Significance
Neurodegenerative diseases such as Alzheimer’s (AD), Parkinson’s (PD), and Huntington’s (HD) present a significant and increasing burden on society. Perturbations in the kynurenine pathway (KP) of tryptophan degradation have been linked to the pathogenesis of these disorders, and thus manipulation of this pathway may have therapeutic relevance. Here we show that genetic inhibition of two KP enzymes—kynurenine-3-monooxygenase and tryptophan-2,3-dioxygenase (TDO)—improved neurodegeneration and other disease symptoms in fruit fly models of AD, PD, and HD, and that alterations in levels of neuroactive KP metabolites likely underlie the beneficial effects. Furthermore, we find that inhibition of TDO using a drug-like compound reverses several disease phenotypes, underscoring the therapeutic promise of targeting this pathway in neurodegenerative disease.
Keywords: neurodegeneration, KMO, TDO, Parkinson’s disease, Alzheimer’s disease
Abstract
Metabolites of the kynurenine pathway (KP) of tryptophan (TRP) degradation have been closely linked to the pathogenesis of several neurodegenerative disorders. Recent work has highlighted the therapeutic potential of inhibiting two critical regulatory enzymes in this pathway—kynurenine-3-monooxygenase (KMO) and tryptophan-2,3-dioxygenase (TDO). Much evidence indicates that the efficacy of KMO inhibition arises from normalizing an imbalance between neurotoxic [3-hydroxykynurenine (3-HK); quinolinic acid (QUIN)] and neuroprotective [kynurenic acid (KYNA)] KP metabolites. However, it is not clear if TDO inhibition is protective via a similar mechanism or if this is instead due to increased levels of TRP—the substrate of TDO. Here, we find that increased levels of KYNA relative to 3-HK are likely central to the protection conferred by TDO inhibition in a fruit fly model of Huntington’s disease and that TRP treatment strongly reduces neurodegeneration by shifting KP flux toward KYNA synthesis. In fly models of Alzheimer’s and Parkinson’s disease, we provide genetic evidence that inhibition of TDO or KMO improves locomotor performance and ameliorates shortened life span, as well as reducing neurodegeneration in Alzheimer's model flies. Critically, we find that treatment with a chemical TDO inhibitor is robustly protective in these models. Consequently, our work strongly supports targeting of the KP as a potential treatment strategy for several major neurodegenerative disorders and suggests that alterations in the levels of neuroactive KP metabolites could underlie several therapeutic benefits.
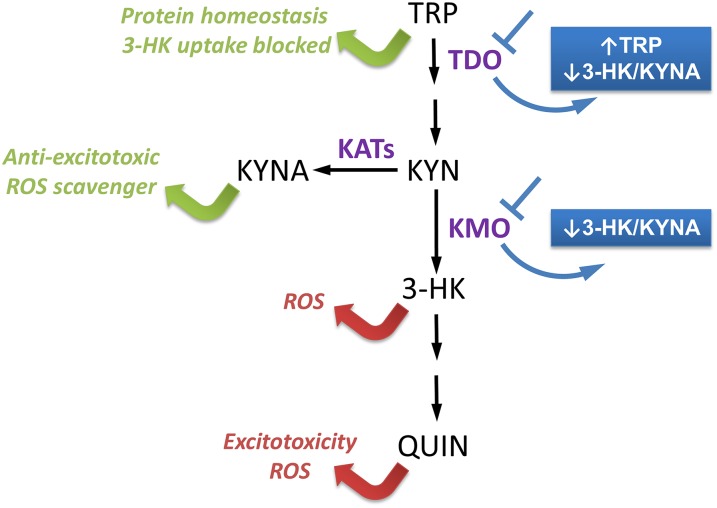
The kynurenine pathway (KP), the major catabolic route of tryptophan (TRP) metabolism in mammals (Fig. 1), has been closely linked to the pathogenesis of several brain disorders (1). This pathway contains several neuroactive metabolites, including 3-hydroxykynurenine (3-HK), quinolinic acid (QUIN) and kynurenic acid (KYNA) (2). QUIN is a well-characterized endogenous neurotoxin that specifically activates N-methyl-D-aspartate (NMDA) receptors, thereby inducing excitotoxicity (3, 4). The metabolites 3-HK and QUIN are also neurotoxic via the generation of free radicals and oxidative stress (5, 6). Conversely, KYNA—synthesized by kynurenine aminotransferases (KATs)—is neuroprotective through its antioxidant properties and antagonism of both the α7 nicotinic acetylcholine receptor and the glycine coagonist site of the NMDA receptor (7–13). Levels of these metabolites are regulated at two critical points in the KP: (i) the initial, rate-limiting conversion of TRP into N-formylkynurenine by either tryptophan-2,3-dioxygenase (TDO) or indoleamine-2,3-dioxygenase 1 and 2 (IDO1 and IDO2); and (ii) synthesis of 3-HK from kynurenine by the flavoprotein kynurenine-3-monoxygenase (KMO) (1).
Fig. 1.
Consequences of KP manipulation. KP metabolites and enzymatic steps are indicated in black, whereas the key KP enzymes TDO, KMO, and KATs are indicated in purple. The metabolites 3-HK and QUIN are neurotoxic (as indicated by red arrows), whereas KYNA and TRP are neuroprotective (as indicated by green arrows). Inhibition of TDO results in increased TRP levels, and either TDO or KMO inhibition leads to a reduction in the 3-HK/KYNA ratio (highlighted in blue). The enzyme 3-hydroxyanthranilic acid dioxygenase is not present in flies, and thus QUIN is not synthesized.
Alterations in levels of the KP metabolites have been observed in a broad range of brain disorders, including both neurodegenerative and psychiatric conditions (14). In neurodegenerative diseases such as Huntington’s (HD), Parkinson’s (PD), and Alzheimer’s (AD), a shift toward increased synthesis of the neurotoxic metabolites QUIN and 3-HK relative to KYNA may contribute to disease (1). Indeed, in patients with HD and HD model mice, 3-HK and QUIN levels are increased in the neostriatum and cortex (15, 16). Moreover, KYNA levels are reduced in the striatum of patients with HD (17). Several studies have also found perturbation in KP metabolites in the blood and cerebrospinal fluid of patients with AD, with decreased levels of KYNA correlating with reduced cognitive performance (18, 19). Similarly, in the basal ganglia of patients with PD, a reduction in KYNA levels combined with increased 3-HK has been observed (20, 21).
Drosophila melanogaster has provided a useful model for interrogation of the KP in both normal physiology and in neurodegenerative disease (22, 23). In fruit flies, TDO and KMO are encoded by vermillion (v) and cinnabar (cn), respectively, and both are implicated in Drosophila eye color pigmentation and brain plasticity (24, 25). In flies, TDO is the sole enzyme that catalyzes the initial step of the KP, as IDO1 and IDO2 are not present (Fig. 1), and so provides a distinctive model for examining the role of this critical step in the pathway. Moreover, we have previously found that downregulating cn and v gene expression significantly reduces neurodegeneration in flies expressing a mutant huntingtin (HTT) fragment—the central causative insult underlying HD (22). We also observed that pharmacological manipulations that reduced the 3-HK/KYNA ratio were always associated with neuroprotection. Notably, reintroduction of physiological levels of 3-HK in HD flies that lacked this metabolite due to KMO inhibition was sufficient to abolish neuroprotection (22). Furthermore, in a Caenorhabditis elegans model of PD, genetic down-regulation of TDO ameliorates α-synuclein (aSyn) toxicity (26). This effect appeared to be independent of changes in the levels of serotonin or KP metabolites but was correlated with increased TRP levels. Supplementing worms with TRP also suppressed aSyn-dependent phenotypes (26). The present study was designed to further define the mechanism(s) that underlies the neuroprotection conferred by TRP treatment and TDO inhibition and to extend our analyses of the neuroprotective potential of the KP to fruit fly models of AD and PD.
Results
TRP Is Neuroprotective in HD Flies via Modulation of Downstream KP Metabolites.
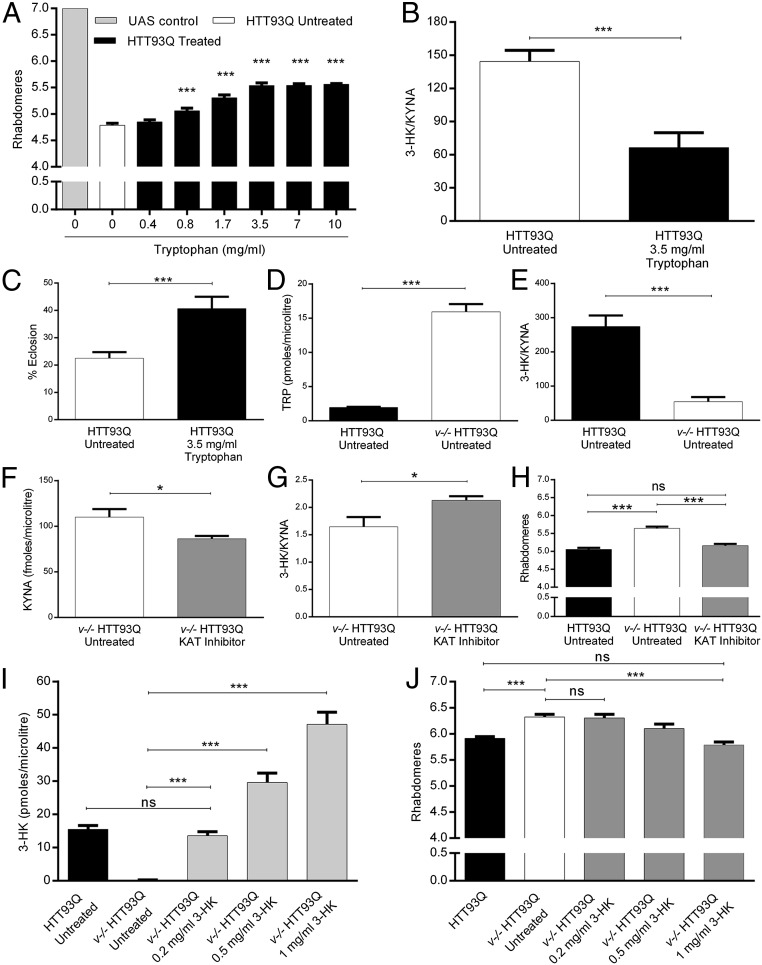
As work in C. elegans suggests that TDO inhibition and TRP treatment may confer protection against toxicity arising from misfolded proteins independent of KP metabolites (26), here we investigated whether alterations in KP metabolites were central to this protection in HD flies. These flies feature the panneuronal expression of a mutant HTT exon 1 encoding fragment (HTT93Q) under control of the elavGAL4 panneuronal driver, and serve as a well-characterized model of HD (27). In particular, degeneration of photoreceptor neurons (rhabdomeres) in the eye serves as a robust and reproducible readout for neurodegeneration, which can easily be scored using the pseudopupil assay. HTT93Q flies were allowed to develop on media supplemented with various concentrations of TRP (from 0.4 to 10 mg/mL), and neurodegeneration was assessed at day 0 on newly emerged flies. TRP supplementation resulted in a dose-dependent amelioration of neurodegeneration compared with untreated controls, with 0.8 mg/mL being the minimum protective concentration (P < 0.001), and the protection saturating at 3.5 mg/mL TRP (P < 0.001; Fig. 2A). To assess whether TRP-induced neuroprotection was dependent upon changes in downstream neuroactive KP metabolites, we next determined the levels of the neurotoxic metabolite 3-HK and the neuroprotective metabolite KYNA. TRP treatment of HTT93Q flies substantially reduced levels of 3-HK relative to KYNA (P < 0.001; Fig. 2B), predominantly driven by increased levels of KYNA (SI Appendix, Fig. S1 A and B). Furthermore, we observed that the low level of emergence of adult HD flies from the pupal case (eclosion; SI Appendix, Fig. S1C) was significantly enhanced by feeding of 3.5 mg/mL TRP (P < 0.001; Fig. 2C). These data suggest that the neuroprotection conferred by TRP treatment is due—at least in part—to increased levels of KYNA in these flies.
Fig. 2.
TRP feeding ameliorates HTT93Q toxicity in fruit flies. (A) Rhabdomere quantification of HD flies treated with different concentrations of TRP during development. TRP concentrations higher than 0.4 mg/mL significantly ameliorate rhabdomere neurodegeneration. n = 6–13 per condition, ***P < 0.001. (B) The 3-HK/KYNA ratio is reduced in TRP-fed HTT93Q flies. n = 5–6 per condition, ***P < 0.001. (C) TRP feeding rescues HTT93Q-dependent eclosion defects. Untreated HD flies: n = 938; TRP-treated HD flies: n = 728, ***P < 0.001. (D) TRP levels are significantly increased in v−/− HTT93Q flies compared with HTT93Q flies. n = 5 per condition, ***P < 0.001. (E) 3-HK/KYNA levels are reduced in v−/− HTT93Q compared with HTT93Q flies. n = 5 flies per condition, ***P < 0.001. (F) Treatment with the KAT inhibitor AOAA (100 μM in the food) reduces the level of KYNA in v−/− HTT93Q flies. n = 5 per condition. *P < 0.05. (G) AOAA treatment leads to a reduction in the 3-HK/KYNA ratio. n = 5 per condition, *P < 0.05. (H) KAT inhibition abrogates the neuroprotection conferred by the v mutation. n = 12–14 per condition, ***P < 0.001; ns, not significant. (I) Feeding of 3-HK leads to increased levels of 3-HK in v−/− HTT93Q flies. n = 5–6 per treatment. ***P < 0.001; ns, not significant. (J) Supplementation of 3-HK in the food of v−/− HTT93Q flies reduces neuroprotection compared with untreated HD flies. n = 8–12 per condition, ***P < 0.001; ns, not significant. Data are the mean ± SEM (one-way ANOVA with Newman–Keuls post hoc test).
We next explored the mechanism(s) by which TDO inhibition leads to neuroprotection. First, HTT93Q flies carrying a strong amorphic allele of v (v36f) were used to assess the role of KYNA. These v−/− HTT93Q flies exhibit a dramatic approximately eightfold increase in TRP levels compared with controls (P < 0.001; Fig. 2D), as well as a significant ∼80% reduction in the 3-HK/KYNA ratio (P < 0.001; Fig. 2E and SI Appendix, Fig. S1 D and E). To reduce levels of KYNA in the HTT93Q v−/− background, we used the nonspecific KAT inhibitor aminooxyacetic acid (AOAA), which effectively reduces KYNA synthesis in rodents in vitro and in vivo (28, 29). Animals administered 100 µM of AOAA in their food exhibited a significant decrease in KYNA levels (P < 0.05; Fig. 2F), which resulted in an increase in the 3-HK/KYNA ratio (P < 0.05; Fig. 2G). Strikingly, these animals showed a complete reversal of the neuroprotection conferred by TDO inhibition (P < 0.001; Fig. 2H). No changes were seen in levels of TRP or 3-HK (SI Appendix, Fig. S1 F and G), so these findings strongly suggest that KYNA is central to the neuroprotection observed.
We next asked whether modulation in 3-HK levels also plays a role in the neuroprotection observed in TDO-deficient flies, which have greatly reduced 3-HK levels (22) (Fig. 2I). In line with our demonstration that reintroduction of 3-HK in KMO-deficient flies (cn−/−) is sufficient to restore neurodegeneration (22), we administered several concentrations of 3-HK (0.2–1 mg/mL) to the flies in the food (Fig. 2 I and J). Surprisingly, we found that—unlike cn−/− HTT93Q flies (22)—restoration of physiological levels of 3-HK in v−/− HTT93Q flies (at the 0.2 mg/mL dose) did not reverse neuroprotection (Fig. 2J). Increasing 3-HK to hyperphysiological levels via administration of 1 mg/mL 3-HK enhanced neurodegeneration in v−/− HTT93Q flies, thereby eliminating the neuroprotection normally observed (P < 0.001; Fig. 2J). In all cases, we found that 3-HK treatment led to significant increases in the 3-HK/KYNA ratio (SI Appendix, Fig. S1 H and I). Thus, restoration of physiological 3-HK levels is not sufficient to abrogate the neuroprotection conferred in TDO-deficient flies.
The Excitotoxin QUIN Promotes Neurodegeneration in Drosophila.
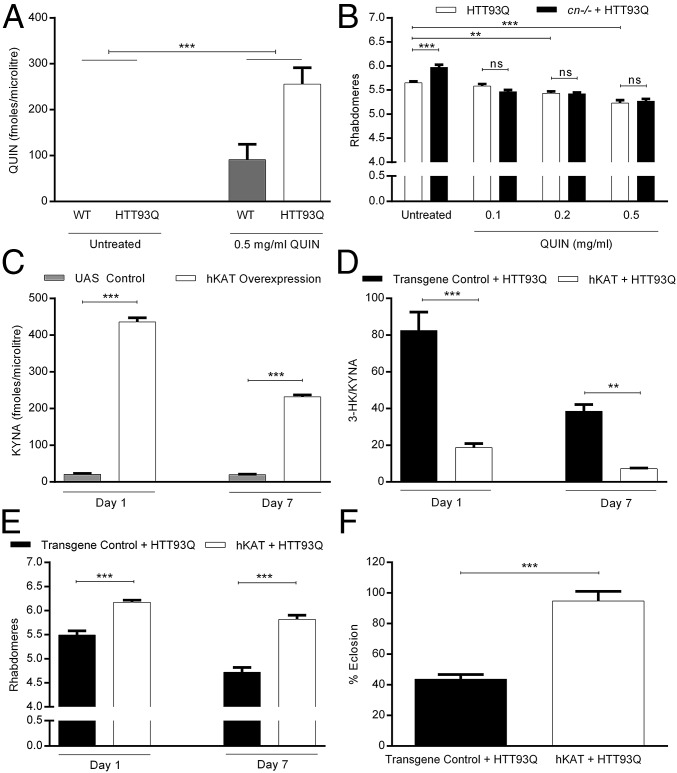
Drosophila do not express the KP enzyme 3-hydroxyanthranilic acid dioxygenase, and thus fruit flies do not synthesize QUIN (30). Therefore, we fed elavGAL4-driven HTT93Q flies with increasing QUIN concentrations during development and assessed neurodegeneration by scoring the number of rhabdomeres at day 0. We first measured QUIN in wild-type (WT) and HD fly heads. As expected, QUIN was detected in flies fed 0.5 mg/mL QUIN, but not in the untreated group (P < 0.001; Fig. 3A). Interestingly, we found that HTT93Q flies accumulate more QUIN than WT flies (P < 0.01). Whereas QUIN feeding did not cause degeneration of rhabdomeres in WT flies (SI Appendix, Fig. S2A), QUIN treatment (0.2 and 0.5 mg/mL) enhanced neurodegeneration in HTT93Q flies in a dose-dependent manner (Fig. 3B). Notably, KMO inhibition did not protect against QUIN-induced neurotoxicity in HTT93Q flies carrying homozygous cn3 mutation, a strong amophic cn allele (Fig. 3B).
Fig. 3.
QUIN exacerbates neurodegeneration in HD flies and overexpression of hKAT is neuroprotective via increased KYNA levels. (A) QUIN levels in WT and HTT93Q-expressing flies. QUIN is detected in flies fed with 0.5 mg/mL of QUIN, but was not measurable in untreated flies. n = 3–5 flies per treatment, ***P < 0.001. (B) HTT93Q and cn−/− HTT93Q flies fed QUIN exhibit increased rhabdomere degeneration compared with untreated flies. Neuroprotection conferred by the cn mutation is abolished by QUIN feeding. n = 11–12 per treatment, **P < 0.01, ***P < 0.001. (C) Panneuronal overexpression of hKAT in a WT background causes an increase in KYNA production compared with controls at both posteclosion ages tested. n = 3–5 per genotype, ***P < 0.001. (D) HTT93Q flies with panneuronal overexpression of hKAT show a significant reduction in the 3-HK/KYNA ratio. The transgene control used in this experiment was a transgenic Drosophila line expressing an empty pJFRC2 vector. n = 4–5 per condition, **P < 0.01, ***P < 0.001. (E) Overexpression of hKAT is neuroprotective in HTT93Q flies at both posteclosion ages tested. n = 9–13 flies per condition, ***P < 0.001. (F) Overexpression of hKAT ameliorates the eclosion phenotype observed in HTT93Q flies. Transgene control + Htt93Q flies: n = 1084; hKAT + Htt93Q flies: n = 1,010, ***P < 0.001; ns, not significant. Data are the mean ± SEM (one-way ANOVA with Newman–Keuls post hoc test).
Endogenous Synthesis of KYNA in Fruit Flies Is Neuroprotective.
We next generated a Drosophila line carrying a transgene encoding hKAT (UAS-hKAT), the KP enzyme that converts kynurenine to KYNA (Fig. 1). In wild-type flies, panneuronal hKAT expression driven by elavGAL4 caused a dramatic increase in KYNA levels at both day 1 and day 7 compared with controls (P < 0.001; Fig. 3C), and in HD flies, dramatically reduced the 3-HK/KYNA ratio at day 1 and day 7 posteclosion (P < 0.01; Fig. 3D and SI Appendix, Fig. S2 B and C). This effect was associated with a significant amelioration of both rhabdomere neurodegeneration (P < 0.001; Fig. 3E) and impaired eclosion in HTT93Q flies (P < 0.001; Fig. 3F).
KMO and TDO Inhibition Ameliorates Disease Phenotypes in Fly Models of PD and AD.
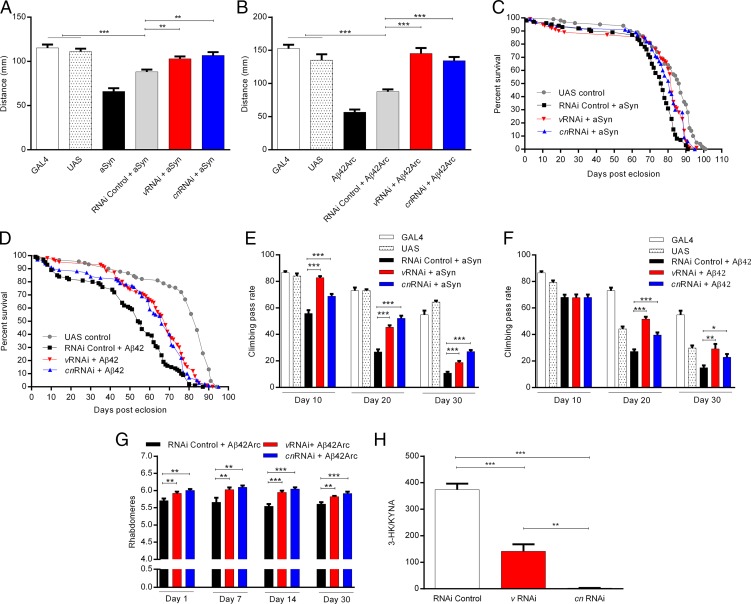
As both KMO and TDO inhibition were robustly protective in HD model fruit flies, we tested the efficacy of these approaches in Drosophila models of PD and AD. For recapitulating these disorders, we used transgenic fly lines expressing human aSyn as a model of PD (31) and the human Aβ42 peptide [either WT or the Arctic mutant form (E693G), which causes autosomal dominant AD, Aβ42Arc] as a model of AD (32, 33). We used RNA interference (RNAi) to down-regulate expression of the genes encoding either TDO (v) or KMO (cn) and found a dramatic reduction in the 3-HK/KYNA ratio, mainly due to increased synthesis of KYNA (P < 0.001; Fig. 4H and SI Appendix, Fig. S3 B and C). TRP levels were not significantly altered by these manipulations (SI Appendix, Fig. S3A), which reduce cn and v expression by ∼80–95% (22).
Fig. 4.
v and cn down-regulation ameliorates PD- and AD-related impairments in Drosophila. Expression of aSyn (A) or Aβ42ARC (B) in motorneurons using the c164GAL4 driver reduces the distance crawled by third instar larvae. The silencing of v or cn significantly ameliorates these locomotor defects. n = 20 larvae per genotype. **P < 0.01 and ***P < 0.001. Panneuronal expression of aSyn (C) or Aβ42 (D) reduces average life span, which is reversed by v and cn silencing. n = 100 per genotype. Median survival in days for aSyn experiments: UAS control = 86; RNAi control + aSyn = 76; vRNAi + aSyn = 82; cnRNAi + aSyn = 81. Median survival in days for Aβ42 experiments: UAS control = 84; RNAi control + Aβ42 = 54; vRNAi + Aβ42 = 68; cnRNAi + Aβ42 = 66. (E and F) Mean climbing pass rate at different posteclosion ages for flies expressing aSyn or Aβ42 panneuronally. Both aSyn (E) and Aβ42 (F) reduce climbing performance, and the effects are reversed by down-regulation of v and cn. n = 50–60 per genotype and condition. *P < 0.05, **P < 0.01, and ***P < 0.001. Panneuronal expression of Aβ42Arc reduces mean rhabdomeres per ommatidium (G); v and cn silencing protects rhabdomere degeneration at all posteclosion ages tested. n = 7–11 per condition. **P < 0.01 and ***P < 0.001. (H) The 3-HK/KYNA ratio is decreased in flies with RNAi down-regulation of v and cn. n = 5 per genotype. **P < 0.01 and ***P < 0.001. Data are the mean ± SEM (one-way ANOVA with Newman–Keuls post hoc test).
For both models, we first assessed larval crawling as an indication of behavioral impairments during early developmental stages (34). Expression of either aSyn or Aβ42Arc in motor neurons using the c164GAL4 driver led to a reduction in the distance crawled by third instar larvae (P < 0.001; Fig. 4 A and B, respectively). The down-regulation of the genes encoding either TDO (v) or KMO (cn) by RNAi significantly enhanced crawling behavior in both models (aSyn, P < 0.01; Aβ42Arc, P < 0.001).
As the expression of aSyn shortens life span compared with control flies (35), we next assessed life span upon panneuronal expression of either aSyn or Aβ42 constructs. We observed a small but significant improvement in the shortening of median life span in aSyn flies in which either TDO (v) or KMO (cn) had been silenced (P < 0.001, Fig. 4C). Notably, silencing of either of the two enzymes also significantly ameliorated shortened median life span in Aβ42 flies from 54 d to 66 d and 68 d, respectively (P < 0.001, Fig. 4D). Complementing this observation, cn down-regulation also significantly reversed shortened life span in the Aβ42Arc model (P < 0.001, SI Appendix, Fig. S4A).
Locomotor behavior in adult flies was assessed by examining negative geotaxis ability (climbing) as a measurement of motor impairment. elavGAL4-driven aSyn, Aβ42,and Aβ42Arc flies exhibited a reduction in climbing at all of the posteclosion ages tested (Fig. 4 E and F and SI Appendix, Fig. S4B). Silencing of either TDO or KMO improved climbing ability in all these models (Fig. 4 E and F and SI Appendix, Fig. S4B). Notably, scoring the number of rhabdomeres per ommatidium revealed that genetic knockdown of either enzyme also consistently reduced neurodegeneration in elavGAL4-driven Aβ42Arc flies at all ages tested (P < 0.01, Fig. 4G).
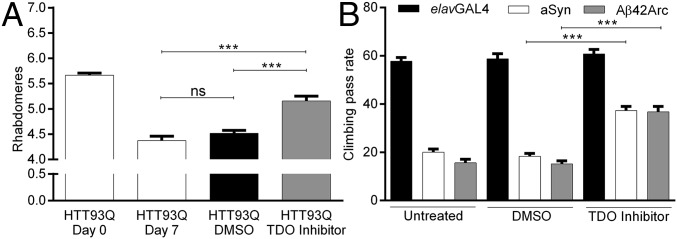
Finally, we interrogated the effect of pharmacological TDO inhibition in the three fly models of neurodegeneration, using the commercially available TDO inhibitor 680C91 (36). Feeding of 680C91 (100 µM) to newly emerged HTT93Q flies resulted in dramatically reduced neurodegeneration 7 d posteclosion compared with flies fed vehicle alone (Fig. 5A, P < 0.001). In PD and AD flies, pharmacological inhibition of TDO with 100 µM 680C91 led to a significant amelioration in climbing performance compared with respective controls 10 d posteclosion (Fig. 5B, P < 0.001).
Fig. 5.
Pharmacological inhibition of TDO is neuroprotective in HD, PD, and AD flies. (A) Reduced neurodegeneration in HTT93Q flies treated with the TDO inhibitor 680C91 (100 µM) 7 d posteclosion. n = 8–17 per condition. (B) aSyn and Aβ42Arc flies treated with 680C91 (100 µM) display improved climbing compared with controls. n = 50–60 per genotype. DMSO, dimethyl sulfoxide. ***P < 0.001; ns, not significant. Data are the mean ± SEM (one-way ANOVA with Newman–Keuls post hoc test).
Discussion
Impairments in KP metabolism have been linked to several neurodegenerative disorders, and in particular to the pathogenesis of HD (37). Notably, increased levels of 3-HK and QUIN have been measured in the neostriatum and cortex of patients with early stage HD (15), and these changes are associated with an up-regulation of IDO1 transcription (38) and a reduction in the activity of KAT, which is critical for KYNA synthesis (17). These data in patients with HD are supported by observations in HD mice, which show increased cerebral KMO activity (39). We previously found that either genetic or pharmacological inhibition of KMO is protective in HD flies and leads to a corresponding increase in KYNA levels relative to 3-HK (22). Furthermore, we reported that KYNA treatment reduced neurodegeneration in these flies. Here, we have extended this work by generating transgenic flies that overexpress hKAT and thereby synthesize ∼20-fold more KYNA than control flies. This increased formation of KYNA reduced neurodegeneration and eclosion defects in HD model flies. Furthermore, KMO inhibition by RNAi revealed beneficial effects in several behavioral and disease-relevant outcome measures, including larval crawling, longevity, climbing, and rhabdomere degeneration, in AD and PD model flies. These results strongly support the notion that KMO inhibition has relevance as a treatment strategy in a broad range of neurodegenerative diseases. In addition, these data also suggest that the design of small molecules capable of increasing KAT activity could have therapeutic relevance for neurodegenerative disorders.
The present results, demonstrating that both genetic and pharmacological inhibition of TDO provides robust neuroprotection in fly models of AD and PD, also confirmed and extended the results of our previous study, which had identified TDO as a candidate drug target in HD flies (22). These protective effects are associated with a decrease in the 3-HK/KYNA ratio, i.e., a shift toward increased KYNA synthesis. Work in C. elegans has revealed that TDO inhibition is also protective in models of proteotoxicity, although amelioration of the phenotypes occurred independently of changes in the levels of KP metabolites and was instead associated with elevated TRP levels (26). Although the underlying mechanism remained unclear, the favorable effects of high TRP levels in the nematode were substantiated by the fact that TRP treatment conferred robust protection from disease-related phenotypes (Fig. 1). In the present study, too, TRP supplementation of the diet was effective, ameliorating rhabdomere degeneration and eclosion defects in HD flies. However, TRP feeding was also associated with a reduction in the 3-HK/KYNA ratio, suggesting that the protective effects of the amino acid may be linked to an increase in the production of the neuroprotective metabolite KYNA (Fig. 1). Indeed, partial inhibition of KYNA synthesis in TDO-deficient flies proved sufficient to completely reverse neuroprotection. In addition, restoration of physiological 3-HK levels in TDO-deficient HD flies did not reverse neuroprotection, in contrast to KMO-deficient flies (22). In primary neurons, 3-HK toxicity is dependent upon its uptake via neutral amino acid transporters, and coapplication of TRP can block this toxicity by competing for the same transporters (6). Thus, it is possible that the vast excess of TRP observed in the heads of HTT93Q v−/− flies (approximately eightfold versus controls) competes with 3-HK for rhabdomere uptake, thereby requiring hyperphysiological levels of 3-HK to reverse TDO-dependent neuroprotection. A similar mechanism may also contribute to the neuroprotection observed with TRP treatment in general. Herein, we have also found that RNAi knockdown of either cn or v does not increase TRP levels, and thus the neuroprotection observed in the AD and PD flies strongly correlates with a decrease in the 3-HK/KYNA ratio. The mechanism causing TRP treatment to favor KYNA synthesis over the formation of 3-HK in Drosophila, as well as the unexpected qualitative differences in the effects of TDO inhibition and TRP administration on KP metabolism between fruit flies and nematodes, clearly requires further investigation.
Interestingly, we found that QUIN—which is not normally synthesized in fruit flies (30)—potentiated neurodegeneration in HD flies, and reversed the protective effects of KMO inhibition. As the same QUIN treatment did not cause neuron loss in wild-type flies, mutant HTT may potentiate vulnerability by enhancing NMDA receptor function (40, 41) and/or by increasing susceptibility to toxic free radicals (42), i.e., by augmenting the two major mechanisms known to be involved in QUIN-induced neurotoxicity (43). If verified in mammals, a reduction in brain QUIN levels—along with a decrease in 3-HK levels—relative to KYNA could therefore be especially promising in the treatment of HD (44). Our observation of increased levels of QUIN in HTT93Q versus WT flies is enigmatic, but may be due to altered feeding behavior, increased permeability of the blood–brain barrier (45, 46), or differences in KP metabolism, and would be interesting to explore in future studies.
In conclusion, the present set of experiments further validates the hypothesis that KP metabolism is causally linked to neuronal viability and that modulation of the KP constitutes a promising therapeutic strategy for a variety of major neurodegenerative disorders. Notably, we provide the first genetic evidence to our knowledge that KMO inhibition is protective in animal models of PD and AD and that pharmacological targeting of TDO is also neuroprotective. We have clarified the mechanism underlying the protective effects of TDO inhibition, which will stimulate efforts to target this step of the KP in neurodegenerative disease. These results, together with supportive studies in flies (47) and rodents (48), raise the possibility that inhibition of TDO and KMO—or combinatorial treatment—may offer therapeutic advantages. The availability of new TDO inhibitors (49, 50), and access to the crystal structures of both TDO (51) and KMO (52), should allow further testing of these hypotheses in the near future.
Materials and Methods
Fruit flies were maintained on standard maize food at 25 °C in a light/dark cycle of 12:12 h. The elavGAL4 [c155], w; +; UASaSyn (8146), w; +; UASAβ42 (32037), w; +; UASAβ42Arc (33774), cn3, and v36f null fly stocks were obtained from the Bloomington Drosophila Stock Center. The c164GAL4 driver line was a gift from Juan Botas, Baylor College of Medicine, Houston. HTT93Q exon 1 flies (27) were a gift from Larry Marsh and Leslie Thompson, University of California, Irvine. cn and v RNAi lines are part of the phiC31 RNAi Library (KK) and were obtained from the Vienna Drosophila RNAi Center (53).
The gene encoding kynurenine aminotransferase (hKAT) was amplified from a human fetal cDNA library (54) and cloned into the pJFRC2 vector (55)—a gift from Gerald Rubin (Addgene plasmid no. 26214)—by standard methods. The resulting construct was injected by BestGene into attP40 Drosophila strains (56).
Pseudopupil analysis, eclosion analysis, feeding experiments, measurement of KP metabolites, behavioral assays, longevity analysis, and statistical analyses are described in detail in SI Appendix, Materials and Methods. Measurement of KP metabolites in treated flies was performed at either 0 or 7 d posteclosion.
Supplementary Material
Acknowledgments
We thank J. Lawrence Marsh, Leslie Thompson, and Juan Botas for their transgenic fly lines and BestGene for the generation of transgenic lines. C.B. was supported by grants from the CHDI Foundation and Parkinson’s UK (to F.G. and C.P.K.). F.G. and C.P.K. also acknowledge grants from the Medical Research Council and the Biotechnology and Biological Sciences Research Council for valuable infrastructure supporting this work. Work in the R.S. laboratory was supported by NIH Grant R01-NS057715.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604453113/-/DCSupplemental.
References
- 1.Amaral M, Outeiro TF, Scrutton NS, Giorgini F. The causative role and therapeutic potential of the kynurenine pathway in neurodegenerative disease. J Mol Med (Berl) 2013;91(6):705–713. doi: 10.1007/s00109-013-1046-9. [DOI] [PubMed] [Google Scholar]
- 2.Thevandavakkam MA, Schwarcz R, Muchowski PJ, Giorgini F. Targeting kynurenine 3-monooxygenase (KMO): Implications for therapy in Huntington’s disease. CNS Neurol Disord Drug Targets. 2010;9(6):791–800. doi: 10.2174/187152710793237430. [DOI] [PubMed] [Google Scholar]
- 3.Stone TW, Perkins MN. Quinolinic acid: A potent endogenous excitant at amino acid receptors in CNS. Eur J Pharmacol. 1981;72(4):411–412. doi: 10.1016/0014-2999(81)90587-2. [DOI] [PubMed] [Google Scholar]
- 4.Schwarcz R, Whetsell WO, Jr, Mangano RM. Quinolinic acid: An endogenous metabolite that produces axon-sparing lesions in rat brain. Science. 1983;219(4582):316–318. doi: 10.1126/science.6849138. [DOI] [PubMed] [Google Scholar]
- 5.Okuda S, Nishiyama N, Saito H, Katsuki H. Hydrogen peroxide-mediated neuronal cell death induced by an endogenous neurotoxin, 3-hydroxykynurenine. Proc Natl Acad Sci USA. 1996;93(22):12553–12558. doi: 10.1073/pnas.93.22.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okuda S, Nishiyama N, Saito H, Katsuki H. 3-Hydroxykynurenine, an endogenous oxidative stress generator, causes neuronal cell death with apoptotic features and region selectivity. J Neurochem. 1998;70(1):299–307. doi: 10.1046/j.1471-4159.1998.70010299.x. [DOI] [PubMed] [Google Scholar]
- 7.Goda K, Hamane Y, Kishimoto R, Ogishi Y. Radical scavenging properties of tryptophan metabolites. Estimation of their radical reactivity. Adv Exp Med Biol. 1999;467:397–402. doi: 10.1007/978-1-4615-4709-9_50. [DOI] [PubMed] [Google Scholar]
- 8.Foster AC, Vezzani A, French ED, Schwarcz R. Kynurenic acid blocks neurotoxicity and seizures induced in rats by the related brain metabolite quinolinic acid. Neurosci Lett. 1984;48(3):273–278. doi: 10.1016/0304-3940(84)90050-8. [DOI] [PubMed] [Google Scholar]
- 9.Carpenedo R, et al. Presynaptic kynurenate-sensitive receptors inhibit glutamate release. Eur J Neurosci. 2001;13(11):2141–2147. doi: 10.1046/j.0953-816x.2001.01592.x. [DOI] [PubMed] [Google Scholar]
- 10.Hilmas C, et al. The brain metabolite kynurenic acid inhibits alpha7 nicotinic receptor activity and increases non-alpha7 nicotinic receptor expression: Physiopathological implications. J Neurosci. 2001;21(19):7463–7473. doi: 10.1523/JNEUROSCI.21-19-07463.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkins MN, Stone TW. An iontophoretic investigation of the actions of convulsant kynurenines and their interaction with the endogenous excitant quinolinic acid. Brain Res. 1982;247(1):184–187. doi: 10.1016/0006-8993(82)91048-4. [DOI] [PubMed] [Google Scholar]
- 12.Hardeland R, et al. Indole-3-pyruvic and -propionic acids, kynurenic acid, and related metabolites as luminophores and free-radical scavengers. Adv Exp Med Biol. 1999;467:389–395. doi: 10.1007/978-1-4615-4709-9_49. [DOI] [PubMed] [Google Scholar]
- 13.Lugo-Huitrón R, et al. On the antioxidant properties of kynurenic acid: Free radical scavenging activity and inhibition of oxidative stress. Neurotoxicol Teratol. 2011;33(5):538–547. doi: 10.1016/j.ntt.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Schwarcz R, Bruno JP, Muchowski PJ, Wu HQ. Kynurenines in the mammalian brain: When physiology meets pathology. Nat Rev Neurosci. 2012;13(7):465–477. doi: 10.1038/nrn3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guidetti P, Luthi-Carter RE, Augood SJ, Schwarcz R. Neostriatal and cortical quinolinate levels are increased in early grade Huntington’s disease. Neurobiol Dis. 2004;17(3):455–461. doi: 10.1016/j.nbd.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Guidetti P, et al. Elevated brain 3-hydroxykynurenine and quinolinate levels in Huntington disease mice. Neurobiol Dis. 2006;23(1):190–197. doi: 10.1016/j.nbd.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Jauch D, et al. Dysfunction of brain kynurenic acid metabolism in Huntington’s disease: Focus on kynurenine aminotransferases. J Neurol Sci. 1995;130(1):39–47. doi: 10.1016/0022-510x(94)00280-2. [DOI] [PubMed] [Google Scholar]
- 18.Hartai Z, et al. Decreased serum and red blood cell kynurenic acid levels in Alzheimer’s disease. Neurochem Int. 2007;50(2):308–313. doi: 10.1016/j.neuint.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Heyes MP, et al. Quinolinic acid and kynurenine pathway metabolism in inflammatory and non-inflammatory neurological disease. Brain. 1992;115(Pt 5):1249–1273. doi: 10.1093/brain/115.5.1249. [DOI] [PubMed] [Google Scholar]
- 20.Knyihár-Csillik E, et al. Effect of 6-hydroxydopamine treatment on kynurenine aminotransferase-I (KAT-I) immunoreactivity of neurons and glial cells in the rat substantia nigra. Acta Neuropathol. 2006;112(2):127–137. doi: 10.1007/s00401-006-0086-4. [DOI] [PubMed] [Google Scholar]
- 21.Ogawa T, et al. Kynurenine pathway abnormalities in Parkinson’s disease. Neurology. 1992;42(9):1702–1706. doi: 10.1212/wnl.42.9.1702. [DOI] [PubMed] [Google Scholar]
- 22.Campesan S, et al. The kynurenine pathway modulates neurodegeneration in a Drosophila model of Huntington’s disease. Curr Biol. 2011;21(11):961–966. doi: 10.1016/j.cub.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green EW, et al. Drosophila eye color mutants as therapeutic tools for Huntington disease. Fly (Austin) 2012;6(2):117–120. doi: 10.4161/fly.19999. [DOI] [PubMed] [Google Scholar]
- 24.Tearle R. Tissue specific effects of ommochrome pathway mutations in Drosophila melanogaster. Genet Res. 1991;57(3):257–266. doi: 10.1017/s0016672300029402. [DOI] [PubMed] [Google Scholar]
- 25.Savvateeva E, et al. Age-dependent memory loss, synaptic pathology and altered brain plasticity in the Drosophila mutant cardinal accumulating 3-hydroxy-kynurenine. J Neural Transm (Vienna) 2000;107(5):581–601. doi: 10.1007/s007020070080. [DOI] [PubMed] [Google Scholar]
- 26.van der Goot AT, et al. Delaying aging and the aging-associated decline in protein homeostasis by inhibition of tryptophan degradation. Proc Natl Acad Sci USA. 2012;109(37):14912–14917. doi: 10.1073/pnas.1203083109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413(6857):739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 28.Turski WA, Gramsbergen JB, Traitler H, Schwarcz R. Rat brain slices produce and liberate kynurenic acid upon exposure to L-kynurenine. J Neurochem. 1989;52(5):1629–1636. doi: 10.1111/j.1471-4159.1989.tb09218.x. [DOI] [PubMed] [Google Scholar]
- 29.Speciale C, et al. Determination of extracellular kynurenic acid in the striatum of unanesthetized rats: Effect of aminooxyacetic acid. Neurosci Lett. 1990;116(1-2):198–203. doi: 10.1016/0304-3940(90)90410-b. [DOI] [PubMed] [Google Scholar]
- 30.Linzen B. Tryptophan ommochrome pathway in insects. Adv Insect Physiol. 1974;10:117–246. [Google Scholar]
- 31.Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404(6776):394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 32.Murakami K, et al. Synthesis, aggregation, neurotoxicity, and secondary structure of various A beta 1-42 mutants of familial Alzheimer’s disease at positions 21-23. Biochem Biophys Res Commun. 2002;294(1):5–10. doi: 10.1016/S0006-291X(02)00430-8. [DOI] [PubMed] [Google Scholar]
- 33.Nilsberth C, et al. The ‘Arctic’ APP mutation (E693G) causes Alzheimer’s disease by enhanced Abeta protofibril formation. Nat Neurosci. 2001;4(9):887–893. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- 34.Nishimura Y, et al. Selection of behaviors and segmental coordination during larval locomotion is disrupted by nuclear polyglutamine inclusions in a new Drosophila Huntington’s disease-like model. J Neurogenet. 2010;24(4):194–206. doi: 10.3109/01677063.2010.514367. [DOI] [PubMed] [Google Scholar]
- 35.Breda C, et al. Rab11 modulates α-synuclein-mediated defects in synaptic transmission and behaviour. Hum Mol Genet. 2015;24(4):1077–1091. doi: 10.1093/hmg/ddu521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salter M, Hazelwood R, Pogson CI, Iyer R, Madge DJ. The effects of a novel and selective inhibitor of tryptophan 2,3-dioxygenase on tryptophan and serotonin metabolism in the rat. Biochem Pharmacol. 1995;49(10):1435–1442. doi: 10.1016/0006-2952(95)00006-l. [DOI] [PubMed] [Google Scholar]
- 37.Maddison DC, Giorgini F. The kynurenine pathway and neurodegenerative disease. Semin Cell Dev Biol. 2015;40:134–141. doi: 10.1016/j.semcdb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Mazarei G, et al. Expression analysis of novel striatal-enriched genes in Huntington disease. Hum Mol Genet. 2010;19(4):609–622. doi: 10.1093/hmg/ddp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sathyasaikumar KV, et al. Dysfunctional kynurenine pathway metabolism in the R6/2 mouse model of Huntington’s disease. J Neurochem. 2010;113(6):1416–1425. doi: 10.1111/j.1471-4159.2010.06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ultsch A, Schuster CM, Laube B, Betz H, Schmitt B. Glutamate receptors of Drosophila melanogaster. Primary structure of a putative NMDA receptor protein expressed in the head of the adult fly. FEBS Lett. 1993;324(2):171–177. doi: 10.1016/0014-5793(93)81387-f. [DOI] [PubMed] [Google Scholar]
- 41.Zachepilo TG, et al. Comparative analysis of the locations of the NR1 and NR2 NMDA receptor subunits in honeybee (Apis mellifera) and fruit fly (Drosophila melanogaster, Canton-S wild-type) cerebral ganglia. Neurosci Behav Physiol. 2008;38(4):369–372. doi: 10.1007/s11055-008-0052-9. [DOI] [PubMed] [Google Scholar]
- 42.Wang CT, et al. Reduced neuronal expression of ribose-5-phosphate isomerase enhances tolerance to oxidative stress, extends lifespan, and attenuates polyglutamine toxicity in Drosophila. Aging Cell. 2012;11(1):93–103. doi: 10.1111/j.1474-9726.2011.00762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pérez-De La Cruz V, Santamaria A. Integrative hypothesis for Huntington’s disease: A brief review of experimental evidence. Physiol Res. 2007;56(5):513–526. doi: 10.33549/physiolres.931049. [DOI] [PubMed] [Google Scholar]
- 44.Guidetti P, Schwarcz R. 3-Hydroxykynurenine potentiates quinolinate but not NMDA toxicity in the rat striatum. Eur J Neurosci. 1999;11(11):3857–3863. doi: 10.1046/j.1460-9568.1999.00806.x. [DOI] [PubMed] [Google Scholar]
- 45.Drouin-Ouellet J, et al. Cerebrovascular and blood-brain barrier impairments in Huntington’s disease: Potential implications for its pathophysiology. Ann Neurol. 2015;78(2):160–177. doi: 10.1002/ana.24406. [DOI] [PubMed] [Google Scholar]
- 46.Schirmeier S, Klämbt C. The Drosophila blood-brain barrier as interface between neurons and hemolymph. Mech Dev. 2015;138(Pt 1):50–55. doi: 10.1016/j.mod.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Oxenkrug GF, Navrotskaya V, Voroboyva L, Summergrad P. Extension of life span of Drosophila melanogaster by the inhibitors of tryptophan-kynurenine metabolism. Fly (Austin) 2011;5(4):307–309. doi: 10.4161/fly.5.4.18414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zwilling D, et al. Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell. 2011;145(6):863–874. doi: 10.1016/j.cell.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reniers J, et al. Synthesis and inhibition study of monoamine oxidase, indoleamine 2,3-dioxygenase and tryptophan 2,3-dioxygenase by 3,8-substituted 5H-indeno[1,2-c]pyridazin-5-one derivatives. Eur J Med Chem. 2011;46(12):6104–6111. doi: 10.1016/j.ejmech.2011.09.042. [DOI] [PubMed] [Google Scholar]
- 50.Dolusić E, et al. Tryptophan 2,3-dioxygenase (TDO) inhibitors. 3-(2-(pyridyl)ethenyl)indoles as potential anticancer immunomodulators. J Med Chem. 2011;54(15):5320–5334. doi: 10.1021/jm2006782. [DOI] [PubMed] [Google Scholar]
- 51.Huang W, Gong Z, Li J, Ding J. Crystal structure of Drosophila melanogaster tryptophan 2,3-dioxygenase reveals insights into substrate recognition and catalytic mechanism. J Struct Biol. 2013;181(3):291–299. doi: 10.1016/j.jsb.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 52.Amaral M, et al. Structural basis of kynurenine 3-monooxygenase inhibition. Nature. 2013;496(7445):382–385. doi: 10.1038/nature12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green EW, Fedele G, Giorgini F, Kyriacou CP. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat Methods. 2014;11(3):222–223. doi: 10.1038/nmeth.2856. [DOI] [PubMed] [Google Scholar]
- 54.Thaminy S, Auerbach D, Arnoldo A, Stagljar I. Identification of novel ErbB3-interacting factors using the split-ubiquitin membrane yeast two-hybrid system. Genome Res. 2003;13(7):1744–1753. doi: 10.1101/gr.1276503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186(2):735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40(4):476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.