and animal imaging originate from the insect family.[1b] These enzymes, including firefly luciferase (Fluc), catalyze the oxidation of D-luciferin (1) and release ~500–600 nm light (Figure 1a).[2b, 3] Wavelengths of this sort can penetrate the skin of small rodents and be detected by sensitive cameras, making insect luciferases attractive for imaging in vivo.[6] Indeed, Fluc and related enzymes have been expressed in a variety of tissue and cell types, and when exposed to D-luciferin, light is produced.[1] D-luciferin is also sufficiently bioavailable in rodents[7] and has been used extensively in preclinical models.[8]
Figure 1.
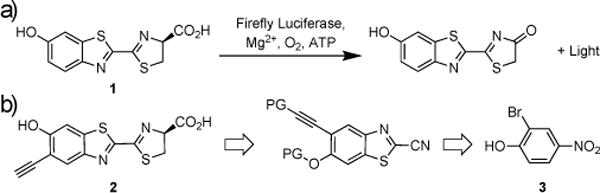
a) The luciferase-catalyzed oxidation of D-luciferin (1) produces visible light. b) Retrosynthetic analysis of alkynyl luciferin (PG= protecting group).
Because of the sensitivity and user-friendly features of bioluminescence, there has been much interest in expanding the scope of the technology.[5d, 9] Several efforts have been directed toward identifying other naturally occuring luciferase-luciferin pairs for multi-component imaging.[1a, 10] The instability and poor tissue penetrance of many luciferins has been prohibitive in many cases. Other attempts have focused on generating luciferases that provide altered emission spectra. For example, several insect luciferases have been engineered to emit different colors of light (ranging from ~500–650 nm) with D-luciferin.[11] While these wavelengths can be adequately resolved in vitro, they cannot be easily discriminated in vivo, where tissue absorption and scatter modulate the color of light that ultimately reaches the detector.[6]
Compared to luciferase engineering efforts, there has been less work invested in crafting new luciferins. Substrate engineering is an obvious strategy to broaden the scope of bioluminescence technology, though, as the luciferin molecules can be modified to emit different colors of light or be selectively utilized by unique luciferases.[12, 13] In some cases, the substrates have proven remarkably cell and tissue permeant and, thus, well suited for in vivo work.[14]
Continued efforts to develop unique bioluminescent tools would benefit from rapid access to diverse collections of light-emitting luciferins. These scaffolds have been notoriously difficult to synthesize owing to their electron-rich and highly substituted cores. Late-stage modifications to luciferin molecules are also complicated. For example, most attempts to derivatize D-luciferin (1) have focused on altering the 6′-position via alkylation or acylation chemistries.[7a, 15] While facile, these strategies have produced scaffolds that are somewhat limited in scope. Electron donation is required for robust emission and, thus, the 6′-position is particularly sensitive to modification.
We aimed to develop a bioluminescent probe modified at an alternative ring position. We were initially drawn to the 5′-alkyne derivative (2) shown in Figure 1b. Previous work established that 5′-fluoro and other small substituents were well tolerated by Fluc and minimally perturbing to the bioluminescent reaction.[12b] Modeling analyses suggested that the alkyne would be similarly accommodated in the luciferase active site (Figure 2a). Furthermore, computational data[16] indicated that 2 would be a viable light emitter (Figure 2b).
Figure 2.
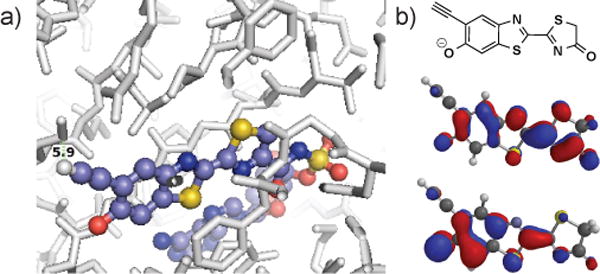
In silico analyses of D-luciferin. a) Overlay of 2 with firefly luciferase (PDB ID: 4G36) suggests that the alkyne motif will be tolerated. b) B3LYP/6-311** MO predictions[16] of the HOMO (middle) and LUMO (bottom) of the oxidzed product (top).
We were further attracted to alkyne 2 as its benzothiazole core could be accessed using C–H activation chemistry previously reported by our group.[17] The functionalized luciferin still presented some synthetic challenges, though. Electron-rich heterocycles like 2 are susceptible to non-specific oxidation and are thus difficult to handle and prepare on scale. Methods to produce highly substituted benzothiazoles are also rare.
To access the desired heterocycle, we began with tri-substituted phenol 3. The hydroxy substituent was first protected with a mesyl group (Scheme 1).[18] Other classic phenol protecting groups (e.g., silyl and methyl) were explored, but most proved either incompatible with subsequent transformations (in the case of bulky silyl groups) or difficult to remove later on in the synthesis (in the case of methyl groups). Mesylate 4 was ultimately subjected to Sonogashira conditions for alkyne installation. Notably, this reaction was readily scalable and provided decagram quantities of 5 (Scheme S1, ESI). The nitro group of 5 was reduced using iron filings and glacial acetic acid[19] to reveal aniline 6 in good yield and purity. Compound 6 was then treated with Appel’s salt 7, and the resulting adduct was fragmented with resin-linked PPh3 to yield thioamide 9 (Scheme 2).[20] It should be noted that while other bulky nucleophiles (e.g, DBU and DBA)[21] can be used for such fragmentations, they resulted in premature deprotection of the mesyl group and reduced overall yields in this case. Subsequent cyclization of thioamide 9 via palladium- and copper-catalyzed C–H activation[22] provided 10 in 61% yield. Attempts to isolate 10 directly from 8 via thermal cyclization resulted in product decomposition and were not further pursued. The desired alkyne luciferin 2 was ultimately isolated following mesyl group removal[23] and cysteine condensation. Importantly, luciferin 2 was stable for weeks as a solid material and in aqueous solution.
Scheme 1.
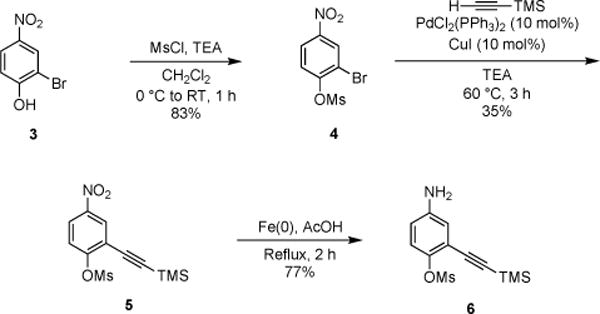
Installation of the alkyne substituent.
Scheme 2.
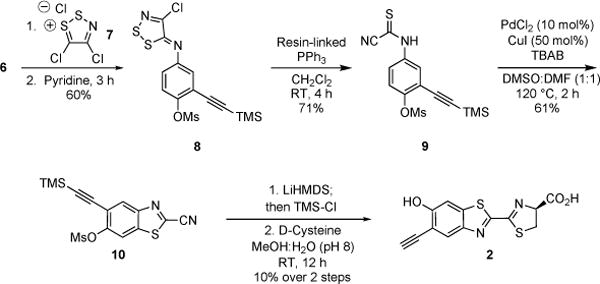
Synthesis of alkyne luciferin 2 using C–H activation chemistry.
Luciferin 2 was also found to be a viable substrate for firefly luciferase (Fluc). When 2 was incubated with Fluc in the presence of ATP, bioluminescent light was observed. As shown in Figure 3, light emission was both concentration-dependent and sustained. The overall photon output from 2 is weaker than that observed with D-luciferin (the native substrate), but on par with other luciferin analogs used in biological assays (Figure S1, ESI).[12b] The measured Km value was 8.5 ± 1 μM, and the apparent Vmax was 130 ± 5 × 106 photons s−1 (Figure S5). Interestingly, the bioluminescence emission spectrum of 2 was substantially red-shifted compared to D-luciferin (λmax = 610 nm at 25 °C, Figure 4). In fact, the alkynyl luciferin spectrum is similar to those of aminoluciferins and other analogs used in BLI.[14, 15b, 24]
Figure 3.
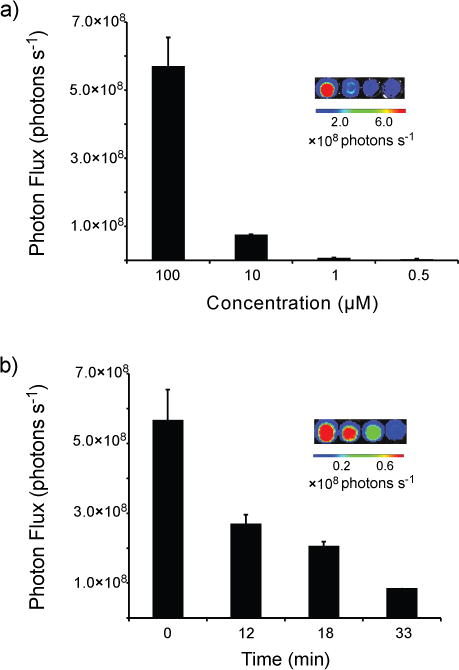
a) Alkynyl luciferin 2 produces light upon incubation with Fluc. Solutions of 2 (0.5–100 μM) were mixed with Fluc, ATP, and CoA in pH 8 buffer in 96-well plates. Light emission was measured using a cooled CCD camera. Sample images are shown in the inset. (b) Analog 2 exhibits sustained light emission. Compound 2 (100 μM) was incubated with Fluc, ATP, and CoA. Light emission was measured over time, and sample images are shown. For a–b, error bars represent the standard deviation of the mean for three replicate experiments.
Figure 4.
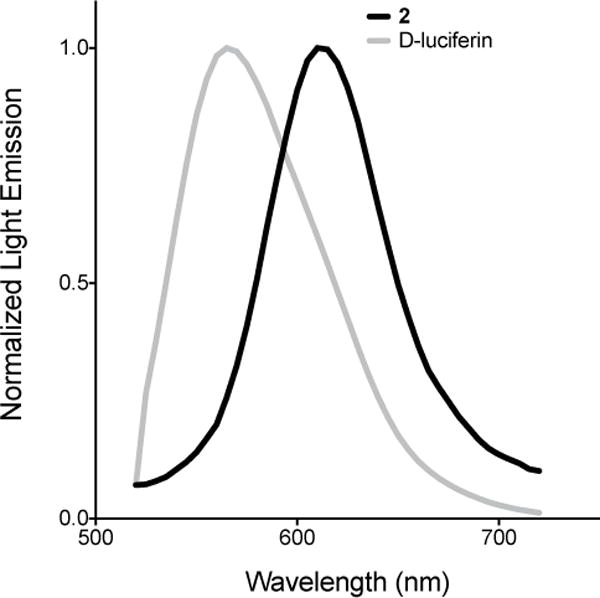
Normalized bioluminescence emission spectra for alkynyl luciferin 2 (λmax 610 nm) and D-luciferin 1 (λmax 565 nm). Samples (100 μM) were combined with Fluc (10 μg) and monitored at 25 °C.
We further evaluated the luciferin analog in live cells. Fluc-expressing HEK293 cells were incubated with 2, and bioluminescent images were acquired. As shown in Figure 5a, dose-dependent light emission was observed, indicating that the alkynyl luciferin is cell permeable. The photon outputs from cultures treated with 2 were weaker than cultures treated with D-luciferin (Figure S7). However, the intensities observed are similar to other luciferin analogs and sufficient for some cellular imaging applications.[12b–d] Importantly, the light emission from cells treated with 2 was also sustained (Figure 5b). Prolonged emission is desirable for routine imaging experiments.
Figure 5.
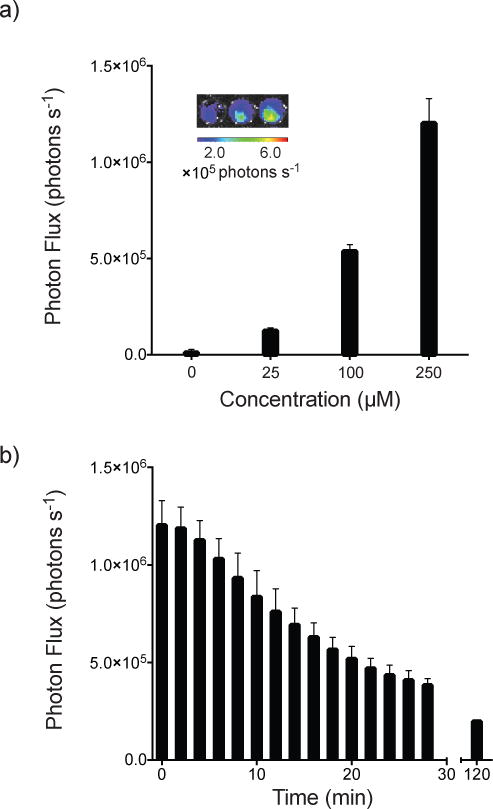
a) Alkynyl luciferin 2 produces light when incubated with HEK293 cells. Analog 2 (25–250 μM in PBS) was added to cells (100,000 cells per well). Sample images are shown (inset). b) Analog 2 exhibits sustained light emssion with HEK293 cells. Analog 2 (250 μM) was incubated with HEK293 cells (100,00 cells per well) and photon production was monitored over time. For a, error bars represent the standard deviation of the mean for 6 replicate experiments. For b, error bars represent the standard deviation of the mean for 3 replicate experiments.
We also recognized that 2 could be further “clicked”[25] with azido appendages via copper-catalyzed azide-alkyne cycloaddition (CuAAC). This transformation could potentially expedite the production of new luciferin analogs, using 2 as a platform for late-stage modification. Model reactions with 2 and various azido compounds suggested that the CuAAC diversification strategy is feasible (Figures S2–S3, and S5, ESI). Notably, the cycloaddition can proceed in aqueous solvents and in the absence of copper chelators (Figure S6, ESI). We envision using CuAAC to produce different classes of luciferins that can be screened for selective processing by mutant luciferases. Recent crystallographic analyses have revealed Fluc amino acids in close proximity to the 5′ carbon of a bound luciferin intermediate.[11h, 27] These amino acids could potentially be mutated to complement more bulky, steric appendages on the luciferin ring, thereby facilitating the development of substrate-specific (i.e., orthogonal) bioluminescent tools.
Conclusions
In conclusion, we identified an alkyne-modified luciferin (2) for use in bioluminescence assays. This scaffold is isolable in reasonable quantities and is a functional light emitter with luciferase. The alkynyl probe can also be selectively modified with azido appendages via CuAAC. Such designer luciferins are applicable to multi-component imaging or biosensing in cells and live organisms.[28] Based on the accessibility and uniqe features of 2, we anticipate that the alkynyl probe will find use in various imaging assays and further expand the scope of bioluminescence technology.
Experimental Section
Experimental details are available in the Supplementary Information.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH, R01GM107630 to J.A.P.). Some experiments were performed in the Laboratory for Fluorescence Dynamics (LFD) at UC Irvine. The LFD is supported jointly by the National Institute of General Medical Sciences of the NIH (8P41GM103540) and UC Irvine. We also thank members of the Jarvo, Chamberlin, and Overman laboratories for providing reagents and experimental advice. We thank Krysten Jones for kindly preparing samples for in vivo analysis. Finally, we thank members of the Prescher lab for assistance with the manuscript.
References
- 1.a) Paley MA, Prescher JA. MedChemComm. 2014;5:255–267. doi: 10.1039/C3MD00288H. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Prescher JA, Contag CH. Curr Opin Chem Biol. 2014;14:80–89. doi: 10.1016/j.cbpa.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 2.a) Viviani VR. Cell Mol Life Sci. 2002;59:1833–1850. doi: 10.1007/PL00012509. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hastings JW. Gene. 1996;173:5–11. doi: 10.1016/0378-1119(95)00676-1. [DOI] [PubMed] [Google Scholar]
- 3.White EH, Miano JD, Umbreit M. J Am Chem Soc. 1975;97:198–200. doi: 10.1021/ja00834a040. [DOI] [PubMed] [Google Scholar]
- 4.Rice BW, Cable MD, Nelson MB. J Biomed Opt. 2001;6:432–440. doi: 10.1117/1.1413210. [DOI] [PubMed] [Google Scholar]
- 5.a) Tinkum KL, White LS, Marpegan L, Herzog E, Piwnica-Worms D, Piwnica-Worms H. J Biol Chem. 2013;288:27999–28008. doi: 10.1074/jbc.M113.494328. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Takakura H, Hattori M, Takeuchi M, Ozawa T. ACS Chem Biol. 2012;7:901–910. doi: 10.1021/cb200360z. [DOI] [PubMed] [Google Scholar]; c) Hattori M, Haga S, Takakura H, Ozaki M, Ozawa T. Proc Natl Acad Sci U S A. 2013;110:9332–9337. doi: 10.1073/pnas.1304056110. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Dothager RS, Flentie K, Moss B, Pan MH, Kesarwala A, Piwnica-Worms D. Curr Opin Biotechnol. 2009;20:45–53. doi: 10.1016/j.copbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Rice BW, Contag CH. Nat Biotechnol. 2009;27:624–625. doi: 10.1038/nbt0709-624. [DOI] [PubMed] [Google Scholar]; b) Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH. J Biomed Opt. 2005;10:41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- 7.a) Shinde R, Perkins J, Contag CH. Biochemistry. 2006;45:11103–11112. doi: 10.1021/bi060475o. [DOI] [PubMed] [Google Scholar]; b) Berger F, Paulmurugan R, Bhaumik S, Gambhir SS. Eur J Nucl Med Mol Imaging. 2008;35:2275–2285. doi: 10.1007/s00259-008-0870-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Contag CH, Bachmann MH. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]; b) Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Olson JA, Zeiser R, Beilhack A, Goldman JJ, Negrin RS. J Immunol. 2009;183:3219–3228. doi: 10.4049/jimmunol.0804268. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, Wen S, Chang YF, Bachmann MH, Shimono Y, Dalerba P, Adorno M, Lobo N, Bueno J, Dirbas FM, Goswami S, Somlo G, Condeelis J, Contag CH, Gambhir SS, Clarke MF. Proc Natl Acad Sci U S A. 2010;107:18115–18120. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Villalobos V, Naik S, Piwnica-Worms D. Annu Rev Biomed Eng. 2007;9:321–349. doi: 10.1146/annurev.bioeng.9.060906.152044. [DOI] [PubMed] [Google Scholar]; b) Sun YQ, Liu J, Wang P, Zhang J, Guo W. Angew Chem Int Ed. 2012;51:8428–8430. doi: 10.1002/anie.201203565. [DOI] [PubMed] [Google Scholar]
- 10.a) Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]; b) Bhaumik S, Gambhir SS. Proc Natl Acad Sci U S A. 2002;99:377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Branchini BR, Ablamsky DM, Davis AL, Southworth TL, Butler B, Fan F, Jathoul AP, Pule MA. Anal Biochem. 2010;396:290–297. doi: 10.1016/j.ab.2009.09.009. [DOI] [PubMed] [Google Scholar]; b) Branchini BR, Ablamsky DM, Murtiashaw MH, Uzasci L, Fraga H, Southworth TL. Anal Biochem. 2007;361:253–262. doi: 10.1016/j.ab.2006.10.043. [DOI] [PubMed] [Google Scholar]; c) Branchini BR, Southworth TL, Khattak NF, Michelini E, Roda A. Anal Biochem. 2005;345:140–148. doi: 10.1016/j.ab.2005.07.015. [DOI] [PubMed] [Google Scholar]; d) Gammon ST, Leevy WM, Gross S, Gokel GW, Piwnica-Worms D. Anal Chem. 2006;78:1520–1527. doi: 10.1021/ac051999h. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Rabinovich BA, Ye Y, Etto T, Chen JQ, Levitsky HI, Overwijk WW, Cooper LJ, Gelovani J, Hwu P. Proc Natl Acad Sci U S A. 2008;105:14342–14346. doi: 10.1073/pnas.0804105105. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Kim JB, Urban K, Cochran E, Lee S, Ang A, Rice B, Bata A, Campbell K, Coffee R, Gorodinsky A, Lu Z, Zhou H, Kishimoto TK, Lassota P. PLoS ONE. 2010;5:e9364. doi: 10.1371/journal.pone.0009364. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Mezzanotte L, Que I, Kaijzel E, Branchini B, Roda A, Lowik C. PLoS ONE. 2011;6:e19277. doi: 10.1371/journal.pone.0019277. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Nakatsu T, Ichiyama S, Hiratake J, Saldanha A, Kobashi N, Sakata K, Kato H. Nature. 2006;440:372–376. doi: 10.1038/nature04542. [DOI] [PubMed] [Google Scholar]
- 12.a) Branchini BR, Hayward MM, Bamford S, Brennan PM, Lajiness EJ. Photochem Photobiol. 1989;49:689–695. doi: 10.1111/j.1751-1097.1989.tb08442.x. [DOI] [PubMed] [Google Scholar]; b) Takakura H, Kojima R, Ozawa T, Nagano T, Urano Y. ChemBioChem. 2012;13:1424–1427. doi: 10.1002/cbic.201200142. [DOI] [PubMed] [Google Scholar]; c) Jathoul AP, Grounds H, Anderson JC, Pule MA. Angew Chem Int Ed. 2014;53:13059–13063. doi: 10.1002/anie.201405955. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kojima R, Takakura H, Ozawa T, Tada Y, Nagano T, Urano Y. Angew Chem Int Ed. 2013;52:1175–1179. doi: 10.1002/anie.201205151. [DOI] [PubMed] [Google Scholar]; e) Mofford DM, Reddy GR, Miller SC. J Am Chem Soc. 2014;136:13277–13282. doi: 10.1021/ja505795s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harwood KR, Mofford DM, Reddy GR, Miller SC. Chem Biol. 2011;18:1649–1657. doi: 10.1016/j.chembiol.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans MS, Chaurette JP, Adams ST, Jr, Reddy GR, Paley MA, Aronin N, Prescher JA, Miller SC. Nat Methods. 2014;11:393–395. doi: 10.1038/nmeth.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Woodroofe CC, Shultz JW, Wood MG, Osterman J, Cali JJ, Daily WJ, Meisenheimer PL, Klaubert DH. Biochemistry. 2008;47:10383–10393. doi: 10.1021/bi800505u. [DOI] [PubMed] [Google Scholar]; b) Reddy GR, Thompson WC, Miller SC. J Am Chem Soc. 2010;132:13586–13587. doi: 10.1021/ja104525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.SPARTAN Student v. 5.0.2 Wavefunction, Inc.
- 17.a) Appel R, Janssen H, Siray M, Knoch F. Chem Ber. 1985;118:1632–1643. [Google Scholar]; b) McCutcheon DC, Paley MA, Steinhardt RC, Prescher JA. J Am Chem Soc. 2014;134:7604–7607. doi: 10.1021/ja301493d. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Besson T, Guillard J, Rees CW. J Chem Soc, Perkin Trans 1. 2000:563–566. [Google Scholar]; d) Koutentis PA, Koyioni M, Michaelidou SS. Org Biomol Chem. 2013;11:621–629. doi: 10.1039/c2ob26993g. [DOI] [PubMed] [Google Scholar]; e) Akhavan-Tafti H, De Silva R, Wang G, Eickholt RA, Gupta R, Kaanumalle LS. 2011/0014599A1. US
- 18.Looker JH, Thatcher DN. J Org Chem. 1954;19:784–788. [Google Scholar]
- 19.Shen M, Driver TG. Org Lett. 2008;10:3367–3370. doi: 10.1021/ol801227f. [DOI] [PubMed] [Google Scholar]
- 20.Besson T, Emayan K, Rees CW. J Chem Soc, Chem Comm. 1995:1419–1420. [Google Scholar]
- 21.Michaelidou SS, Koutentis PA. Synthesis. 2009:4167–4174. [Google Scholar]
- 22.Inamoto K, Hasegawa C, Hiroya K, Doi T. Org Lett. 2008;10:5147–5150. doi: 10.1021/ol802033p. [DOI] [PubMed] [Google Scholar]
- 23.Ritter T, Stanek K, Larrosa I, Carreira EM. Org Lett. 2004;6:1513–1514. doi: 10.1021/ol049514j. [DOI] [PubMed] [Google Scholar]
- 24.Mofford DM, Reddy GR, Miller SC. J Am Chem Soc. 2014;136:13277–13282. doi: 10.1021/ja505795s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.a) Kolb HC, Finn MG, Sharpless KB. Angew Chem, Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]; b) Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Proc Natl Acad Sci U S A. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Patterson DM, Nazarova LA, Prescher JA. ACS Chem Biol. 2014;9:592–605. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- 26.Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. Org Lett. 2004;6:4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- 27.Sundlov JA, Fontaine DM, Southworth TL, Branchini BR, Gulick AM. Biochemistry. 2012;51:6493–6495. doi: 10.1021/bi300934s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.a) Li J, Chen L, Du L, Li M. Chem Soc Rev. 2013;42:662–676. doi: 10.1039/c2cs35249d. [DOI] [PubMed] [Google Scholar]; b) Yao H, So MK, Rao J. Angew Chem Int Ed Engl. 2007;46:7031–7034. doi: 10.1002/anie.200701931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


