Understanding the development of the human trophoblast lineage and placentation is a challenge. Ethical issues prevent the most direct experimentation. Animal models have provided considerable insight, but species differences exist and knowledge of key regulatory events in human development is missing, which precludes an appreciation of conserved processes. In the mouse, it is possible to capture trophoblast stem (TS) cells from the early embryo and propagate them ex vivo and investigate the regulation of their differentiation (1); however, these conditions are not conducive to establishing TS cell cultures from human embryos (2). At this juncture, there does not seem to be a consensus on the identity of factors required for maintaining self-renewal of human TS cells. Serendipitous observations over a decade ago offered a potential alternative strategy for investigating human trophoblast development. Bone morphogenetic protein (BMP) treatment of human ES cells yielded a cell population possessing a trophoblast cell-like phenotype (3). Over the ensuing years, modifications have been made to this differentiation procedure for facilitating trophoblast-like conversion of human ES cells, including the addition of BMP along with inhibitors for Activin/Nodal (A83-01) and FGF2 (PD173074) signaling (BAP; 4–6). As these protocols have been elaborated, concerns have been noted about the identity of the cell derivatives and the physiological relevance of the BMP-directed process (7, 8). A recent study reported in PNAS using a simple yet elegant strategy for capturing subpopulations of cells from BAP-treated human ES cells provides further evidence of the trophoblastic potency of pluripotent stem cells (9).
One of the challenges in analyzing differentiated cells from manipulated stem cell cultures is the resulting complex and often heterogeneous mixture of cell types. This heterogeneity could represent distinct differentiated cell types as well as cells at different stages of differentiation. Purification of specific subpopulations is a strategy to address cell mixtures and is sometimes achieved through recognition of a unique surface antigen or by engineering a reporter that is exclusively activated in the differentiated cell population (10). Single-cell analyses of differentiated cells have also become a popular approach (11). Yabe et al. (9) took advantage of a specialized property of one type of differentiated trophoblast cell that arises through the fusion of progenitors generating multinucleated cells referred to as syncytiotrophoblasts. These cells are known to possess a range of specific functions, including hormonogenesis and bidirectional solute transport, features vital to the placenta and essential for in utero growth and survival of the fetus (12). In the PNAS report, human ES cells were treated with the BAP mixture for 8 d, dissociated, and then passed through nylon cell strainers of different pore sizes to fractionate cells on the basis of size. BAP-treated cells exhibited size heterogeneity (9). Transcript profiles of different cell size fractions were generated and each exhibited a trophoblast signature. Cells larger than 70 μm composed less than 0.2% of the total cell population, but ∼5% of total cellular DNA, and were primarily multinucleated. Transcriptomes of these large multinucleated cells were compared with transcriptomes of syncytiotrophoblast produced in vitro from term human progenitor trophoblast. The latter profiles exhibited many similarities, including transcripts encoding proteins involved in hormonogenesis and bidirectional solute transport. Some differences were also evident and may reflect gestation stage-dependent trophoblast behavior. Yabe et al. (9) assert that human ES cell-derived multinucleated cells bear greater similarity to early-gestation syncytiotrophoblast, especially syncytiotrophoblast arising immediately postimplantation. This represents an attractive hypothesis and if further supported would make their model especially advantageous for investigating early events in human trophoblast lineage development.
Although the authors noted that all BAP-treated cell-size subpopulations possessed a trophoblast signature, each exhibited some transcriptomic differences when compared against the others and with primary term human syncytiotrophoblast. Pluripotent stem cells are dynamic and culture systems are not amenable to fixing a stem cell population in a unitary developmental state. Consequently, it should not be a surprise that the outcome following BAP treatment was heterogeneous. Heterogeneity in the initial stem-cell state may influence the capacity of a human ES cell to respond to BAP and the direction of its response. BAP treatment of human ES cells results in cells activated to differentiate into trophoblast cells, but evidently not all cells progress to the final end-stage syncytiotrophoblast. Pooling mixed cell populations for analyses generates a composite phenotype, which complicates many analyses and interpretations and has led some to discredit the model system. Thus, investigating single cells or fractionating cell subpopulations based on strict criteria following the induction of differentiation, as demonstrated in the authors’ sieving technique, allows for a more detailed analysis of the resulting cells. Additional advances in isolating human stem cells for trophoblast derivation may be on the horizon. Fluorescence-activated cell sorting of subsets of ES cells with enriched capacity to differentiate along a trophoblast lineage has been demonstrated (13). Additionally, pluripotent stem cells derived from single eight-cell human blastomeres exhibit less heterogeneity and a greater proclivity to advance to a trophoblast phenotype (14).
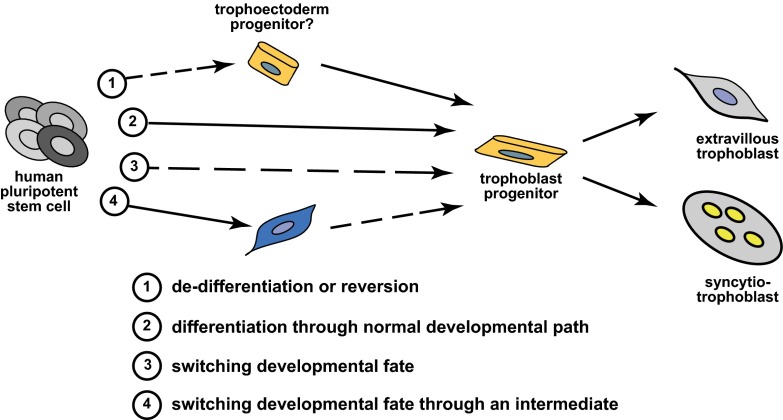
At face value, the work of Yabe et al. (9) would seem to represent a straightforward report of separating and analyzing subpopulations of differentiated cells. However, the differentiation paradigm of human pluripotent stem cells to trophoblast lineages has been controversial. We can acknowledge that human pluripotent stem cells can be converted into specialized syncytiotrophoblast cells, but how did this process transpire? A few potential mechanisms for such a differentiation pathway can be envisioned (Fig. 1).
Fig. 1.
Potential mechanisms for the derivation of trophoblast from human pluripotent stem cells. Four potential scenarios are depicted. Solid arrows indicate differentiation pathways that recapitulate normal developmental processes that occur during embryogenesis. Dashed arrows indicate differentiation pathways that are a result of vitro culture conditions and are not associated with in vivo developmental events.
One possibility is that this differentiation process recapitulates an actual developmental event. Human ES cells possess features similar to the epiblast, the portion of the embryo that gives rise to all adult tissues, but are considered a “synthetic” cell type that does not exist in an actual embryo (15). The notion that epiblast could possess trophoblast differentiative potential would be considered heresy for a developmental biologist trained in the “school of mouse embryogenesis.” Trophoblast lineages are specified in early preimplantation development, well before the existence of the epiblast (16). The gold standard assay for assessing potency of a stem cell population is to place it into a native in vivo environment and monitor differentiative capacity, in this case the early embryo. Ethical concerns prevent introducing human pluripotent stem cells into human embryos and interspecies chimerism has not provided compelling support for mouse embryonic signals directing human pluripotent stem cells to a trophoblast fate (17, 18). Experiential factors may be dictating the outcome of these latter experiments and cues present in human embryos may not exist in the mouse. Consequently, we either assume that the human epiblast has the distinctive capacity for differentiation to trophoblast and wait for supportive information, or we look to other possible mechanisms.
An epiblast origin of trophoblast does not have to be a requirement for the utility of human pluripotent stem cells as a model system for investigating events in trophoblast development. In vitro experiments reflect potential and not necessarily a precise developmental process. The BAP treatment may be returning the pluripotent stem cell to a more primitive state, reminiscent of an early embryonic cell with a trophectoderm progenitor
A recent study reported in PNAS using a simple yet elegant strategy for capturing subpopulations of cells from BAP-treated human ES cells provides further evidence of the trophoblastic potency of pluripotent stem cells.
phenotype, which is then allowed to progress to more mature trophoblast lineages. An alternative mechanism may be that the culture conditions are promoting a reprogramming event and a switching of developmental fate, either directly from pluripotent stem cells to TS cells and trophoblast progenitors and on to terminally differentiated trophoblast cell types or possibly through an intermediate cell type. These alternative ideas posit that BAP-induced differentiation of human pluripotent stem cells to trophoblast does not entirely mimic normal development but nonetheless represents a potentially useful phenomenon that can be exploited. The study of the early events associated with BAP-induced trophoblast differentiation in vitro may distinguish between these mechanisms. However, perhaps the exact details do not matter. Once the trophoblast lineage is activated then the ensuing events can be experimentally dissected, including the establishment of strategies for capturing and expanding human TS cells and trophoblast progenitors and exploring pathways regulating their differentiation.
The fate of a stem cell is dependent upon its environment. Culture systems are artificial and an approximation of an in vivo cellular environment and can be manipulated to promote the requisite epigenetic remodeling to acquire a specific cell fate. The challenge is acquiring more knowledge of how to further manipulate the conditions for efficient differentiation to a trophoblast cell fate. This is an iterative process that benefits from an in vivo reference point. Derivation of mouse TS cells was built on an understanding of signaling events in the early mouse embryo (1). The paucity of information on early events of human development represents a fundamental issue and has challenged the use of human pluripotent stem cells as a model to investigate trophoblast lineage development. Given that BAP-directed human pluripotent stem cells differentiate into syncytiotrophoblast, then a molecular understanding of the early differentiation events that give rise to this and other specialized trophoblast cell types adds significantly to our toolbox for investigating human development and diseases of the placenta.
Acknowledgments
The authors’ research is supported by NIH Grants HD020676 and HD079363.
Footnotes
The authors declare no conflict of interest.
See companion article on page E2598.
References
- 1.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282(5396):2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 2.Kunath T, et al. Developmental differences in the expression of FGF receptors between human and mouse embryos. Placenta. 2014;35(12):1079–1088. doi: 10.1016/j.placenta.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Xu RH, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20(12):1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 4.Ezashi T, Telugu BP, Roberts RM. Model systems for studying trophoblast differentiation from human pluripotent stem cells. Cell Tissue Res. 2012;349(3):809–824. doi: 10.1007/s00441-012-1371-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amita M, et al. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci USA. 2013;110(13):E1212–E1221. doi: 10.1073/pnas.1303094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Parast MM. BMP4 regulation of human trophoblast development. Int J Dev Biol. 2014;58(2-4):239–246. doi: 10.1387/ijdb.130341mp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernardo AS, et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9(2):144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CQE, et al. What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Rep. 2016;6(2):257–272. doi: 10.1016/j.stemcr.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yabe S, et al. Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc Natl Acad Sci USA. 2016;113:E2598–E2607. doi: 10.1073/pnas.1601630113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irion S, Nostro MC, Kattman SJ, Keller GM. Directed differentiation of pluripotent stem cells: From developmental biology to therapeutic applications. Cold Spring Harb Symp Quant Biol. 2008;73:101–110. doi: 10.1101/sqb.2008.73.065. [DOI] [PubMed] [Google Scholar]
- 11.Hoppe PS, Coutu DL, Schroeder T. Single-cell technologies sharpen up mammalian stem cell research. Nat Cell Biol. 2014;16(10):919–927. doi: 10.1038/ncb3042. [DOI] [PubMed] [Google Scholar]
- 12.Maltepe E, Fisher SJ. Placenta: The forgotten organ. Annu Rev Cell Dev Biol. 2015;31:523–552. doi: 10.1146/annurev-cellbio-100814-125620. [DOI] [PubMed] [Google Scholar]
- 13.Hölzenspies J, Dela Cruz G, M Brickman J. Resolving heterogeneity: Fluorescence-activated cell sorting of dynamic cell population from feeder-free mouse embryonic stem cell culture. Methods Mol Biol. 2016;1341:25–40. doi: 10.1007/7651_2015_254. [DOI] [PubMed] [Google Scholar]
- 14.Zdravkovic T, et al. Human stem cells from single blastomeres reveal pathways of embryonic or trophoblast fate specification. Development. 2015;142(23):4010–4025. doi: 10.1242/dev.122846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith AG. Embryo-derived stem cells: Of mice and men. Annu Rev Cell Dev Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- 16.Rossant J, Tam PPL. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136(5):701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 17.James D, Noggle SA, Swigut T, Brivanlou AH. Contribution of human embryonic stem cells to mouse blastocysts. Dev Biol. 2006;295(1):90–102. doi: 10.1016/j.ydbio.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 18.Masaki H, et al. Interspecific in vitro assay for the chimera-forming ability of human pluripotent stem cells. Development. 2015;142(18):3222–3230. doi: 10.1242/dev.124016. [DOI] [PubMed] [Google Scholar]