Significance
At the molecular level, most processes in living systems are mediated by multisubunit protein complexes. Recombinant forms of these complexes are essential for analyzing their structure and function. Multigene expression constructs greatly improve recombinant protein complex preparations, but the generation of such constructs can be a rate-limiting step. To overcome this limitation, we have adapted Gibson assembly reactions for the rapid, efficient, and fast generation of numerous expression constructs in parallel and used the resulting biGBac method for expression of different cell-cycle complexes, composed of up to 17 different subunits. The biGBac technique enables the analyses of large protein complexes by systematic mutagenesis approaches that were not feasible before.
Keywords: protein complexes, baculovirus-insect cell expression, Gibson assembly
Abstract
Analyses of protein complexes are facilitated by methods that enable the generation of recombinant complexes via coexpression of their subunits from multigene DNA constructs. However, low experimental throughput limits the generation of such constructs in parallel. Here we describe a method that allows up to 25 cDNAs to be assembled into a single baculoviral expression vector in only two steps. This method, called biGBac, uses computationally optimized DNA linker sequences that enable the efficient assembly of linear DNA fragments, using reactions developed by Gibson for the generation of synthetic genomes. The biGBac method uses a flexible and modular “mix and match” approach and enables the generation of baculoviruses from DNA constructs at any assembly stage. Importantly, it is simple, efficient, and fast enough to allow the manual generation of many multigene expression constructs in parallel. We have used this method to generate and characterize recombinant forms of the anaphase-promoting complex/cyclosome, cohesin, and kinetochore complexes.
Most cellular processes are mediated by multisubunit protein complexes (1). Structural and functional analyses of such complexes are facilitated by technologies that permit coexpression of their subunits in homologous or heterologous host cells, such as bacteria, yeast, insect, or mammalian cells. The ectopic coexpression of subunits often leads to the assembly of the corresponding protein complexes in the host cells, enabling the purification and subsequent use of these complexes in structural, biochemical, and biophysical studies. The yield, homogeneity, reproducibility, and biological activity of such protein complex preparations can be improved if the genes or cDNAs encoding individual complex subunits are combined in one DNA expression vector, rather than being introduced into the host cells as individual constructs (reviewed in ref. 2). A particularly useful method for the generation of eukaryotic protein complexes are the MultiBac and OmniBac systems, which enable coexpression of multiple subunits from a single baculoviral DNA construct in insect cells, either as individual proteins (3–6) or as a polyprotein that can be proteolytically cleaved into individual subunits (5, 7). The ability of multigene expression constructs to improve the quality and quantity of recombinant protein complexes is presumably due to the fact that in their case only a single DNA construct has to be introduced into the host cells to enable expression of all subunits, whereas otherwise different host cells might receive different numbers and subsets of single-gene expression constructs, possibly resulting in incompletely or wrongly assembled protein complexes.
Protein expression approaches are particularly useful if they are combined with recombinant DNA technologies, which allow the generation of tagged and mutated proteins. These approaches typically require the generation of large sets of DNA constructs to identify tagged proteins that are suitable for purification or localization studies without perturbing protein function or to identify mutants that are defective in specific properties. For example, a comprehensive structure–function analysis of the ubiquitin-conjugating enzyme UBE2S required the generation of 135 mutants, even though this monomeric protein is only composed of 222 amino acid residues (8). The application of tagging and mutagenesis approaches for the generation and analysis of multisubunit protein complexes is more difficult. This is partly due to the large number of differently tagged or mutated versions that can be obtained for multisubunit protein complexes. Moreover, the generation of the corresponding multigene expression constructs is often also rate-limiting, as the efficiency with which multiple DNA fragments can be assembled correlates inversely with their number and size. If sequence-independent methods are used that are based on homology sequences, gene assembly efficiency can also vary strongly depending on the suitability of the chosen homology sequences. As a result, extensive screening efforts are often required to identify correctly assembled multigene expression constructs.
To overcome this limitation, we have developed a gene sequence-independent cloning method that relies on computationally optimized DNA linker sequences for the rapid and efficient recombination of up to 25 individual protein-coding sequences into a single DNA construct in only two sequential reactions. These are based on the one-step isothermal DNA assembly method described by Gibson et al. (9), which was originally developed for the assembly of synthetic genomes. In these “Gibson assembly reactions,” 3′ “overhangs” are created in linear double-stranded DNA molecules by an exonuclease, complementary sequences within these overhangs anneal, gaps between the annealed DNA molecules are filled by a DNA polymerase, and the annealed DNA molecules are covalently linked to each other by a DNA ligase (9). These processes all occur in a single reaction mixture. We have developed this approach for the generation of baculoviral expression constructs. We call the resulting method “biGBac,” with biG referring both to the size of the expression constructs and the use of Gibson assembly reactions (biG being the reverse of the initial three letters of Gibson) and Bac referring to its use for the generation of baculoviral vectors, the backbones of which are derived from the MultiBac system (3, 4, 10). The biGBac method uses a flexible and modular “mix and match” approach, enables the generation of baculoviruses from DNA constructs at any assembly stage, can be carried out in 6 d, and is efficient enough to allow the generation of many multigene expression constructs in parallel. Principally, the biGBac method should also be adaptable for use in other host cells, such as bacteria, yeast, or mammalian cells.
Results
biGBac Enables the Rapid Assembly of Up to 25 cDNAs into a Single Expression Vector.
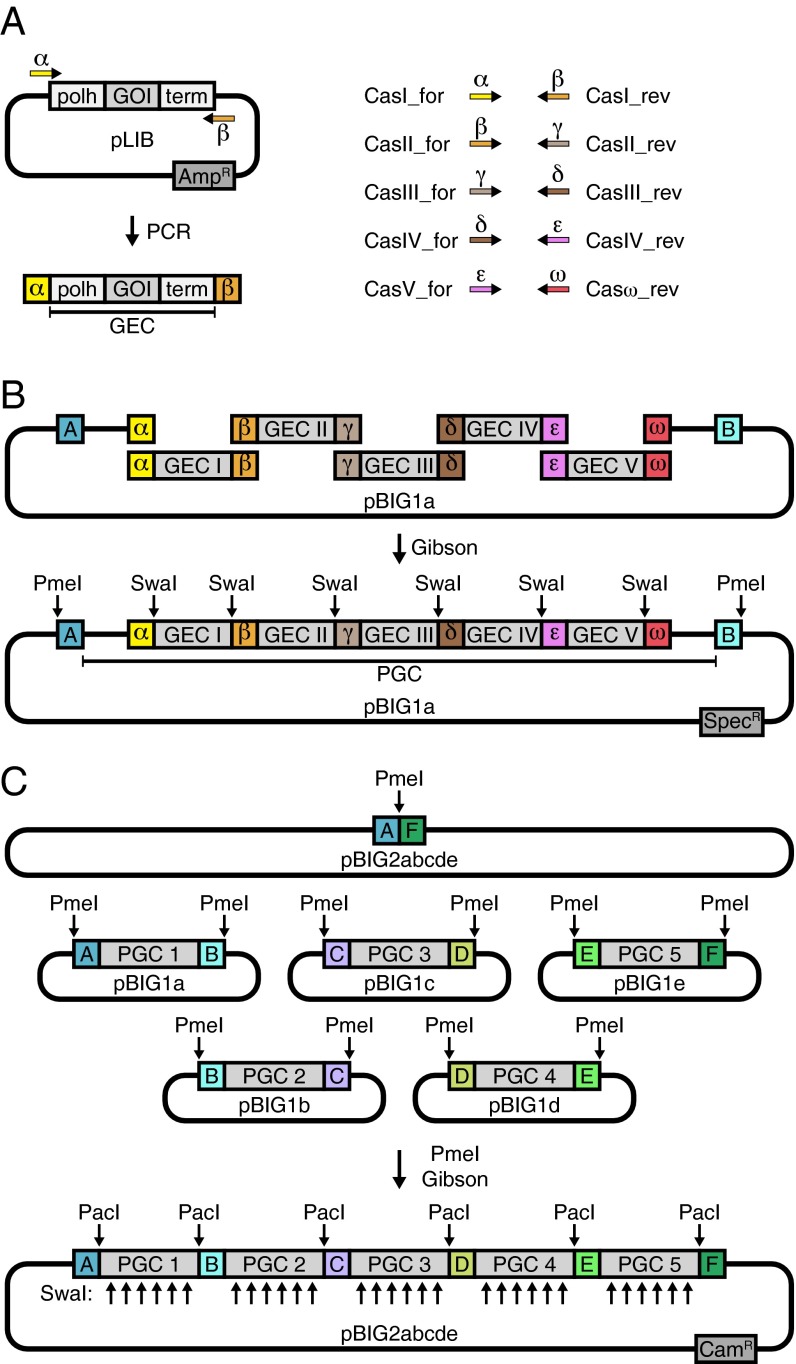
To increase the efficiency of DNA assembly reactions, we computationally identified a set of linker sequences that based on their predicted melting temperatures, propensity to form secondary structures, and lack of cross-complementarity should be well suited for participation in Gibson assembly reactions and should minimize the formation of incorrectly assembled byproducts (Fig. S1). Next, we designed a set of cloning vectors, a set of predefined oligonucleotides, and a protocol in which the optimized linker sequences can be used to assemble up to 25 DNA fragments into one baculoviral expression construct (Fig. 1). Initially, cDNAs or genes encoding individual subunits of a protein complex are cloned using Gibson assembly reactions or conventional techniques into a vector that contains an expression cassette with a polyhedrin promotor (polh), which will later enable expression of the gene of interest (GOI) in insect cells, resulting in a collection of library vectors (pLIBs; Fig. S2). From these, gene expression cassettes (GECs) are amplified by polymerase chain reactions (PCRs) using sets of predefined oligonucleotides that decorate the DNA fragment ends with the optimized linker sequences α, β, γ, δ, ε, and ω (Fig. 1A and Table S1). In the first assembly step (pBIG1 vector level), up to five GECs are combined in a Gibson assembly reaction with one of five pBIG1 vectors, called pBIG1a–pBIG1e. Importantly, the “last” GEC—that is, the one that will occupy the most 3′ position in the pBIG1 construct—has to carry the ω linker sequence (introduced by oligonucleotide Casω_rev). These reactions lead to the assembly of polygene cassettes (PGCs) in circular pBIG1 constructs, which are amplified in Escherichia coli. The procedure introduces SwaI restriction sites flanking individual GECs and PmeI restriction sites flanking the PGCs (Fig. 1B and Fig. S3). The SwaI sites can be used to analyze if all GECs are present in the pBIG1 constructs, whereas the PmeI sites can be used to release the PGCs from the pBIG1 constructs. Importantly, PGCs that are released by PmeI digestion also carry optimized linker sequences at their ends, in this case called A–F (Table S1). These linkers can be used in a second Gibson assembly reaction (pBIG2 vector level) to assemble up to five PGCs from different pBIG1 constructs into one of four pBIG2 vectors, called pBIG2ab, pBIG2abc, pBIG2abcd, and pBIG2abcde. The pBIG2 vector name indicates which and how many of the pBIG1 constructs can be combined. For example, pBIG2ab can assemble the PGCs released from pBIG1a and pBIG1b. The resulting pBIG2 constructs can be analyzed by SwaI or PacI digestion followed by gel electrophoresis (Fig. 1C and Fig. S4). This protocol can be used to assemble different multigene expression constructs and can be completed in 6 d (Table S2). All biGBac vectors—that is, pLIB, pBIG1, and pBIG2 constructs—are designed so that they can be directly used for the generation of baculoviruses using Tn7 transposition (11) to allow protein expression in insect cells.
Fig. S1.
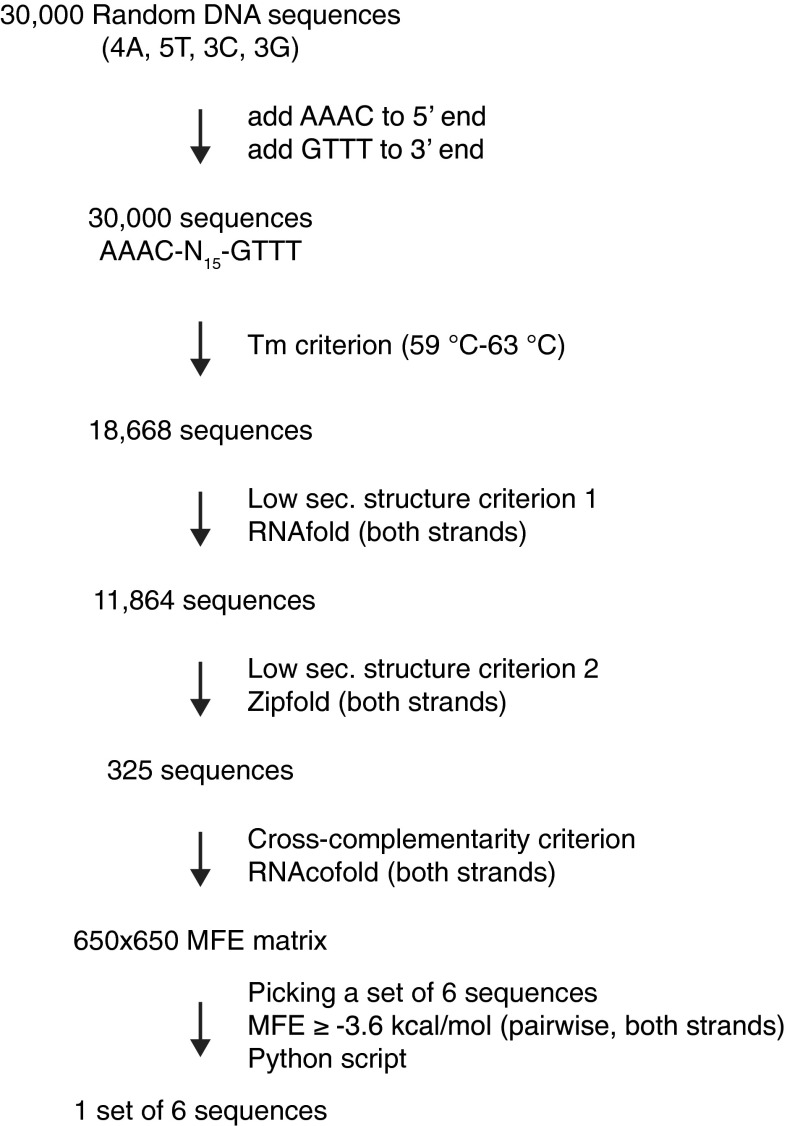
Generation of optimized DNA linker sequences. To generate optimized homology sequences for a six-fragment Gibson assembly reaction, 30,000 random DNA sequences of 15 nt in length and defined base composition were generated, and the sequences AAAC and GTTT required for compatibility with PmeI digestion were added to their 5′ and 3′ ends, respectively. Melting temperatures were predicted, and only sequences within the melting temperature range of 59–63 °C were included in further analyses (18,668 sequences). Sequences with a high probability of secondary structure formation on either strand were excluded using RNAfold (11,864 sequences left). The sequence set was further analyzed for secondary structure formation with Zipfold using parameters that fit with Gibson reaction conditions. Only sequences for which both strands were very unlikely to form secondary structures in Gibson assembly reactions [minimum free energy (MFE) ≥ +1.5 kcal/mol] remained in the sequence set (325 sequences). To pick a set of sequences that provides highest specificity in Gibson reactions (i.e., no false annealing), hybrid structures for each combination of two sequences out of the set of 325 sequences and their reverse complements were predicted using RNAcofold. Free energy values of the MFE structure of each hybrid were entered into a 650 × 650 MFE matrix. An algorithm was written in python to build sequence sets in which no pairwise interactions violate an MFE threshold. When setting the threshold to MFE ≥ –3.6 kcal/mol, it was possible to pick exactly one set of six sequences. This set is used as linker sequences in the second assembly step (linkers A, B, C, D, E, and F). This set was manually slightly modified to generate a set that is applicable in the first assembly step (linearization with SwaI), while maintaining all thermodynamic criteria (linkers α, β, γ, δ, ε, and ω).
Fig. 1.
Schematic representation of the biGBac assembly procedure. (A) A GEC consisting of polyhedrin promoter (polh), cDNA of a GOI, and SV40-terminator (term) is amplified by PCR from pLIB using predefined oligonucleotide sets (Cas_for/rev) to introduce specific linker sequences (Greek letters). (B) First assembly step shown for pBIG1a and five GECs. Linearized vector and PCR products are recombined in a Gibson assembly reaction to a circular product containing a PGC. The positions of SwaI and PmeI restriction sites, which can be used for the analysis of DNA constructs in conjunction with gel electrophoresis, are indicated. (C) Second assembly step shown for pBIG2abcde and five PGCs in pBIG1 vectors. The six plasmids are mixed, digested with PmeI, and recombined in a Gibson assembly reaction. AmpR, ampicillin resistance; CamR, chloramphenicol resistance; SpecR, spectinomycin resistance.
Fig. S2.
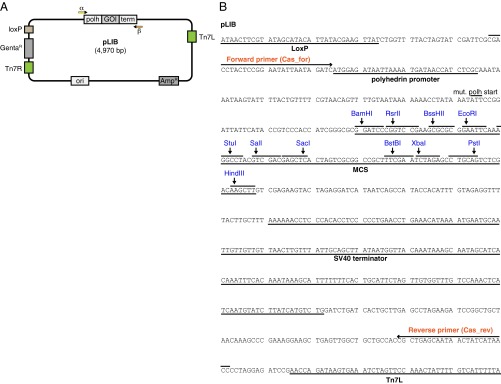
pLIB vector. (A) Schematic representation of pLIB. The cDNA of a GOI is inserted between the polyhedrin promoter (polh) and the SV40 terminator sequence (term). pLIB can be maintained with Ampicillin (AmpR resistance gene). All biGBac vectors (pLIB, pBIG1, pBIG2) contain Tn7 elements (Tn7L, Tn7R) and a gentamicin resistance gene (GentaR) for generation of baculoviruses and a LoxP site for compatibility with Multibac donor plasmids. (B) DNA sequence of empty pLIB from LoxP site to Tn7L element. For generation of library constructs using Gibson reactions, a stock of linearized pLIB can be generated by BamHI/HindIII digestion and a cDNA can be inserted via a Gibson reaction (see Materials and Methods for good homology sequences). Alternatively the MCS can be used to insert a cDNA by conventional restriction/ligation cloning. It is recommended to not clone ORFs in-frame with the mutated polyhedrin start codon (mut. polh start) to avoid the possibility of leaky expression that might lead to N-terminal extensions. The binding sites of the predefined oligonucleotide set (Table S1) for amplification of GECs are indicated (Cas_for/Cas_rev).
Table S1.
Linker sequences and predefined oligonucleotide set
| Identifier | Sequence |
| Linker sequences for second assembly step | |
| A | AAACGCTCTATGGTCTAAAGTTT |
| B | AAACTATATCTCAATCGGGGTTT |
| C | AAACTGGATACTATTGCACGTTT |
| D | AAACCTAATGATGCCTGATGTTT |
| E | AAACGGTTCACATAGCTTAGTTT |
| F | AAACACTGACATTGACTTGGTTT |
| Linker sequences for first assembly step | |
| α (=mod. A) | AACGCTCTATGGTCTAAAGATTT |
| β (=C) | AAACTGGATACTATTGCACGTTT |
| γ (=D) | AAACCTAATGATGCCTGATGTTT |
| δ (=E) | AAACGGTTCACATAGCTTAGTTT |
| ε (=F) | AAACACTGACATTGACTTGGTTT |
| ω (=mod. B) | AAATCTATATCTCAATCGGGGTT |
| Predefined oligonucleotide set | |
| CasI_for | AACGCTCTATGGTCTAAAGATTTAAATCGACCTACTCCGGAATATTAATAGATC |
| CasI_rev | AAACGTGCAATAGTATCCAGTTTATTTAAATGGTTATGATAGTTATTGCTCAGCG |
| CasII_for | AAACTGGATACTATTGCACGTTTAAATCGACCTACTCCGGAATATTAATAGATC |
| CasII_rev | AAACATCAGGCATCATTAGGTTTATTTAAATGGTTATGATAGTTATTGCTCAGCG |
| CasIII_for | AAACCTAATGATGCCTGATGTTTAAATCGACCTACTCCGGAATATTAATAGATC |
| CasIII_rev | AAACTAAGCTATGTGAACCGTTTATTTAAATGGTTATGATAGTTATTGCTCAGCG |
| CasIV_for | AAACGGTTCACATAGCTTAGTTTAAATCGACCTACTCCGGAATATTAATAGATC |
| CasIV_rev | AAACCAAGTCAATGTCAGTGTTTATTTAAATGGTTATGATAGTTATTGCTCAGCG |
| CasV_for | AAACACTGACATTGACTTGGTTTAAATCGACCTACTCCGGAATATTAATAGATC |
| Casω_rev | AACCCCGATTGAGATATAGATTTATTTAAATGGTTATGATAGTTATTGCTCAGCG |
Fig. S3.
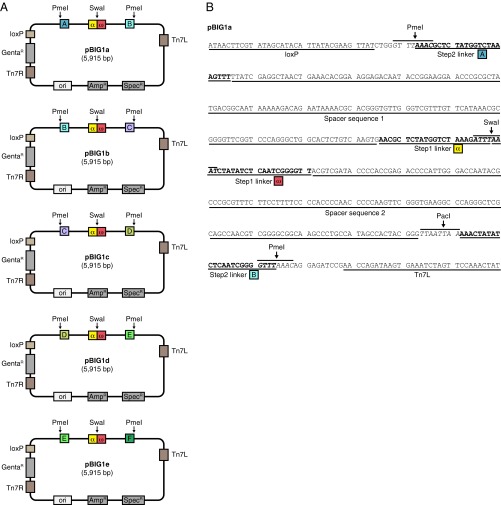
pBIG1 vectors. (A) Schematic representation of pBIG1 vectors. pBIG1 vectors can be maintained with Spectinomycin (SpecR resistance gene) or Ampicillin (AmpR resistance gene). For the selection in the first assembly step, Spectinomycin is used. Stocks of linearized pBIG1 cloning vectors are generated by SwaI digestion. This results in linear vector backbone with linker sequences α and ω at the fragment ends. After the first assembly step, the generated PGCs can be released from the vector backbone by PmeI digestion. The five pBIG1 vectors differ only in the linker sequences (A, B, C, D, E, and F) next to the PmeI sites as indicated. All biGBac vectors (pLIB, pBIG1, pBIG2) contain Tn7 elements (Tn7L, Tn7R) and a Gentamicin resistance gene (GentaR) for generation of baculoviruses and a LoxP site for compatibility with Multibac donor plasmids. (B) DNA sequence of pBIG1a shown from the LoxP site to Tn7L element. The positions of α, ω and A, B linker sequences as well as of the restriction sites SwaI, PmeI, and PacI are shown. The linker sequences of the first and the second assembly step are separated by spacer sequences (derived from HSVtk terminator sequence in pFL) to avoid interference of the two linker sequence sets in Gibson reactions.
Fig. S4.
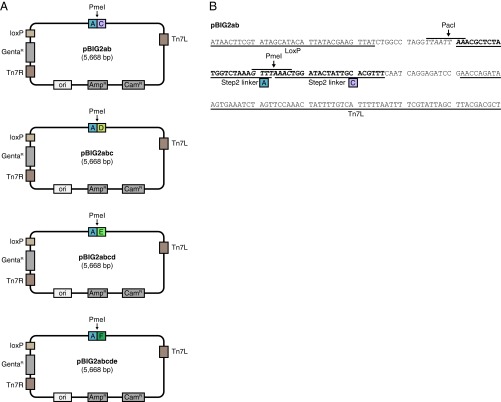
pBIG2 vectors. (A) Schematic representation of pBIG2 vectors. pBIG2 vectors are maintained with Chloramphenicol (CamR resistance gene) or Ampicillin (AmpR resistance gene). For the selection in the second assembly step, Chloramphenicol is used. Stocks of linearized pBIG2 cloning vectors are generated by PmeI digestion. This results in linear vector backbone with linker sequences on both ends. The four pBIG2 vectors contain linker sequence A on one end and differ only in the linker sequence on the other side (C, D, E, or F as indicated). All biGBac vectors (pLIB, pBIG1, pBIG2) contain Tn7 elements (Tn7L, Tn7R) and a Gentamicin resistance gene (GentaR) for generation of baculoviruses and a LoxP site for compatibility with Multibac donor plasmids. (B) DNA sequence of pBIG2ab shown from the LoxP site to Tn7L element. The positions of linker sequences A and C and the restriction sites PmeI and PacI are shown.
Table S2.
Timetable for generation of biGBac expression constructs
| Day | Experimental steps | Time* |
| Day 1 | PCR GECs | ∼8–10 h |
| PCR “cleanup,” determine DNA concentration, agarose gel electrophoresis | ||
| Step 1 Gibson assembly reaction | ||
| Transformation of E. coli | ||
| Day 2 | Inoculate E. coli overnight cultures | ∼20 min |
| Day 3 | “Miniprep” DNA isolation | ∼6–8 h |
| SwaI/PmeI digests, agarose gel electrophoresis | ||
| Submit for DNA Sanger sequencing | ||
| Day 4 | Analyze sequencing results | ∼6 h |
| Step 2 PmeI digest and Gibson assembly reaction | ||
| Transformation of E. coli | ||
| Day 5 | Inoculate E. coli overnight cultures | ∼10 min |
| Day 6 | Miniprep DNA isolation | ∼4 h |
| SwaI/PacI digests, agarose gel electrophoresis |
Times are estimated for the generation of five pBIG1 and one pBIG2 construct. The generation of multiple constructs in parallel can require longer periods of time.
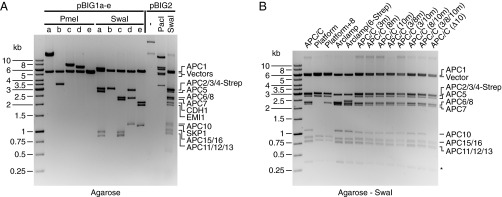
To test biGBac, we assembled an expression construct containing 17 cDNAs encoding the subunits of the 1.2-MDa human anaphase promoting complex/cyclosome (APC/C) bound to CDC20 homolog-1 (CDH1), early mitotic inhibitor-1 (EMI1), and S-phase kinase-associated protein-1 (SKP1). APC/C is a ubiquitin ligase essential for chromosome segregation, mitotic exit, and neuronal differentiation (reviewed in ref. 12); CDH1 is an APC/C coactivator required to recruit substrates to the APC/C during mitotic exit and the G1 and G0 phase (13); and EMI1 and SKP1 form a heterodimeric inhibitor of APC/C–CDH1 (14). Earlier studies relied on the coexpression of human recombinant APC/C subunits from several baculoviruses (14–16). Using biGBac, we were able to assemble all 17 cDNAs previously cloned into pLIB vectors in a defined order into five different pBIG1 constructs and assembled the PGCs from these into one pBIG2 construct (FW_II-1; all pBIG1 and pBIG2 expression constructs used in this study are listed in Tables S3 and S4). During this procedure, we analyzed in total 34 E. coli colonies transformed either with DNA from the step 1 or step 2 assembly reactions. Of these, 17 contained the correctly assembled constructs (Tables S3 and S4). Selected examples of these pBIG1 and pBIG2 constructs, analyzed by PmeI, SwaI, or PacI restriction digest and agarose gel electrophoresis, are shown in Fig. 2A. Multigene expression constructs can therefore be assembled with biGBac rapidly and efficiently.
Table S3.
pBIG1 constructs in this study
| pBIG1 construct | Alias | Vector | CasI | CasII | CasIII | CasIV | CasV | Construct size, kb | Shown in figure | Clones correct/analyzed |
| FW_I-1 | pBIG1a | APC1 | APC2 | APC5 | APC11 | APC15 | 20.0 | Fig. 2A | 1/3 | |
| FW_I-2 | pBIG1b | APC4-Strep | — | — | — | — | 8.9 | Fig. 2A | 3/3 | |
| FW_I-3 | pBIG1c | APC6 | APC7 | APC12(CDC26) | APC13 | APC16 | 13.2 | Fig. 2A | 1/7 | |
| FW_I-4 | pBIG1d | APC3 | APC8 | APC10 | — | — | 12.5 | Fig. 2A | 2/3 | |
| FW_I-5 | pBIG1e | CDH1 | EMI1 | SKP1 | — | — | 10.9 | Fig. 2A | 4/12 | |
| FW_I-6 | pBIG1b | APC4-Strep | APC8 | — | — | — | 11.4 | — | 2/7 | |
| FW_I-7 | pBIG1c | APC6-Strep | APC7 | APC12(CDC26) | APC13 | APC16 | 13.3 | — | 2/13 | |
| FW_I-8 | pBIG1d | APC3m | APC8 | APC10 | — | — | 12.5 | — | 1/3 | |
| FW_I-9 | pBIG1d | APC3 | APC8m | APC10 | — | — | 12.5 | — | 1/3 | |
| FW_I-10 | pBIG1d | APC3 | APC8 | APC10m | — | — | 12.5 | — | 1/3 | |
| FW_I-11 | pBIG1d | APC3m | APC8m | APC10 | — | — | 12.5 | — | 1/3 | |
| FW_I-12 | pBIG1d | APC3m | APC8 | APC10m | — | — | 12.5 | — | 2/3 | |
| FW_I-13 | pBIG1d | APC3 | APC8m | APC10m | — | — | 12.5 | — | 1/3 | |
| FW_I-14 | pBIG1d | APC3m | APC8m | APC10m | — | — | 12.5 | — | 2/3 | |
| FW_I-15 | pBIG1d | APC3 | APC8 | — | — | — | 11.3 | — | 2/3 | |
| FW_I-16 | Cohesin wt | pBIG1c | SMC1A | SMC3-Flag | SCC1 | His-SA1 | — | 21.2 | Fig. 3F | 1/6* |
| FW_I-17 | Cohesin KA | pBIG1c | SMC1A(KA) | SMC3(KA)-Flag | SCC1 | His-SA1 | — | 21.2 | Fig. 3F | 1/42* |
| FW_I-18 | Cohesin EQ | pBIG1c | SMC1A(EQ) | SMC3(EQ)-Flag | SCC1 | His-SA1 | — | 21.2 | Fig. 3F | 4/30* |
| FW_I-19 | Kinetochore N | pBIG1a | Ndc80-His | Nuf2 | Spc24 | Spc25 | — | 12.9 | Fig. 3K | 2/12 |
| FW_I-20 | Kinetochore M | pBIG1b | His-Dsn1 | Mtw1 | Nnf1 | Nsl1 | — | 12.0 | Fig. 3K | 1/6 |
| FW_I-21 | pBIG1b | Dsn1 | Mtw1 | Nnf1 | Nsl1 | — | 12.0 | — | 3/6 | |
| FW_I-22 | pBIG1a | Ndc80-His | Nuf2 | Spc24 | Spc25 | Kre28 | 14.6 | — | 2/10 | |
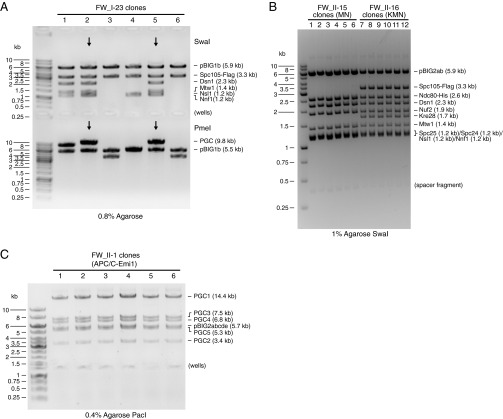
| FW_I-23 | pBIG1b | Dsn1 | Mtw1 | Nnf1 | Nsl1 | Spc105-Flag | 15.3 | Fig. S5A | 2/12 |
Although the assembly of APC/C and kinetochore biGBac constructs occurred with high efficiency, more transformed E. coli clones needed to be analyzed when we assembled the four cohesin core subunits into pBIG1 vectors, possibly due to the relatively large size of the cohesin cDNAs, resulting in pBIG1 constructs of 21.2 kb. We assume this is due to difficulties generating pure PCR products of large GECs. We therefore also generated cohesin expression vectors in two steps with the two SMC subunits on pBIG1a and SCC1 and SA1 on pBIG1b. In the second step, we combined the four subunits into a pBIG2 expression vector. Using this strategy, most analyzed clones were correct. If several subunits of a protein complex are large, it may therefore be advantageous to distribute the cDNAs encoding the large subunits onto more than one pBIG1 vector.
Table S4.
pBIG2 constructs in this study
| pBIG2 construct | Alias | Vector | PGC1 | PGC2 | PGC3 | PGC4 | PGC5 | Construct size, kb | Shown in figure | Clones correct/analyzed |
| FW_II-1 | APC/C–Emi1 | pBIG2abcde | FW_I-1 | FW_I-2 | FW_I-3 | FW_I-4 | FW_I-5 | 43.6 | Fig. 2A and Fig. S5C | 6/6 |
| FW_II-2 | APC/C | pBIG2abcd | FW_I-1 | FW_I-2 | FW_I-3 | FW_I-4 | — | 38.3 | Fig. 2B | 1/4 |
| FW_II-3 | Platform | pBIG2ab | FW_I-1 | FW_I-2 | — | — | — | 23.7 | Fig. 2B | 2/4 |
| FW_II-4 | Platform+8 | pBIG2ab | FW_I-1 | FW_I-6 | — | — | — | 26.0 | Fig. 2B | 2/4 |
| FW_II-5 | Arclamp | pBIG2abcd | pBIG1a(empty) | pBIG1b(empty) | FW_I-3 | FW_I-4 | — | 21.0 | Fig. 2B | 1/1 |
| FW_II-6 | Arclamp(6-Strep) | pBIG2abcd | pBIG1a(empty) | pBIG1b(empty) | FW_I-7 | FW_I-4 | — | 21.1 | Fig. 2B | 1/1 |
| FW_II-7 | APC/C(3m) | pBIG2abcd | FW_I-1 | FW_I-2 | FW_I-3 | FW_I-8 | — | 38.3 | Fig. 2B | 2/4 |
| FW_II-8 | APC/C(8m) | pBIG2abcd | FW_I-1 | FW_I-2 | FW_I-3 | FW_I-9 | — | 38.3 | Fig. 2B | 1/1 |
| FW_II-9 | APC/C(10m) | pBIG2abcd | FW_I-1 | FW_I-2 | FW_I-3 | FW_I-10 | — | 38.3 | Fig. 2B | 1/1 |
| FW_II-10 | APC/C(3/8m) | pBIG2abcd | FW_I-1 | FW_I-2 | FW_I-3 | FW_I-11 | — | 38.3 | Fig. 2B | 5/10 |
| FW_II-11 | APC/C(3/10m) | pBIG2abcd | FW_I-1 | FW_I-2 | FW_I-3 | FW_I-12 | — | 38.3 | Fig. 2B | 1/1 |
| FW_II-12 | APC/C(8/10m) | pBIG2abcd | FW_I-1 | FW_I-2 | FW_I-3 | FW_I-13 | — | 38.3 | Fig. 2B | 1/4 |
| FW_II-13 | APC/C(3/8/10m) | pBIG2abcd | FW_I-1 | FW_I-2 | FW_I-3 | FW_I-14 | — | 38.3 | Fig. 2B | 1/1 |
| FW_II-14 | APC/C(Δ10) | pBIG2abcd | FW_I-1 | FW_I-2 | FW_I-3 | FW_I-15 | — | 37.1 | Fig. 2B | 1/1 |
| FW_II-15 | Kinetochore MN | pBIG2ab | FW_I-19 | FW_I-21 | — | — | — | 19.5 | Fig. 3K and Fig. S5B | 6/6 |
| FW_II-16 | Kinetochore KMN | pBIG2ab | FW_I-22 | FW_I-23 | — | — | — | 24.5 | Fig. 3K and Fig. S5B | 6/6 |
Fig. 2.
Analysis of biGBac constructs. (A) Analysis of DNA constructs for the expression of APC/C–CDH1–EMI1–SKP1 by restriction digest and gel electrophoresis. The 17 GECs were distributed over the five pBIG1 vectors a–e (FW_I-1/2/3/4/5; Table S3). Correct constructs were analyzed by PmeI or SwaI digestion, releasing PGCs or GECs from the vector “backbone,” respectively, followed by gel electrophoresis and ethidium bromide staining. The five PGCs were recombined with a pBIG2abcde vector (FW_II-1), and a correctly assembled construct is shown before (–) and after digestion with PacI or SwaI, releasing PGCs or GECs, respectively. (B) SwaI digestion of 13 pBIG2 constructs coding for APC/C, subcomplexes, or mutant complexes (FW_II-2–FW_II-14; Table S4). *SwaI digestion of pBIG2 constructs generates additional fragments of 0.3 kb containing spacer sequences between PGCs.
biGBac Allows the Assembly of Many Expression Constructs in Parallel.
To address if biGBac enables the generation of multigene constructs in parallel, we next assembled 13 different expression constructs side by side (Fig. 2B). We decided to assemble constructs encoding wild-type APC/C, subcomplexes, and mutants of APC/C, as this experiment also allowed us to test if existing biGBac constructs can be flexibly recombined to generate new constructs. For example, by combining four of the five pBIG1 constructs that we had used for the APC/C–CDH1–EMI1–SKP1 construct (all except pBIG1e FW_I-5, which contains cDNAs encoding CDH1, EMI1, and SKP1; Table S3), we assembled a construct (pBIG2abcd FW_II-2) encoding the 14 core subunits of APC/C. By combining pBIG1a FW_I-1 and pBIG1b FW_I-2, we generated a construct encoding the six subunits of an APC/C subcomplex known as the “platform” (17–19). By combining pBIG1c and pBIG1d constructs FW_I-3 and FW_I-4 with “empty” pBIG1a and pBIG1b vectors, we assembled a construct encoding APC/C’s “arc lamp” subcomplex (17–19). To generate mutated forms of the APC/C that are predicted to have deficiencies in coactivator and substrate binding (20, 21), we generated pLIB vectors encoding APC3N575A,L606A, APC8N339A, and APC10N144A,H145A,R149A,D150A and generated pBIG1d and pBIG2abcd constructs with combinations of these mutated subunits, as well as a version lacking the cDNA for APC10. In this experiment, we analyzed 81 E. coli colonies transformed either with DNA from the step 1 or step 2 assembly reactions. Of these, 35 contained the correctly assembled DNA constructs. Examples of these constructs, analyzed by SwaI restriction digest and agarose gel electrophoresis, are shown in Fig. 2B. The biGBac method is therefore suitable for the generation of multigene expression constructs in parallel and can assemble vectors in different combinations, enabling the generation of constructs that encode subcomplexes, tagged versions, and mutants of protein complexes.
Functional and Structural Characterization of Complexes Expressed from biGBac Constructs.
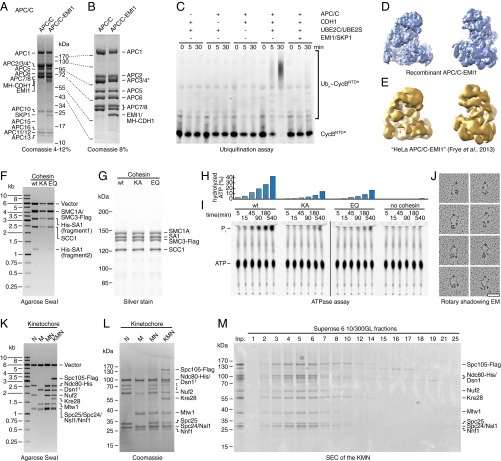
To test if the constructs assembled by biGBac can be used to generate baculoviruses that produce functional protein complexes in insect cells, we used biGBac vectors to express three different types of protein complexes (Fig. 3). First, we coexpressed the 14 core subunits of APC/C from construct FW_II-2 (Table S4). During the course of these experiments, we observed that higher protein yields could be obtained when insect cells were in addition coinfected with a baculovirus containing the platform construct FW_II-3, possibly because otherwise some of the platform subunits were expressed at substoichiometric ratios. Proteins expressed from these constructs assembled into complexes that could be purified by affinity, anion exchange, and size exclusion chromatography (SEC) and that contained all 14 different types of subunits, as seen by SDS–polyacrylamide gel electrophoresis (SDS/PAGE) followed by Coomassie staining (Fig. 3 A and B). Binding of coactivator CDH1 and inhibitor EMI1–SKP1 to these complexes and purification by tandem affinity chromatography led to the formation of complexes containing all 17 subunits (Fig. 3 A and B). In the presence of the ubiquitin-activating enzyme UBA1, the ubiquitin-conjugating enzymes UBE2C (also known as UBCH10) and UBE2S, ubiquitin, ATP, and CDH1, the 14-subunit APC/C, was able to ubiquitinate an N-terminal fragment of cyclin B1, whereas addition of EMI1–SKP1 inhibited this reaction (Fig. 3C). Cryoelectron microscopy (cryo-EM) was used to determine the structure of recombinant APC/C that was expressed from biGBac constructs and bound to CDH1, EMI1, and SKP1 (Fig. 3D). This reconstruction resembled that of endogenous APC/C purified from HeLa cells to which recombinant CDH1, EMI1, and SKP1 had been bound (Fig. 3E) (14). Importantly, we determined the structure of APC/C–CDH1–EMI1–SKP1 expressed from biGBac constructs at a resolution of 7 Å by cryo-EM, whereas we had only obtained a resolution of 20 Å when using endogenous HeLa APC/C and negative-stain EM (14). These results indicate that biGBac vectors can be used to express fully assembled enzymatically active recombinant human APC/C, which is sensitive to inhibition by EMI1–SKP1 and whose structural homogeneity allowed us to study its structure by cryo-EM. This conclusion is further supported by the accompanying manuscript by Qiao et al. (22), which describes the generation and functional characterization of 47 different APC/C variants mutated in up to 68 mitotic phosphorylation sites. Furthermore, the recombinant forms of APC/C described in Brown et al. (8, 23) were also expressed from biGBac constructs.
Fig. 3.
Functional and structural characterization of protein complexes expressed from biGBac constructs. (A) Coomassie-stained 4–12% gradient SDS/PAGE gel of purified APC/C alone and bound to CDH1/EMI1/SKP1 (“APC/C-EMI1”). (B) Coomassie-stained 8% SDS/PAGE gel of samples shown in A. *The Strep-tag on APC4 is cleaved in the APC/C sample but not in the APC/C-EMI1 sample. (C) Ubiquitination assay with fluorescein-labeled Cyclin BNTD (CycBNTD*) as substrate. APC/C, coactivator CDH1, E2 ubiquitin-conjugating enzymes UBE2C/UBE2S, and inhibitor EMI1–SKP1 were added as indicated. (D) Single-particle reconstruction by cryo-EM of recombinant APC/C–CDH1–EMI1–SKP1 (“APC/C–EMI1”). (E) HeLa cell APC/C bound to recombinant CDH1 and EMI1-SKP1 (“APC/C-EMI1”) negative stain EM structure data from ref. 14 for comparison. (F) SwaI digestion of pBIG1 constructs coding for wild-type cohesin tetramer (wt; FW_I-16), Walker A double mutant (KA; FW_I-17), and Walker B double mutant (EQ; FW_I-18). (G) SDS/PAGE and silver staining of purified cohesin complexes. (H and I) ATPase activity of cohesin. Thin-layer chromatography (I) and quantification (H) of [γ-32P]-ATP hydrolysis. (J) Representative micrographs of wild-type cohesin tetramers after rotary-shadowing EM. (K) SwaI digestion of yeast kinetochore constructs N (FW_I-19), M (FW_I-20), MN (FW_II-15), and KMN (FW_II-16). †Dsn1 contains a His-tag in M (FW_I-20) but not in MN (FW_II-15) and KMN (FW_II-16). (L) Coomassie-stained SDS/PAGE gel of purified yeast kinetochore subcomplexes. ‡Marked bands were found in samples containing Ndc80-His but not in its absence, suggesting that these represent degradation products of Ndc80-His. (M) Coomassie-stained SDS/PAGE gel of SEC fractions of the KMN complex using a Superose 6 10/300GL column.
Second, we assembled pBIG1 vectors encoding the four core subunits (SMC1A, SMC3, SCC1, and SA1) of human cohesin (Fig. 3F), a ring-shaped 500-kDa complex that mediates sister chromatid cohesion, higher order chromatin structure, and DNA damage repair (24). We generated expression constructs encoding either wild-type cohesin or versions in which the subunits SMC1 and SMC3 carry mutations in amino acid residues required for ATP binding (KA) or hydrolysis (EQ) (25). Subunits expressed from these constructs assembled into complexes that could be purified by two-step affinity chromatography and contained all four subunits in stoichiometric amounts, as judged by SDS/PAGE and silver staining (Fig. 3G). Samples containing wild-type cohesin hydrolyzed ATP more effectively than samples containing similar amounts of cohesin containing the KA or EQ mutations in SMC1 and SMC3 (Fig. 3 H and I). The long coiled-coils of SMC1 and SMC3 result in cohesin’s characteristic ring-like appearance, as visualized by rotary shadowing EM (Fig. 3J). These results indicate that biGBac vectors can also be used to express recombinant human cohesin that is indistinguishable from previously generated recombinant cohesin regarding its subunit stoichiometry, ATPase activity, and structure (25–27).
Third, we assembled biGBac constructs encoding four subcomplexes of the budding yeast kinetochore, a macromolecular complex that mediates the attachment of chromosomes to microtubules of the mitotic or meiotic spindle (reviewed in ref. 28). Within the kinetochore, the conserved Knl1–Mis12–Ndc80 (KMN) complex, composed of the heterodimeric Spc105–Kre28 complex (KNL1-ZWINT in humans), the four-subunit Ndc80 complex and the four-subunit Mis12 complex, is of particular interest, as it constitutes the evolutionary conserved microtubule-binding site of the outer kinetochore and is required for recruitment of spindle assembly checkpoint proteins, which control APC/C activity (29). A biochemical analysis of the KMN has remained challenging, as full-length yeast Spc105 and human KNL1 are largely refractory to recombinant expression in significant yield, and therefore, truncation mutants of these proteins had to be used so far (30). We tested if this limitation could be overcome by coexpression of KMN subunits from biGBac constructs. First, we generated constructs either encoding the four subunits of the yeast Ndc80 complex (N) or the four subunits of the yeast Mis12 complex (M), or both (MN). By including Spc105 and Kre28, we also generated a construct encoding all 10 full-length subunits of the yeast KMN, which had not been reconstituted from recombinant subunits so far (Fig. 3K). Infection of insect cells with baculoviruses containing these expression constructs led to the assembly of all four kinetochore complexes, as judged by SDS/PAGE and Coomassie staining of proteins purified by affinity and SEC (Fig. 3 L and M). We conclude that our approach allows reconstitution of the budding yeast KMN with full-length proteins, generating an assembly that contains the Spc105–Kre28 “platform” for spindle assembly checkpoint proteins as well as the microtubule-binding interface in the Ndc80 complex.
Discussion
Multigene expression systems like MultiBac (3, 4, 6) have proven to be invaluable for the generation of recombinant protein complexes. Although robust standardized protocols, principally suitable for robotic automatization, have been developed for the generation of large expression constructs (5, 31), the generation of such constructs can still represent a technical challenge, in particular if multiple constructs containing many cDNAs or genes are needed and robotic support is not available. We have therefore adapted single-step DNA assembly reactions initially developed for the assembly of synthetic genomes (9) for the generation of multigene expression constructs. By identifying DNA sequences that are particularly well suited for the efficient and specific annealing of multiple linear DNA fragments via complementary base pairing and that can be linked to any linear DNA fragment, we have been able to optimize Gibson assembly reactions to a degree that enables the rapid and efficient generation of multigene expression constructs within a few days. Our experience is that a single person can use the resulting biGBac method to generate up to 10 constructs in only 6 to 7 d, whereas 2 wk are typically needed using biGBac for the parallel generation of 30–40 constructs (Table S2). Importantly, biGBac can be carried out with simple techniques and does not require robotics—that is, should be usable in any molecular biology laboratory. Unlike other methods for the generation of multigene expression constructs, biGBac does not depend on combining “donor” and “acceptor” vectors. Any biGBac construct can therefore be used for the generation of baculoviruses and protein expression in insect cells—for example, for the generation of subcomplexes at the pBIG1 level. Finally, biGBac vectors are designed in a way that enables a flexible mix and match utilization of genes, cDNAs, or cassettes containing multiples of these. As is illustrated by our generation of APC/C expression constructs in this and the accompanying manuscript by Qiao et al. (22), this flexibility is particularly useful for the generation of constructs that encode subcomplexes, mutants, or differently tagged versions of a given protein complex. Based on these advantageous features of biGBac (summarized in Table S5), we expect that this method will further advance structural and functional studies of large protein complexes.
Table S5.
Advantageous features of biGBac
| Simple—can be used in standard molecular biology laboratories, no robotics needed |
| Efficient—cloning efficiency for APC/C pBIG1 and pBIG2 on average ∼33% and ∼80%, respectively |
| Fast—final pBIG2 multigene expression constructs can be generated in 6 d |
| Scalable—can be used to generate up to 40 constructs in parallel |
| Flexible—any biGBac construct can be used for baculovirus generation (pLIB, pBIG1, pBIG2), does not rely on combinations of “donor” and “acceptor” vectors |
| Modular—by allowing multiple combinations of different biGBac constructs (mix and match) |
Materials and Methods
Generation of pLIB Constructs.
For adding a gene to the pLIB library, the pLIB vector was linearized by digestion with BamHI and HindIII. The linearized vector fragment was purified by gel extraction (QIAquick Gel Extraction Kit, Qiagen). cDNAs were amplified by PCR. When using Gibson assembly for pLIB construct generation, it is recommendable to use forward primers that carry the sequence overhang 5′-CCACCATCGGGCGCGGATCCA (followed by the start codon and gene-specific sequences) and reverse primers that carry the sequence overhang 5′-TCCTCTAGTACTTCTCGACAAGCTT (followed by the reverse complement of stop codon and gene-specific sequences). The PCR products were purified by gel extraction (QIAquick Gel Extraction Kit, Qiagen). We mixed 100 ng of the linearized pLIB vector with the PCR product at a molar ratio of 1:5 in a Gibson assembly reaction. The Gibson assembly reaction was performed as described below. Alternatively, cDNAs can be cloned via conventional restriction–ligation cloning using the multiple cloning site (MCS) of the pLIB vector (Fig. S2). In this case, it is recommendable to clone cDNAs not in frame with the mutated polyhedrin gene start codon from which sporadic leaky expression was reported (6, 32). If PmeI sites are present in the coding sequences (which is rare), they should be removed at this stage by constructing the pLIB construct from two PCR products introducing a silent mutation.
Generation of pBIG1 Constructs.
To linearize the pBIG1 vectors, 10 µg of pBIG1a, pBIG1b, pBIG1c, pBIG1d, or pBIG1e were digested using 1 µL SwaI (New England Biolabs) in NEBuffer 3.1 at 25 °C. After overnight incubation, another 2 µL of SwaI were added, and incubation was continued for 2 h. SwaI was heat-inactivated at 65 °C for 20 min, and the linearized vector was purified using the QIAquick PCR purification kit (Qiagen). Complete SwaI digestion of the pBIG1 vectors is crucial to avoid colonies containing empty pBIG1. GECs were generated by PCR on pLIB templates using oligonucleotides of the predefined oligonucleotide sets (PAGE-purified quality, Microsynth; Table S1). The oligonucleotide pair that is used for the generation of the GEC defines its position in the pBIG1 construct. If less than five GECs are to be assembled, the Casω_rev oligonucleotide is used as reverse primer of the last GEC to create an overlap with the linearized pBIG1 vector. PCR was performed using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) with an annealing temperature of 65 °C. PCR products were purified with PureLink PCR Purification Kit (Invitrogen) using the provided high-cutoff binding buffer B3 and elution in 30 µL elution buffer E1 that was preheated to 70 °C. Purity of the DNA fragments was confirmed by agarose gel electrophoresis.
To perform the first assembly step, 100 ng of a linearized pBIG1 vector was mixed with up to five PCR-amplified GECs at a fivefold molar excess of each GEC over the vector. The Gibson assembly reaction was performed as described below. Up to 5 µL of the assembly reaction was transformed into 50 µL chemically competent DH5alpha cells. After 1 h of recovery in lysogeny broth (LB) at 37 °C, cells were plated onto LB agar plates containing 50 µg/mL spectinomycin. Single colonies were picked and grown in 5 mL LB medium containing 50 µg/mL spectinomycin at 37 °C overnight. Plasmid DNA was purified using the QIAprep Spin Miniprep Kit (Qiagen) and analyzed by digestion at 25 °C with SwaI to release individual GECs or with PmeI to release the PGC (Fig. S5). Clones that displayed the correct restriction patterns were analyzed by Sanger sequencing using gene-specific oligonucleotides. PCR errors were found to be rare.
Fig. S5.
Examples for construct analysis. (A) Analytical digest of a pBIG1 construct. Six clones of FW_I-23 (pBIG1b:Dsn1/Mtw1/Nnf1/Nsl1/Spc105-Flag) were digested by SwaI or PmeI and analyzed on a 0.8% agarose gel. The expected sizes of pBIG1 vector backbone and GECs after SwaI digestion and of pBIG1 vector backbone and the PGC after PmeI digestion are indicated. Note that spacer sequences (see Fig. S3) stay with the vector backbone after SwaI digestion (5.9 kb) and with the PGC after PmeI digestion (backbone 5.5 kb). Clones 2 and 5 show the correct restriction pattern. (B) Analytical SwaI digest of pBIG2 constructs. Six clones of FW_II-15 (Kinetochore MN) and six clones of FW_II-16 (Kinetochore KMN) were digested with SwaI and analyzed on a 1% agarose gel. The expected sizes of pBIG2 vector backbone and GECs are indicated. Note that SwaI digestion of pBIG2 constructs yields an additional fragment of 0.3 kb consisting mainly of spacer sequences between PGCs. All clones show the correct restriction pattern. (C) Analytical PacI digest of a pBIG2 construct. Six clones of FW_II-1 (APC/C-Emi1) were digested with PacI and analyzed on a 0.4% agarose gel. The expected sizes of pBIG2 vector backbone and PGCs are indicated. All clones show the correct restriction pattern.
Generation of pBIG2 Constructs.
To linearize the pBIG2 vectors, 10 µg of pBIG2ab, pBIG2abc, pBIG2abcd, or pBIG2abcde were digested using 1 µL of PmeI (New England Biolabs) in CutSmart Buffer at 25 °C. After overnight incubation, another 2 µL of PmeI was added, and incubation was continued for 2 h at 37 °C. The DNA was purified using the QIAquick PCR purification kit (Qiagen).
To perform the second assembly step, 33 ng of a linearized pBIG2 vector was mixed with up to five compatible pBIG1 constructs at a fivefold molar excess of each pBIG1 construct over the pBIG2 vector, and water was added to a total volume of 12.5 µL. After addition of 4 µL of 5× isothermal reaction buffer (IRB) buffer (see Gibson Assembly Reactions), 1 µL of PmeI was added and the reaction was incubated at 37 °C for 90 min to release the PGCs from the pBIG1 constructs. The reaction was transferred to 4 °C. The Gibson assembly enzymes were added on ice, and Gibson assembly was performed as described below. When generating constructs with a size >20 kb, electroporation was used. Up to 0.4 µL of the assembly reaction were transformed into 40 µL of electrocompetent DH10B cells using a 0.1-cm cuvette at 1,800 V (Eppendorf Electroporator 2510). Cells were recovered in LB at 37 °C for 1 h and plated onto LB agar plates containing 34 µg/mL chloramphenicol. Colonies were picked and grown in 5 mL LB medium containing 34 µg/mL chloramphenicol at 37 °C overnight. Plasmids were purified using the QIAprep Spin Miniprep Kit (Qiagen). Plasmid DNA was analyzed by digestion with SwaI to release individual GECs or with PacI to release the pBIG1-derived PGCs (Fig. S5).
Gibson Assembly Reactions.
One-step isothermal DNA assembly reactions were performed as described (9). The DNA fragments, 4 µL 5× IRB [25% (wt/vol) PEG-8000, 500 mM Tris∙HCl pH 7.5, 50 mM MgCl2, 50 mM DTT, 1 mM each of the four dNTPs, 5 mM NAD] (9), 2 µL Taq DNA Ligase (M0208, New England Biolabs), 0.25 µL Phusion HF DNA Polymerase (F-530, Thermo Scientific), and 0.25 µL T5 Exonuclease (prediluted 1:30 in 1× IRB) (T5E4111K, Epicentre) were mixed on ice in a total volume of 20 µL and immediately transferred to a 50 °C preheated thermocycler block and incubated for 60 min.
Additional details can be found in SI Materials and Methods.
SI Materials and Methods
Generation of Optimized Linker Sequences.
Random DNA sequences of defined base composition were generated using the FaBox online toolbox (33). Melting temperatures were predicted using Thermo Scientific’s Multiple Primer Analyzer tool for Phusion polymerase. Single-stranded DNA secondary structure formation probabilities were estimated using RNAfold (v2.0.7; parameters, −noGU −noPS -T 50; ViennaRNA Package) (34) and Zipfold (parameters, DNA, 50 °C, 0.1 M Na+, 0.01 M Mg2+; DINAMelt Web Server) (35, 36). Minimum free energy hybrid structures were predicted using RNAcofold (v2.0.7; parameters, −noGU −noPS -T 50; ViennaRNA Package) (34). The optimized sequence set was determined using a python script.
biGBac Vector Generation.
The biGBac vectors described here are derived from the Multibac pFL vector (4, 10). The pLIB vector was generated by deletion of the p10 expression cassette from pFL by digestion with PmeI and BstZ17I followed by blunt end ligation (Fig. S2). The pBIG1 vectors were generated from pFL by replacing the p10 expression cassette, the multiplication module, and the polh expression cassette with the optimized DNA linker sequences for both assembly steps separated by spacer sequences of about 150 bp (derived from HSVtk terminator sequence in pFL) that contain SwaI, PmeI, and PacI endonuclease recognition sites (Fig. S3) using Gibson assembly reactions. In addition, the gene conferring spectinomycin resistance was inserted between the ampicillin resistance gene and the Tn7L sequence. The pBIG2 vectors were generated from pFL by replacing the p10 expression cassette, the multiplication module, and the polh expression cassette with DNA linker sequences and recognition sites of the endonucleases PmeI and PacI (Fig. S4) using Gibson assembly reactions. The gene conferring chloramphenicol resistance was inserted between the ampicillin resistance gene and the Tn7L sequence.
Baculovirus Generation and Protein Expression.
pLIB, pBIG1, or pBIG2 constructs were used to generate recombinant baculoviral genomes by Tn7 transposition in DH10EMBacY cells (5). Viruses were generated by transfection of Sf9 insect cells (Thermo Scientific) with the recombinant baculoviral genome using Fugene 6 reagent (Promega). After 3 d, viruses were amplified by adding transfection supernatant to Sf9 suspension cultures. Protein complexes were expressed in Sf9 or HighFive (Thermo Scientific) insect cell suspension cultures.
Protein Purification.
Recombinant APC/C and APC/C–CDH1–EMI1–SKP1 were expressed using the baculovirus expression system in HighFive cells (Thermo Scientific) and purified for biochemical analysis or EM essentially as described (8). In brief, APC/C, which is expressed with a Twin-Strep(II)-tag on the C terminus of APC4, was purified by affinity to Strep-Tactin Sepharose (IBA), then by ion exchange, and finally by SEC. APC/C–CDH1–EMI1–SKP1 for EM was prepared by mixing the lysate from HighFive cells expressing APC/C or Myc-6xHis-CDH1. APC/C–CDH1 was captured on Strep-Tactin Sepharose and then incubated with purified FLAG-EMI1/SKP1 (14). APC/C–CDH1–EMI1–SKP1 eluted from the Strep-Tactin Sepharose was affinity purified again using anti-FLAG M2 affinity gel (Sigma). The equivalent of 250 μg of APC/C–CDH1–EMI1–SKP1 was further processed through GraFix (37). Recombinant UBA1, UBE2C, UBE2S, CDH1, EMI1/SKP1, and ubiquitin were all expressed and purified as described previously (8, 14, 23, 38). Cyclin B NTD* (1–95) was purified and C-terminally labeled with fluorescein 5-maleimide, as denoted by the asterisk (38).
Recombinant cohesin complexes were expressed in Sf9 cells and purified essentially as described (26). Cohesin tetramers, which were expressed with a His-tag on SA1 and a Flag-tag on SMC3, were purified by affinity to Ni-NTA agarose beads (Qiagen), eluted with 150 mM imidazole, and then purified by affinity to anti-FLAG M2 affinity gel (Sigma) and eluted with 0.5 mg/mL Flag peptide.
Recombinant yeast kinetochore subcomplexes were expressed in Sf9 cells. The complexes were purified by affinity and SEC. The Ndc80 (N) complex and theMN complex were expressed with a His-tag on Ndc80 and were purified by affinity to Ni-NTA agarose beads (Qiagen). The Mis12 (M) complex was expressed with a His-tag on Dsn1 and was purified by affinity to Ni-NTA agarose beads (Qiagen). The KMN complex was expressed with a Flag-tag on Spc105 and was purified by affinity to anti-FLAG M2 agarose beads (Sigma). The Ndc80 (N) and Mis12 (M) complexes were further purified by SEC using a Superdex 200 10/300GL column (GE Healthcare). The MN and KMN complexes were further purified by SEC using a Superose 6 10/300GL column.
Protein Complex Characterization.
APC/C-mediated ubiquitination assays were performed as previously described (8, 14, 23). In summary, 30 nM APC/C, 1 μM CDH1, 200 nM UBE2C, 200 nM UBE2S, 1 μM EMI1, 0.5 mg/mL BSA, 100 nM UBA1, 5 mM MgCl2, 5 mM ATP, and 500 nM fluorescein-labeled CycBNTD* were mixed on ice. The reactions were then equilibrated to room temperature and initiated by addition of 250 µM Ub. Following a defined incubation period, the reactions were quenched and then separated by SDS/PAGE. Ubiquitinated CycBNTD* was visualized using a Typhoon FLA 9500 PhosphorImager. The cryo-EM structure of APC/C–CDH1–EMI1–SKP1 was determined essentially as previously described (8, 23). Recombinant cohesin ATPase assays and rotary shadowing EM were performed as previously described (26).
Acknowledgments
We thank Imre Berger for the pFL vector and Daniela Götz, Marc Jarvis, Maciej Zaczek, and Roman Stocsits for assistance. For funding, we thank Boehringer Ingelheim, the Austrian Research Promotion Agency (FFG Laura Bassi Centre for Optimized Structural Studies), the European Union (Seventh Framework Programme Grant 227764 MitoSys), and the Austrian Science Fund (SFB-F34 and Wittgenstein award) (to J.-M.P.); Deutsche Forschungsgemeinschaft Sonderforschungsbereich 860 (to H.S.); Jane Coffin Childs Foundation, Leukemia & Lymphoma Society (N.G.B.); American Lebanese Syrian Associated Charities, NIH Grants R37GM065930 and P30CA021765, and Howard Hughes Medical Institute (to B.A.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604935113/-/DCSupplemental.
References
- 1.Alberts B. The cell as a collection of protein machines: Preparing the next generation of molecular biologists. Cell. 1998;92(3):291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Bieniossek C, Imasaki T, Takagi Y, Berger I. MultiBac: Expanding the research toolbox for multiprotein complexes. Trends Biochem Sci. 2012;37(2):49–57. doi: 10.1016/j.tibs.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol. 2004;22(12):1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald DJ, et al. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006;3(12):1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 5.Vijayachandran LS, et al. Robots, pipelines, polyproteins: Enabling multiprotein expression in prokaryotic and eukaryotic cells. J Struct Biol. 2011;175(2):198–208. doi: 10.1016/j.jsb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Yang J, Barford D. Recombinant expression and reconstitution of multiprotein complexes by the USER cloning method in the insect cell-baculovirus expression system. Methods. 2016;95:13–25. doi: 10.1016/j.ymeth.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Nie Y, Bellon-Echeverria I, Trowitzsch S, Bieniossek C, Berger I. Multiprotein complex production in insect cells by using polyproteins. Methods Mol Biol. 2014;1091:131–141. doi: 10.1007/978-1-62703-691-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown NG, et al. Mechanism of polyubiquitination by human anaphase-promoting complex: RING repurposing for ubiquitin chain assembly. Mol Cell. 2014;56(2):246–260. doi: 10.1016/j.molcel.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald DJ, et al. Multiprotein expression strategy for structural biology of eukaryotic complexes. Structure. 2007;15(3):275–279. doi: 10.1016/j.str.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Luckow VA, Lee SC, Barry GF, Olins PO. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol. 1993;67(8):4566–4579. doi: 10.1128/jvi.67.8.4566-4579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Primorac I, Musacchio A. Panta rhei: The APC/C at steady state. J Cell Biol. 2013;201(2):177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol Cell. 2005;18(5):543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Frye JJ, et al. Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nat Struct Mol Biol. 2013;20(7):827–835. doi: 10.1038/nsmb.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uzunova K, et al. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol. 2012;19(11):1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, et al. Recombinant expression, reconstitution and structure of human anaphase-promoting complex (APC/C) Biochem J. 2013;449(2):365–371. doi: 10.1042/BJ20121374. [DOI] [PubMed] [Google Scholar]
- 17.Dube P, et al. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol Cell. 2005;20(6):867–879. doi: 10.1016/j.molcel.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Herzog F, et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323(5920):1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreiber A, et al. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470(7333):227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 20.Carroll CW, Enquist-Newman M, Morgan DO. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr Biol. 2005;15(1):11–18. doi: 10.1016/j.cub.2004.12.066. [DOI] [PubMed] [Google Scholar]
- 21.Matyskiela ME, Morgan DO. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol Cell. 2009;34(1):68–80. doi: 10.1016/j.molcel.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiao R, et al. Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc Natl Acad Sci USA. 2016;113:E2570–E2578. doi: 10.1073/pnas.1604929113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown NG, et al. RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase-promoting complex. Proc Natl Acad Sci USA. 2015;112(17):5272–5279. doi: 10.1073/pnas.1504161112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22(22):3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 25.Ladurner R, et al. Cohesin’s ATPase activity couples cohesin loading onto DNA with Smc3 acetylation. Curr Biol. 2014;24(19):2228–2237. doi: 10.1016/j.cub.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huis in ’t Veld PJ, et al. Characterization of a DNA exit gate in the human cohesin ring. Science. 2014;346(6212):968–972. doi: 10.1126/science.1256904. [DOI] [PubMed] [Google Scholar]
- 27.Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156(3):419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLuca JG, Musacchio A. Structural organization of the kinetochore-microtubule interface. Curr Opin Cell Biol. 2012;24(1):48–56. doi: 10.1016/j.ceb.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127(5):983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 30.Petrovic A, et al. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol Cell. 2014;53(4):591–605. doi: 10.1016/j.molcel.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 31.Berger I, et al. The multiBac protein complex production platform at the EMBL. J Vis Exp. 2013;(77):e50159. doi: 10.3791/50159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feil R, Müller S, Hofmann F. High-level expression of functional cGMP-dependent protein kinase using the baculovirus system. FEBS Lett. 1993;336(1):163–167. doi: 10.1016/0014-5793(93)81632-a. [DOI] [PubMed] [Google Scholar]
- 33.Villesen P. FaBox: An online toolbox for fasta sequences. Mol Ecol Notes. 2007;7(6):965–968. [Google Scholar]
- 34.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6(1):26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33(Web Server issue) suppl 2:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markham NR, Zuker M. UNAFold: Software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 37.Kastner B, et al. GraFix: Sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5(1):53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi M, et al. Structure of an APC3-APC16 complex: Insights into assembly of the anaphase-promoting complex/cyclosome. J Mol Biol. 2015;427(8):1748–1764. doi: 10.1016/j.jmb.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]