Significance
For plant viruses a successful infection correlates with the ability to access the vascular phloem and move systemically into distal tissues. However, how viruses gain access to and usurp vascular tissues is poorly understood. Here we show how tobacco mosaic virus (TMV) enhances its access to the phloem of mature plant tissues through the targeted disruption of auxin/indole acetic acid (Aux/IAA) transcriptional regulators that control expression of host genes involved in virus cell-to-cell movement, plasmodesmata gating, and defense. TMV’s ability to disrupt Aux/IAA function successfully confers a significant advantage in the systemic spread of this virus, allowing it to outcompete nondisrupting viruses. In summary, TMV interacts with Aux/IAA proteins to reprogram the vascular phloem, making it more conducive to systemic movement.
Keywords: pathogen defense, plant hormone signaling, virus movement, plasmodesmata gating, age-related resistance
Abstract
Vascular phloem loading has long been recognized as an essential step in the establishment of a systemic virus infection. In this study, an interaction between the replication protein of tobacco mosaic virus (TMV) and phloem-specific auxin/indole acetic acid (Aux/IAA) transcriptional regulators was found to modulate virus phloem loading in an age-dependent manner. Promoter expression studies show that in mature tissues TMV 126/183-kDa–interacting Aux/IAAs predominantly express and accumulate within the nuclei of phloem companion cells (CCs). Furthermore, CC Aux/IAA nuclear localization is disrupted upon infection with an interacting virus. In situ analysis of virus spread shows that the inability to disrupt Aux/IAA CC nuclear localization correlates with a reduced ability to load into the vascular tissue. Subsequent systemic movement assays also demonstrate that a virus capable of disrupting Aux/IAA localization is significantly more competitive at moving out of older plant tissues than a noninteracting virus. Similarly, CC expression and overaccumulation of a degradation-resistant Aux/IAA-interacting protein was found to inhibit TMV accumulation and phloem loading selectively in flowering plants. Transcriptional expression studies demonstrate a role for Aux/IAA-interacting proteins in the regulation of salicylic and jasmonic acid host defense responses as well as virus-specific movement factors, including pectin methylesterase, that are involved in regulating plasmodesmata size-exclusion limits and promoting virus cell-to-cell movement. Combined, these findings indicate that TMV directs the reprogramming of auxin-regulated gene expression within the vascular phloem of mature tissues as a means to enhance phloem loading and systemic spread.
To establish a systemic infection, plant viruses must access their host’s vascular phloem. The first step in this process involves cell-to-cell movement through intercellular cytoplasmic and endo-membrane connections called “plasmodesmata” (PD). Virus movement through the PD is facilitated by viral movement proteins (MP) that function to modulate the size-exclusion limits of the PD, allowing virus-transport forms composed of either nucleoprotein complexes or virions to pass between cells (1, 2). For systemic movement viruses must “load” into the vascular phloem. Phloem loading requires passage through specialized branched PD connections known as “pore units” that occur between companion cells (CCs) and phloem sieve elements (SE) (3). Once in the anucleate SEs, viruses move following the source-to-sink path of photoassimilates to distal plant tissues (4). In addition to the transport of photoassimilates, the vascular phloem also serves as a conduit for the movement of numerous host components including proteins, mRNA, microRNAs, and small molecules involved in a range of plant responses including development and flowering as well as abiotic and biotic stress responses (5–7). It is clear that the vascular phloem functions as a gatekeeper between distal plant tissues, controlling the passage of numerous molecules that affect many aspects of plant physiology as well as responses to outside stimuli. Thus, to establish a systemic infection, plant viruses must usurp this gateway. However, how plant viruses appropriate the vascular phloem remains a fundamental question in plant virology.
A number of virus and host components have been shown to impact virus systemic movement (2, 8, 9). For tobacco mosaic virus (TMV), systemic transport via the phloem involves the viral coat protein (CP), P30 MP, and 126/183-kDa replication proteins. Both the MP and replication protein are required for cell-to-cell movement via the PD (10). Phloem loading and systemic movement also involve the MP and replication protein, because transit via PD is necessary to access the SEs. In addition, a functional virus CP is required for TMV systemic movement. Mutations that disrupt CP expression or its ability to form virions are known to inhibit systemic movement (11–13). Furthermore, chimeric TMV recombinants encoding the MP or CP of the orchid-infecting Tobamovirus Ondontoglossum ringspot virus display impaired systemic movement, and similar chimeric recombinants derived from sun-hemp mosaic virus suggest a role for the viral replication proteins in vascular movement (14–16). Combined, these findings suggest a host-selective gating mechanism involving multiple virus–host interactions as essential factors in TMV phloem loading.
Several host components also have been found to impact the systemic spread of TMV. Many of these host factors affect PD gating and have been reviewed in detail (2, 8, 10). However, there is an emerging link between virus systemic movement and transcriptional reprograming. For example, the crucifer strain of TMV-cg has been shown to modulate the expression of a WRKY8 transcription factor, and the P30 protein from tomato mosaic virus (ToMV) associates in vivo with the transcriptional coactivator KELP (17, 18). WRKY transcription factors are associated with basal defense responses, and suppression of WRKY8 during infection correlates with enhanced TMV-cg systemic accumulation (18). Similarly, overexpression of KELP resulted in the partial relocation of P30 to the nucleus and prevented ToMV cell-to-cell and systemic movement. Furthermore, the P30 protein from the Tobamovirus turnip vein clearing virus (TVCV) encodes a nuclear localization signal required for both cell-to-cell and systemic virus movement (19). Association of the TVCV P30 protein with nuclear F-actin filaments was hypothesized to alter gene expression and promote virus infection (19). These studies provide evidence that virus interactions with host-associated transcriptional regulators can modulate systemic virus movement.
In our previous studies we identified an interaction between the 126/183-kDa replication protein of TMV and specific auxin/indole acetic acid (Aux/IAA) host transcriptional regulators (20–22). These studies show that the helicase domain present within the virus replication protein interacts strongly with Aux/IAA member IAA26 and more weakly with IAA27 and IAA18 in Arabidopsis thaliana as well as an IAA26 homolog in tomato. Members of the Aux/IAA family encode short-lived nuclear proteins that mediate auxin-dependent gene expression (23–25). Based on current evidence, Aux/IAA proteins interact with auxin-responsive transcription factors (ARFs) that in turn regulate numerous auxin-responsive genes (26, 27). Within Arabidopsis there are 29 Aux/IAA family members and 23 ARF members capable of forming an array of hetero- and homo-complexes from which auxin signaling can be orchestrated (28–30). Within the plant, auxin binds TIR1/AFB F-box proteins of the SCF E3 ubiquitin ligase complex, promoting their association with Aux/IAA proteins (24, 27, 31, 32). Ubiquitination of Aux/IAA proteins results in their targeted degradation via the 26S proteasome (27, 33). The plant’s auxin gradient thus provides a spatially sensitive means to regulate Aux/IAA activity via proteolysis.
Cellular localization studies demonstrate that the nuclear localization of interacting Aux/IAA proteins is disrupted during TMV infection (20, 22). In contrast, Aux/IAA proteins remain localized to the nucleus during infection by noninteracting viruses. Subsequent analysis indicates that viruses with reduced ability to interact with Aux/IAA proteins are compromised in their ability to accumulate and move in inoculated tissue (21). Interestingly, effects on virus accumulation are observed only in mature tissues and not in younger, immature tissues (21). This developmental relationship corresponds to the accumulation of Aux/IAA proteins in mature tissues and is consistent with the lower auxin levels and reduced auxin-mediated degradation found in older plant tissues (21, 34).
In this study we investigated the mechanism through which the TMV–Aux/IAA interactions affect virus accumulation and spread. Results indicate that interacting Aux/IAA proteins are expressed predominantly in the CC of mature vascular phloem tissues. The ability to disrupt the nuclear localization of these Aux/IAAs was found to correlate with enhanced virus phloem loading and movement within the vascular tissues. As a result, TMV, with the ability to interact with Aux/IAA proteins, gains a significant advantage in systemic movement over a virus defective in this interaction. Transcriptomic analysis of plant genes under the control of an interacting Aux/IAA protein identified an array of host genes linked to plant defense responses, virus cell-to-cell movement, and PD regulation. Interestingly, overaccumulation of Aux/IAA proteins in CCs before virus infection led to decreased accumulation and phloem loading of TMV only in mature flowering plants. This correlation with flowering has similarities to age-related resistance (ARR) and suggests that interactions with the identified Aux/IAA proteins may provide TMV with a mechanism to overcome this form of host resistance (35). Based on these studies, we propose that within mature plant tissues TMV selectively targets phloem-expressed Aux/IAA proteins to reprogram functions of the CC–SE complex that contribute to phloem loading.
Results
Interacting Aux/IAA Factors Are Expressed and Localize Within the Nucleus of Phloem CCs.
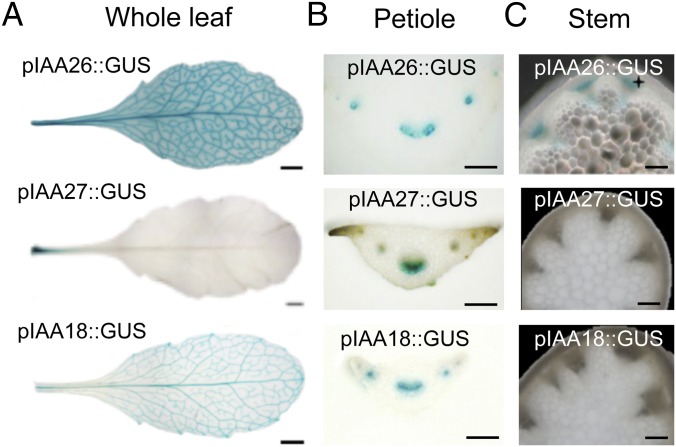
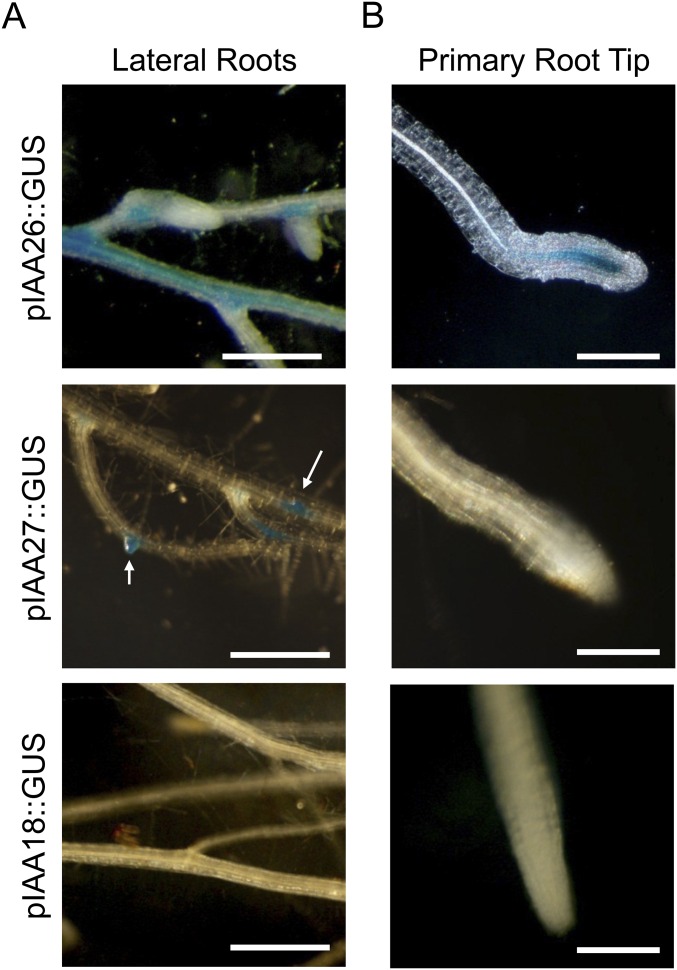
To define the importance of the Aux/IAA interaction on TMV accumulation, we first examined the expression patterns of three known interacting Arabidopsis Aux/IAA proteins. Previously IAA26, IAA27, and IAA18 were identified as interacting with and displaying varying levels of cytoplasmic colocalization with the TMV 126-kDa replication protein (22). The strength of the interaction corresponded to the level of cytoplasmic colocalization. IAA26 confers the strongest interaction with the virus 126-kDa protein and shows the greatest disruption in nuclear localization, followed by IAA27 and IAA18. These past studies used only the cauliflower mosaic virus 35S constitutive promoter to drive the expression of GFP-tagged IAA proteins in whole-leaf tissues. However, IAA family members are known to be differentially expressed and to confer tissue-specific functions (24, 36). To determine the expression pattern of the three TMV 126/183-kDa–interacting IAAs, promoter sequences upstream of their translation start codons (2,000 nt for IAA26, 1,500 nt for IAA27, and 2,000 nt for IAA18) were all cloned in front of the β-glucuronidase (GUS) ORF to create pIAA26::GUS, pIAA27::GUS, and pIAA18::GUS. All three promoter–reporter constructs were transformed into the systemic TMV host A. thaliana ecotype Shahdara (37). Fully expanded leaves from T2-generation plants were histochemically stained for GUS activity. Results from three or four independent plant lines for each of the three interacting IAAs consistently showed the predominant GUS activity in vascular tissues, but with distinct expression patterns (Fig. 1). pIAA26::GUS displayed the most robust expression with strong levels of GUS staining in all vein classes (I, II, III, and IV) of the leaves. Conversely, pIAA27::GUS was expressed predominately in vein class I of the petiole. Like pIAA26::GUS, pIAA18::GUS expression was observed in all vein classes, but at a markedly lower level, indicating that in leaf tissue IAA18 is not as highly expressed as IAA26 (Fig. 1A). Petiole cross-sections of pIAA26::GUS, pIAA27::GUS, and pIAA18::GUS lines showed GUS staining predominantly localized to the phloem for all three interacting IAA family members (Fig. 1B). pIAA26::GUS expression also was observed in stem and root vascular phloem (Fig. 1C and Fig. S1). In contrast pIAA27::GUS expression was not observed in stem or root vascular tissues but was observed at the sites of lateral root formation (Fig. S1). pIAA18::GUS expression was not observed in either stem or roots (Fig. 1C and Fig. S1).
Fig. 1.
Histochemical analysis of pIAA26::GUS, pIAA27::GUS, and pIAA18::GUS expression in A. thaliana Shahdara leaves, petioles, and stems. (A) GUS expression in 7-wk-old leaves after staining overnight. (Scale bars, 2 mm.) (B) Petiole cross-sections. (Scale bars, 200 μm.) (C) Stem cross-sections. (Scale bars, 100 μm.)
Fig. S1.
Histochemical analysis of pIAA26::GUS, pIAA27::GUS, and pIAA18::GUS expression in A. thaliana roots. (A) GUS expression in lateral roots after staining overnight. (Scale bars, 500 μm.) (B) Primary root tip. (Scale bars, 200 μm.)
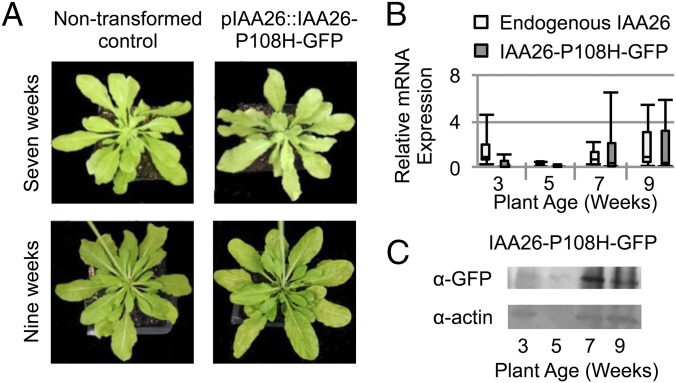
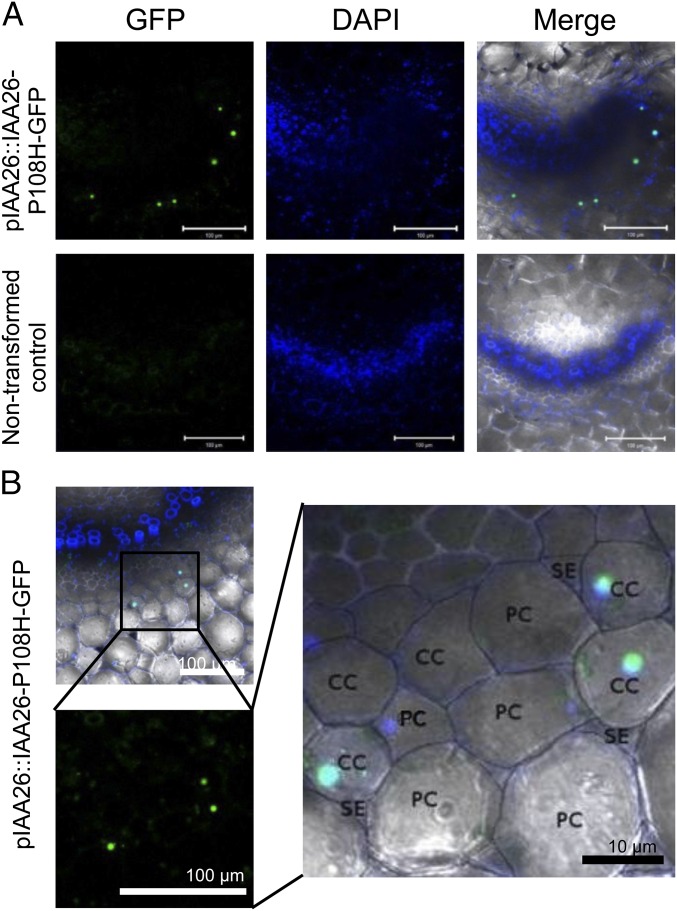
To investigate the localization of Aux/IAA proteins further, we focused on IAA26 because it displays by far the strongest interaction with the TMV 126-kDa protein and, as described above, is the most abundantly expressed in leaf and root vascular tissues. However, the rapid turnover of Aux/IAA proteins can make detecting or visualizing Aux/IAA proteins difficult. To address this issue, we used a previously generated IAA26 mutant allele, IAA26-P108H, which is resistant to auxin-mediated degradation but retains the ability to interact with the TMV 126/183-kDa protein (22). The native IAA26 promoter (2,000 nt upstream of the start codon) was cloned in front of the IAA26-P108H ORF and fused to GFP, creating pIAA26::IAA26-P108H-GFP. This construct was used to transform A. thaliana ecotype Shahdara. Transgenic pIAA26::IAA26-P108H-GFP plant lines grew and developed similarly to nontransformed Shahdara (Fig. 2A). In addition, quantitative RT-PCR (qRT-PCR) analysis for the endogenous IAA26 and transgene IAA26-P108H-GFP mRNAs show a similar expression pattern of increasing transcripts in older plant tissues (Fig. 2B). Thus, transgenic pIAA26::IAA26-P108H-GFP plants express IAA26-P108H-GFP mRNA in a manner similar to the endogenous IAA26 gene. Consistent with the mRNA expression analysis, the IAA26-P108H-GFP protein is predominantly detectable by Western immunoblot in 7- to 9-wk-old tissues (Fig. 2C). Subsequent petiole cross-sections from T2-generation plant lines showed IAA26-P108H-GFP fluorescence specifically expressed and localized to the nuclei of phloem-associated CCs (Fig. 3). Similar to immunoblot studies, IAA26-P108H-GFP fluorescence was detected only in 7- to 11-wk-old tissues. From these studies it is clear that interacting IAA26 is expressed primarily within older leaf and stem phloem CCs.
Fig. 2.
Characterization of pIAA26::IAA26-P108H-GFP transgenic plant lines. (A) Representative images of 7- and 9-wk-old nontransformed control Shahdara plants and pIAA26::IAA26-P108H-GFP transgenic Shahdara plants. (B) Relative mRNA expression of endogenous IAA26 mRNA in control plants or IAA26-P108H-GFP mRNA expressed from the pIAA26::IAA26-P108H-GFP transgene in 3- to 9-wk-old plants as measured by qRT-PCR. Total RNA was isolated from the leaves of four plants from two independent pIAA26::IAA26-P108H-GFP plant lines or from nontransformed controls and was pooled for each of three biological replicates. 18S RNA was used as an internal control for normalization. (C) Western immunoblot detection of IAA26-P108H-GFP protein levels in 3- to 9-wk-old pIAA26::IAA26-P108H-GFP transgenic plants as probed with anti-GFP antibody. Actin levels were used as a loading control.
Fig. 3.
Localization of IAA26-P108H-GFP expressed from the IAA26 native promoter in A. thaliana Shahdara petioles. (A) Representative fluorescent images of cells expressing IAA26-P108H-GFP from the IAA26 native promoter or nontransformed controls. Petioles from 9- to 11-wk-old T2-generation plants were hand sectioned for imaging. Green, GFP; blue, DAPI. (Scale bars, 100 μm.) (B) Localization of IAA26-P108H-GFP in CC nuclei. PC, phloem parenchyma cells.
TMV Disrupts the Nuclear Localization of Interacting IAA26 Within Phloem CCs.
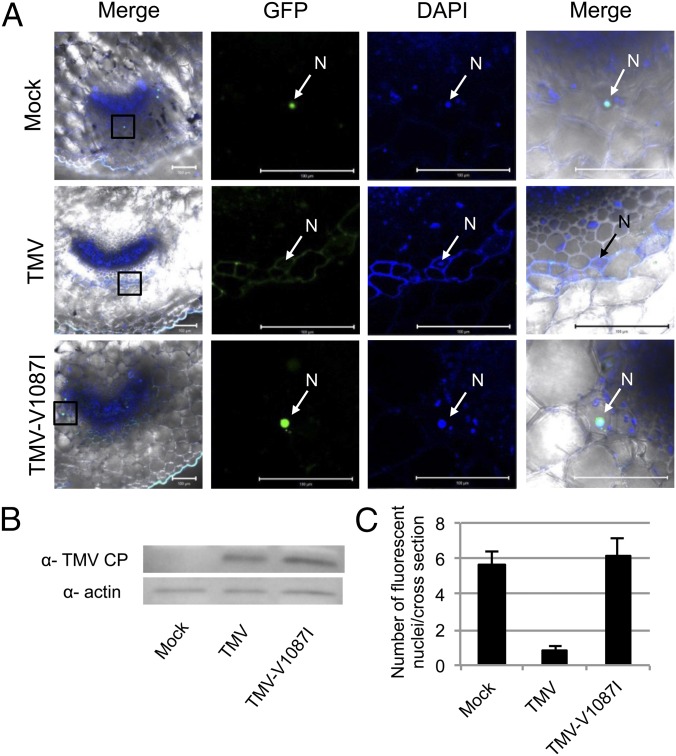
To examine pIAA26::IAA26-P108H-GFP nuclear localization in response to TMV infection, 9- to 11-wk-old T2-generation plants from two independent lines were infected with TMV and observed for IAA26-P108H-GFP fluorescence at 12 d post inoculation (dpi). Within TMV-infected tissues the number of phloem cells displaying IAA26-P108H-GFP–derived nuclear fluorescence was reduced sevenfold in comparison with mock-inoculated tissues (Fig. 4 A and C). In contrast, tissues infected with TMV-V1087I, a mutant virus that is able to replicate in protoplasts at levels similar to TMV but that does not interact with IAA26, did not disrupt the nuclear localization of IAA26-P108H-GFP in CCs (Fig. 4 A and B) (20, 38). Thus, TMV disrupts the nuclear localization of an interacting Aux/IAA protein expressed within its native CC environment.
Fig. 4.
Localization of IAA26-P108H-GFP in virus-infected tissue. (A) Representative fluorescent images of cells expressing IAA26-P108H-GFP in mock-infected, TMV-infected, or TMV-V1087I–infected phloem tissue. Inoculated leaf petioles were hand sectioned and imaged 12 dpi. Green, GFP; blue, DAPI. (Scale bars, 100 μm.) N, nucleus. (B) Western immunoblot for the detection of the TMV CP present in the investigated cross-sections of pIAA26::IAA26-P108H-GFP plants at 12 dpi using actin levels as a loading control. (C) Quantification of cell nuclei with GFP fluorescence reported as the mean of 10 petiole cross-sections ± SE.
Stabilization of IAA26 Expressed from its Native Phloem Promoter Inhibits TMV Accumulation.
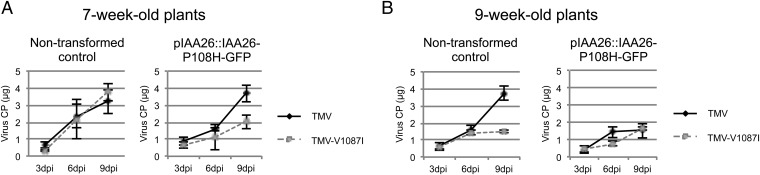
A. thaliana ecotype Shahdara lines transformed with pIAA26::IAA26-P108H-GFP were used to assess the effect of IAA26 accumulation on TMV infection. We hypothesized that stabilization of IAA26 within the phloem CCs, resulting in greater accumulation of the protein before virus infection, would significantly impact the ability of both TMV and the noninteracting TMV-V1087I to accumulate in leaf tissues. The studies described above indicated that IAA26-P108H-GFP protein accumulation is consistently detectable by 7 wk in transformed Shahdara (Fig. 2C). TMV and TMV-V1087I infections were subsequently monitored in both 7- and 9-wk-old plants using two independent lines expressing IAA26-P108H-GFP from the native IAA26 promoter or in nontransformed control plants at 3, 6, and 9 dpi. Virus accumulation was measured by Western immunoblot analysis using an antibody specific for the TMV CP. TMV and TMV-V1087I accumulate to similar levels in 7-wk-old nontransformed plants. Conversely, TMV-V1087I accumulated to significantly lower levels than TMV (P = 0.014, Student’s t test) in 7-wk-old pIAA26::IAA26-P108H-GFP plants (Fig. 5A). Thus, stabilization and overaccumulation of IAA26 in the younger, 7-wk-old tissues significantly impacts the ability of this non–Aux/IAA-interacting virus to accumulate.
Fig. 5.
Accumulation of TMV and TMV-V1087I in pIAA26::IAA26-P108H-GFP and nontransformed control A. thaliana Shahdara leaf tissue. (A) Virus accumulation in 7-wk-old inoculated leaves at 3, 6, and 9 dpi. (B) Virus accumulation in 9-wk-old inoculated leaves at 3, 6, and 9 dpi. Each data point represents the mean and SE derived from 18 independent leaves, each inoculated with 10 μg of TMV or TMV-V1087I. Virus infection was monitored for the accumulation of TMV CP by Western immunoblotting and was quantified using known CP standards. Actin was used as a loading control.
In 9-wk-old nontransformed plants that have bolted and begun to flower, TMV-V1087I accumulated to significantly lower levels than TMV (P = 0.006, Student’s t test) (Fig. 5B). This result is consistent with our previous findings that reduced accumulation of the non–Aux/IAA-interacting TMV-V1087I occurred only in older leaf tissues (21). However, in 9-wk-old pIAA26::IAA26-P108H-GFP plants TMV accumulated at significantly lower levels than in nontransformed control plants (P = 0.046, Student’s t test), at a level similar to the noninteracting TMV-V1087I (Fig. 5B). Together these findings indicate that the accumulation of IAA26-P108H-GFP before infection creates a cellular environment that is not conducive to infection and cannot be overcome by the TMV–Aux/IAA interaction.
Aux/IAA Interaction Specifically Enhances TMV Phloem Loading and Accumulation.
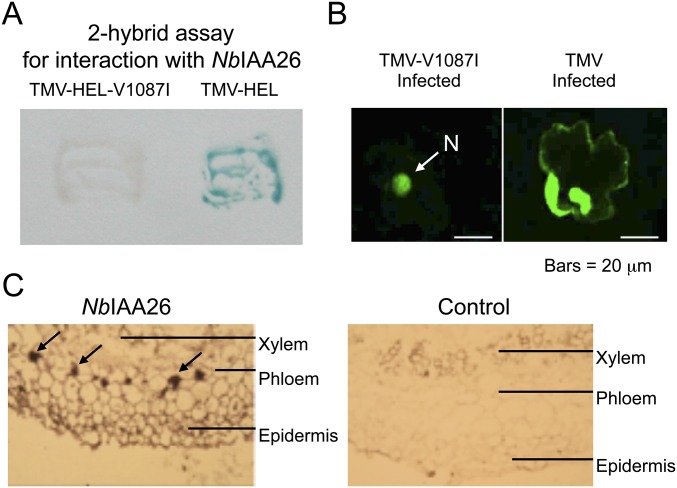
To identify the mechanism through which IAAs disrupt TMV accumulation, we investigated the ability of both interacting TMV and noninteracting TMV-V1087I to spread within inoculated leaves. To do so, we first developed a tobacco system for the in situ detection of TMV and tobacco IAA26. Previously we identified TMV 126/183-kDa–interacting Aux/IAA genes in both Arabidopsis and tomato (20, 21). In this study we cloned the corresponding Nicotiana benthamiana NbIAA26 and found it shares 96% sequence identity and 97% similarity with Nicotiana tabacum NtIAA26 and 51% sequence identity and 64% similarity with A. thaliana AtIAA26 (Fig. S2). Similar to our previous studies with the Arabidopsis and tomato IAA26 homologs, yeast two-hybrid analysis demonstrated the NbIAA26 protein interacts with the TMV 126/183-kDa helicase domain but not with the TMV-V1087I helicase domain (Fig. S3A). In addition, localization studies using a transiently expressed NbIAA26 fused to GFP demonstrated that, in the presence of the interacting TMV, NbIAA26 relocalizes from the nucleus to the cytoplasm but is unaffected by infection with the noninteracting TMV-V1087I (Fig. S3B). We also examined the cellular expression pattern of NbIAA26 mRNA using in situ hybridization. These experiments were performed on fixed stem tissue using a digoxigenin-labeled probe specific for the NbIAA26 sequence. Results clearly indicate that NbIAA26 mRNA is expressed predominantly in the vascular tissues of N. benthamiana stem sections (Fig. S3C). Thus, as in Arabidopsis, TMV appears to target similar vascular-expressed tobacco Aux/IAAs.
Fig. S2.
Sequence alignment of NbIAA26, NtIAA26, and AtIAA26. Similar residues are displayed in the same color. The four conserved Aux/IAA domains are indicated by horizontal black bars.
Fig. S3.
Characterization of NbIAA26. (A) Yeast two-hybrid assay showing the interaction of the TMV-helicase-bait protein and NbIAA26-prey protein and the absence of interaction between TMV-V1087I-helicase-bait protein and NbIAA26-prey protein. (B) TMV infection interferes in the nuclear localization of NbIAA26-GFP. Fluorescent images of N. benthamiana leaf tissue transiently expressing NbIAA26-GFP fusion protein in TMV-V1087I–infected or TMV-infected cells. Yeast two-hybrid and transient localization assays were done as previously described (20). (Scale bars, 20 μm.) (C) In situ immunolocalization of NbIAA26 in N. benthamiana stem cross-sections. Dark brown color indicates NbIAA26 mRNA accumulation. Arrows denote phloem location.
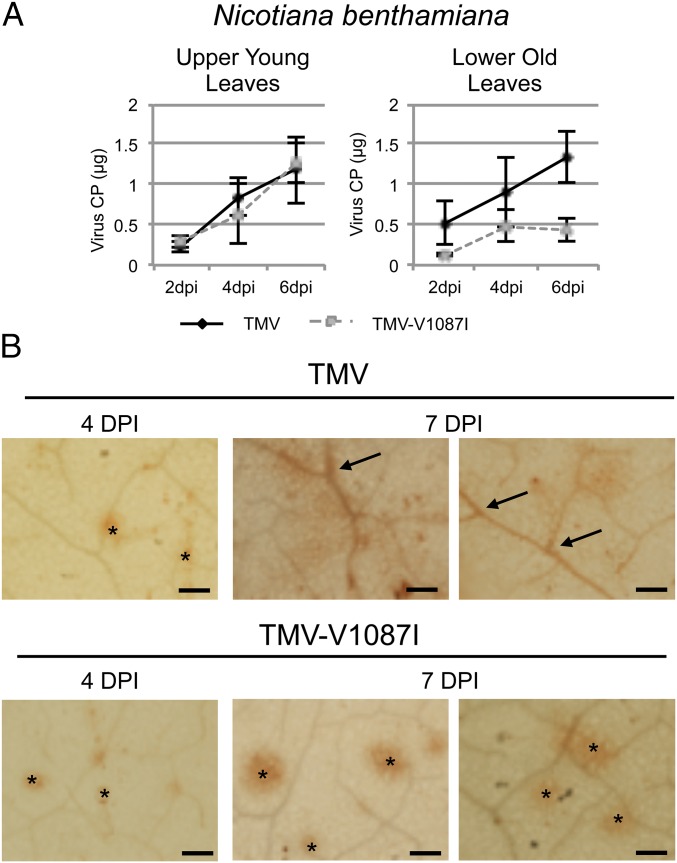
To examine the effect of the NbIAA26 interaction on the cellular movement of TMV, in situ hybridization was used to analyze the accumulation of TMV and TMV-V1087I (non–IAA-interacting) in inoculated leaves of N. benthamiana. Previous protoplasts studies comparing TMV and TMV-V1087I revealed no significant differences in the replication of these viruses in single cells (38). However, as is consistent with our previous findings in Arabidopsis, the noninteracting TMV-V1087I virus accumulates to significantly lower levels (P = 0.004, Student’s t test) in mature tissues than those observed for TMV (Fig. 6A). For this study mature N. benthamiana leaves were inoculated with either TMV or TMV-V1087I, were harvested at 4 and 7 dpi, and were subjected to in situ immunodetection for the TMV CP. Results indicate that at 4 dpi both TMV and TMV-V1087I show defined infection foci that are similar in size and appearance (Fig. 6B). In contrast, at 7 dpi TMV shows significant accumulation along the veins and vascular networks of the leaf, whereas TMV-V1087I shows continued cell-to-cell spread but limited accumulation within the vascular networks. These findings indicate that a non–Aux/IAA-interacting virus is less able to load into the vascular network.
Fig. 6.
Accumulation and phloem loading of TMV and TMV-V1087I in N. benthamiana leaves. (A) Virus accumulation in upper (young) and lower (old) inoculated leaves at 2, 4, and 6 dpi. Each data point represents the mean and SE derived from six independent leaves inoculated with 5 μg of TMV or TMV-V1087I. Virus infection was monitored for the accumulation of TMV CP by Western immunoblotting and was quantified using CP standards. (B) In situ immunolocalization for the accumulation of TMV CP using anti-CP antibody. A positive response for the TMV CP is visualized as a brown stain in the cleared N. benthamiana leaves. Arrows denote TMV accumulation in vascular tissue. Asterisks denote TMV accumulation in mesophyll. (Scale bars, 3 mm.)
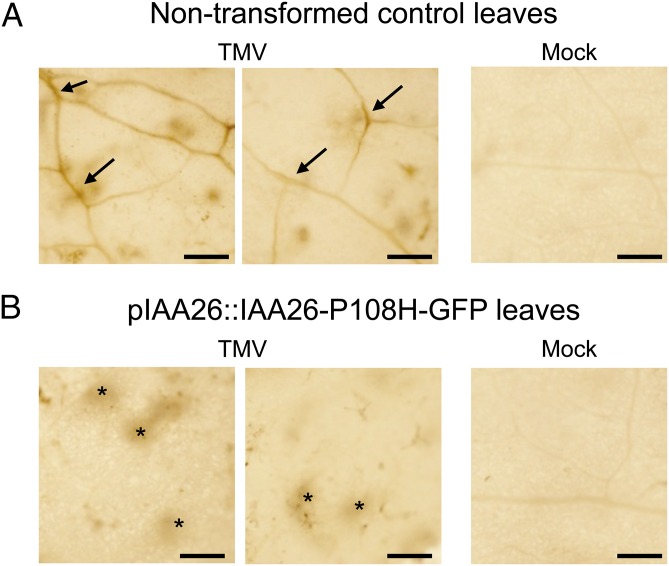
To investigate further the role of the interacting Aux/IAA proteins on TMV phloem loading, we performed similar in situ immunodetection for TMV in transgenic A. thaliana Shahdara pIAA26::IAA26-P108H-GFP plants. We reasoned that age-related factors combined with IAA26-P108H-GFP overaccumulation would produce alterations in vascular traits that restrict TMV phloem loading. For these experiments leaves from 9-wk-old plants that had flowered were harvested for analysis at 9 dpi. Results show significant accumulation of TMV within the vascular tissues of nontransformed control plants (Fig. 7A). In contrast, in plants expressing IAA26-P108H-GFP, TMV produces detectable viral accumulation at initial infection foci but little accumulation within the vascular tissues (Fig. 7B). This finding confirms that accumulation of IAA26-P108H-GFP before infection leads to reduced vascular loading of TMV in flowering plants. These findings also are consistent with the observed reduced accumulation of TMV in the older 9-wk-old pIAA26::IAA26-P108H-GFP plants (Fig. 5B). Combined, these findings suggest that within mature tissues additional factors such as ARR impact TMV phloem loading.
Fig. 7.
In situ immunolocalization for the TMV CP in pIAA26::IAA26-P108H-GFP A. thaliana Shahdara leaves infected with 10 μg of TMV at 9 dpi. (A) Immunolocalization in 9-wk-old nontransformed control leaf tissue infected with TMV (Left and Center) or mock infected (Right). (B) Similar immunolocalization panels for TMV CP in 9-wk-old leaf tissue from pIAA26::IAA26-P108H-GFP plants similarly infected with TMV or mock infected. Arrows denote TMV accumulation in the veins visualized as a brown stain. Asterisks denote TMV accumulation in mesophyll. (Scale bars, 1 mm.)
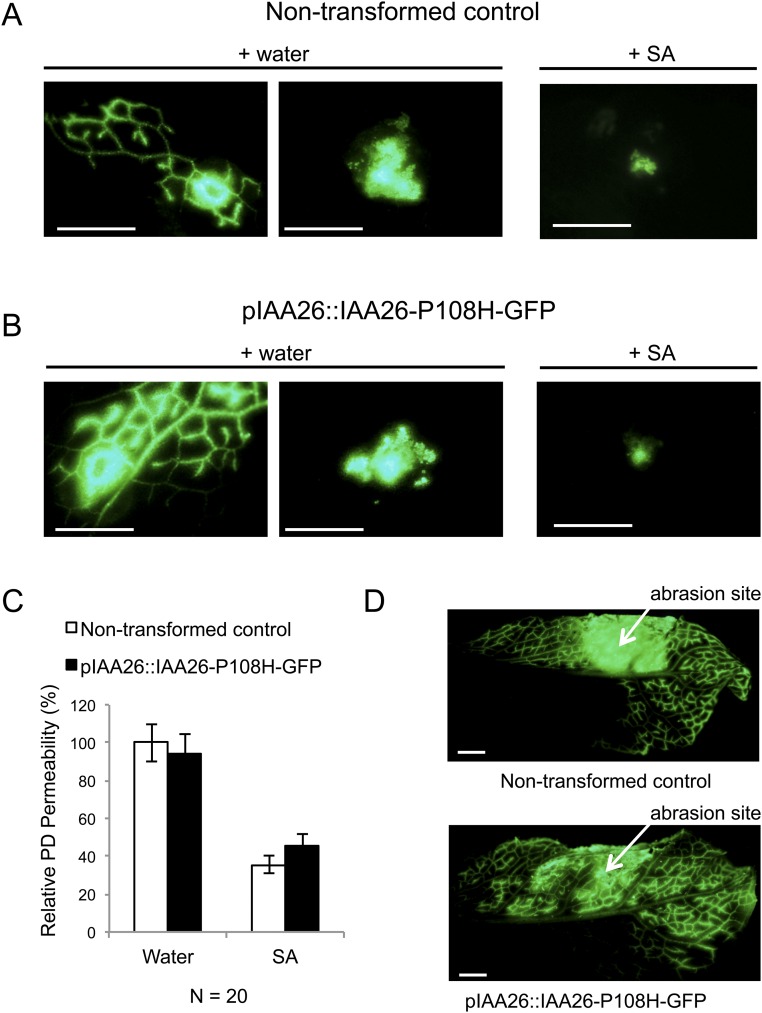
To address the specificity of observed phloem-loading restrictions, we used a fluorescent dye-based cell-to-cell movement and phloem-loading assay. Several studies have demonstrated that changes in PD size-exclusion limit and vascular movement can be monitored using 5(6)-carboxyfluorescein diacetate (CFDA), a membrane-permeable nonfluorescent dye that upon cell entry is cleaved by esterases to produce 5(6)-carboxyfluorescein (CF), a non–membrane-permeable fluorescent dye (39–41). In these experiments we compared CF mobility in 9-wk-old nontransformed control and pIAA26::IAA26-P108H-GFP plants. CF mobility was found to be similar in pIAA26::IAA26-P108H-GFP and the control plants (Fig. S4). Consistent with previous studies, treatment of control or pIAA26::IAA26-P108H-GFP plants with SA 24 h before CF application produced significant reductions in CF fluorescence movement (Fig. S4 A and B). Reduced CF movement after SA treatment has been attributed to the deposition of callose at the PD (41). The inability of pIAA26::IAA26-P108H-GFP plants to affect CF movement suggests that CC expression of IAA26-P108H-GFP is insufficient to impact whole-leaf PD closure. The effects of IAA26-P108H-GFP on PD gaiting thus are likely to be specifically directed toward disrupting virus movement into the phloem.
Fig. S4.
Analysis of PD size-exclusion limits in nontransformed control and pIAA26::IAA26-P108H-GFP plants. PD permeability and phloem loading were examined by CFDA-based dye-loading assays. (A) Nine-week-old control plants treated with water (Left and Center) or SA (Right). (B) Nine-week-old pIAA26::IAA26-P108H-GFP plants treated with water (Left and Center) or SA (Right). (C) Quantification of CF movement. Each assay represents 20 CFDA spot treatments done on the leaves of 10 different plants. The extent of dye diffusion was quantified by measuring the area and was normalized to nontransformed control plants. Error bars represent mean ± SE. (D) Phloem loading of CF after abrasion with fine sandpaper. (Scale bars, 5 mm.)
Aux/IAA Interactions Confer a Competitive Advantage.
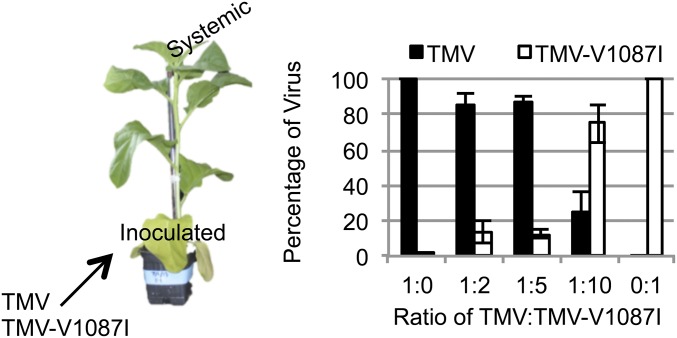
To determine the impact of Aux/IAA interactions on TMV systemic movement, we performed mixed infection assays for the accumulation of TMV and TMV-V1087I in systemic noninoculated leaf tissues. For these assays a single lower leaf of N. tabacum plants with at least six fully expanded leaves was inoculated with TMV, TMV-V1087I, or TMV and TMV-V1087I at a 1:1, 1:5 or 1:10 ratio of TMV to TMV-V1087I. At 10 dpi symptomatic systemic leaf tissue was harvested, and cDNA was generated from total RNA. Quantitative sequencing for the V1087I mutation was used to determine the relative concentrations of each virus in the systemic tissue (42). Results indicated that inoculation ratios of greater than five times TMV-V1087I to TMV were required for TMV-V1087I to be the predominant systemic virus (Fig. 8). As a control, plants inoculated with only TMV-V1087I did not yield reversions to the TMV sequence, demonstrating that this mutation is stable. Taken together, these results suggest that interaction with Aux/IAA proteins significantly enhances the ability of TMV to load into the vascular tissue and move systemically.
Fig. 8.
Competitive accumulation of Aux/IAA-interacting TMV vs. the non–Aux/IAA-interacting TMV-V1087I. In this assay the lower leaves of N. tabacum cv. Xanthi plants were inoculated with one of five inoculation treatments: single inoculation with either TMV or TMV-V1087I or simultaneous inoculation with TMV and TMV-V1087I at a 1:1, 1:5, or 1:10 ratio of TMV to TMV-V1087I. The infectivity of TMV and TMV-V1087I inoculum was determined by local lesion assays on N. tabacum cv. Xanthi-NC and was standardized to ensure an equal infection potential. RNA was isolated from systemic leaves upon the appearance of mosaic disease symptoms at 10 dpi. The relative amount of each virus was determined by quantitative sequencing (42). Each bar represents the mean of three independent plants ± SE.
IAA26-Directed Transcriptional Reprogramming.
To identify genes regulated by IAA26, we compared the transcriptomes of 7-wk-old pIAA26::IAA26-P108H-GFP transgenic plants and nontransformed control plants. These plants were selected for analysis because at this age IAA26-P108H-GFP protein accumulates to high levels and induces a significant reduction in the accumulation of TMV-V1087I (Figs. 2C and 5A). For transcriptomic analysis, mRNA from three biological replicates, each composed of leaves from four to six plants combined from two independent T2-generation pIAA26::IAA26-P108H-GFP plant lines or from similarly grown nontransformed control plants were used to construct cDNA libraries for RNA sequencing (RNAseq) analysis. Results showed 1,228 genes were significantly altered [a greater than twofold change and false discovery rate (FDR) P value < 0.05] in pIAA26::IAA26-P108H-GFP plants compared with nontransformed control plants (Dataset S1). Of these genes, 236 were altered more than 10-fold, and 28 genes were reduced more than 100-fold. Overall we observed that the majority of the transcripts (78.5%) were reduced in IAA26-stabilized plants, whereas only 21.5% were up-regulated. Additionally, 70% of the identified transcriptionally altered genes contain at least one ARF-binding auxin-responsive element (AuxRE) in their promoter region within 1,500 bp upstream of the start codon, and 35% of identified genes contain an AuxRE within 500 bp upstream of the start codon, indicating that a significant portion of these genes could be directly impacted by the excess accumulation of IAA26-P108H-GFP.
Gene ontology (GO) annotations were incorporated into our gene list, and hypergeometric distributions were used to identify GO categories that occur more frequently in our subset of differentially expressed genes than would be expected by chance (43, 44). We found that genes annotated as involved in “response to stress” were overrepresented and accounted for 21% of the 1,228 genes displaying altered expression levels in auxin-resistant pIAA26::IAA26-P108H-GFP plants (Dataset S1). More specific categories of overrepresented genes included 54 genes involved in “response to wounding,” 46 genes involved in “systemic acquired resistance,” 43 genes involved in “response to salicylic acid,” and 65 genes involved in “response to jasmonic acid” (Table S1 and Dataset S1). In addition, genes that respond to auxin were found to be overrepresented in the transcriptomic analysis, a result that is consistent with IAA26-P108H-GFP functioning in auxin regulation (Dataset S1). Additional differentially expressed groups of genes that were not found to be overrepresented but were known to have an impact on virus biology were identified, including those involved in callose deposition (14 genes) and virus movement (three genes) (Table S1).
Table S1.
Subset of genes differentially expressed in IAA26-stabilized plants with known links to virus movement or defense responses
| AGI identifier* | Gene name | Fold-change† | AuxRE‡ 500 bp | AuxRE‡ 1,500 bp | Description§ |
| Callose | |||||
| AT2G02310 | PP2-B6 | 15.86 | 0 | 2 | Defense response by callose deposition |
| AT1G72520 | LOX4 | 9.21 | 0 | 1 | Defense response by callose deposition |
| AT4G17615 | CBL1 | 3.75 | 0 | 0 | Defense response by callose deposition |
| AT3G04010 | O-Glycosyl hydrolases family 17 | 2.75 | 0 | 0 | β-1,3-Glucanase |
| AT4G39950 | CYP79B2 | 2.61 | 1 | 1 | Defense response by callose deposition |
| AT4G18010 | AT5PTASE2 | 2.50 | 1 | 3 | Defense response by callose deposition |
| AT4G31500 | CYP38B1 | 2.48 | 0 | 3 | Defense response by callose deposition |
| AT5G62570 | CBP60A | 2.36 | 1 | 1 | Defense response by callose deposition |
| AT1G74360 | LRR protein kinase family | 2.25 | 0 | 1 | Defense response by callose deposition |
| AT1G18650 | PCBP3 | −2.20 | 1 | 2 | Callose binding |
| AT1G06490 | CalS7, GSL7 | −2.14 | 0 | 0 | Callose synthase |
| AT4G29360 | O-Glycosyl hydrolases family 17 | −2.68 | 0 | 1 | β-1,3-Glucanase |
| AT5G67460 | O-Glycosyl hydrolases family 17 | −3.57 | 0 | 2 | β-1,3-Glucanase |
| AT5G08000 | PCBP2 | −6.45 | 0 | 0 | Callose binding |
| Virus movement | |||||
| At2G33330 | PDLP3 | −2.22 | 1 | 2 | PD-mediated intercellular transport |
| AT3G47690 | EB1A | −2.49 | 0 | 1 | Viral process |
| AT5G47500 | PME5 | −12.07 | 1 | 1 | Cell wall modification |
| SA-mediated defense | |||||
| At2G35980 | NHL10 | 30.42 | 1 | 1 | SA-mediated signaling pathway |
| AT1G71390 | RLP11 | 22.18 | 0 | 0 | SA mediated signaling pathway |
| At2g14610 | PR1 | 10.57 | 0 | 1 | Response to sa, marker for sar |
| AT3G57260 | PR2 | 8.42 | 0 | 1 | Response to sa, marker for sar |
| AT1G28480 | GRX480, ROXY19 | 6.63 | 1 | 1 | SA-inducible, SA/JA cross-talk |
| AT4G27410 | ANAC072 | 5.76 | 0 | 0 | Inhibition of SA synthesis |
| AT4G04490 | CRK36 | 5.18 | 0 | 2 | SA-mediated signaling pathway |
| AT1G52890 | ANAC019 | 4.89 | 0 | 0 | Inhibition of SA synthesis |
| AT4G14365 | XBAT34 | 4.24 | 0 | 0 | SA-mediated signaling pathway |
| AT2G32680 | RLP23 | 4.19 | 0 | 1 | SA-mediated signaling pathway |
| AT1G75040 | PR5 | 3.89 | 0 | 4 | Response to SA, marker for SAR |
| AT5G60900 | RLK1 | 2.31 | 0 | 0 | SA-mediated signaling pathway |
| AT1G74360 | LRR protein kinase family | 2.25 | 0 | 1 | SA-mediated signaling pathway |
| AT2G17290 | CPK6 | 2.16 | 1 | 2 | SA-mediated signaling pathway |
| AT3G05660 | RLP33 | 2.15 | 2 | 2 | SA-mediated signaling pathway |
| AT5G10380 | RING1 | 2.04 | 0 | 2 | SA-mediated signaling pathway |
| AT4G33070 | PDC1 | −6.21 | 0 | 1 | SA-mediated signaling pathway |
| AT3G02550 | LBD41 | −6.68 | 1 | 4 | SA-mediated signaling pathway |
| AT5G15120 | PCO1 | −9.79 | 0 | 0 | SA-mediated signaling pathway |
| AT1G43800 | SAD6 | −156.34 | 0 | 0 | SA-mediated signaling pathway |
| JA signaling | |||||
| AT2G44810 | DAD1 | 32.97 | 2 | 2 | JA biosynthesis |
| AT2G34600 | JAZ7 | 30.96 | 0 | 1 | Negative regulator of JA signaling |
| AT5G63450 | CYP94B1 | 21.47 | 1 | 3 | Oxidative turnover of JA-Ile |
| AT2G27690 | CYP94C1 | 17.95 | 0 | 0 | Oxidative turnover of JA-Ile |
| AT1G17420 | LOX3 | 15.49 | 0 | 1 | JA biosynthetic process |
| At3G22275 | JAZ13 | 13.67 | 1 | 3 | Negative regulator of JA signaling |
| AT1G30135 | JAZ8 | 11.53 | 0 | 0 | Negative regulator of JA signaling |
| AT1G17380 | JAZ5 | 9.96 | 0 | 3 | Negative regulator of JA signaling |
| AT1G72520 | LOX4 | 9.21 | 0 | 1 | JA biosynthetic process |
| AT5G13220 | JAZ10 | 9.05 | 1 | 1 | Negative regulator of JA signaling |
| AT1G19180 | JAZ1 | 6.81 | 0 | 1 | Negative regulator of JA signaling |
| AT3G25780 | AOC3 | 6.42 | 1 | 4 | JA biosynthesis |
| AT2G06050 | ORP3 | 5.80 | 0 | 1 | JA biosynthesis |
| AT1G74950 | JAZ2 | 3.04 | 0 | 1 | Negative regulator of JA signaling |
| At1G32640 | MYC2 | 2.60 | 0 | 0 | bHLH transcriptional activator |
| AT2G46510 | JAM1 | 2.45 | 0 | 2 | bHLH transcriptional activator |
| AT1G01260 | JAM2 | 2.10 | 0 | 1 | bHLH transcriptional activator |
| WRKY transcription factors | |||||
| At5g24110 | WRKY30 | 7.46 | 0 | 0 | WRKY transcription factor |
| At2g40740 | WRKY55 | 5.53 | 1 | 3 | WRKY transcription factor |
| At5g64810 | WRKY51 | 3.38 | 1 | 1 | WRKY transcription factor |
| At5g22570 | WRKY38 | 3.23 | 0 | 1 | WRKY transcription factor |
| At3g01080 | WRKY58 | 2.93 | 1 | 3 | WRKY transcription factor |
| At2g37260 | WRKY44 | −2.85 | 0 | 1 | WRKY transcription factor |
| At2g34830 | WRKY35 | −14.01 | 0 | 0 | WRKY transcription factor |
| At2g44745 | WRKY12 | −15.67 | 1 | 2 | WRKY transcription factor |
| At1g30650 | WRKY14 | −25.84 | 2 | 2 | WRKY transcription factor |
AGI Identifier, unique locus identifier that is part of the Arabidopsis genome sequence annotation at TAIR (90).
RNAseq fold-change expression values were determined after empirical analysis of digital gene expression using the CLC Genomics Workbench (89). The RNAseq experiment compares three biological replicates of A. thaliana Shahdara pIAA26:IAA26-P108H-GFP plants to three biological replicates of nontransformed A. thaliana Shahdara control plants.
Number of AuxRE found in the upstream promoter region of each gene. AuxRE is defined as a pattern match to TGTCTC, TGTCGG, or TGTSTSBC on either strand within 500 bp or 1,500 bp upstream of the start codon.
Description of the predicted biological process in which each gene is involved collected from TAIR (90).
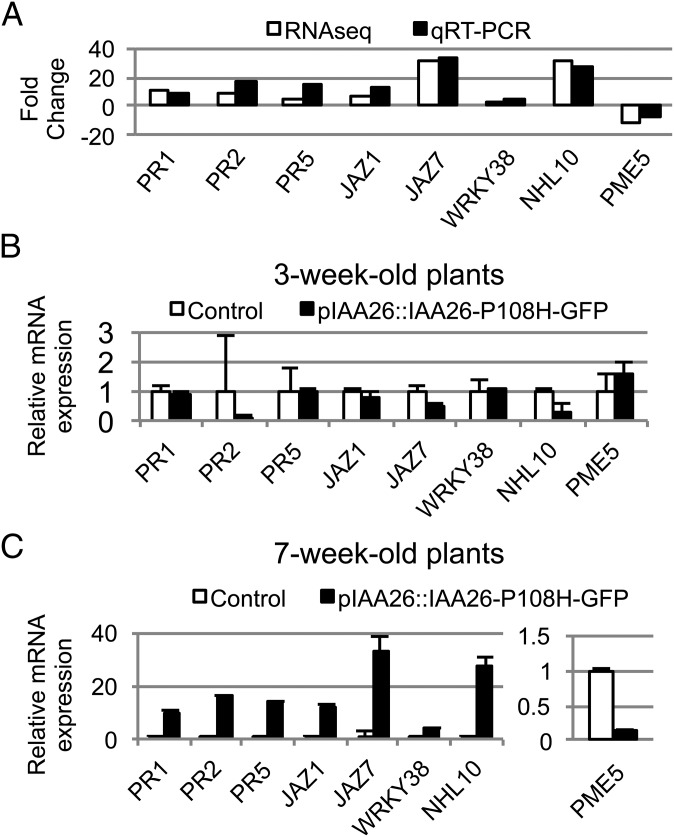
To validate our transcriptomic analysis, we selected a set of eight genes that displayed varying levels of regulation in response to the overaccumulation of IAA26-P108H-GFP for further qRT-PCR analysis. Total RNA from two additional 7-wk-old biological replicates was isolated from the leaves of four to six plants from two independent pIAA26::IAA26-P108H-GFP plant lines. qRT-PCR results clearly demonstrated that the mRNA levels for each of the eight selected genes mirrored those obtained in our RNAseq studies (Fig. 9A). In addition, we also compared mRNA levels from the leaf tissue of both 3- and 7-wk-old pIAA26::IAA26-P108H-GFP plants. We speculated that if these genes are altered by the overaccumulation of IAA26-P108H-GFP, then this regulation should occur primarily in older tissues, where the IAA26-P108H-GFP protein was found to accumulate, and not in younger tissues, where IAA26-P108H-GFP does not accumulate significantly (Fig. 2 B and C). Results indicated that in 3-wk-old tissue none of the eight selected genes show a significant alteration in expression in comparison with a nontransformed control plant (Fig. 9B). In contrast, in 7-wk-old plants all eight genes display altered expression levels that roughly correspond to their alterations within our transcriptomic analysis (Fig. 9C). These findings are consistent with the accumulation of IAA26-P108H-GFP protein only in older leaf tissues, supporting a role for IAA26-P108H-GFP in the regulation of these genes.
Fig. 9.
Validation of IAA26-P108H-GFP–affected gene expression. qRT-PCR expression analysis for eight selected genes that displayed significant transcriptional alterations within pIAA26::IAA26-P108H-GFP A. thaliana Shahdara plants. (A) Fold-change comparisons between qRT-PCR (black bars) and RNAseq (white bars) studies. The qRT-PCR fold-change represents the mean from two biological replicates. The RNAseq fold-change was determined by empirical analysis of digital gene expression for three biological replicates (91). (B and C) Fold-change comparisons for the eight selected genes in 3- and 7-wk-old pIAA26::IAA26-P108H-GFP plants (black bars) versus nontransformed control plants (white bars). (B) None of the eight selected genes shows a significant alteration in expression in 3-wk-old tissue. (C) In contrast, all eight genes display significantly altered expression levels in 7-wk-old tissue. Each bar represents the mean ± SE from two biological replicates composed of leaf tissue from four to six plants from two independent plant lines. The 18S RNA gene was chosen as an internal control for normalization.
Discussion
To maintain physiological functions, plants must tightly regulate the movement of molecules into and out of the vascular network. The gatekeeper function of the vascular phloem thus represents a significant obstacle through which viruses must navigate to achieve a systemic infection. For TMV, our studies indicate that in mature tissue access to the vascular gateway is enhanced significantly by the disruption of select phloem-expressed Aux/IAA transcriptional regulators. Auxin, an essential plant hormone, is involved in many processes, including the development and regulation of the vascular network (45). Aux/IAA proteins are a key component in this regulatory system, playing a central role in converting auxin concentrations into gene expression (24, 46). The targeted disruption of specific phloem-localized Aux/IAA proteins results in the transcriptional reprograming of the vascular tissue. This reprogramming corresponds with enhanced phloem loading and viral systemic movement and confers a significant advantage to TMV over viruses that are unable to disrupt these Aux/IAAs.
Aux/IAA proteins are transcriptional regulators that interact with and control ARF transcription factors (23, 24). ARFs target the AuxRE that include TGTCTC, TGTCGG, and TGTSTSBC located in the promoters of auxin-responsive genes (47). Within Arabidopsis 71% of the mapped genes contain at least one AuxRE within 1,500 bp upstream of the start codon, and 31% contain at least one AuxRE within 500 bp upstream of the start codon. Thus, Aux/IAA proteins have the potential to impact the regulation of a significant portion of the plant’s genome. In previous studies we found that only 3 of 10 tested Arabidopsis Aux/IAA proteins were capable of interacting with the TMV replication protein (22). In this study we show that all three interacting Aux/IAA family members are expressed predominately in the phloem, and in the case of the strongest interactor, IAA26, in the phloem CCs. Consistent with our results, IAA26, IAA27, and IAA18 have been reported to be three of the six Aux/IAA family members enriched in vascular tissue isolated from mature Arabidopsis leaves (48). Furthermore, in translatome-profiling studies IAA26 was found to be enriched in root and shoot CCs (49). It also is interesting that in micrografting experiments the mRNA of IAA18 has been shown to be phloem mobile and transported to the roots, where it negatively regulates lateral root development (48). Taken together, these findings confirm that Aux/IAA proteins targeted by TMV are predominantly phloem expressed.
Within the CC–SE complex SEs provide the transport conduit between distal tissues and, lacking a nucleus, are dependent upon CCs for RNA and protein synthesis. For viruses such as TMV that require an assembly-functional CP to move systemically, it seems likely that viral genomic RNA and CP are transported separately through the specialized PD pore units that connect CCs and SEs, with virion assembly occurring within the SEs (11, 50). Thus, TMV loading from CCs to the SEs is likely to function differently from the cell-to-cell movement that occurs between other cell types where CP is not required. Enhancing a virus’s ability to move through this unique gateway thus would confer a significant advantage. TMV clearly has this advantage within mature tissues, where phloem loading is very efficient for TMV but delayed for the non–Aux/IAA-interacting virus (Fig. 6B), or when IAA26 is stabilized and over accumulates in the phloem (Fig. 7). Additionally, the advantage conferred through the disruption of phloem Aux/IAA functions is of particular importance for a mechanically inoculated virus such as TMV that does not have a vector and therefore has no control over the types or age of the tissue it infects.
The targeting of these specific Aux/IAA proteins indicates that they function in the transcriptional regulation of genes that directly impact TMV phloem loading. This possibility is strengthened by the inability of IAA26-P108H-GFP to affect CF dye movement in whole leaves, suggesting that observed effects on phloem loading are restricted to the CC–SE complex and are specific to virus movement (Fig. S4). Transcriptomic analysis of plants expressing a stabilized auxin-resistant version of IAA26 from its native promoter identified a number of host genes known to impact the transport of molecules across PD or to modulate host defense signals as affected by IAA26 (Table S1 and Dataset S1). Additionally, as expected, several auxin-responsive genes are altered when IAA26 is stabilized (Dataset S1). The known function of these IAA26-altered genes defines several potential mechanisms through which disruption of phloem-specific Aux/IAA proteins could enhance the ability of TMV to move systemically.
At the local level, genes affected by IAA26 may be involved directly in regulating the cell-to-cell movement of TMV. Three genes with established connections to virus movement that displayed altered expression in IAA26-stabilized plants included pectin methylesterase 5 (PME5), microtubule end-binding 1a (EB1a), and PD-located protein 3 (PDLP3) (Table S1). PME5 is of particular interest because it displays a 12-fold reduction in pIAA26::IAA26-P108H-GFP plants. Pectin methylesterases (PMEs) catalyze the demethylesterification of pectin, releasing both protons and methanol. PME-dependent methanol emission has been shown to trigger PD dilation (51). In addition, PMEs have been shown to be involved with Tobamovirus local cell-to-cell movement through PD via an interaction with the TMV MP (52). When TMV MP is mutated so it no longer binds PME, the local spread of virus is reduced. Additionally, knockout of PMEs reduce local viral movement (53, 54). PMEs also may play a role in systemic viral movement, because TMV systemic movement is delayed in PME knockdown plants (52). Overexpression of PME inhibitors in tobacco and Arabidopsis also limited Tobamovirus systemic movement (55). Furthermore, PME5 contains an AuxRE in its promoter region, suggesting it could be directly regulated by IAA26. Reduction of PME5 expression in IAA26-stabilized plants could explain the reduced phloem loading of TMV in these plants.
Both EB1a and PDLP3 display greater than twofold reductions in expression when IAA26 is stabilized. EB1 proteins are evolutionarily conserved and localize to growing microtubule plus ends where they regulate microtubule dynamics. TMV MP has been shown to colocalize and interact with GFP-tagged EB1a in Arabidopsis (56). It has been suggested that this EB1a/TMV-MP association could assist in the movement of TMV replication complexes to the PD. Consistent with the role of microtubule polymerization in cell-to-cell viral movement, tobacco mutants that have reduced microtubule dynamics also have reduced TMV cell-to-cell movement (57). Thus, altering the expression of EB1a may be advantageous for virus movement. PDLPs are another group of proteins involved in the movement of viruses through PD. PDLPs interact with tubule-forming MPs, and disruption of this interaction leads to delayed infection and attenuated symptoms (58). Although TMV does not form tubules, down-regulation of PDLPs suggests IAA26 may regulate host pathways involved in the general movement of macromolecules within the vascular phloem.
Additional genes linked to virus movement include members of the β-1,3-glucanase family, two of which show two- to threefold reductions in pIAA26::IAA26-P108H-GFP plants (Table S1). β-1,3-glucanases are enzymes that degrade callose. Several studies have shown that callose deposition at PD leads to decreased cell-to-cell movement, whereas treatments inhibiting callose deposition lead to increased size-exclusion limits of PD (59–61). Reduction of β-1,3-glucanases could lead to the accumulation of callose at PD and decreased cell-to-cell movement when IAA26 accumulates. Additionally, two PD callose-binding proteins, PDCB2 and PDCB3, were significantly down-regulated in pIAA26::IAA26-P108H-GFP plants, by 6.4- and 2.2-fold, respectively (Table S1). PDCBs specifically bind to callose in vitro and have been proposed to be involved in callose-mediated regulation within the PD (62). Taken together, the observed down-regulation of genes that positively impact virus movement suggests that IAA26 functions to regulate these genes negatively.
Host defense pathways, including SA and jasmonic acid (JA), have been shown to be key factors in the development of systemic resistance against TMV (63–65). Transcriptomic analysis of pIAA26::IAA26-P108H-GFP plants showed marked changes in genes associated with these defense pathways. Of particular interest were genes related to SA defense responses, including the SA defense markers pathogenesis-related (PR) genes PR1, PR2, and PR5, which are up-regulated 10.5-, 8.4-, and 3.8-fold, respectively, upon IAA26 stabilization in the phloem (Table S1) (66). The up-regulation of SA-mediated defense genes is consistent with previously published results showing that, when the AUX/IAA protein AXR2/IAA7 is stabilized, AvrRpt2, a Pseudomonas syringae effector, is no longer able to suppress the induction of SA-mediated defense genes (67). Additional genes involved in SA biosynthesis, including ANAC019 and ANAC072, and genes involved in SA-mediated signaling also were up-regulated in pIAA26::IAA26-P108H-GFP plants (Table S1 and Dataset S1). Thus, Aux/IAA proteins appear to play important roles in the regulation of SA-mediated defense response.
We also observed transcriptional alterations in several key genes associated with cross-communication between the SA and JA defense pathways. In particular, we identified nine WRKY transcription factors that were altered in IAA26-stabilized plants (Table S1). WRKY transcription factors function as regulators between the SA and JA defense pathways (68). Five WRKY family members were up-regulated (WRKY30, -38, -51, -55, and -58), and four were down-regulated (WRKY12, -14, -35, and -44). Interestingly, the five up-regulated WRKYs have all been shown previously to be induced in response to SA treatment, whereas the four that were down-regulated were all undetectable (WRKY12, -14, and -35) or repressed (WRKY44) by SA treatment (69). WRKY38 expression has been linked to the degradation of NPR1 and in combination with WRKY62 is required for systemic acquired resistance (70), and WRKY51 has been shown to mediate SA-dependent repression of JA signaling (71). Another important regulator affecting the antagonism between SA- and JA-mediated signaling is GRX480, which showed a sixfold increase in IAA26-P108H-GFP plants (72). Additionally, seven JASMONATE ZIM-DOMAIN (JAZ) repressor proteins were up-regulated (Table S1). JAZ proteins negatively regulate the JA-signaling pathway by repressing transcription factors that control JA-regulated genes (73). It has been proposed that stabilization of JAZ proteins may be one way that SA exerts an antagonistic effect on JA signaling (74). Taken together, these results indicate that IAA26 likely functions as an important regulator of these host defense pathways within the phloem of mature tissues.
There is growing evidence for auxin involvement in disease resistance through cross-talk with the SA and JA signaling pathways. Treatment of Arabidopsis with the SA analog benzothiadiazole S-methylester (BTH) resulted in an overall reduction of auxin responses (75). One mechanism by which SA might inhibit auxin signaling is through transcriptional repression of the auxin-receptor genes, leading to reduced degradation of Aux/IAA proteins and thus to the repression of auxin responses. Support for this mechanism comes from observations that auxin-receptor genes are down-regulated in response to BTH treatment and that detection of Aux/IAA proteins by Western blot is increased after SA treatment (75). Conversely, activation of auxin signaling leads to the suppression of SA biosynthesis and signaling (76). It also has been reported that auxin triggers the induction of genes involved in JA biosynthesis (77). Consistent with these findings are studies that have linked the stabilization of Aux/IAA proteins to bacterial resistance. For example, when axr2-1 plants, which produce a nondegradable form of AXR2/IAA7, were infected with the bacterial pathogen P. syringae pv. maculicola, there was a 10-fold reduction in bacterial growth (75). Consistent with these findings, our results also demonstrate a role for auxin as a negative regulator of the SA pathway, perhaps via activation of JA signaling.
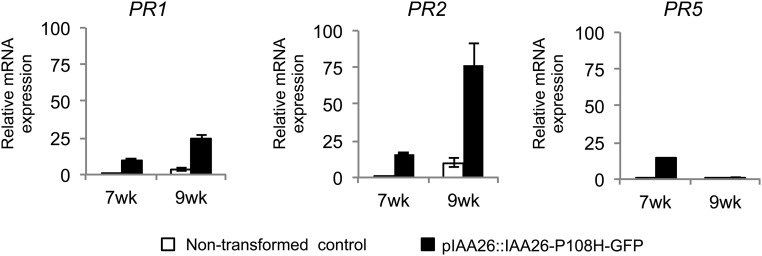
Interestingly, the biological advantage of the TMV–Aux/IAA interaction was observed only in older plant tissues (Figs. 5 and 6). In particular, inhibition of TMV phloem loading occurred in 9-wk-old and not in 7-wk-old pIAA26::IAA26-P108H-GFP plants, even though both developmental stages expressed and accumulated similar levels of IAA26-P108H-GFP mRNA and protein (Fig. 2 B and C). This finding suggests that additional factors can impact the ability of IAA26 to affect TMV phloem loading. Although no obvious phenotypic differences were observed between pIAA26::IAA26-P108H-GFP plants and nontransformed control plants, we did find that bolting and the initiation of flowering occurred between week 7 and week 9. This finding is of interest, because previous studies have noted that the development of ARR, defined as the development of resistance in mature tissues, is often associated with transition to flowering (35, 78). Within Arabidopsis, ARR has been shown to impact the virulence of P. syringae and the systemic movement of cauliflower mosaic virus (79, 80). In both systems, mutations in genes associated with the induction and maintenance of flowering were shown to influence the development of this resistance. Resistance against P. syringae has been linked to the accumulation of SA (81). To examine ARR in pIAA26::IAA26-P108H-GFP plants, we determined whether the expression of SA-associated genes changed between week 7 and week 9. Results indicated that PR1, PR2 and PR5 expression is greater in pIAA26::IAA26-P108H-GFP plants than in nontransformed controls at both week 7 and week 9. In addition, PR1 and PR2, but not PR5, show significantly increased expression in flowering 9-wk-old plants (Fig. S5). These findings indicated that the developmental age of the plant significantly influences the effect of IAA26-P108H-GFP accumulation on the expression of these SA-associated genes. Although additional studies are needed, these findings suggest that IAA26 is a factor in the regulation of ARR.
Fig. S5.
PR1, PR2, and PR5 gene expression in 7- and 9-wk-old plants. Fold-change comparisons from qRT-PCR expression analysis between nontransformed control (white bars) and pIAA26::IAA26-P108H-GFP (black bars) plants. Each bar represents the mean ± SE from three biological replicates composed of leaf tissue from four to six plants from two independent plant lines. The 18S RNA gene was chosen as an internal control for normalization.
In summary, IAA26 plays an important role in regulating the phloem environment within mature plant tissues and appears to be a contributor in ARR. The diverse array of genes impacted through the stabilization of IAA26 suggests that this auxin-regulated protein has a significant role in modulating both the local environment of the CC–SE complex and surrounding and distal tissues. The regulation of genes that impact PD function and the inability of TMV to move into the phloem in older IAA26-stabilized plants suggest that this protein regulates the transport of macromolecules within the phloem. Impacts on SA and JA defense pathways provide another mechanism whereby IAA26 could produce wider cellular responses both in tissues surrounding the vascular phloem and systemically throughout the plant, providing a means for TMV to modulate responses such as ARR and systemic acquired resistance. Additional studies directed at understanding the precise impact of this TMV-directed vascular reprogramming should provide greater details about the role these mechanisms play in promoting virus phloem loading and systemic movement.
Experimental Procedures
Promoter Constructs and GUS Assays.
Promoter fragments of 2 kb for IAA26 and IAA18 and 1.5 kb for IAA27 were amplified from genomic DNA extracted from A. thaliana ecotype Shahdara using promoter-specific primers (Table S2). Cloned promoter fragments were moved into pBI101.1 (Clontech) upstream of the GUS reporter ORF via primer-generated restriction sites. Promoter pIAA::GUS constructs were introduced into the Agrobacterium tumefaciens strain GV3101 (82), and a floral dip method was used for plant transformation (83). All plants were maintained in growth chambers for a 12-h photoperiod at 24 °C. Histochemical staining for GUS activity was performed as previously described (84). After staining, whole leaves or hand-sectioned petioles or stems were imaged using an Olympus Stereo MVX10 or BX60 Microscope.
Table S2.
Primers used for promoter amplification and qRT-PCR
| Symbol | Name | Forward primer | Reverse primer |
| pIAA26 | IAA26 promoter (2 kb) | ATCGATCTCCTTTTTTAGTTCACTAA | GGATCCTGATCAACCCAAGATTCC |
| pIAA27 | IAA27 promoter (1.5 kb) | TCTAGATTTTGTATGTCTTCTTGCTATGTAT | CCCGGGTTTTTTCAAATATAATTGTGGAC |
| pIAA18 | IAA18 promoter (2 kb) | GTCGACTATCGATCTAAAGTATAAGATGAC | GGATCCTAGGATTTTTTTTAGAGGAACTACA |
| PR1 | Pathogenesis-related gene 1 | CCCACAAGATTATCTAAGGGTTCAC | TCCCACTGCATGGGACCTA |
| PR2 | Pathogenesis-related protein 2 | TCAACCAACAGCTGGACAAA | CCGATGGACTTGGCAAGGTA |
| PR5 | Pathogenesis-related gene 5 | AACGGCGGCGGCGTTC | GCCGCCATCGCCTACTAGA |
| JAZ1 | Jasmonate-ZIM-domain protein 1 | GCCTGATGTCASATGGAACTTTAG | CAGATTCGTCGGTTTAACATCTTG |
| JAZ7 | Jasmonate-ZIM-domain protein 7 | GCGACAAGCCTTACTCAATTT | TGATTCGTCCAACGAGCTATG |
| WRKY38 | WRKY DNA-binding protein 38 | TCTAGGGATTCTTCTCCTCCTC | GTGAATCGTCCCTCCAATTCT |
| NHL10 | NDR1/HIN1-like 10 | CAAGTTCAGGCTTAGGGTTAGG | AGGTAGTTGTAGTTCCGTTTGAG |
| PME5 | Pectin methylesterase 5 | AGTCATCACCGTCTCACTTAAC | TAGCTGGAACCACCACTTTC |
pIAA26::IAA26-P108H-GFP Construction and Characterization.
To create pIAA26::IAA26-P108H-GFP, the ORF of our previously described 35S::IAA26-P108H-GFP (22) construct was removed by BamHI and SacI digestion and ligated into a similarly cut pIAA26::GUS (described above), replacing the GUS ORF with that of IAA26-P108H-GFP. A. thaliana ecotype Shahdara plants were transformed with pIAA26::IAA26-P108H-GFP as described above (85). Plants were maintained as described above, and 3- to 9-wk-old T2-generation plants from two independent transgenic lines were analyzed for IAA26-P108H-GFP mRNA and protein expression as described in SI Experimental Procedures. GFP fluorescence localization for IAA26-P108H-GFP was done by hand-sectioning petioles. Sectioned tissues were stained with 1 μg/mL DAPI and were imaged using a Zeiss LSM700 lase- scanning confocal microscope (Carl Zeiss, Inc.).
NbIAA26 Cloning and Characterization.
Total leaf RNA from N. benthamiana plants was used to amplify the NbIAA26 ORF as described in SI Experimental Procedures. Yeast two-hybrid and transient NbIAA26-GFP localization studies were performed as previously described (20–22). NbIAA26 in situ hybridizations were done following previously published methods (86). Details for this protocol can be found in the SI Experimental Procedures.
Virus Accumulation, in Situ Localizations, and Systemic Movement Assays.
Seven- and nine-week-old Shahdara control and transgenic pIAA26::IAA26-P108H-GFP plants were used for virus accumulation studies. Rosette leaves were dusted with carborundum and mechanically inoculated with 10 μg of purified TMV or TMV-V1087I. Leaf punches were collected from 18 independently inoculated rosette leaves at 3, 6, and 9 dpi. For studies in N. benthamiana, plants were inoculated with 5 μg of TMV or TMV-V1087I, and leaf punches were collected from six inoculated leaves at 2, 4, and 6 dpi. All samples were ground in Laemmli sample buffer (87) and analyzed by Western immunoblot analysis using TMV-specific antibodies as previously reported (21, 37). Whole-leaf in situ localizations for the detection of the TMV CP were done as previously described (88).
For systemic movement assays lower leaves of N. tabacum cv. Xanthi plants were inoculated with 10 ng of virus. Three plants were used for each inoculation treatment. Plants were randomly assigned to one of five inoculation treatments: single inoculation with either TMV or TMV-V1087I or simultaneous inoculation with TMV and TMV-V1087I at a 1:1, 1:5, or 1:10 ratio of TMV to TMV-V1087I. The infectivity of TMV and TMV-V1087I inoculum was determined by local lesion assays on N. tabacum cv. Xanthi-NC and was standardized to ensure an equal infection potential. RNA was isolated from systemic leaves at 10 dpi, when mosaic disease symptoms were first observed, and cDNA was prepared as described above. Quantitative sequencing for the detection and quantification of the G-to-A TMV-V1087I point mutation was done as described in ref. 42. See SI Experimental Procedures for details.
Transcriptomic Analysis and Validation.
Total RNA was isolated from 7-wk-old pIAA26::IAA26-P108H-GFP transgenic plants and nontransformed control Shahdara plants using the RNeasy kit (Qiagen). Four fully expanded leaves were collected from each test plant, and total RNA from four to six individual test plants was pooled for each of the three biological replicates. RNAseq library construction and sequencing was done by the University of Maryland Institute for Bioscience and Biotechnology Research (UM-IBBR) Sequencing Core. RNAseq analysis was done using CLC Bio genomics workbench as described in SI Experimental Procedures (89, 90). The RNAseq data have been deposited in NCBI’s Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo) and are accessible through GEO series accession number GSE75983.
To validate RNAseq findings, total RNA from 3- or 7-wk-old pIAA26::IAA26-P108H-GFP plants or nontransformed control plants was isolated and used to generate cDNA. qRT-PCR was performed as described in SI Experimental Procedures for two additional biological replicates, each containing three technical replicates. Primer sequences used for the amplification of selected genes are provided in Table S2. The 18S RNA gene was chosen as an internal control for normalization.
SI Experimental Procedures
Characterization of pIAA26::IAA26-P108H-GFP Plant Lines.
For mRNA expression total RNA was isolated from 3- to 9-wk-old pIAA26::IAA26-P108H-GFP plants or from nontransformed control plants using the RNeasy Plant Mini Kit (Qiagen). One microgram of isolated RNA was pretreated with RQ1 DNase (Promega), followed by reverse transcription using the SuperScript III First-Strand cDNA Synthesis System and random hexamer primers (Invitrogen). SYBR Green real-time qRT-PCR was performed in 96-well reaction plates in an ABI Prism 7100 (Applied Biosystems). Experiments were done in three biological replicates, each containing three technical replicates. The 18S RNA gene was chosen as an internal control for normalization and was amplified using the primers 5′-CGTCCCTGCCCTTTGTACAC and 3′-CGAACACTTCACCGGATCATT. Primers 5′-TCAAGAAGGAAACAGAAGACAAATC and 3′-TGTGAGATGTGAGGAACAGAAG were used to detect endogenous IAA26 transcripts in control plants. Primers 5′-GATGCAGTAAGCAAGAGAAGATG and 3′-CTGAACTTGTGGCGTTTAC that span the IAA26-P108H to GFP fusion were used to detect the IAA26-P108H-GFP transgene mRNA. For protein expression, leaf tissue from 3- to 9-wk-old plants was collected and ground in Laemmli sample buffer (87). Samples were loaded onto denaturing polyacrylamide gels and blotted onto nitrocellulose membranes. Blotted proteins were probed with anti-GFP antibodies (Santa Cruz Biotechnology, Inc.) or anti-actin antibodies (Sigma) as a loading control.
NbIAA26 Isolation and Analysis.
Whole-leaf RNA from N. benthamiana isolated via RNeasy minipreps (Qiagen) was used to generate cDNA via the SuperScript First-Strand Synthesis System (Invitrogen by Life Technologies). NbIAA26-specific 5′-CGGATCCAATGGAGGGTTATTCAAG and 3′-GCTCGAGTCAGGTCAGCTTATTTCC primers containing BamHI and XhoI restriction sites were used to amplify the 828-bp NbIAA26 ORF. The amplified ORF was cloned into pCRII-TOPO (Invitrogen) and sequenced. Protein sequence alignments for NbIAA26, NtIAA26, and AtIAA26 were performed using CLC Genomics Workbench v 7.5.1 (CLC Bio).
In situ analysis of NbIAA26 mRNA localization was done using plant stem tissues fixed in methanol-glacial acetic acid (3:1) for 45 min, followed by chlorophyll removal in methanol and in absolute ethanol. Fixed tissue samples were rehydrated (95%, 75%, 50%, 25% ethanol), refixed, and washed in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) with 0.1% Tween-20 at room temperature. Prehybridization occurred at 55 °C for 1 h in buffer (0.1% Tween-20, 50% formamide, 2× saline sodium citrate buffer, pH 7.0), and a digoxigenin-labeled probe specific for NbIAA26 was hybridized for 16 h. Probed tissues were washed and incubated with anti–digoxigenin-AP Fab fragments. Incubated tissues were subsequently stained for hybridization via alkaline phosphatase. Stained leaves were imaged using an Olympus Stereo MVX10 microscope.
Quantitative Sequencing for TMV–TMV-V1087I in Systemically Infected Tissues.
A 500-bp region of the 126-kDa gene that contains the point mutation G-to-A for TMV-V1087I was amplified using PCR primers 5′-CTTTGTCATGAGCACTTCTTCG and 3′-TTGTGTTCCTGCATCGACC. The PCR products were purified using GeneJet PCR purification kit (Fermentas, Thermo Fisher Scientific) and sequenced using the 5′ amplification primer. The relative quantification of TMV and TMV-V1087I was determined using sequencing chromatograms as previously published (42). Briefly, the peak height ratio at the polymorphic site (height of A peak)/(height of A peak + height of G peak) was determined for each sample and compared with a standard curve developed by sequencing nine known ratios (1:0, 1:1, 1:2, 1:5, 1:10, 2:1, 5:1, 10:1, and 0:1) of TMV and TMV-V1087I.
RNAseq Analysis.
For RNAseq analysis RNA isolated from pIAA26::IAA26-P108H-GFP plants was run on a 2100 Bioanalyzer Eukaryotic Total RNA Nanochip (Agilent Technologies) to check RNA quality. RNA integrity numbers for all samples were between 7.2 and 8.0. Library preparation with the Illumina TruSeq RNA sample preparation kit and sequencing with the Illumina HiSeq1500 rapid run was done by the UM-IBBR Sequencing Core (https://www.ibbr.umd.edu/facilities/sequencing). Each cDNA library generated between 21 and 64 million 50-bp single-end sequence reads. Reads were mapped to the A. thaliana Col-0 genome reference TAIR10 using the CLC Genomics Workbench v 7.5.1 RNA-seq analysis tool and default parameters (mismatch cost 2, insertion cost 3, deletion cost 3, length fraction 0.8, similarity fraction 0.8, maximum hits for a read 10) (CLC Bio) (89, 90). Approximately 93.9–98.5% of total reads mapped for each cDNA library, and 96.6–98.0% of mapped reads aligned to exons (Dataset S1). Reads aligned to exons were used in all subsequent analyses. Statistical analysis to identify differentially expressed genes was performed using the CLC Genomics Workbench Empirical Analysis of DGE (digital gene expression) tool with default dispersion estimation parameters (total count filter cutoff 5.0) (91). Results were filtered with a cut of value of fold-change >2.0 and the FDR-corrected P value <0.05. Gene symbols, gene model descriptions, and GO annotations were retrieved from TAIR on August 4, 2015 (90). The generic GO term-finder tool developed at the Lewis–Sigler Institute at Princeton University (44) was used to identify GO annotation terms that occurred more frequently than expected by chance in the list of differentially regulated genes. A Bonferroni-corrected P value of <0.01 was used as a cutoff for significantly enriched GO terms. Gene promoter regions for AuxREs were analyzed using the PatMatch tool (92). AuxRE was defined as a pattern match to TGTCTC, TGTCGG, or TGTSTSBC on either strand within 1,500 bp upstream of the start codon.
Dye-Loading Assays.
To detect alterations to the size-exclusion limits of PD, we used CFDA as previously described, with slight modifications (41). Briefly, 5 μL of 1 mM CFDA was applied to the adaxial surface of a mature source leaf of an intact 9-wk-old nontransformed control or pIAA26::IAA26-P108H-GFP plant. After 1 h any remaining dye was removed, and leaves were washed with sterile water. The abaxial surface of the leaf was imaged using an Olympus Stereo MVX10 microscope. The area of CF spread was measured using ImageJ (93). For SA treatments, plants were sprayed with 100 μM of SA 24 h before CFDA application.
To examine phloem loading, the adaxial surface of a mature source leaf of an intact 9-wk-old nontransformed control or pIAA26::IAA26-P108H-GFP plant was gently abraded with fine sandpaper. CFDA (20 μL; 60 μg/mL) was applied to the adaxial leaf surface as previously described (94). The abaxial side of the leaf was imaged after translocation in the light for 2 h.
qRT-PCR Analysis of PR1, PR2, and PR5 Expression in 7- and 9-Wk-Old Plants.
Total RNA from 7- or 9-wk-old pIAA26::IAA26-P108H-GFP plants or nontransformed control plants was isolated and used to generate cDNA. qRT-PCR was performed as described above for characterization of pIAA26::IAA26-P108H-GFP plant lines using three biological replicates, each containing three technical replicates. Primer sequences used for the amplification of PR1, PR2, and PR5 are provided in Table S2. The 18S RNA gene was chosen as an internal control for normalization.
Supplementary Material
Acknowledgments
This work was supported in part by US Department of Agriculture National Research Initiative Competitive Grant 2008-35319-19168 and National Science Foundation Grant ISO-1120044. T.D.C. was additionally supported by NIH Institutional Training Grant 5T32AI051967 awarded to the University of Maryland.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE75983).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524390113/-/DCSupplemental.
References
- 1.Heinlein M. Plasmodesmata: Channels for viruses on the move. Methods Mol Biol. 2015;1217:25–52. doi: 10.1007/978-1-4939-1523-1_2. [DOI] [PubMed] [Google Scholar]
- 2.Solovyev AG, Savenkov EI. Factors involved in the systemic transport of plant RNA viruses: The emerging role of the nucleus. J Exp Bot. 2014;65(7):1689–1697. doi: 10.1093/jxb/ert449. [DOI] [PubMed] [Google Scholar]
- 3.Stadler R, et al. Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. Plant J. 2005;41(2):319–331. doi: 10.1111/j.1365-313X.2004.02298.x. [DOI] [PubMed] [Google Scholar]
- 4.Leisner SM, Turgeon R. Movement of virus and photoassimilate in the phloem: A comparative analysis. BioEssays. 1993;15(11):741–748. doi: 10.1002/bies.950151107. [DOI] [PubMed] [Google Scholar]
- 5.Ham BK, Lucas WJ. The angiosperm phloem sieve tube system: A role in mediating traits important to modern agriculture. J Exp Bot. 2014;65(7):1799–1816. doi: 10.1093/jxb/ert417. [DOI] [PubMed] [Google Scholar]
- 6.Kehr J, Buhtz A. Long distance transport and movement of RNA through the phloem. J Exp Bot. 2008;59(1):85–92. doi: 10.1093/jxb/erm176. [DOI] [PubMed] [Google Scholar]
- 7.Turgeon R, Wolf S. Phloem transport: Cellular pathways and molecular trafficking. Annu Rev Plant Biol. 2009;60:207–221. doi: 10.1146/annurev.arplant.043008.092045. [DOI] [PubMed] [Google Scholar]
- 8.Harries P, Ding B. Cellular factors in plant virus movement: At the leading edge of macromolecular trafficking in plants. Virology. 2011;411(2):237–243. doi: 10.1016/j.virol.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Chisholm ST, Parra MA, Anderberg RJ, Carrington JC. Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long-distance movement of tobacco etch virus. Plant Physiol. 2001;127(4):1667–1675. [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C, Nelson RS. The cell biology of Tobacco mosaic virus replication and movement. Front Plant Sci. 2013;4:12. doi: 10.3389/fpls.2013.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding X, Shintaku MH, Carter SA, Nelson RS. Invasion of minor veins of tobacco leaves inoculated with tobacco mosaic virus mutants defective in phloem-dependent movement. Proc Natl Acad Sci USA. 1996;93(20):11155–11160. doi: 10.1073/pnas.93.20.11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Culver JN, Dawson WO. Tobacco mosaic virus coat protein: An elicitor of the hypersensitive reaction but not required for the development of mosaic symptoms in Nicotiana sylvestris. Virology. 1989;173(2):755–758. doi: 10.1016/0042-6822(89)90592-8. [DOI] [PubMed] [Google Scholar]
- 13.Dawson WO, Bubrick P, Grantham GL. Modifications of the Tobacco Mosaic-Virus Coat Protein Gene Affecting Replication, Movement, and Symptomatology. Phytopathology. 1988;78(6):783–789. [Google Scholar]
- 14.Hilf ME, Dawson WO. The tobamovirus capsid protein functions as a host-specific determinant of long-distance movement. Virology. 1993;193(1):106–114. doi: 10.1006/viro.1993.1107. [DOI] [PubMed] [Google Scholar]
- 15.Fenczik CA, Padgett HS, Holt CA, Casper SJ, Beachy RN. Mutational analysis of the movement protein of odontoglossum ringspot virus to identify a host-range determinant. Mol Plant Microbe Interact. 1995;8(5):666–673. doi: 10.1094/mpmi-8-0666. [DOI] [PubMed] [Google Scholar]
- 16.Deom CM, Quan S, He XZ. Replicase proteins as determinants of phloem-dependent long-distance movement of tobamoviruses in tobacco. Protoplasma. 1997;199(1-2):1–8. [Google Scholar]
- 17.Sasaki N, et al. Over-expression of putative transcriptional coactivator KELP interferes with Tomato mosaic virus cell-to-cell movement. Mol Plant Pathol. 2009;10(2):161–173. doi: 10.1111/j.1364-3703.2008.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Zhang L, Li D, Wang F, Yu D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA. 2013;110(21):E1963–E1971. doi: 10.1073/pnas.1221347110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy A, Zheng JY, Lazarowitz SG. The tobamovirus Turnip Vein Clearing Virus 30-kilodalton movement protein localizes to novel nuclear filaments to enhance virus infection. J Virol. 2013;87(11):6428–6440. doi: 10.1128/JVI.03390-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padmanabhan MS, Goregaoker SP, Golem S, Shiferaw H, Culver JN. Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. J Virol. 2005;79(4):2549–2558. doi: 10.1128/JVI.79.4.2549-2558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padmanabhan MS, Kramer SR, Wang X, Culver JN. Tobacco mosaic virus replicase-auxin/indole acetic acid protein interactions: Reprogramming the auxin response pathway to enhance virus infection. J Virol. 2008;82(5):2477–2485. doi: 10.1128/JVI.01865-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padmanabhan MS, Shiferaw H, Culver JN. The Tobacco mosaic virus replicase protein disrupts the localization and function of interacting Aux/IAA proteins. Mol Plant Microbe Interact. 2006;19(8):864–873. doi: 10.1094/MPMI-19-0864. [DOI] [PubMed] [Google Scholar]
- 23.Calderon-Villalobos LI, Tan X, Zheng N, Estelle M. Auxin perception--structural insights. Cold Spring Harb Perspect Biol. 2010;2(7):a005546. doi: 10.1101/cshperspect.a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salehin M, Bagchi R, Estelle M. SCFTIR1/AFB-based auxin perception: Mechanism and role in plant growth and development. Plant Cell. 2015;27(1):9–19. doi: 10.1105/tpc.114.133744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reed JW. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 2001;6(9):420–425. doi: 10.1016/s1360-1385(01)02042-8. [DOI] [PubMed] [Google Scholar]
- 26.Guilfoyle TJ. The PB1 domain in auxin response factor and Aux/IAA proteins: A versatile protein interaction module in the auxin response. Plant Cell. 2015;27(1):33–43. doi: 10.1105/tpc.114.132753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- 28.Piya S, Shrestha SK, Binder B, Stewart CN, Jr, Hewezi T. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci. 2014;5:744. doi: 10.3389/fpls.2014.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nanao MH, et al. Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun. 2014;5:3617. doi: 10.1038/ncomms4617. [DOI] [PubMed] [Google Scholar]
- 30.Korasick DA, et al. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci USA. 2014;111(14):5427–5432. doi: 10.1073/pnas.1400074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 32.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 33.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414(6861):271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 34.Fleming AJ. Plant signalling: The inexorable rise of auxin. Trends Cell Biol. 2006;16(8):397–402. doi: 10.1016/j.tcb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Carella P, Wilson DC, Cameron RK. Some things get better with age: Differences in salicylic acid accumulation and defense signaling in young and mature Arabidopsis. Front Plant Sci. 2015;5:775. doi: 10.3389/fpls.2014.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayashi K. The interaction and integration of auxin signaling components. Plant Cell Physiol. 2012;53(6):965–975. doi: 10.1093/pcp/pcs035. [DOI] [PubMed] [Google Scholar]
- 37.Dardick CD, Golem S, Culver JN. Susceptibility and symptom development in Arabidopsis thaliana to Tobacco mosaic virus is influenced by virus cell-to-cell movement. Mol Plant Microbe Interact. 2000;13(10):1139–1144. doi: 10.1094/MPMI.2000.13.10.1139. [DOI] [PubMed] [Google Scholar]
- 38.Goregaoker SP, Lewandowski DJ, Culver JN. Identification and functional analysis of an interaction between domains of the 126/183-kDa replicase-associated proteins of tobacco mosaic virus. Virology. 2001;282(2):320–328. doi: 10.1006/viro.2001.0831. [DOI] [PubMed] [Google Scholar]
- 39.Oparka K, Duckett C, Prior D, Fisher D. Real‐time imaging of phloem unloading in the root tip of Arabidopsis. Plant J. 1994;6(5):759–766. [Google Scholar]
- 40.Knoblauch M, van Bel AJ. Sieve tubes in action. Plant Cell. 1998;10(1):35–50. [Google Scholar]
- 41.Wang X, et al. Salicylic acid regulates Plasmodesmata closure during innate immune responses in Arabidopsis. Plant Cell. 2013;25(6):2315–2329. doi: 10.1105/tpc.113.110676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall GS, Little DP. Relative quantitation of virus population size in mixed genotype infections using sequencing chromatograms. J Virol Methods. 2007;146(1-2):22–28. doi: 10.1016/j.jviromet.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berardini TZ, et al. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 2004;135(2):745–755. doi: 10.1104/pp.104.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle EI, et al. GO:TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20(18):3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattsson J, Ckurshumova W, Berleth T. Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 2003;131(3):1327–1339. doi: 10.1104/pp.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimizu-Mitao Y, Kakimoto T. Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB. Plant Cell Physiol. 2014;55(8):1450–1459. doi: 10.1093/pcp/pcu077. [DOI] [PubMed] [Google Scholar]
- 47.Mironova VV, Omelyanchuk NA, Wiebe DS, Levitsky VG. Computational analysis of auxin responsive elements in the Arabidopsis thaliana L. genome. BMC Genomics. 2014;15(Suppl 12):S4. doi: 10.1186/1471-2164-15-S12-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Notaguchi M, Wolf S, Lucas WJ. Phloem-mobile Aux/IAA transcripts target to the root tip and modify root architecture. J Integr Plant Biol. 2012;54(10):760–772. doi: 10.1111/j.1744-7909.2012.01155.x. [DOI] [PubMed] [Google Scholar]
- 49.Mustroph A, et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA. 2009;106(44):18843–18848. doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito T, Yamanaka K, Okada Y. Long-distance movement and viral assembly of tobacco mosaic virus mutants. Virology. 1990;176(2):329–336. doi: 10.1016/0042-6822(90)90002-9. [DOI] [PubMed] [Google Scholar]
- 51.Dorokhov YL, et al. Airborne signals from a wounded leaf facilitate viral spreading and induce antibacterial resistance in neighboring plants. PLoS Pathog. 2012;8(4):e1002640–e1002640. doi: 10.1371/journal.ppat.1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen MH, Citovsky V. Systemic movement of a tobamovirus requires host cell pectin methylesterase. Plant J. 2003;35(3):386–392. doi: 10.1046/j.1365-313x.2003.01818.x. [DOI] [PubMed] [Google Scholar]
- 53.Heinlein M. The spread of tobacco mosaic virus infection: Insights into the cellular mechanism of RNA transport. Cell Mol Life Sci. 2002;59(1):58–82. doi: 10.1007/s00018-002-8406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lucas WJ, Gilbertson RL. Plasmodesmata in relation to viral movement within leaf tissues. Annu Rev Phytopathol. 1994;32(1):387–415. [Google Scholar]
- 55.Lionetti V, Raiola A, Cervone F, Bellincampi D. Transgenic expression of pectin methylesterase inhibitors limits tobamovirus spread in tobacco and Arabidopsis. Mol Plant Pathol. 2014;15(3):265–274. doi: 10.1111/mpp.12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brandner K, et al. Tobacco mosaic virus movement protein interacts with green fluorescent protein-tagged microtubule end-binding protein 1. Plant Physiol. 2008;147(2):611–623. doi: 10.1104/pp.108.117481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ouko MO, et al. Tobacco mutants with reduced microtubule dynamics are less susceptible to TMV. Plant J. 2010;62(5):829–839. doi: 10.1111/j.1365-313X.2010.04195.x. [DOI] [PubMed] [Google Scholar]
- 58.Amari K, et al. A family of plasmodesmal proteins with receptor-like properties for plant viral movement proteins. PLoS Pathog. 2010;6(9):e1001119–e1001119. doi: 10.1371/journal.ppat.1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beffa RS, Hofer R-M, Thomas M, Meins F., Jr Decreased Susceptibility to Viral Disease of [beta]-1,3-Glucanase-Deficient Plants Generated by Antisense Transformation. Plant Cell. 1996;8(6):1001–1011. doi: 10.1105/tpc.8.6.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iglesias VA, Meins F., Jr Movement of plant viruses is delayed in a β-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J. 2000;21(2):157–166. doi: 10.1046/j.1365-313x.2000.00658.x. [DOI] [PubMed] [Google Scholar]
- 61.Lee J-Y, et al. A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell. 2011;23(9):3353–3373. doi: 10.1105/tpc.111.087742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simpson C, Thomas C, Findlay K, Bayer E, Maule AJ. An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell. 2009;21(2):581–594. doi: 10.1105/tpc.108.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White RF. Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology. 1979;99(2):410–412. doi: 10.1016/0042-6822(79)90019-9. [DOI] [PubMed] [Google Scholar]
- 64.Malamy J, Carr JP, Klessig DF, Raskin I. Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science. 1990;250(4983):1002–1004. doi: 10.1126/science.250.4983.1002. [DOI] [PubMed] [Google Scholar]
- 65.Zhu F, et al. Salicylic acid and jasmonic acid are essential for systemic resistance against tobacco mosaic virus in Nicotiana benthamiana. Mol Plant Microbe Interact. 2014;27(6):567–577. doi: 10.1094/MPMI-11-13-0349-R. [DOI] [PubMed] [Google Scholar]
- 66.Fu ZQ, Dong X. Systemic acquired resistance: Turning local infection into global defense. Annu Rev Plant Biol. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 67.Cui F, et al. The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol. 2013;162(2):1018–1029. doi: 10.1104/pp.113.219659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caarls L, Pieterse CM, Van Wees SC. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci. 2015;6:170. doi: 10.3389/fpls.2015.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol. 2003;51(1):21–37. doi: 10.1023/a:1020780022549. [DOI] [PubMed] [Google Scholar]
- 70.Spoel SH, et al. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137(5):860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao Q-M, Venugopal S, Navarre D, Kachroo A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 2011;155(1):464–476. doi: 10.1104/pp.110.166876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ndamukong I, et al. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007;50(1):128–139. doi: 10.1111/j.1365-313X.2007.03039.x. [DOI] [PubMed] [Google Scholar]
- 73.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448(7154):666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 74.Leon-Reyes A, et al. Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta. 2010;232(6):1423–1432. doi: 10.1007/s00425-010-1265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol. 2007;17(20):1784–1790. doi: 10.1016/j.cub.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 76.Robert-Seilaniantz A, Navarro L, Bari R, Jones JD. Pathological hormone imbalances. Curr Opin Plant Biol. 2007;10(4):372–379. doi: 10.1016/j.pbi.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 77.Tiryaki I, Staswick PE. An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 2002;130(2):887–894. doi: 10.1104/pp.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Develey-Rivière MP, Galiana E. Resistance to pathogens and host developmental stage: A multifaceted relationship within the plant kingdom. New Phytol. 2007;175(3):405–416. doi: 10.1111/j.1469-8137.2007.02130.x. [DOI] [PubMed] [Google Scholar]
- 79.Rusterucci C, et al. Age-related resistance to Pseudomonas syringae pv. tomato is associated with the transition to flowering in Arabidopsis and is effective against Peronospora parasitica. Physiol Mol Plant Pathol. 2005;66(6):222–231. [Google Scholar]
- 80.Leisner SM, Turgeon R, Howell SH. Effects of host plant development and genetic determinants on the long-distance movement of cauliflower mosaic virus in Arabidopsis. Plant Cell. 1993;5(2):191–202. doi: 10.1105/tpc.5.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cameron RK, Zaton K. Intercellular salicylic acid accumulation is important for age-related resistance in Arabidopsis to Pseudomonas syringae. Physiol Mol Plant Pathol. 2004;65(4):197–209. [Google Scholar]
- 82.Holsters M, et al. In vivo transfer of the ti-plasmid of Agrobacterium tumefaciens to Escherichia coli. Mol Gen Genet. 1978;163(3):335–338. doi: 10.1007/BF00271963. [DOI] [PubMed] [Google Scholar]
- 83.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 84.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Holsters M, et al. Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet. 1978;163(2):181–187. doi: 10.1007/BF00267408. [DOI] [PubMed] [Google Scholar]
- 86.Raikhel NV, Bednarek SY, Lerner DR. Plant Molecularl Biology Manual. Springer; New York: 1989. In situ RNA hybridization in plant tissues; pp. 371–402. [Google Scholar]
- 87.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 88.Zachgo S, Perbal MC, Saedler H, Schwarz-Sommer Z. In situ analysis of RNA and protein expression in whole mounts facilitates detection of floral gene expression dynamics. Plant J. 2000;23(5):697–702. doi: 10.1046/j.1365-313x.2000.00826.x. [DOI] [PubMed] [Google Scholar]
- 89.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 90.Lamesch P, et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012;40(Database issue) D1:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9(2):321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 92.Yan T, et al. PatMatch: A program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res. 2005;33(Web Server issue) suppl 2:W262-6. doi: 10.1093/nar/gki368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roberts AG, et al. Phloem unloading in sink leaves of Nicotiana benthamiana: Comparison of a fluorescent solute with a fluorescent virus. Plant Cell. 1997;9(8):1381–1396. doi: 10.1105/tpc.9.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.