Significance
Mycoplasmas are minimal pathogenic bacteria able to infect humans and a wide range of economically important animals; as such, they are major causes of concern in the medical and veterinary fields. These pathogens often lead to chronic infections, and their mechanisms of immunity evasion are poorly known. Here we describe a two-protein system from the ruminant pathogen Mycoplasma mycoides subspecies capri that is involved in the capture and cleavage of antibodies. MIB is able to capture the antibodies and to subsequently recruit MIP, a protease that is able to cleave the antibody heavy chain. The MIB–MIP system appears to be widespread among pathogenic mycoplasmas and is potentially a key player for the virulence and immunity evasion mechanisms of these bacteria.
Keywords: mycoplasmas, immunoglobulin, protease
Abstract
Mycoplasmas are “minimal” bacteria able to infect humans, wildlife, and a large number of economically important livestock species. Mycoplasma infections include a spectrum of clinical manifestations ranging from simple fever to fulminant inflammatory diseases with high mortality rates. These infections are mostly chronic, suggesting that mycoplasmas have developed means to evade the host immune response. Here we present and functionally characterize a two-protein system from Mycoplasma mycoides subspecies capri that is involved in the capture and cleavage of IgG. The first component, Mycoplasma Ig binding protein (MIB), is an 83-kDa protein that is able to tightly bind to the Fv region of a wide range of IgG. The second component, Mycoplasma Ig protease (MIP), is a 97-kDa serine protease that is able to cleave off the VH domain of IgG. We demonstrate that MIB is necessary for the proteolytic activity of MIP. Cleavage of IgG requires a sequential interaction of the different partners of the system: first MIB captures the IgG, and then MIP is recruited to the MIB–IgG complex, enabling protease activity. MIB and MIP are encoded by two genes organized in tandem, with homologs found in the majority of pathogenic mycoplasmas and often in multiple copies. Phylogenetic studies suggest that genes encoding the MIB–MIP system are specific to mycoplasmas and have been disseminated by horizontal gene transfer. These results highlight an original and complex system targeting the host immunoglobulins, playing a potentially key role in the immunity evasion by mycoplasmas.
Mycoplasmas are small-sized bacteria belonging to the class Mollicutes (1). These organisms are characterized by a fast evolution that has been marked by a drastic genome reduction, leading to current mycoplasmas having a reduced coding capacity, no cell wall, and a limited number of metabolic pathways (2, 3). As a result, they are obligate parasites and are found in vertebrate animals including mammalian, avian, reptilian, and piscine hosts (4).
Despite their apparent simplicity, many mycoplasmas are pathogenic for humans and a wide range of animals, and are major causes of concern in both the medical and veterinary fields (5). In humans, two of the most significant pathogens are Mycoplasma pneumoniae and Mycoplasma genitalium, infecting the respiratory and urogenital tracts, respectively; they are mostly associated with diseases characterized by high morbidity and low mortality, like interstitial pneumonia or urethritis (6, 7). In farm animals, mycoplasmas have a high economic impact, thus warranting intensive veterinary surveillance and regulation. In cattle, one of the diseases listed by the World Organization for Animal Health is the contagious bovine pleuropneumonia (CBPP) caused by Mycoplasma mycoides subspecies (subsp.) mycoides. CBPP is still highly prevalent in Africa, and its mortality rate can reach up to 50% when introduced into naive herds (8). In goats and sheep, mycoplasma infections are predominantly caused by Mycoplasma capricolum subsp. capricolum, Mycoplasma capricolum subsp. capripneumoniae, and Mycoplasma mycoides subsp. capri. These pathogens cause acute to peracute diseases, often lethal, including contagious caprine pleuropneumonia (9). Mycoplasmas also infect swine (Mycoplasma hyopneumoniae, Mycoplasma hyorhinis) (10) and economically important Galliformes, such as chicken and turkey (Mycoplasma gallisepticum, Mycoplasma synoviae, and Mycoplasma iowae) (11).
Overall, the genetic bases of mycoplasma pathogenicity remain unclear, but they appear to lack the effectors exported through specialized secretion systems found in numerous bacterial pathogens (12). In addition, the number of toxins or other toxic compounds produced by mycoplasmas is limited. Among the known exceptions is the capacity of several mycoplasmas to produce hydrogen peroxide (H2O2) as a by-product of glycerol metabolism (13, 14). For a small number of Mycoplasma species, the secretion of virulence-mediating proteins was demonstrated. For instance, the rodent pathogen Mycoplasma arthritidis secretes the effector Mycoplasma arthritidis mitogen, an immunomodulatory protein that demonstrates classical superantigenic properties, resulting in an excessive activation of the immune system (15). Another example is the production by M. pneumoniae of the community acquired respiratory distress syndrome toxin, an ADP ribosylating toxin with vacuolating activity (16).
Though some mycoplasma infections can lead to acute inflammatory episodes, most are chronic, suggesting that these bacteria have developed strategies to face and to evade the sophisticated immune system of mammals (17). The most widespread mechanism involves the antigenic variation of surface proteins, including immunodominant lipoproteins. In many cases, the antigens that are subject to variation are associated with cytoadherence (18, 19). Indirect evidences also point toward the existence of a molecular mimicry mechanism in M. pneumoniae, because infections by this mycoplasma have been followed in some cases by acute autoimmune diseases (20).
It has been suggested that some mycoplasmas can directly target the host’s immunoglobulins as a means to evade the immune system. In Ureaplasma, the cleavage of the human IgA1 at the hinge region, releasing intact Fab and Fc fragments, has been reported. It was also shown that isolates of Ureaplasma could cleave the IgA1 of their host species, but not IgA1 of other species (21–23). This activity appears to be linked to a serine protease, but the source of this activity remains cryptic, because no gene candidate could be identified. In the poultry pathogens M. synoviae and M. gallisepticum, a similar activity was attributed to the cysteine protease CysP (24). More recently, a new type of antibody-binding protein found in M. genitalium was described. This protein, named protein M, is extracellular, membrane-anchored, and exhibits an antibody-binding mechanism distinct from other known antibody-binding proteins like protein A from Staphylococcus aureus and protein G from streptococcal bacteria (25). Protein M binds with a high affinity to multiple types of human and nonhuman IgG, through attachment to conserved portions of the VL domain of k and λ light chains. As a result, protein M has the ability to prevent the antibody–antigen union, and could be part of an immunity evasion system based on antibody neutralization. Clear homologs to protein M were only identified in the closely related species M. pneumoniae, suggesting that the system is restricted to a few species.
In this study, we have identified a gene of M. mycoides subsp. capri GM12 encoding a putative Mycoplasma Ig binding protein (MIB) presenting an internal domain with a strongly predicted structural similarity to protein M despite very low sequence conservation. Consistently, we show that MIB acts as a high-affinity IgG binding protein. In addition, we have identified a gene encoding a putative Mycoplasma Ig protease (MIP) immediately downstream to the MIB encoding gene. We demonstrate that MIP specifically cleaves the IgG heavy chain at an unconventional site, located between the VH and CH3 domains. Our data also reveal that prior binding of MIB to the IgG substrate is required for MIP activity. The MIB–MIP system is encoded by a pair of genes often found in several copies in a wide range of mycoplasmas that infect various hosts. Our bioinformatics analysis predicts horizontal gene transfer of the MIB–MIP system between mycoplasmas sharing the same host, suggesting that MIB–MIP is a shared system involved in a global strategy to escape the host immune system. The identification of the MIB–MIP system opens a new avenue of research to better understand the strategies used by minimal bacteria to evade the sophisticated immune system of complex hosts.
Results
MMCAP2_0583 Encodes an IgG Binding Protein.
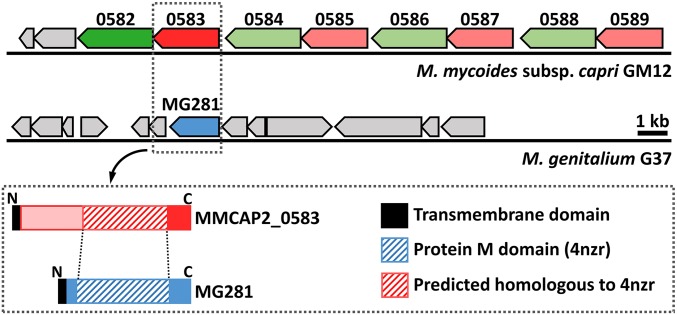
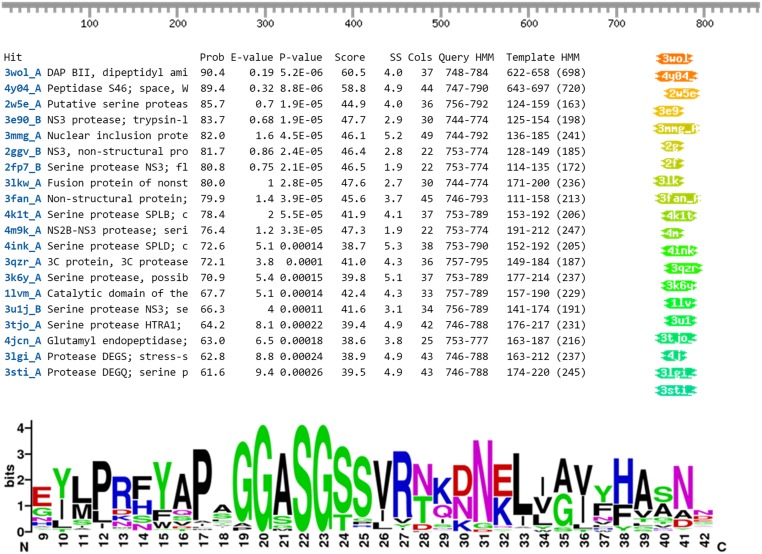
M. mycoides subsp. capri (Mmc) is a pathogenic mycoplasma associated with high mortality outbreaks of respiratory mycoplasmosis in small ruminants (26). We have identified in Mmc an atypically organized cluster of genes, comprised of four tandems (Fig. 1). The tandems all encode homologs of two putative proteins: a 97-kDa predicted serine protease (MMCAP2_0582, 0584, 0586, and 0588) and an 83-kDa protein of unknown function (MMCAP2_0583, 0585, 0587, and 0589). These proteins bear a transmembrane domain at their N-terminal extremity and are predicted to be located at the cell surface. A RT-PCR experiment showed that all of the genes of this cluster are expressed in Mmc during growth on serum-based media (Fig. S1). To functionally characterize these two protein groups, we focused our study on two genes: MMCAP2_0582 and MMCAP2_0583. Analysis of MMCAP2_0583 using HHpred (27) to find remote homologs predicted a very strong structural similarity with the functional domain MTD of protein M from M. genitalium despite very limited sequence similarities (Fig. S2). This domain homologous to protein M is located on the C-terminal side of MMCAP2_0583, whereas the N-terminal side corresponded to a 250-aa domain with no homolog (Fig. 1).
Fig. 1.
Genome context of protein M homologs coding sequences. (Upper) Schematic diagram of the genomic regions surrounding the genes MG281 in M. genitalium (blue) and MMCAP2_0583 (red) and MMCAP2_0582 (dark green) in M. mycoides subsp. capri. Genes homologous to MMCAP2_0583 are colored in light red, and genes homologous to MMCAP2_0582 are colored in light green. (Lower) Schematic representation of the domain architecture of protein M and MMCAP_0583.
Fig. S1.
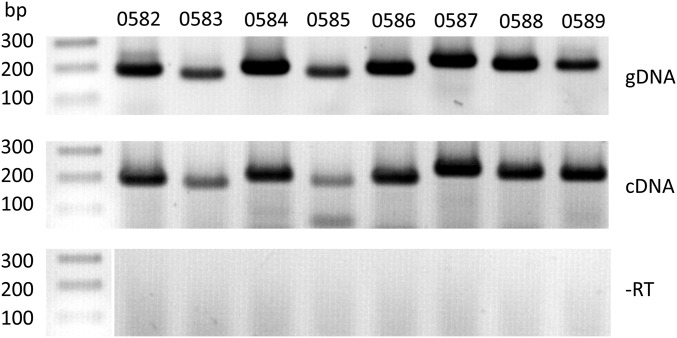
RT-PCR analysis of the expression of the atypical gene cluster of Mmc GM12. To assess whether the eight genes of the cluster were expressed, a RT-PCR experiment was performed. The eight primer pairs designed to specifically amplify a fragment of each of the genes were first validate using genomic DNA (gDNA) as a PCR template. The presence of cDNA reverse transcribed from mRNA was then assessed (cDNA). As a control, an identical PCR was performed using non–retrotranscribed RNA (−RT) to validate the lack of DNA contamination in the RNA samples.
Fig. S2.
Homology detection between MMCAP2_0583 and MG281. (A) The amino acids sequence of the putative protein encoded by the gene MMCAP2_0583 was analyzed using HHpred software, available at toolkit.tuebingen.mpg.de/hhpred. Parameters were set to default. The figure depicts the results obtained by comparing the query sequence to the PDB database updated on November 23, 2015. 4nzr_M is the PDB entry corresponding to the core domain of protein M (dubbed protein MTD). This domain is necessary and sufficient for the Ig binding function of protein M. (B) The amino acid sequences of MMCAP2_0583 and MG281 were aligned using MUSCLE with default parameters. The alignment file was then processed using ESPript (espript.ibcp.fr/) to generate the figure. Gaps are indicated as dots (“.”), fully conserved residues are highlighted in red, and conservation between groups of strongly similar properties is indicated in red letters and framed in blue.
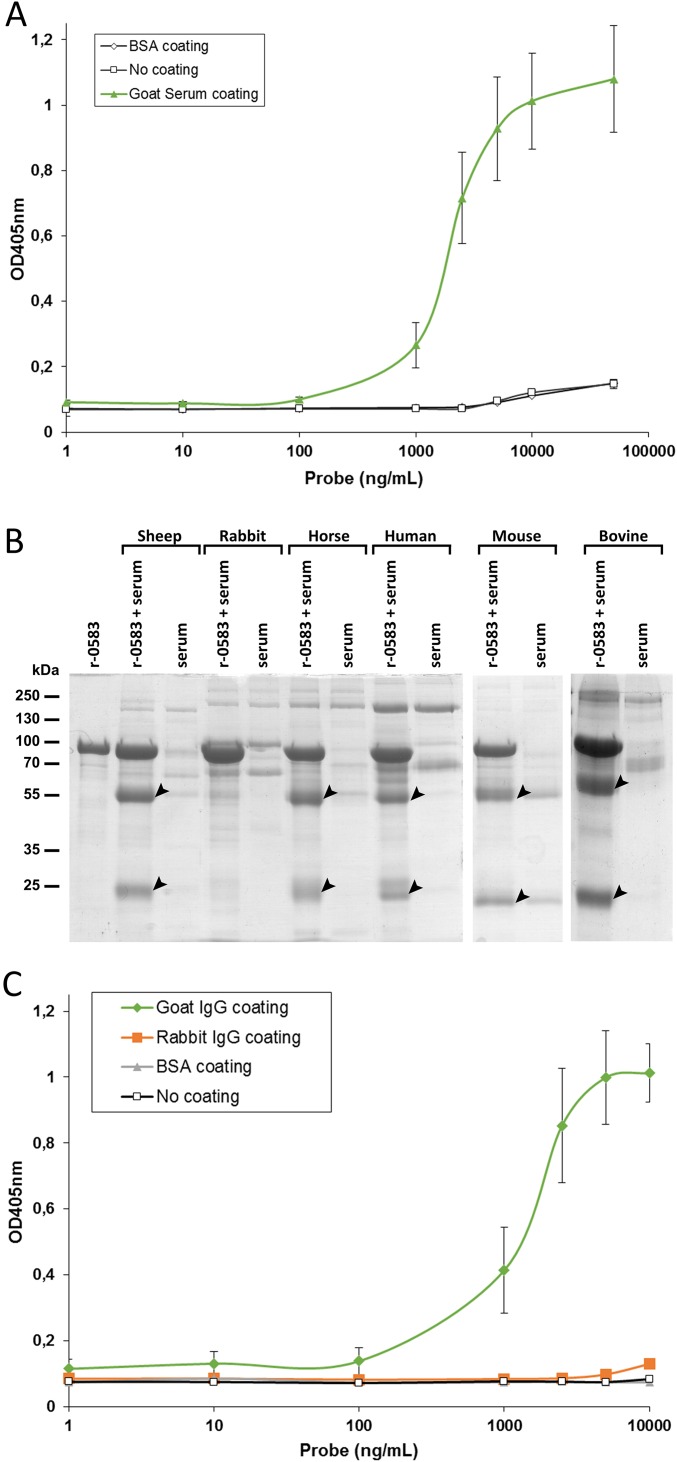
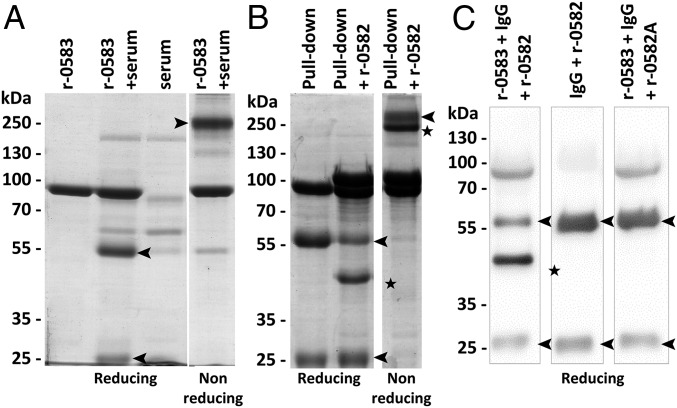
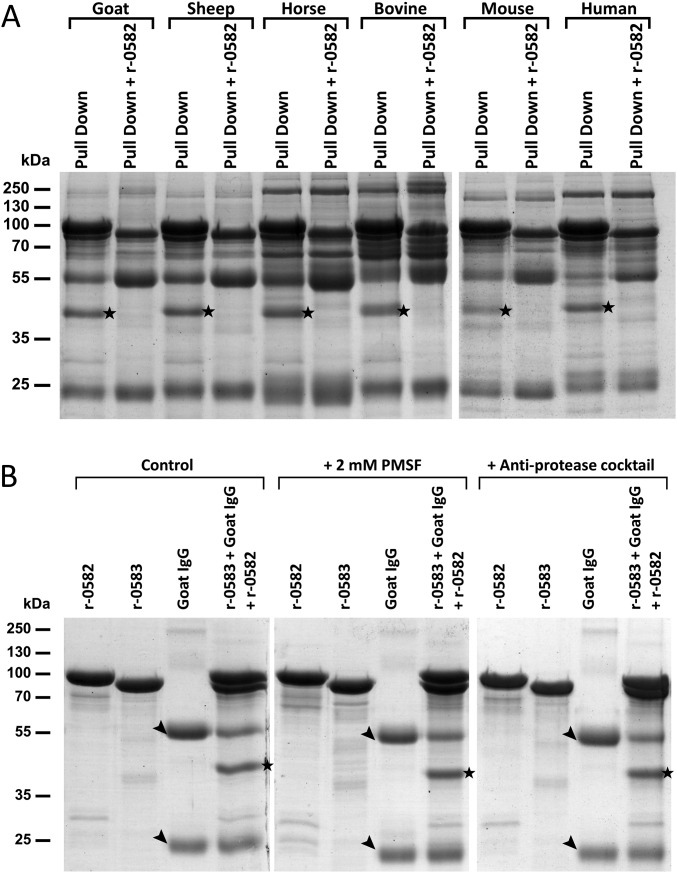
To study the function of MMCAP2_0583, the recombinant protein r-0583 was produced by replacing the transmembrane domain with a histidine tag. The protein r-0583 was expressed in Escherichia coli and purified to homogeneity. An initial ELISA experiment demonstrated that r-0583 is able to interact with a component of naive goat serum (Fig. S3). To identify this serum component, a pull-down experiment was performed using r-0583 as bait and naive goat serum as prey (Fig. 2A). Analysis of the eluted fractions showed that two proteins were purified from the serum through binding to r-0583. These proteins were identified by liquid chromatography–tandem mass spectrometry (LC-MS/MS) as the heavy and light chain of IgG, in accordance with their apparent molecular weights of 55 kDa and 25 kDa, respectively. The same experiment was performed using sera from various mammals, and showed that r-0583 is also able to interact with IgG from sheep, bovine, horse, mouse, and human sources (Fig. S3). Interestingly, r-0583 is not able to bind to immunoglobulins from rabbit serum. This result was confirmed by an ELISA comparing the interaction between r-0583 and purified IgG from goat or rabbit (Fig. S3). Overall, these results indicate that r-0583 shares the function of protein M and acts as a broad spectrum IgG binding protein, hereafter termed MIB.
Fig. S3.
ELISA and pull-down assay of the interactions between r-0583 and various mammalian sera, or rabbit and goat IgG. (A) ELISA was used to assess the ability of purified r-0583 to bind to serum components. Sera from healthy goats (i.e., goats that have never been in contact with any mycoplasma strains) were coated on an ELISA plate at 50 µg/mL in bicarbonate/carbonate 0.1 M pH 9.6 buffer. Binding to the coating was detected using a mouse anti-polyhistidine primary antibody and an anti-mouse AP-conjugated secondary antibody. Alkaline phosphatase activity was measured using pNPP substrate in diethanolamine buffer pH 9.8 at 37 °C. Error bars for the goat serum coating samples correspond to the SD from eight biological samples. Error bars for the BSA coating and no-coating samples correspond to the SD of three technical replicates each. (B) Purified r-0583 is bound to a Ni-NTA resin via its histidine tag, and incubated with normal serum from sheep, rabbit, horse, human, mouse, and bovine. After extensive washing, proteins bound to the resin are eluted with a pulse of imidazole and analyzed by SDS/PAGE. The control experiments include the bait protein r-0583 without serum (lane: r-0583) and the naked resin incubated with the serum (lane: serum). Black arrows indicate the heavy and light chains of immunoglobulins (55 kDa and 25 kDa, respectively). Identification of the proteins was confirmed by LC/MS-MS analysis. (C) ELISA was used to assess the ability of purified r-0583 to bind to IgG. Goat or rabbit purified IgG were coated on an ELISA plate at 50 µg/mL in bicarbonate/carbonate 0.1 M pH 9.6 buffer. Binding to the coating was detected using a mouse anti-polyhistidine primary antibody and an anti-mouse AP-conjugated secondary antibody. Alkaline phosphatase activity was measured using pNPP substrate in diethanolamine buffer pH 9.8 at 37 °C. Error bars correspond to the SD of six technical replicates.
Fig. 2.
Functional analysis of r-0583 and r-0582. (A) Pull-down interaction assay between r-0583 and naïve goat serum. r-0583 was bound to a resin and incubated with serum. After extensive washing, bound proteins were eluted and analyzed by SDS/PAGE in reducing (Left) and nonreducing conditions (Right). Control experiments included the bait protein r-0583 without serum (lane: r-0583) and the naked resin incubated with the serum (lane: serum). Black arrow, IgG chains (55 kDa, heavy; 25 kDa, light; ∼250 kDa, whole IgG). (B) Effect of r-0582 on IgG pulled down from goat serum. Proteins eluted from the pull-down experiments were mixed with purified r-0582 (lane: pull down + r-0582) and incubated for 15 min at 30 °C. Samples were checked by SDS/PAGE in reducing (Left) and nonreducing conditions (Right). Black star, cleaved IgG heavy chain. (C) Cleavage of purified goat IgG by r-0582. Samples were checked by SDS/PAGE and the IgG fragments were detected by Western blot. Nonspecific detection of r-0583 was caused by its nonimmunological binding of the primary antibody.
MMCAP2_0582 Encodes an IgG Protease.
The gene in tandem with the MIB-encoding MMCAP2_0583 was annotated as a putative serine protease, bearing the predicted domain of unknown function 31 (DUF31). Because the MMCAP2_0582 gene is genetically linked to an IgG binding protein, we hypothesized that it could act as an IgG protease. To validate this hypothesis, the recombinant protein r-0582 was produced by replacing the N-terminal transmembrane domain with a histidine tag. The protein r-0582 was expressed in E. coli, and subsequently purified to homogeneity. To test its activity, r-0582 was added to the fractions eluted from the pull-down experiment described above. After incubation, the integrity of the IgG chains was checked by electrophoresis (Fig. 2B). The results showed that the addition of r-0582 lead to a cleavage of the heavy chain of the IgG, producing a fragment of ∼44 kDa. This result was confirmed by performing the cleavage experiment with purified goat IgG, and detecting the Ig fragments with a specific antibody (Fig. 2C). Interestingly, this experiment also showed that combining r-0582 with the IgG alone is not sufficient for cleavage to occur, and that the presence MIB is necessary for the protease activity. The proteolytic cleavage of pulled-down IgG was also performed with serum samples from various mammals (Fig. S4). This experiment showed that, in the presence of MIB, r-0582 is also able to cleave the IgG heavy chain from sheep, bovine, horse, mouse, and human, resulting in a 44-kDa fragment. Overall, these results point toward an IgG protease function for r-0582, hereafter termed MIP.
Fig. S4.
Functional characterization of r-0582. (A) Samples eluted from the sera pull-down experiment described in Fig. S4B (lane: pull-down) were mixed with purified r-0582 (lane: pull-down + r-0582) and incubated for 15 min at 37 °C. After incubation, the samples were analyzed by SDS/PAGE. Black star indicates large fragment resulting from the cleavage of Ig heavy chain by r-0582 (∼44 kDa). (B) The impact of various protease inhibitors on the proteolytic activity of r-0582 was assessed. The cleavage reaction was set up by mixing r-0583 and goat IgG first, then adding r-0582 preincubated for 10 min with the protease inhibitor. Once all of the components were mixed, the samples were incubated for 15 min. After incubation, the samples were analyzed by SDS/PAGE. Black stars indicate the large fragment resulting from the cleavage of Ig heavy chain by r-0582 (∼44 kDa). Black arrows indicate the heavy and light chains of immunoglobulins (55 kDa and 25 kDa, respectively). PMSF is a serine protease inhibitor. The anti-protease mixture was added at a 1/100th dilution as recommended by the manufacturer protocol. The mixture contains the following protease inhibitors: AEBSF (serine protease inhibitor), phosphoramidon (thermolysine and collagenase inhibitor), pepstatin A (acid protease inhibitor), bestatin (aminopeptidase inhibitor), and E-64 (cysteine protease inhibitor).
MIP Is a Serine Protease.
Predictions based on sequence similarity suggested that MIP is a serine protease. To validate this hypothesis, we performed the IgG cleavage assay described above using MIP preincubated with PMSF, a serine protease inhibitor that reacts specifically with the active catalytic serine (Fig. S4). Unexpectedly, MIP was still able to cleave IgG in the presence of the inhibitor. The same result was obtained when using a commercial mixture of protease inhibitors targeting the majority of protease types. Nonetheless, the serine protease annotation for MMCAP2_0582 appeared to be confirmed by an HHpred analysis, because a small C-terminal domain of the protein had multiple hits in serine proteases (Fig. S5). Interestingly, the catalytic serines of the hits were systematically aligned with the Serine727 of MIP. Furthermore, analysis of the homologs of MIP in multiple mycoplasma species showed that Serine727 and its immediate surroundings were highly conserved (Fig. S5). Consequently, a Ser727Ala mutant (r-0582A) was produced, expressed in E. coli and purified. This mutant was used in an IgG cleavage assay and showed no proteolytic activity (Fig. 2C), confirming that MIP is indeed a serine protease.
Fig. S5.
Identification of the putative catalytic serine of MMCAP2_0582. (Upper) HHpred analysis of the amino acid sequence of MMCAP2_0582. Parameters were set to default. The results were obtained by comparing the query sequence to the PDB database updated on November 23, 2015. Several low-confidence hits were obtained at the C-terminal end of MMCAP2_0582. The majority of the hits are annotated as serine protease. The catalytic serine of the hits is always located inside the matched sequence, and always corresponds to Ser727 of MMCAP2_0582. (Lower) WebLogo graphical representation (weblogo.berkeley.edu/) of the amino acid conservation of the region surrounding Ser727 in MMCAP2_0582 and its homologs.
The IgG Cleavage Site by MIP Is Located Between the VH and CH3 Domains.
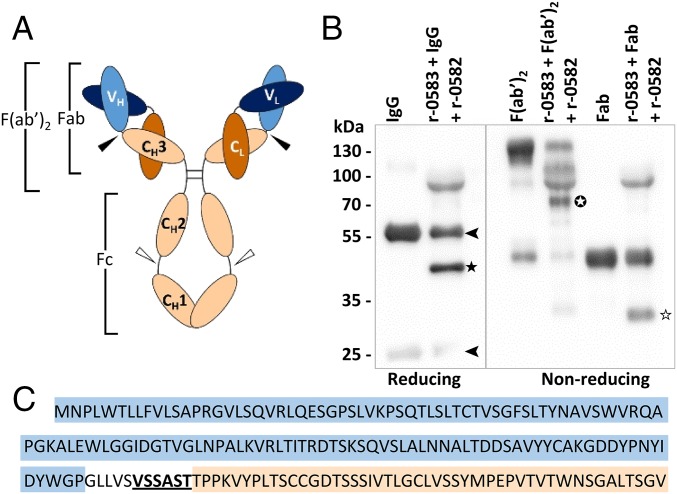
Digestion of IgG by MIP led to a 44-kDa fragment, resulting from the cleavage of the heavy chain. Based on the size of this polypeptide, two possible cleavage sites can be deduced: the first located between the CH1 and CH2 domains, and the second between VH and CH3 (Fig. 3A). We performed a cleavage assay using IgG fragments F(ab′)2 and Fab instead of whole IgG (Fig. 3B), because these fragments bore only one of the possible cleavage sites. The results of this experiment showed that MIP is able to cleave both F(ab′)2 and Fab, confirming that the cleavage site is located between the VH and CH3 domains. The exact cleavage site was determined by sequencing the N-terminal amino acids of the 44-kDa fragment. The sequence obtained was VSSAST, and was identified at the N-terminal extremity of the CH3 domain (Fig. 3C). Interestingly, the triplet VSS is also present on the IgG of sheep, bovine, horse, human, and mouse (Fig. S6), which can all be cleaved by MIP.
Fig. 3.
Identification of r-0582 proteolytic site. (A) Schematic representation of the domain architecture of IgG, F(ab′)2 and Fab. CH, conserved domain heavy chain (beige); VH, variable domain heavy chain (orange); CL, conserved domain light chain (light blue); VL, variable domain light chain (dark blue). Arrows, possible cleavage sites. (B) Proteolytic cleavage of IgG fragments by r-0582. Samples were checked by SDS/PAGE and the IgG fragments were detected by Western blot. Black arrow, heavy and light chains of immunoglobulins; black star, cleaved IgG heavy chain; circled star, cleaved F(ab′)2 fragment; white star, cleaved Fab fragment. (C) Cleavage site of r-0582. The N-terminal sequence of the large cleaved fragment of IgG heavy chain (black star) was determined by Edman degradation (bold and underlined). This sequence is located between the VH domain (light blue) and CH3 domain (beige) of goat IgG.
Fig. S6.
Sequence alignment of the VH domain of Ig gamma-1. Sequences for the VH domain of IgG1 of goat (Capra hircus), sheep (Ovis aries), bovine (Bos taurus), horse (Equus caballus), mouse (Mus musculus), and human (Homo sapiens) were retrieved from the Abysis database. The amino acid sequences were aligned using MUSCLE with default parameters. The alignment file was then processed using ESPript to generate the displayed image. Gaps are indicated as dots (“.”), fully conserved residues are highlighted in red, and conservation between groups of strongly similar properties is indicated in red letters.
The MIB–MIP System Requires a Sequential Assembly.
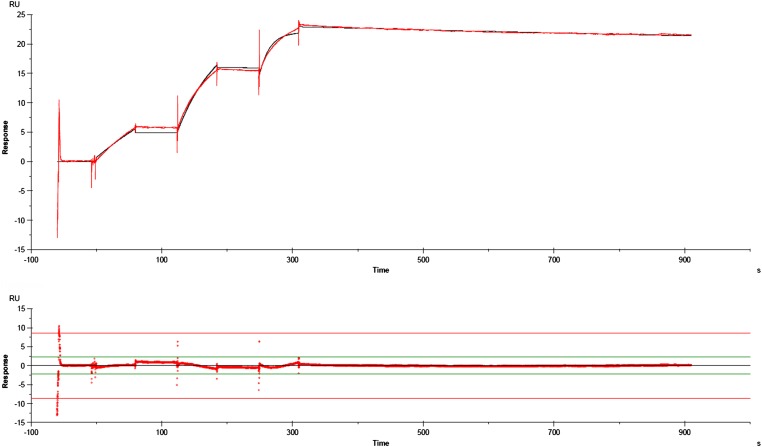
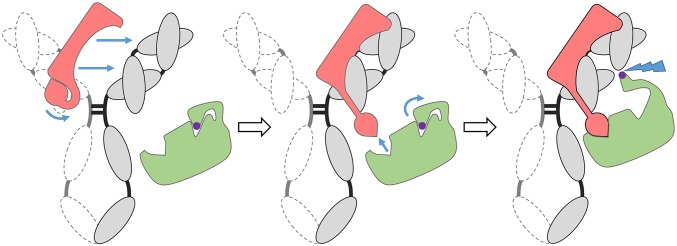
To better understand the interplay between MIB and MIP, and in particular to comprehend the inability of the protease to cleave the IgG alone, we studied the protein–protein interactions between the different partners. Using surface plasmon resonance (SPR) in a qualitative approach (Fig. 4A), we showed that r-0582 is unable to interact with either r-0583 or with IgG. Interestingly, when an r-0583/IgG complex is preassembled, r-0582 is able to bind very tightly to this compound. The kinetic parameters for this interaction were determined using the inactive variant r-0582A (Fig. S7). We elected to use the inactive form of MIP to remove from the system the loss of mass potentially caused by the cleavage of the VH domain, which could interfere with the SPR signal. This experiment determined that KD = 0.37 nM, with ka = 3.19 × 105 s−1⋅M−1 and kd = 1.18 × 10−4⋅s−1, indicative of a very strong interaction between MIP and the MIB–IgG complex. To determine the stoichiometry of the interaction, we measured the absolute molar mass of the MIB–IgG–MIP complex using size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS). After evaluating the mass of the individual components and confirming that their basic states are monomeric (Fig. S8), we analyzed the complexes IgG/r-0583 and IgG/r-0583/r-0582 (Fig. 4B). The absolute molar mass of the IgG/r-0583 complex was determined at 303 kDa, which is consistent with a 1 × IgG – 2 × r-0583 stoichiometry [expected molar mass ∼140 + (2 × 83) = 306 kDa]. For the IgG/r-0583/r-0582 complex, the absolute molar mass was determined at 495 kDa, which is consistent with a 1 × IgG – 2 × r-0583 − 2 × r0582 stoichiometry [expected molar mass ∼140 + (2 × 83) + (2 × 97) = 500 kDa]. Overall, these results indicate that one MIB is able to bind to each of the two Fab domains of IgG. The resulting structure is able to recruit two MIP, one for each MIB–Fab pair (Fig. 5). The resulting complex appears to be stable over time.
Fig. 4.
Assembly of the IgG–MIB–MIP complex. (A) Interactions among MIB, MIP, and goat IgG. SPR was used to analyze the interactions among IgG, r-0583, and r-0582. In a first experiment (Left), r-0583 was immobilized on the sensor chip and probed with r-0582 (1 µM), goat IgG (0.5 µM), and again with r-0582 (1 µM). In a second experiment (Right), goat IgG was immobilized on the sensor chip and probed with r-0582 (1 µM), r-0583 (1 µM), and again with r-0582 (1 µM). (B) Molecular weight of the IgG–MIB and IgG–MIB–MIP complexes. Absolute molecular masses of the IgG/r-0583 complex (blue line) and IgG/r-0582/r-0583 complex (black line) were determined by SEC-MALS. The horizontal lines correspond to the MALS calculated masses. Average mass values were calculated over the half-height width of the elution peaks.
Fig. S7.
Kinetic analysis of the interaction of r-0582 with a preformed IgG/r-0583 complex. (Upper) Sensorgrams (red line) and fitting curve (black line) of the single-cycle kinetics analysis. Before the experiment, goat IgG was captured on the sensor chip and allowed to interact with r-0583. This stable complex was subsequently probed with three injections of r-0582A at 12.5 nM, 50 nM, and 200 nM. The experiment temperature was set at 30 °C, and two technical replicates were performed. A flow cell left blank was used for double-referencing of the sensorgrams. The global data were fitted to a 1:1 Langmuir model of interaction to determine the association and dissociation rate constants, Ka and Kd, respectively, and to calculate the dissociation equilibrium constant Kd = Ka/Kd. (Lower) Residual plot showing the difference between experimental and fitted data.
Fig. S8.
Molar mass and oligomeric state of goat IgG, r-0583, and r-0582. SEC-MALS analysis was used to determine the absolute molecular mass of the proteins in solution, and infer their oligomerization status. The blue line corresponds to the UV elution profile at 280 nm. The horizontal red lines correspond to the MALS calculated masses. Average mass values are calculated over the half-height width of the elution peaks.
Fig. 5.
Model of the assembly of the MIB–IgG–MIP complex. MIB (red) is able to bind tightly to the FV region of the IgG (gray). Upon formation of the IgG–MIB complex, MIP (green) is recruited. Upon binding, the catalytic serine of MIP is exposed (purple) and the cleavage of the heavy chain occurs (blue lightning bolt).
The MIB–MIP System Is Widespread in Mollicutes.
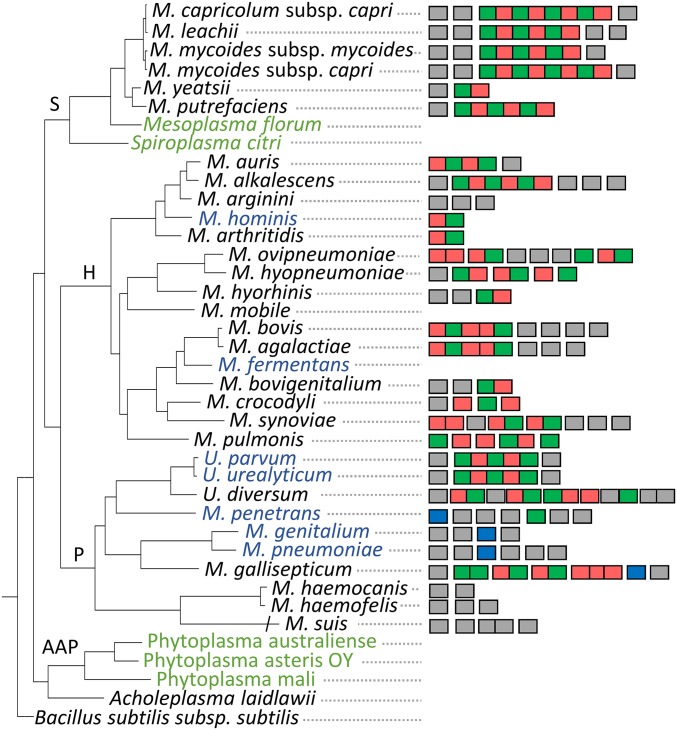
After the identification of MIB and MIP, we searched for homologous genes in the genomes of 39 Mollicutes representative of the diversity of this class, both in terms of taxonomy and host tropism (Fig. 6). Our data showed that, in contrast to protein M, which is only found in a small number of pathogens, MIB and MIP homologs are found in the majority of animal pathogenic mycoplasmas. The tandem architecture was found in most of the genomes containing the MIB–MIP system, and was often complemented by isolated MIB or MIP homologous genes at remote loci. The system was noticeably absent from plant pathogens such as Mesoplasma florum and Spiroplasma citri and the acholeplasma/phytoplasma group. It is also interesting to note that the MIB–MIP system and protein M appear to be mutually exclusive because they are not found in the same genomes. The only exception is the presence of a remote homolog of protein M in M. gallisepticum alongside clear homologs of the MIB–MIP system. Furthermore, we scanned the different genomes for the presence of genes annotated as DUF31 that are not homologs of MIP-encoding genes. This Pfam domain was the only annotation initially available for MMCAP2_0582, and all of the MIP predicted homologs shared this domain annotation. This domain appears to be found almost exclusively in the Mollicutes class. DUF31-annotated genes are widespread among animal and human pathogenic mycoplasmas, and interestingly absent from plant pathogenic species. It is noteworthy that, in some species like M. hemocanis or M. genitalium, DUF31-annotated genes are found in multiple copies, whereas the MIB–MIP system is absent. The exact function of these DUF31 genes is still elusive, although amino acid conservation suggests a serine–protease function.
Fig. 6.
Distribution of the MIB and MIP homologs in Mollicutes. The occurrence of a gene homolog of MIB, MIP, protein M, or a gene annotated DUF31 is indicated by rectangles colored in red, green, dark blue, and light gray, respectively. Adjoining rectangles denote genes located on the same loci. Vertical alignment of the symbols does not infer synteny. The host specificity of the Mollicutes species is indicated as follows: black, animal host; cyan, human host; green, plant host. The phylogenetic tree was inferred using the maximum-likelihood method from the concatenated multiple alignments of 79 proteins encoded by genes present at one copy in each genome. Main phylogenetic groups are indicated: S, spiroplasma; H, hominis; P, pneumoniae; and AAP, acholeplasma/phytoplasma.
Discussion
In this study, we identified and functionally characterized a two-protein system from the small-ruminant pathogen Mycoplasma mycoides subsp. capri, involved in the capture and cleavage of IgG. The first component of the system is the protein MIB, which is able to bind tightly to IgG. Given that MIB has a very strong predicted structural similarity with protein M from M. genitalium, and a similar function, we assumed that its binding mechanism was similar. Protein M binds to immunoglobulins predominantly through attachment to the conserved portions of the VL domain of the light chain (25). This model is consistent with the observation that two MIB molecules can bind to a single IgG. Furthermore, MIB was able to bind to the Fab and F(ab′)2 fragments, confirming its interaction with the FV regions of the immunoglobulins.
The second element of the system is the serine protease MIP, which is able to cleave the heavy chain of IgG. Our data showed that MIP is unable to interact with either isolated MIB or IgG, but can bind very strongly to the complex they form. The exact assembly mechanism of the MIB–IgG-MIP complex is not yet resolved, and it is unclear whether MIP binds directly to a domain of MIB, to a domain of IgG, or to both. Interestingly, we showed that MIP alone is not able to cleave the IgG heavy chain, suggesting that it is inactive by itself. MIP is insensitive to PMSF, despite the fact that this inhibitor inactivates virtually all serine protease and is a diagnostic for this protease class. PMSF inhibits serine proteases by reacting specifically with the catalytic serine and thus blocking the active site (28). We therefore hypothesized that the catalytic residue Ser727 is either not exposed to the solvent or not in the activated state before the interaction of MIP with its cognate partners. Upon binding to the MIB–IgG complex it is probable that significant conformational changes occur in MIP, leading to the activation of the catalytic serine or to its positioning in the vicinity of the IgG cleavage site. Further studies on the 3D structure of the different partners within and outside of the complex should provide us with a better understanding of this intricate mechanism. The data we gathered also show that the protease is bound very tightly to its partners, and is not released after the cleavage occurs. This observation could be linked to the in vitro experimental set-up used in this study. The MIB–MIP system might be missing one or more partners involved in further processing of the antibody before its release. Potential candidates include other proteases such as the three homologs of MMCAP2_0582, the three other DUF31 annotated genes, or the gene MMCAP2_0490, which encodes a distant homolog of the IgG protease CysP (24). Combined activities of these proteases might further degrade immunoglobulins and lead to a final release of the MIB–MIP components.
Another idiosyncrasy of the MIP protease is the location of its cleavage site on the IgG. All of the bacterial Ig proteases characterized to date have a cleavage site located in the hinge region (29, 30), leading to the release of the Fc and Fab fragments. It is surmised that the role of these proteases is to protect the bacteria from the host immune system by either preventing the binding of the complement to Fc or masking the cell with a cloak of host-like components (31). In the case of MIP, it is not clear whether the VH domain would be released from the antibody after cleavage or not. The VH domain is bound noncovalently to the VL domain (32), but with relatively low dissociation constants (10−5 − 10−8 M) that are not sufficient to keep the domains associated at low concentrations of protein or under mildly destabilizing conditions (33). Loss of the VH domain would be highly detrimental for the function of the antibody as this domain bears three of the six complementarity determining regions (CDRs) involved in antigen recognition, including CDR-H3 which is the key player in the antigen affinity (34–36). Therefore, the role of the MIB–MIP system could be to render the IgG nonfunctional, unable to target their antigens, thus limiting the efficiency of the immune response.
In the genome of Mmc, four pairs of genes encoding MIB and MIP proteins were found in tandem (Fig. 1). The intraspecies diversity between these multiple variants appeared to be quite high (Tables S1–S4). Amino acids conservation between MIB and MIB paralogs was limited to 78.4–83.5% similarity and 63.5–70% identity, with most of the sequence differences located in the N-terminal domain of unknown function. For MIP paralogs, the dissimilarities were even more important (75.8–79.1% similarity and 58.4–64% identity) but spread throughout the protein. The specific roles and interplay of the different homologs and paralogs of MIB and MIP, as well as their regulation, remains to be explored. Nonetheless, data gathered from two recent publications provided initial evidence that the MIB–MIP system is expressed in vivo. The proteome of M. mycoides subsp. mycoides (Mmm), a species closely related to Mmc, was analyzed in vitro and in vivo. These studies showed that five of six homologs of MIB and MIP could be detected in Mmm (37) and confirmed that these proteins were located at the surface of the bacteria (38). Based on sequence similarity values, the MIP and MIB homologs expressed in Mmm correspond to the proteins 0582, 0583, 0584, 0587, and 0588 from Mmc.
Table S1.
Similarity percentages between MIB homologs
| MMCAP2_0582 | MMCAP2_0584 | MMCAP2_0586 | MMCAP2_0588 | |
| MMCAP2_0582 | 100 | 75.8 | 77.1 | 77.5 |
| MMCAP2_0584 | 75.8 | 100 | 79.1 | 76.0 |
| MMCAP2_0586 | 77.1 | 79.1 | 100 | 77.4 |
| MMCAP2_0588 | 77.5 | 76.0 | 77.4 | 100 |
Identity and similarity percentages were calculated based on pairwise Needleman-Wunsch alignment of protein sequences.
Table S4.
Identity percentages between MIB homologs
| MMCAP2_0583 | MMCAP2_0585 | MMCAP2_0587 | MMCAP2_0589 | |
| MMCAP2_0583 | 100 | 65.0 | 68.0 | 63.7 |
| MMCAP2_0585 | 65.0 | 100 | 66.1 | 63.5 |
| MMCAP2_0587 | 68.0 | 66.1 | 100 | 70.0 |
| MMCAP2_0589 | 63.7 | 63.5 | 70.0 | 100 |
Identity and similarity percentages were calculated based on pairwise Needleman-Wunsch alignment of protein sequences.
Table S2.
Identity percentages between MIB homologs
| MMCAP2_0582 | MMCAP2_0584 | MMCAP2_0586 | MMCAP2_0588 | |
| MMCAP2_0582 | 100 | 60.9 | 58.4 | 59.0 |
| MMCAP2_0584 | 60.9 | 100 | 64 | 59.1 |
| MMCAP2_0586 | 58.4 | 64.0 | 100 | 62.6 |
| MMCAP2_0588 | 59.0 | 59.1 | 62.6 | 100 |
Identity and similarity percentages were calculated based on pairwise Needleman-Wunsch alignment of protein sequences.
Table S3.
Similarity percentages between MIP homologs
| MMCAP2_0583 | MMCAP2_0585 | MMCAP2_0587 | MMCAP2_0589 | |
| MMCAP2_0583 | 100 | 80.8 | 81.8 | 80.4 |
| MMCAP2_0585 | 80.8 | 100 | 80.6 | 78.4 |
| MMCAP2_0587 | 81.8 | 80.6 | 100 | 83.4 |
| MMCAP2_0589 | 80.4 | 78.4 | 83.4 | 100 |
Identity and similarity percentages were calculated based on pairwise Needleman-Wunsch alignment of protein sequences.
MIB–MIP system homologs were found in the majority of animal pathogenic Mollicutes genomes, including the human pathogens M. hominis, Ureaplasma parvum, and Ureaplasma urealyticum (Fig. 6). By contrast, no MIB–MIP system could be predicted in any plant pathogenic phytoplasmas and spiroplasmas. The system was also missing in several mycoplasmas including the two human pathogenic species M. genitalium and M. pneumoniae. Interestingly, these two organisms possess the protein M that is a structurally related to MIB. Despite a clear structural relatedness, the amino acid sequences of MIB homologs and protein M are only very weakly similar, which suggests a fast and specific evolution of the protein M encoding gene in the few species where it is found. Moreover, no protein homologous to MIP could be identified for these two species, but multiple genes encoding proteins with a predicted DUF31 domain were predicted, which could potentially be functional homologs of MIP—namely, IgG proteases working in pair with protein M.
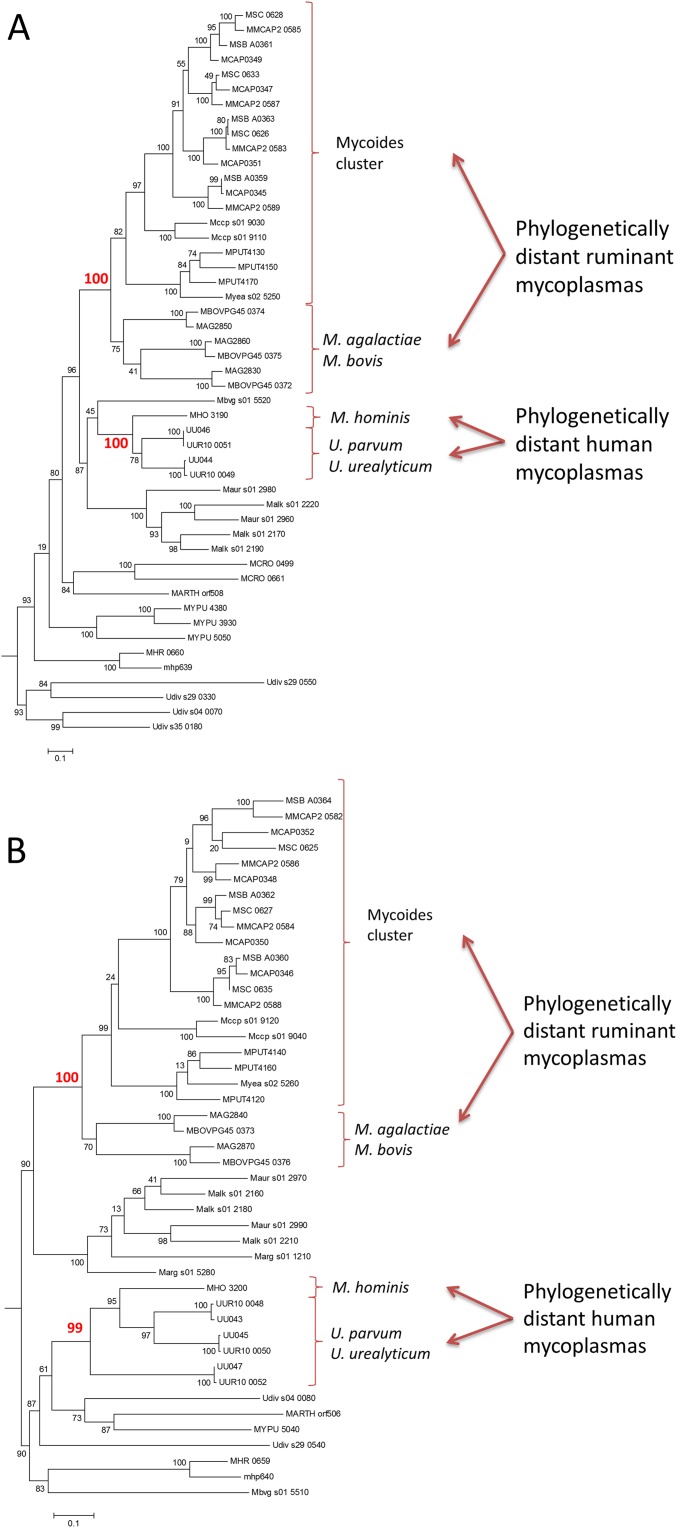
The wide distribution of the MIB–MIP system in mycoplasmas infecting different animals and belonging to different phylogenetic groups suggested a biological importance of this system regarding host–pathogen interactions. Phylogenetic analyses of the MIB and MIP (Fig. S9) proteins strongly suggested that horizontal gene transfers (HGTs) occurred several times during the Mollicutes’ evolution. In particular, HGT were predicted among ruminant mycoplasmas between the ancestor of the M. agalactiae/M. bovis group and the ancestor of the mycoides cluster M. yeatsii/M. putrefaciens group. Other HGTs of potential virulence factors have already been described among these groups (39). In urogenital human mycoplasmas, HGTs were predicted between M. hominis and the ancestor of the U. parvum/U. urealyticum group, in accordance with previous study (40). These genetic exchanges of the MIB–MIP system among mycoplasmas colonizing the same host are in complete agreement with the potential significance of this system in the evolutionary process leading to the adaptation to a new host.
Fig. S9.
Phylogenetic analysis of MIB and MIP homologs. (A) The evolutionary history of MIB proteins was inferred by using the pipeline available at www.phylogeny.fr with the following parameters: alignment with MUSCLE; curation with Gblock 0.91b; phylogenetic tree reconstruction with PhyML (substitution model: WAG; gamma shape parameter: 0.981; number of categories: four; proportion of invariant: 0.039), statistical tests for branch support aLRT (SH-like). Protein sequences are indicated by their locus tag. Strong phylogenetic associations supporting HGT between evolutionary distant species sharing a same host are indicated. (B) The evolutionary history of MIP proteins was inferred by using the pipeline available at www.phylogeny.fr with the following parameters: alignment with MUSCLE; curation with Gblock 0.91b; phylogenetic tree reconstruction with PhyML (substitution model: WAG; gamma shape parameter: 1.427; number of categories: four; proportion of invariant: 0.138), statistical tests for branch support aLRT (SH-like). Protein sequences are indicated by their locus tag. Strong phylogenetic associations supporting HGT between evolutionary distant species sharing a same host are indicated.
Finally, we noted that the MIB–MIP system homologs were found exclusively in Mollicutes. It is interesting to find that such an intricate mechanism evolved in so-called “minimal” bacteria, which lack some of the most basic cell functions. Although further studies are necessary to completely elucidate this system and its in vivo implications, the identification of MIB and MIP opens a new avenue of research to better understand the pathogenicity of mycoplasmas and their immunity evasion mechanisms.
Materials and Methods
Cloning.
MMCAP2_0582 and MMCAP2_0583 coding sequences were codon optimized for expression in E. coli. Synthetic genes were ordered from Eurofins Genomics and cloned in the pET28a(+) vector (Novagen). Details for this and subsequent methods can be found in SI Materials and Methods.
Recombinant Proteins Expression and Purification.
Recombinant protein were expressed in E. coli BL21(DE3) (Novagen) grown in autoinduction medium at 20 °C. Protein purification was performed on a Nickel-NTA column (GE Healthcare Life Sciences) followed by polishing and buffer exchange by gel filtration on a Superdex 200 column (GE Healthcare Life Sciences).
Pull-Down Assays.
Animal sera were obtained either from commercial vendors (Sigma) or as gifts from academic research laboratories. Nickel-NTA agarose beads (GE Healthcare Life Sciences) coated with purified r-0583 were incubated with serum for 2 h at room temperature. Unbound proteins were thoroughly washed with buffer containing 20 mM imidazole, and bound proteins were subsequently eluted using buffer containing 250 mM imidazole.
MS Identification.
Protein identity was determined using a LC-MS/MS analysis pipeline. Proteins were first reduced then alkylated before undergoing tryptic digestion overnight at 37 °C. Extracted peptides were analyzed on an Ultimate 3000 nanoLC system (Dionex) coupled to a nanospray LTQ-Orbitrap XL mass spectrometer (Thermo Finnigan). Data were searched by SEQUEST through Proteome Discoverer 1.4 (Thermo Fisher Scientific Inc.) against databases containing amino acid sequences of Ig extracted from the abYsis database (www.bioinf.org.uk/abysis).
Ig Cleavage Assays.
Purified goat IgG was purchased from Sigma, and purified goat F(ab′)2 and Fab were purchased from Jackson ImmunoResearch. All cleavage assays were performed at 30 °C by mixing all of the components in PBS buffer pH 7.4. The protease was always the last element added to the reaction mixture.
Western Blots.
Proteins separated by SDS/PAGE were transferred on a nitrocellulose membrane using a TE-77 semidry transfer system (GE Healthcare Life Sciences). Detection was performed using a primary antibody (mouse anti-goat IgG H+L; Jackson ImmunoResearch) and a secondary antibody (goat anti-mouse Fc HRP coupled) and the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
Protein–Protein Interaction Analysis.
Analysis of the interactions was performed by SPR on a Biacore T200 instrument using CM5 S Series sensor chips (GE Healthcare Life Sciences). Ligands were coupled to the chip surface using the Biacore amine coupling protocol, and analytes were injected at a flow of 20 µL⋅min−1. All of the analyses were performed at 30 °C, in PBS pH 7.4 Tween 20 0.05% buffer. Kinetic parameters were determined by performing a single-cycle kinetic experiment, and data were fitted to a 1:1 Langmuir model.
SEC-MALS.
Absolute molecular masses were determined using SEC coupled to a MALS detector. SEC was performed using a HPLCy system and a Superose 6 10/300 GL column (GE) thermostated at 30 °C, at a flow rate of 0.5 mL⋅min−1. The elution profiles were followed by refractive index, and by static light scattering using an Optilab rEX, a miniDAWN TREOS, respectively (Wyatt Technology). Molar masses are given as mean over the half-height width of the elution peaks.
SI Materials and Methods
Gene Expression Analysis.
RNA was purified from cells grown in SP5 media until late exponential phase using the Qiagen miRNeasy kit according to the manufacturer’s recommendation. The purified total RNA was subsequently treated with RQ1 DNase (Promega) to remove any contaminant DNA. Single-stranded cDNA were synthesized from the treated RNA using the SuperScript II reverse transcriptase using random hexamers (Invitrogen) for priming. A control experiment without reverse transcriptase was performed to assess the efficiency of the DNase step. The cDNA were amplified by PCR using eight pairs of primers, designed to each amplify a fragment of the different genes of the cluster (0582: 191 bp; 0583: 178 bp; 0584: 201 bp; 0585: 181 bp; 0586: 195 bp; 0587: 218 bp; 0588: 210 bp; 0589: 209 bp). The amplicons were checked by electrophoresis on a 2% (wt/vol) agarose gel in Tris-borate-EDTA buffer.
Cloning.
MMCAP2_0582 and MMCAP2_0583 coding sequences were retrieved from the MolliGen database (services.cbib.u-bordeaux2.fr/molligen/), and subsequently codon-optimized for expression in E. coli. The optimization process included conversion of the opal codon TGA to a tryptophan-encoding TGG, removal of the rho-independent terminators and maximization of the Codon Adaptation Index. Synthetic genes for the optimized MMCAP2_0582 and MMCAP2_0583 were purchased from Eurofins Genomics. The sequences were further modified by removing the N-terminal transmembrane domain of both proteins. The resulting coding sequences were cloned in the pET28a(+) vector (Novagen) using the In-Fusion HD Cloning kit (Clontech) according to the manufacturer’s protocol. Briefly, the coding sequences were amplified and flanked with 15-bp regions homologous to the cloning site in the vector. The vector was linearized using PCR. All of the PCR steps were performed on a C1000 thermal cycler (BioRad) using the Q5 Polymerase (NEB) according to the manufacturer’s protocols. The Ser727Ala mutant of MMCAP2_0582 (r-0582A) was constructed by modifying 1 bp in the codon 727, using the Q5 Site-directed Mutagenesis Kit (NEB) according to the manufacturer’s protocol. All primers were ordered from Eurogentec. All constructs were designed to contain a His tag at the N-terminal extremity for subsequent purification. Plasmid were maintained and propagated in E. coli DH5α.
Expression and Purification.
Recombinant MMCAP2_0582 and MMCAP2_0583 (respectively, r-0582, r-0582A and r-0583) were expressed in E. coli BL21(DE3) grown in Studier autoinduction medium at 24 °C, supplemented with the appropriate antibiotics (kanamycin: 100 µg⋅mL−1) in UltraYield flasks (Thomson Instrument Co.). Cells were harvested by centrifugation and resuspended in TBS buffer (140 mM NaCl, 3 mM KCl, 25 mM Tris, pH 7.4) with 10 mM imidazole. Cell lysis was achieved by sonication and the lysate was subsequently clarified by centrifugation. Protein purification was performed by immobilized metal ion affinity chromatography on a 1- mL HisTrap HP Nickel-NTA column (GE). Bound proteins were eluted with a gradient of imidazole in TBS buffer, and fractions were collected and analyzed by SDS/PAGE before pooling. Eluted proteins were concentrated by ultrafiltration (Vivaspin; 10-kDa molecular mass cutoff, PES membrane; Sartorius) and further purified and buffer-exchanged by gel filtration on a Superdex 200 column (GE) in PBS buffer pH 7.4. All of the purification steps were performed on an ÄKTApurifier FPLC system (GE) under the control of the UNICORN 4.0 software. Protein concentration was determined spectrophotometrically (OD280) with an Epoch plate reader coupled to a Take3 microvolume quantification plate (BioTek) using the extinction coefficient of each protein computed with the ProtParam tool (web.expasy.org/protparam/). Purified proteins were snap-frozen in liquid nitrogen and stored in aliquots at −20 °C until use.
Pull-Down Assays.
Animal sera were obtained from either Sigma or as gifts from academic research laboratories. His SpinTrap Nickel-NTA agarose beads (GE) were equilibrated with 10-column volume of PBS buffer (NaCl 137 mM, KCl 2.7 mM, Na2HPO4 10 mM, KH2PO4 1.8 mM, pH 7.4). The beads were coated with purified r-0583 in PBS buffer for 1 h, then washed with 10-column volume of PBS. Subsequently, the coated beads were incubated with animal serum diluted in PBS buffer for 2 h at room temperature. Unbound proteins were washed using 3 × 10-column volume of PBS, 20 mM imidazole. Bound proteins were eluted from the beads using 0.5-column volume of PBS, 250 mM imidazole. Eluted samples were checked by SDS/PAGE analysis.
Mass Spectrometry Identification.
The identity of the proteins present in the SDS/PAGE gel bands was determined using a LC-MS/MS analysis pipeline. Bands of interest were cut out from the gel and subsequently broken up in 1 × 1-mm gel pieces. Gel pieces were destained in 25 mM ammonium bicarbonate 50% (vol/vol) acetonitrile (ACN), rinsed twice in ultrapure water and shrunk in ACN for 10 min. Proteins were first reduced in 10 mM DTT, 100 mM NH4HCO3 for 30 min at 56 °C, then alkylated in 100 mM iodoacetamide, 100 mM NH4HCO3 for 30 min at room temperature and shrunk in ACN for 10 min. After ACN removal, gel pieces were rehydrated with 100 mM NH4HCO3 for 10 min at room temperature and shrunk again in ACN. After ACN removal, gel pieces were dried at room temperature, covered with the trypsin solution [10 ng/µL in 40 mM NH4HCO3 and 10% (vol/vol) ACN], rehydrated at 4 °C for 10 min, and finally incubated overnight at 37 °C. Gel pieces were incubated for 15 min in 40 mM NH4HCO3 and 10% (vol/vol) ACN at room temperature. The supernatant was collected, and an H2O/ACN/HCOOH (47.5:47.5:5) extraction solution was added onto gel slices for 15 min. The extraction step was repeated twice. Supernatants were pooled and dried in a vacuum centrifuge. Finally samples were suspended in 30 µL H2O/HCOOH (100:0.1). Peptide mixture was analyzed on an Ultimate 3000 nanoLC system (Dionex) coupled to a nanospray LTQ-Orbitrap XL mass spectrometer (Thermo Finnigan). Ten microliters of peptide digests were loaded onto a 300 µm (inner diameter) × 5 mm C18 PepMap trap column (LC Packings) at a flow rate of 20 µL/min. The peptides were eluted from the trap column onto an analytical 75 mm (inner diameter) × 15 cm C18 Pep-Map column (LC Packings) with a 2–40% linear gradient of solvent B in 48 min [solvent A was 0.1% formic acid in 5% (vol/vol) ACN, and solvent B was 0.1% formic acid in 80% (vol/vol) ACN]. The separation flow rate was set at 200 nL/min. The mass spectrometer operated in positive ion mode at a 1.8-kV needle voltage and a 42-V capillary voltage. Data were acquired in a data-dependent mode. MS scans (m/z 300–1,700) were recorded at a resolution of R = 60,000 (m/z 400) and an AGC target of 5 × 105 ions collected within 500 ms. Dynamic exclusion was set to 30 s and the top six ions were selected from fragmentation in collision-induced dissociation mode. MS/MS scans with a target value of 1 × 104 ions were collected with a maximum fill time of 200 ms. Data were searched by SEQUEST through Proteome Discoverer 1.4 (Thermo Fisher Scientific Inc.) against house-made databases containing amino acid sequences of Ig extracted from the abYsis database (www.bioinf.org.uk/abysis). Spectra from peptides higher than 5,000 Da or lower than 350 Da were rejected. The search parameters were as follows: mass accuracy of the monoisotopic peptide precursor and peptide fragments was set to 10 ppm and 0.6 Da, respectively. Only b and y ions were considered for mass calculation. Oxidation of methionines (+16 Da) and carbamidomethylation of cysteines (+57 Da) were considered as variable modifications. Two missed trypsin cleavages were allowed. Peptide validation was performed using Percolator algorithm and only “high-confidence” peptides were retained corresponding to a 1% false positive rate at peptide level.
Ig Cleavage Assay.
Purified goat IgG were purchased from Sigma, and F(ab′)2 and Fab fragments were purchased from Jackson ImmunoResearch. All Ig cleavage assays were performed at room temperature, by mixing all of the components in PBS buffer pH 7.4. The r-0582 was always added last to the mixture. Reaction products were checked by analyzing the samples by SDS/PAGE.
Western Blots.
Proteins separated by SDS/PAGE were transferred on a Protran BA85 nitrocellulose membrane (Whatman) using a TE-77 semidry transfer system (GE). After transfer, the membrane was saturated with blocking buffer [2% (wt/vol) BSA fraction V, Sigma, in PBS Tween 20 0.1%] for 1 h at room temperature. Primary antibody (mouse anti-goat IgG H+L; Jackson ImmunoResearch) diluted at 1:2,000 in blocking buffer was incubated with the membrane for 1 h. Unbound primary antibody was washed away with 3 × 30 mL of PBS Tween 20 0.1%, before incubation of the membrane with the secondary antibody (goat anti-mouse Fc HRP coupled) diluted at 1:20,000 in blocking buffer for 1 h. Unbound secondary antibody was washed away with 3 × 30 mL of PBS Tween 20 0.1%, before revelation. Antibody-coupled peroxidase activity was detected using the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) according to the manufacturer’s indications. Membrane imaging was performed on a Gel Doc XR+ system (BioRad).
Edman Sequencing.
The r-0582 cleavage site on goat IgG was determined by sequencing the N-terminal amino acids of the large heavy-chain fragment. The SDS/PAGE gel band containing the fragment was excised and incubated in a buffer to passively extract the protein from the acrylamide gel. After overnight incubation under shaking, the solution was eluted on a ProSorb Filter (Applied Biosystems) to fixate the protein on a PVDF disk. The N-terminal sequence of the protein was determined by introducing the PVDF disk into an Applied Biosystems 494 automated protein sequencer (Applied Biosystems). Runs of Edman degradation, 10 cycles of pulsed-liquid PVDF chemistry, were carried out.
Protein–Protein Interaction Analysis.
Qualitative and quantitative analysis of the interactions among r-0582A, r-0583, and goat IgG was performed by SPR on a Biacore T200 instrument, using CM5 S Series sensor chips (GE). Ligands were coupled to the chip surface using the Biacore amine coupling protocol, and analytes were injected at various concentrations at a flow of 20 µL/min. All of the analyses were performed at 30 °C in PBS (pH 7.4) Tween 20 0.05% buffer. For qualitative analysis, r-0583 or goat IgG were immobilized directly on the sensor chip. Weakly bound proteins were washed off with pulses of NaOH 20 mM, NaCl 1 M. Interactions were assessed by injecting the putative ligands 60 s. For quantitative analysis, a rabbit anti-goat IgG, Fc fragment specific (Jackson ImmunoResearch), was immobilized on the sensor chip. Weakly bound proteins were washed off with pulses of NaOH 20 mM, NaCl 1 M. Goat IgG was subsequently captured by the immobilized rabbit antibody. This capture method allowed efficient regeneration of the immobilized capture antibody using 60-s pulses of glycine 10 mM pH 1.5. Experimental controls were performed to validate that the r-0583 and r-0582 did not interact with the rabbit antibody. Kinetic parameters were determined by performing a three-step single-cycle kinetic analysis (41). The sensorgrams were process using Biacore T200 Evaluation Software 2.0 (Biacore). The association and dissociation rate constants, Ka and Kd, respectively, were determined by direct curve fitting of the sensorgrams to a Langmuir 1:1 model of interaction. The dissociation equilibrium constant, Kd, was calculated as Ka/Kd.
SEC-MALS.
The molecular weight of the various recombinant proteins and protein–protein complexes was determined using SEC coupled to a multiple-angle light-scattering detector. All experiments were performed in PBS pH 7.4 buffer. SEC was performed into a HPLCy system equipped with a LC-20AD pump, an autosampler SIL20-ACHT storing the samples at 4 °C, a communication interface CBM-20A (Shimadzu), using a Superose 6 10/300 GL column (GE), thermostated at 20 °C in an oven XL-Therm (WynSep), and equilibrated with PBS buffer pH 7.4 (ChemSolute Bio). Volumes of 40–90 µL were injected at a flow rate of 0.5 mL/min. The elution profiles were followed on-line at 280 nm (SPD-M20A Shimadzu) by refractive index and static light scattering using a laser emitting at 658 nm (Optilab rEX and miniDAWN TREOS detectors, respectively; Wyatt Technology). Data were analyzed using ASTRA V 5.3.4.20 software (Wyatt Technology). The basic equation for light scattering, restricted to diluted solutions of small particles (<20 nm), reduces to I = KcM × (∂n/∂c)2, where I is the excess intensity of scattered light, K is an optical parameter, c is the weight concentration, and ∂n/∂c is the refractive index increment. The concentration is evaluated online from the refractive index signal. From their amino acid composition and using Sedfit (https://sedfitsedphat.nibib.nih.gov/), we obtained ∂n/∂c of 0.186, 0.188, and 0.190 mL⋅g−1 for IgG, r-0583, and r-0582, respectively. Molar masses are given as mean over the half-height width of the elution peaks.
Acknowledgments
The authors thank Sybille Duret and Dr. Jan Naessens for their assistance, and Dr. Maude Ele-Deverttus for the discussions. Goat sera were kindly provided by Dr. Tardy. We thank the structural biophysico-chemistry facility (UMS 3033/US001) of the Institut Européen de Chimie et Biologie (Pessac, France) for access to the Biacore T200 instrument that was acquired with the support of the Conseil Régional d'Aquitaine, the Groupement d'Intérêt Scientifique–Infrastrutures en Biologie Sante et Agronomie, and the Cellule Hôtels à Projets of the CNRS. The platform of the Grenoble Instruct Center (Integrated Structural Biology Grenoble; UMS 3518 CNRS-CEA-UJF-EMBL) is supported by the French Infrastructure for Integrated Structural Biology (ANR-10-INSB-05-02) and Grenoble Alliance for Integrated Structural Cell Biology (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology. This work was funded by the National Science Foundation Basic Research to Enable Agricultural Development Program Grant 1110151. L.M. is supported by the SIRIC BRIO.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600546113/-/DCSupplemental.
References
- 1.Brown DR, Whitcomb RF, Bradbury JM. Revised minimal standards for description of new species of the class Mollicutes (division Tenericutes) Int J Syst Evol Microbiol. 2007;57(Pt 11):2703–2719. doi: 10.1099/ijs.0.64722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.May M, Balish MF, Blanchard A. 2014. The Prokaryotes: Firmicutes and Tenericutes, eds Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (Springer, Berlin), pp 515–550.
- 3.Sirand-Pugnet P, Citti C, Barré A, Blanchard A. Evolution of mollicutes: Down a bumpy road with twists and turns. Res Microbiol. 2007;158(10):754–766. doi: 10.1016/j.resmic.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Brown DR, et al. 2011. Genus I. mycoplasma. Bergey’s Manual of Systematic Bacteriology, ed Krieg NR, et al. (Springer, New York), 2nd Ed, Vol 4, pp 575–613.
- 5.Citti C, Blanchard A. Mycoplasmas and their host: Emerging and re-emerging minimal pathogens. Trends Microbiol. 2013;21(4):196–203. doi: 10.1016/j.tim.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Atkinson TP, Balish MF, Waites KB. Epidemiology, clinical manifestations, pathogenesis and laboratory detection of Mycoplasma pneumoniae infections. FEMS Microbiol Rev. 2008;32(6):956–973. doi: 10.1111/j.1574-6976.2008.00129.x. [DOI] [PubMed] [Google Scholar]
- 7.Taylor-Robinson D, Jensen JS. Mycoplasma genitalium: From chrysalis to multicolored butterfly. Clin Microbiol Rev. 2011;24(3):498–514. doi: 10.1128/CMR.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiaucourt F, Aboubakar Y, Wesonga H, Manso-Silvan L, Blanchard A. Contagious bovine pleuropneumonia vaccines and control strategies: recent data. Dev Biol (Basel) 2004;119:99–111. [PubMed] [Google Scholar]
- 9.Manso-Silván L, Dupuy V, Chu Y, Thiaucourt F. Multi-locus sequence analysis of Mycoplasma capricolum subsp. capripneumoniae for the molecular epidemiology of contagious caprine pleuropneumonia. Vet Res (Faisalabad) 2011;42:86. doi: 10.1186/1297-9716-42-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simionatto S, Marchioro SB, Maes D, Dellagostin OA. Mycoplasma hyopneumoniae: From disease to vaccine development. Vet Microbiol. 2013;165(3-4):234–242. doi: 10.1016/j.vetmic.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 11.Stipkovits L, Kempf I. Mycoplasmoses in poultry. Rev Sci Tech. 1996;15(4):1495–1525. doi: 10.20506/rst.15.4.986. [DOI] [PubMed] [Google Scholar]
- 12.Mattoo S, Lee YM, Dixon JE. Interactions of bacterial effector proteins with host proteins. Curr Opin Immunol. 2007;19(4):392–401. doi: 10.1016/j.coi.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins C, Samudrala R, Geary SJ, Djordjevic SP. Structural and functional characterization of an organic hydroperoxide resistance protein from Mycoplasma gallisepticum. J Bacteriol. 2008;190(6):2206–2216. doi: 10.1128/JB.01685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidl SR, et al. A trigger enzyme in Mycoplasma pneumoniae: Impact of the glycerophosphodiesterase GlpQ on virulence and gene expression. PLoS Pathog. 2011;7(9):e1002263. doi: 10.1371/journal.ppat.1002263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo W, et al. Association of Mycoplasma arthritidis mitogen with lethal toxicity but not with arthritis in mice. Infect Immun. 2008;76(11):4989–4998. doi: 10.1128/IAI.00667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker A, et al. Structure of CARDS toxin, a unique ADP-ribosylating and vacuolating cytotoxin from Mycoplasma pneumoniae. Proc Natl Acad Sci USA. 2015;112(16):5165–5170. doi: 10.1073/pnas.1420308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons WL, Dybvig K. How some mycoplasmas evade host immune responses. Microbe Mag. 2007;2(11):537–543. [Google Scholar]
- 18.Rosengarten R, Wise KS. Phenotypic switching in mycoplasmas: Phase variation of diverse surface lipoproteins. Science. 1990;247(4940):315–318. doi: 10.1126/science.1688663. [DOI] [PubMed] [Google Scholar]
- 19.Ron Y, Flitman-Tene R, Dybvig K, Yogev D. Identification and characterization of a site-specific tyrosine recombinase within the variable loci of Mycoplasma bovis, Mycoplasma pulmonis and Mycoplasma agalactiae. Gene. 2002;292(1-2):205–211. doi: 10.1016/s0378-1119(02)00679-0. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs E, Bartl A, Oberle K, Schiltz E. Molecular mimicry by Mycoplasma pneumoniae to evade the induction of adherence inhibiting antibodies. J Med Microbiol. 1995;43(6):422–429. doi: 10.1099/00222615-43-6-422. [DOI] [PubMed] [Google Scholar]
- 21.Spooner RK, Russell WC, Thirkell D. Characterization of the immunoglobulin A protease of Ureaplasma urealyticum. Infect Immun. 1992;60(6):2544–2546. doi: 10.1128/iai.60.6.2544-2546.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paralanov V, et al. Comparative genome analysis of 19 Ureaplasma urealyticum and Ureaplasma parvum strains. BMC Microbiol. 2012;12:88. doi: 10.1186/1471-2180-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robertson JA, Stemler ME, Stemke GW. Immunoglobulin A protease activity of Ureaplasma urealyticum. J Clin Microbiol. 1984;19(2):255–258. doi: 10.1128/jcm.19.2.255-258.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cizelj I, et al. Mycoplasma gallisepticum and Mycoplasma synoviae express a cysteine protease CysP, which can cleave chicken IgG into Fab and Fc. Microbiology. 2011;157(Pt 2):362–372. doi: 10.1099/mic.0.045641-0. [DOI] [PubMed] [Google Scholar]
- 25.Grover RK, et al. A structurally distinct human mycoplasma protein that generically blocks antigen-antibody union. Science. 2014;343(6171):656–661. doi: 10.1126/science.1246135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez L, et al. Mycoplasma mycoides subsp. capri associated with goat respiratory disease and high flock mortality. Can Vet J. 2006;47(4):366–369. [PMC free article] [PubMed] [Google Scholar]
- 27.Remmert M, Biegert A, Hauser A, Söding J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9(2):173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 28.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102(12):4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 29.Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J Innate Immun. 2009;1(2):70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mistry D, Stockley RA. IgA1 protease. Int J Biochem Cell Biol. 2006;38(8):1244–1248. doi: 10.1016/j.biocel.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brezski RJ, Jordan RE. Cleavage of IgGs by proteases associated with invasive diseases: An evasion tactic against host immunity? MAbs. 2010;2(3):212–220. doi: 10.4161/mabs.2.3.11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horne C, Klein M, Polidoulis I, Dorrington KJ. Noncovalent association of heavy and light chains of human immunoglobulins. III. Specific interactions between VH and VL. J Immunol. 1982;129(2):660–664. [PubMed] [Google Scholar]
- 33.Röthlisberger D, Honegger A, Plückthun A. Domain interactions in the Fab fragment: A comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J Mol Biol. 2005;347(4):773–789. doi: 10.1016/j.jmb.2005.01.053. [DOI] [PubMed] [Google Scholar]
- 34.Bujotzek A, et al. Prediction of VH-VL domain orientation for antibody variable domain modeling. Proteins. 2015;83(4):681–695. doi: 10.1002/prot.24756. [DOI] [PubMed] [Google Scholar]
- 35.Ohno S, Mori N, Matsunaga T. Antigen-binding specificities of antibodies are primarily determined by seven residues of VH. Proc Natl Acad Sci USA. 1985;82(9):2945–2949. doi: 10.1073/pnas.82.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirai H, Kidera A, Nakamura H. H3-rules: Identification of CDR-H3 structures in antibodies. FEBS Lett. 1999;455(1-2):188–197. doi: 10.1016/s0014-5793(99)00821-2. [DOI] [PubMed] [Google Scholar]
- 37.Weldearegay YB, et al. Proteomic characterization of pleural effusion, a specific host niche of Mycoplasma mycoides subsp. mycoides from cattle with contagious bovine pleuropneumonia (CBPP) J Proteomics. 2016;131:93–103. doi: 10.1016/j.jprot.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 38.Krasteva I, et al. Characterization of the in vitro core surface proteome of Mycoplasma mycoides subsp. mycoides, the causative agent of contagious bovine pleuropneumonia. Vet Microbiol. 2014;168(1):116–123. doi: 10.1016/j.vetmic.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 39.Sirand-Pugnet P, et al. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet. 2007;3(5):e75. doi: 10.1371/journal.pgen.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pereyre S, et al. Life on arginine for Mycoplasma hominis: Clues from its minimal genome and comparison with other human urogenital mycoplasmas. PLoS Genet. 2009;5(10):e1000677. doi: 10.1371/journal.pgen.1000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palau W, Di Primo C. Simulated single-cycle kinetics improves the design of surface plasmon resonance assays. Talanta. 2013;114:211–216. doi: 10.1016/j.talanta.2013.04.022. [DOI] [PubMed] [Google Scholar]