We welcome the additional data and analyses of Wang et al. (1), but believe there are some misunderstandings regarding the methods and findings of Shannon et al. (2). First, although we merged Nepal and Mongolia when plotting linkage disequilibrium (LD) decay in figure 5B of ref. 2 for legibility, we did not assume Nepal and Mongolia represented a single, interbreeding population, and indeed computed separate LD scores for each population (figure 5A of ref. 2), matching Wang et al.’s (1) observation of slightly lower LD in Nepal than Mongolia. Although Nepal (along with India) is commonly considered part of South Asia, Nepal borders Central Asia. Dog populations in two Central Asian countries, Mongolia and Afghanistan, both have lower LD than India. Nepal does not border Southeast Asia. Because we cannot, given the resolution of current sampling data, say the center of diversity is precisely within Nepal, the LD pattern observed is most consistent with an origin of dogs in or near Central Asia.
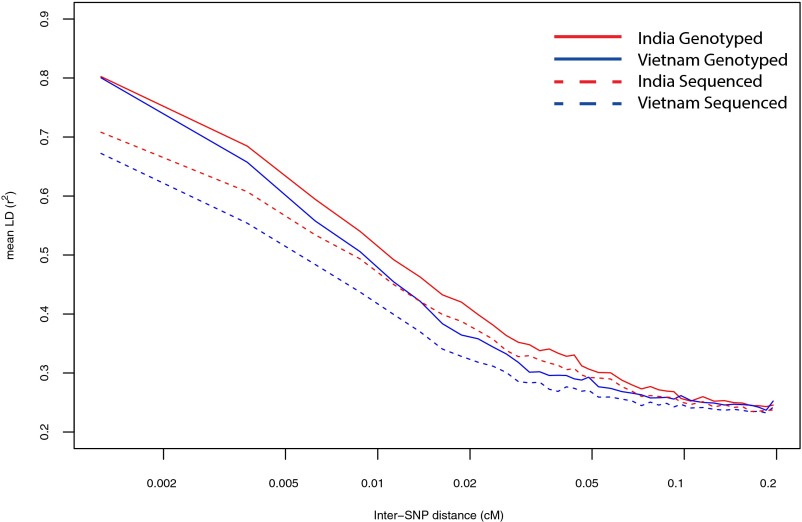
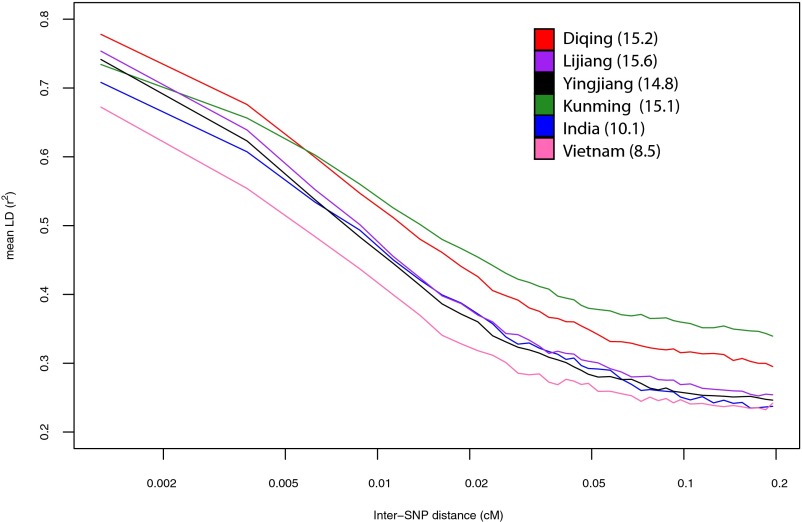
Second, comparison of our LD curves with published resequencing data is problematic due to the low coverage of existing village dog genomes (3). Genotyping error at this coverage depth using the Samtools pipeline is ∼1% in human datasets (4); in our laboratory, we see a similar level of genotyping discordance between the Illumina CanineHD array (2) and variants called from whole-genome sequencing using the Genome Analysis Toolkit (GATK) (5). As a consequence, LD curves computed from sequencing data are lower than LD curves computed from array data, even when the same dogs are considered at the same markers (Fig. 1). Indeed, we see even lower LD in sequenced Vietnamese or Indian village dogs (5) than in the Chinese village dogs reported by Wang et al. (1) (Fig. 2), but this is likely a reflection of the lower coverage depth of the non-Chinese populations (8–10× vs. 15×) and not representative of the true geographic patterning of genetic variation.
Fig. 1.
Decay of LD curves computed from variant calls from whole-genome sequencing [Auton et al. (5), dashed lines] and from Illumina array genotypes [Shannon et al. (2), solid lines]. For each population, the same six dogs were used for sequencing and genotyping, and curves are computed based on the 156,000 autosomal markers passing quality control in both datasets. Thus, differences in LD decay estimates for the same population are due solely to genotyping discordances between the sequenced and genotyped data.
Fig. 2.
Decay of LD in village dog populations based on six resequenced samples based on variant calls only at positions on the Illumina array. Mean depth of sequencing coverage is given in parentheses.
Last, Wang et al. (1) are correct that Y haplogroup 8 (HG8) was previously observed by Natanaelsson et al. (6) in a single Southeast Asian sample (a Thai Ridgeback). Genome-wide studies of Thai Ridgebacks are lacking, so it is unclear whether this HG8 haplogroup is likely present in indigenous Southeast Asian village dog populations or was brought to Southeast Asia by the common ancestor of Thai and African Ridgeback dogs. Even accepting the presence of HG8 in Southeast Asia, African village dogs still contain more Y haplogroups than Southeast Asian dogs because HG9 is present in Africa (and Central Asia and Siberia) but not Southeast Asia.
We appreciate the reanalysis and incorporation of new data by Wang et al. (1), and agree that consistent genetic data generated from additional samples from Central Asia (including western China) and neighboring regions are needed for better characterization of patterns of genetic diversity in Asian dogs. However, without stringent controls for the biases created by errors in low- and intermediate-coverage sequencing, comparisons of population genetic statistics like LD between array-based and sequence-based data can lead to spurious conclusions and should be interpreted with great caution.
Footnotes
Conflict of interest statement: A.R.B. and R.H.B. are cofounders and officers of Embark Veterinary, Inc., a canine genetics testing company.
References
- 1.Wang G-D, Peng M-S, Yang H-C, Savolainen P, Zhang Y-P. Questioning the evidence for a Central Asian domestication origin of dogs. Proc Natl Acad Sci USA. 2016;113:E2554–E2555. doi: 10.1073/pnas.1600225113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shannon LM, et al. Genetic structure in village dogs reveals a Central Asian domestication origin. Proc Natl Acad Sci USA. 2015;112(44):13639–13644. doi: 10.1073/pnas.1516215112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gou X, et al. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 2014;24(8):1308–1315. doi: 10.1101/gr.171876.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng AY, Teo YY, Ong RT. Assessing single nucleotide variant detection and genotype calling on whole-genome sequenced individuals. Bioinformatics. 2014;30(12):1707–1713. doi: 10.1093/bioinformatics/btu067. [DOI] [PubMed] [Google Scholar]
- 5.Auton A, et al. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 2013;9(12):e1003984. doi: 10.1371/journal.pgen.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Natanaelsson C, et al. Dog Y chromosomal DNA sequence: identification, sequencing and SNP discovery. BMC Genet. 2006;7:45. doi: 10.1186/1471-2156-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]