Significance
Adoptive transfer of allogeneic natural killer (NK) cells into leukemia patients can lead to remission; however, therapies are hindered by inefficient expansion and limited persistence of these lymphocytes. We now report that Kruppel-like factor 2 (KLF2) regulates both NK cell proliferation and survival. KLF2 limits homeostatic expansion of NK cells in a cell-intrinsic manner. In addition, KLF2 instructs mature NK cells to home to IL-15–rich niches, which is necessary for continued survival under homeostatic conditions. Therefore, targeting KLF2 while providing rate-limiting survival factors such as transpresented IL-15 may improve NK cell engraftment and sustainability in cancer patients.
Keywords: NK cell, KLF2, NK cell proliferation, NK cell homeostasis, IL-15
Abstract
Natural killer (NK) cells are innate lymphocytes that recognize and lyse virally infected or transformed cells. This latter property is being pursued in clinics to treat leukemia with the hope that further breakthroughs in NK cell biology can extend treatments to other cancers. At issue is the ability to expand transferred NK cells and prolong their functionality within the context of a tumor. In terms of NK cell expansion and survival, we now report that Kruppel-like factor 2 (KLF2) is a key transcription factor that underpins both of these events. Excision of Klf2 using gene-targeted mouse models promotes spontaneous proliferation of immature NK cells in peripheral tissues, a phenotype that is replicated under ex vivo conditions. Moreover, KLF2 imprints a homeostatic migration pattern on mature NK cells that allows these cells to access IL-15–rich microenvironments. KLF2 accomplishes this feat within the mature NK cell lineage via regulation of a subset of homing receptors that respond to homeostatic ligands while leaving constitutively expressed receptors that recognize inflammatory cytokines unperturbed. Under steady-state conditions, KLF2-deficient NK cells alter their expression of homeostatic homing receptors and subsequently undergo apoptosis due to IL-15 starvation. This novel mechanism has implications regarding NK cell contraction following the termination of immune responses including the possibility that retention of an IL-15 transpresenting support system is key to extending NK cell activity in a tumor environment.
Natural killer (NK) cells are a subset of group 1 innate lymphoid cells (ILCs) that participate in viral and tumor clearance by directly lysing stressed cells and producing cytokines that recruit and activate effector leukocytes (1). Humans and mice that lack NK cells have increased incidence of cancer (2), and clinical trials have demonstrated that adoptively transferred allogeneic NK cells can improve patient outcome without contributing to graft-versus-host disease (3). Moreover, in vivo expansion and persistence of donor NK cells correlates with tumor clearance (4), which suggests that therapeutic efficacy can be enhanced by augmenting NK cell survival. Therefore, understanding basic mechanisms that support NK cell homeostasis has clinical implications in terms of cancer therapy.
Following the establishment of a diverse NK cell receptor repertoire, NK cells exit the bone marrow and circulate throughout peripheral tissues including the lungs, liver, gut, lymph nodes, blood, and splenic red pulp (5, 6). In mice, peripheral NK cell differentiation is further described in relation to CD11b and CD27 surface expression, progressing in maturity from CD27+CD11b− (stage 1) to CD27+CD11b+ (stage 2) to CD27−CD11b+ (stage 3) (7). With regard to peripheral homeostasis, early CD27+ NK cell stages are associated with IL-15–dependent proliferation (8, 9), whereas later CD11b+ stages require IL-15 for survival (10). As such, these two IL-15–dependent events are prime targets for controlling NK cell expansion and in vivo persistence.
To better understand how NK cell homeostasis is regulated, we investigated the potential role of transcription factor Kruppel-like factor 2 (KLF2) within the NK cell compartment by using Klf2 gene-targeted mice. The rational for this study was threefold: (i) KLF2 maintains homeostasis in other lymphocyte compartments, including quiescent B (11–13) and T cells (14, 15); (ii) NK cell proliferation is regulated by a P13K-PDK1-Akt-mTOR signaling pathway (9, 16–18), which terminates KLF2 expression in other lymphocyte populations (19, 20; and (iii) Foxo1, which regulates Klf2 transcription in T cells (21, 22), inhibits late stage NK cell differentiation (23). Based on these reports, we predicted that Klf2 gene-targeted mice would exhibit mature NK cell hyperplasia because of dysregulated proliferation and relaxed maturation checkpoints. Indeed, Klf2 excision promoted CD27+ NK cell cycling in a cell-intrinsic manner. However, instead of a preponderance of late-stage NK cells, we found that KLF2 was necessary for CD11b+ effector cell survival. Under steady-state conditions, KLF2-deficient NK cells altered expression of homeostatic homing receptors, thereby preventing these cells from accessing IL-15–rich microenvironments. Importantly, aberrant migration proceeded KLF2-deficient NK cell death, which was confined to an in vivo setting. Therefore, we conclude that KLF2 regulates mature NK cell homeostasis by limiting production of newly differentiated effector cells while simultaneously supporting their survival by guiding these cells toward transpresented IL-15. This latter event may represent a novel form of tolerance that terminates unwarranted NK cell activity.
Results
KLF2 Is Necessary for Conventional NK Cell Homeostasis.
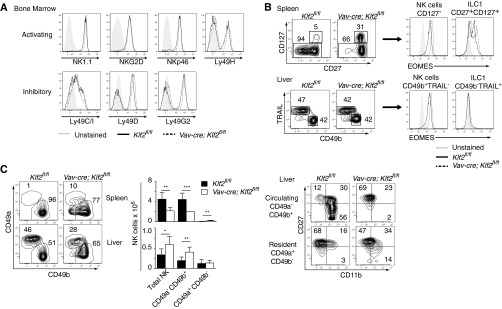
KLF2 is necessary to maintain B and T-cell homeostasis (11–15). To determine whether this transcription factor played a similar role in NK cells, we first verified that KLF2 was expressed under steady-state conditions. Following lineage commitment and initial development in the bone marrow, NK cells home to peripheral tissues, where they continue a differentiation program that is characterized by the surface expression of CD27 and CD11b (7). Isolating individual populations (CD27+CD11b−, CD27+CD11b+, and CD27−CD11b+), mRNA and protein analysis revealed that KLF2 is expressed early during NK cell development and increases with maturation (Fig. 1A). Next, we assessed NK cell homeostasis in Klf2 gene-targeted mice. To ensure KLF2 was depleted from the entire NK cell compartment (Fig. 1B), Vav-cre transgenic animals were used to excise floxed alleles of Klf2 (Klf2fl/fl) in hematopoietic stem cells. Early NK cell development and MHC licensing was intact in Vav-cre; Klf2fl/fl animals, as reflected by normal frequencies of bone marrow-derived NK cells expressing activating (NK1.1, NKG2D, NKp46, Ly49H) and inhibitory (Ly49C/I, Ly49D, Ly49G2) receptors (Fig. S1A). In contrast, loss of KLF2 resulted in increased CD27+CD11b− NK cell frequencies in all tissues, with the exception of mesenteric lymph nodes (Fig. 1C). Absolute CD27+CD11b− NK cell numbers were also increased in the spleen, liver, and bone marrow, the latter two tissues being sites that preferentially harbor immature NK cell populations. These cells expressed high levels of transcription factor EOMES and were either CD127− (spleen) or CD49b+TRAIL− (liver), indicating that they were NK cells and not misidentified NK1.1+ ILC1 cells (24) (Fig. S1B). Instead, ILC1 numbers remained constant in Vav-cre; Klf2fl/fl animals. Likewise, loss of KLF2 did not affect CD49a+CD49b− tissue-resident NK cells in the liver (25) (Fig. S1C). Collectively, these data suggest that KLF2 limits proliferation associated with CD27+CD11b− NK cell differentiation but does not affect homeostasis of neighboring lineages, including ILC1 cells.
Fig. 1.
KLF2 is necessary for NK cell homeostasis. (A) Klf2 mRNA and KLF2 protein levels in NK cell subsets. Splenic CD122+Lin− (CD3, CD8, CD19, Gr-1, TCRβ) NK cells were FACS sorted into maturing NK cell subsets (R1, CD27+CD11b−; R2, CD27+CD11b+; R3, CD27−CD11b+) from C57BL/6 mice. Klf2 mRNA and KLF2 protein levels were normalized to gapdh and tubulin, respectively. This experiment was repeated twice. (B) Klf2 mRNA levels expressed in MACS-sorted NK cells harvested from Klf2fl/fl versus Vav-cre; Klf2fl/fl mice (normalized to gapdh). (C) Contour plots of CD122+Lin− NK cell populations harvested from Klf2fl/fl (black) versus Vav-cre; Klf2fl/fl (white) littermates. Frequencies and absolute cell numbers are graphed. BM, bone marrow; MsLN, mesenteric lymph nodes. Data are pooled from three independent experiments (n = 10 mice per group). (D) Alternate analysis of CD122+Lin− NK cell populations, using CD43 and CD11b as maturity markers. n = 10 mice per cohort. (E) IL-15R surface expression on splenic NK cells harvested from Klf2fl/fl versus Vav-cre; Klf2fl/fl mice. n = 3 experiments. (F) IL-15R signaling capacity of control (Top) versus KLF2-deficient NK cell populations (Lower). NK cells were cultured ± IL-15 (1 h) before intracellular staining for phosphorylated S6, a downstream target of mTOR activity. This experiment was repeated three times. *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. S1.
NK cell MHC licensing, ILC1 homeostasis, and liver-resident NK cell homeostasis is intact in Vav-cre; Klf2fl/fl mice. (A) Histogram overlays of activating and inhibitory receptors expressed on the surface of NK cells (CD122+ Lin− NK1.1+) isolated from the bone marrow of Klf2fl/fl versus Vav-cre; Klf2fl/fl animals. This experiment was repeated twice. (B) Conventional lineage markers were used to identify ILC1 cells in the spleen (CD27+ CD127+) and liver (CD49b− TRAIL+) of Klf2fl/fl and Vav-cre; Klf2fl/fl mice after gating on CD122+ Lin− NK1.1+ cells. Consistent with the literature (24), EOMES+ ILC1 cells are confined to the spleen. (C) Liver-resident NK cells, which are distinct from conventional NK cells (25) were found at normal numbers in Vav-cre; Klf2fl/fl mice. CD49a+ CD49b− NK cells (gated on CD122+ Lin− NK1.1+ cells) were primarily confined to the liver in both sets of animals. Increased CD49a− CD49b+ NK cells found in the liver of Vav-cre; Klf2fl/fl mice were CD27+CD11b− (R1). This experiment was repeated twice by using three mice per cohort. *P < 0.05; **P < 0.01; ***P < 0.001.
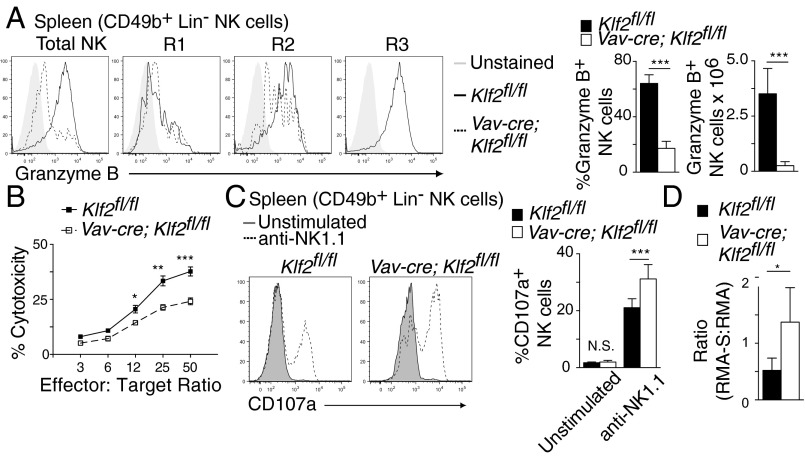
Despite the increase in early stage NK cells, Vav-cre; Klf2fl/fl mice had significantly fewer CD27+CD11b+ and CD27−CD11b+ NK cells relative to littermate controls (Fig. 1C). Costaining for CD43+ and CD11b+ (markers for late-stage NK cells) confirmed that KLF2 gene-targeted mice lacked mature CD43+CD11b+ populations in all examined tissues (Fig. 1D). Given that IL-15 is essential for NK cell survival (26–28), we hypothesized that KLF2-deficient NK cells were unable to respond to this cytokine. However, KLF2-deficient NK cells expressed normal surface levels of CD122 (IL-2Rβ) and CD132 (γc) (Fig. 1E) that conveyed IL-15–mediated signaling events when stimulated ex vivo (Fig. 1F), which suggested that defective IL-15R signaling was not responsible for mature NK cell depletion. [Increased baseline expression of phospho-S6 in immature NK cells reflects elevated metabolism, as noted (17).] Instead, it was possible that mature NK cells were present in Vav-cre; Klf2fl/fl mice but that they were either misidentified (i.e., KLF2 regulates CD11b and CD43 expression) or located in alternate tissues. To address the former option, we used cytolytic activity as a surrogate marker to identify mature KLF2-deficient NK cells. Reflective of their immature status, total NK cells harvested from Vav-cre; Klf2fl/fl mice expressed low levels of granzyme B (Fig. 2A) and had a reduced ability to lyse YAC-1 target cells ex vivo (Fig. 2B). At the same time, KLF2-deficient NK cells responded to anti-NK1.1 stimulation by increasing surface expression of the degranulation marker CD107a (Fig. 2C), which indicated that cell-intrinsic effector functions were not directly regulated by KLF2. Together, these data suggest that mature NK cells are not present in the spleens of Vav-cre; Klf2fl/fl mice. To extend this finding to additional tissues, in vivo tumor clearance was analyzed in animal cohorts following coinjection of NK cell-sensitive (RMA-S) and NK cell-resistant (RMA) tumor cells. Compared with littermate controls, Vav-cre; Klf2fl/fl animals were unable to effectively clear RMA-S target cells (Fig. 2D). Therefore, we conclude that effector NK cells are absent in Vav-cre; Klf2fl/fl mice and that KLF2 is necessary to support mature NK cell homeostasis.
Fig. 2.
Mature cytolytic NK cells are absent in Vav-cre; Klf2fl/fl mice. (A) Histogram overlays (Left) and quantification (Right) of granzyme B expression following (PMA + ionomycin)-simulation of splenic NK cells harvested from Klf2fl/fl versus Vav-cre; Klf2fl/fl mice. Histograms display individual subsets, whereas columns are total NK cells. n = 9–11 mice per cohort, pooled from three independent experiments. (B) Ex vivo cytolytic activity of IL-2–primed splenocytes cultured with Yac-1 target cells for 4 h in an LDH release assay. This experiment was performed once in quadruplicate. (C) CD107a surface expression on NK cells cultured for 6 h ± plate-bound NK1.1 antibody. n = 6 mice per cohort, pooled from two independent experiments. (D) RMA control and RMA-S target cells were coinjected at a 1:1.5 ratio into Klf2fl/fl versus Vav-cre; Klf2fl/fl mice and RMA/RMA-S survival was assessed 48 h later. This experiment was repeated twice by using three mice per group. *P < 0.05; **P < 0.01; ***P < 0.001.
KLF2-Regulated NK Cell Homeostasis Is Cell Intrinsic.
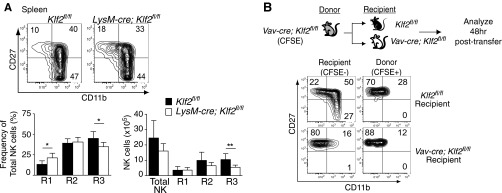
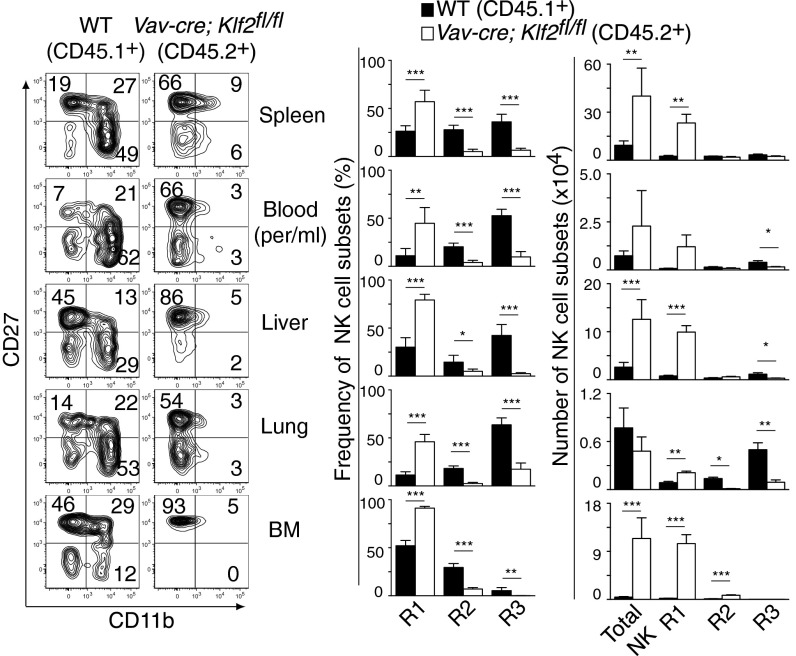
Mature NK cell survival depends on IL-15 that is typically transpresented as an IL-15/IL-15Rα complex on the surface of mesenchymal and myeloid cells (28–31). Because Vav-cre; Klf2fl/fl mice excised Klf2 within the myeloid compartment, NK cell homeostasis may have been altered because of compromised IL-15 presentation. To test this hypothesis, NK cell populations were analyzed in LysM-cre; Klf2fl/fl mice that excised Klf2 in a myeloid-specific manner (Fig. S2A). Using this genetic model, we found normal numbers of CD27+CD11b− and CD11b+CD27+ cells, and a slight but statistical decrease in CD27−CD11b+ NK cells. These results suggest that major NK cell phenotypes observed in Vav-cre; Klf2fl/fl mice were independent of myeloid cells. Additionally, a prior study demonstrated that transferring mutant NK cells into wild-type animals could overcome myeloid-specific defects in NK cell development (32); however, KLF2-deficient NK cell survival was not rescued under these circumstances (Fig. S2B). To confirm that KLF2 intrinsically regulates NK cell homeostasis, lethally irradiated wild-type CD45.2+ mice were reconstituted with a 1:1 ratio of wild type (CD45.1+) versus Vav-cre; Klf2fl/fl (CD45.2+) bone marrow and analyzed 8 wk later. As shown in Fig. 3, CD27+CD11b+ and CD27−CD11b+ NK cells were primarily derived from wild-type recipients, whereas the majority of CD27+CD11b− NK cells were KLF2-deficient. Collectively, these data indicate that KLF2 maintains late-stage NK cell homeostasis in a cell-intrinsic manner and may a play a role in suppressing early stage NK cell proliferation.
Fig. S2.
Defective NK cell homeostasis is consistent with a cell-intrinsic mechanism. (A) Flow cytometric analysis of CD122+ Lin− NK cells harvested from the spleens of 8-wk-old Klf2fl/fl versus LysM-cre; Klf2fl/fl littermates. n = 7 mice per cohort. (B) CFSE-labeled CD19-depleted Vav-cre; Klf2fl/fl splenocytes (2.5 × 107) were adoptively transferred into Klf2fl/fl or Vav-cre; Klf2fl/fl recipients. CFSE+ CD122+ Lin− NK cells were analyzed 48 h after transfer to determine whether neighboring cells could rescue KLF2-deficient NK cell differentiation. This experiment was repeated twice. *P < 0.05; **P < 0.01.
Fig. 3.
KLF2-mediated NK cell homeostasis is cell intrinsic. Analysis of mixed bone marrow chimeras that were generated by reconstituting lethally irradiated Klf2fl/fl (CD45.2+) mice with wild-type (CD45.1+) and Vav-cre; Klf2fl/fl (CD45.2+) bone marrow. Flow cytometric analysis was performed 8 wk after transfer. Representative contour plots, frequencies, and cell numbers of gated (CD122+Lin−) KLF2-sufficient (black) and KLF2-deficient (white) populations are shown. This experiment was performed once by using five recipient animals. *P < 0.05; **P < 0.01; ***P < 0.001.
KLF2 Suppresses Homeostatic Proliferation of NK Cells.
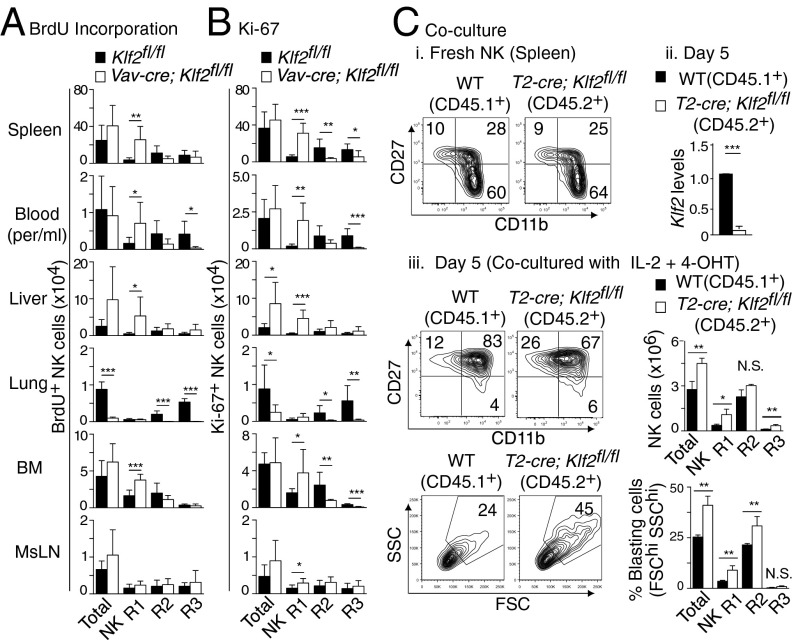
KLF2 was originally reported to prevent the spontaneous proliferation of quiescent T cells (33, 34); however, subsequent work demonstrated that this event was not a cell-intrinsic effect (14, 15). To better characterize KLF2’s role in early stage NK cell cycling, 5-bromo-2′-deoxyuridine (BrdU) incorporation was quantified as a measure of steady-state NK cell proliferation (Fig. 4A). Compared with control littermates, significantly more BrdU+ NK cells were present in the bone marrow of Vav-cre; Klf2fl/fl animals, the majority of which were CD27+CD11b−. Nuclear staining for Ki-67, which identifies proliferating cells, confirmed that immature NK cells were undergoing increased cell cycling in Vav-cre; Klf2fl/fl mice (Fig. 4B). To determine whether KLF2-deficient NK cells had increased access to proliferation-inducing factors in vivo, NK cell cycling was examined under controlled ex vivo conditions. Using a tamoxifen-inducible Cre system, NK cells harvested from T2-cre; Klf2fl/fl versus littermate control mice were cocultured (1:1 ratio) in media supplemented with 4-hydroxytamoxifen (4-OHT) and IL-2, the latter cytokine added to support cell cycling. After 5 d in culture, increased numbers of CD45.2+ cells from T2-cre; Klf2fl/fl mice were recovered relative to CD45.1+ control cells (Fig. 4C), primarily due to an expansion of CD27+CD11b− NK cells. Flow cytometric analysis also demonstrated that more KLF2-deficient NK cells exhibited a blast morphology compared with KLF2-sufficent cells, consistent with KLF2 limiting early stage NK cell proliferation.
Fig. 4.
KLF2 suppresses proliferation in immature NK cells. (A) BrdU incorporation over 5 d was used to assess NK cell (CD122+Lin−) proliferation in various tissues harvested from Klf2fl/fl versus Vav-cre; Klf2fl/fl littermates. n = 6 mice per cohort, pooled from two independent experiments. (B) The percentage of CD122+Lin− NK cells actively proliferating was quantified by Ki-67 expression. n = 7 mice per group (two pooled experiments). (C) Equal numbers of MACS-sorted NK cells from wild type (CD45.1+) versus T2-cre; Klf2fl/fl (CD45.2+) mice were cocultured in 4-OHT and IL-2 to induce Klf2 excision and support proliferation, respectively. NK cells were analyzed by flow cytometry before (i) and after (iii) excision and Klf2 expression was assessed by RT-PCR at day 5 (ii). This experiment was repeated twice by using three biological replicates per group. *P < 0.05; **P < 0.01; ***P < 0.001; N.S., not significant.
Mature NK Cells Require KLF2 To Access Transpresented IL-15.
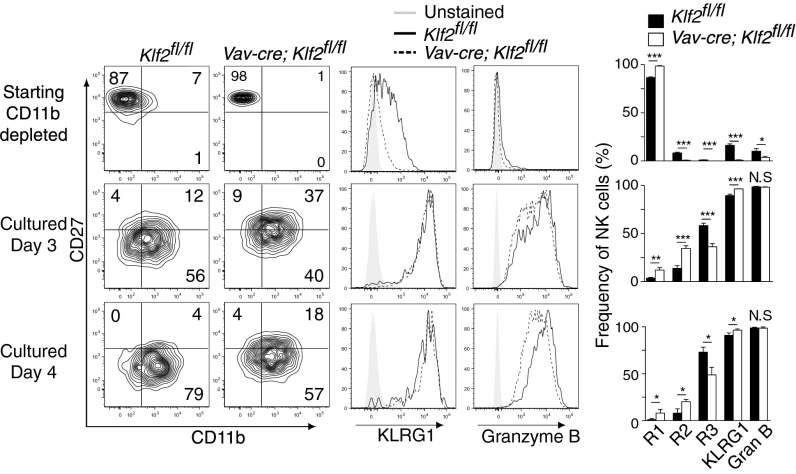
Lymphocyte proliferation and differentiation are typically exclusionary events, which raised the possibility that KLF2 was necessary for mature NK cell differentiation. To test this hypothesis, we performed ex vivo differentiation assays by using NK cells harvested from control versus Vav-cre; Klf2fl/fl mice. To aid in our analysis, input populations were depleted of CD11b+ cells to offset the increased frequency of mature NK cells present in wild-type animals. Following 72 h and 96 h in stromal culture supplemented with IL-15, IL-12, and IL-18, KLF2-deficient NK cells differentiated into CD27+CD11b+ (R2) and CD27−CD11b+ (R3) cells that expressed the maturity markers, KLRG1 and granzyme B (Fig. 5). From this result, we conclude that KLF2 is not necessary for late-stage NK cell differentiation.
Fig. 5.
KLF2-deficient NK cell differentiation is reestablished in culture. CD11b-depleted bone marrow from Klf2fl/fl and Vav-cre; Klf2fl/fl mice was plated on wild-type bone marrow (CD45.1+) supplemented with IL-15, IL-12, and IL-18. Starting material and cells cultured for 3–4 d were initially gated (CD45.2+CD122+Lin−) then analyzed for NK cell differentiation (CD27, CD11b contour plots) and maturity markers (KLRG1, granzyme B) by flow cytometry. Differentiation experiments were performed twice in triplicate, generating similar results. *P < 0.05; **P < 0.01; ***P < 0.001; N.S., not significant.
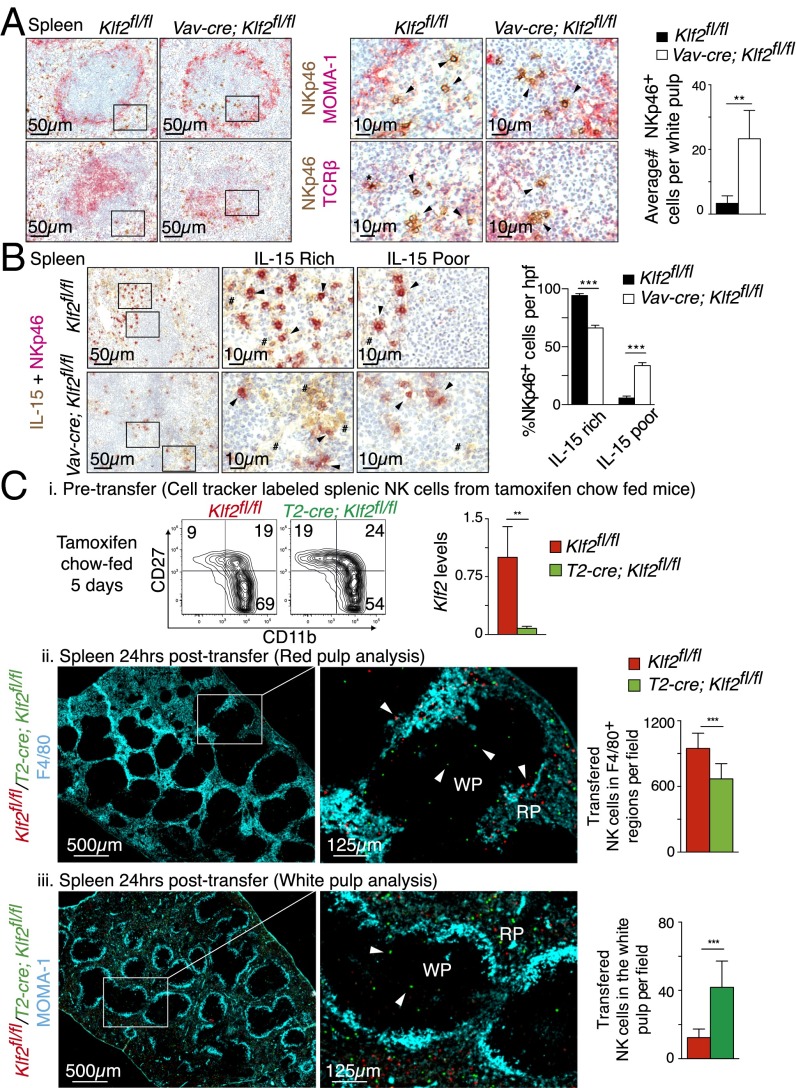
Given that KLF2-deficient NK cells can fully differentiate ex vivo but not in vivo, we hypothesized that NK cells were unable to access a limited survival niche in Vav-cre; Klf2fl/fl mice. Transpresented IL-15 is necessary for late-stage NK cell survival, which is rate limiting under steady-state conditions (35). For this reason, we performed immunohistochemistry to document NK cell localization relative to IL-15 in the spleens of Vav-cre; Klf2fl/fl versus littermate controls. Using a metallophilic macrophage antibody (MOMA-1) to delineate the marginal sinus that forms a ring around the white pulp, we found that NK cells predominately localized in the red pulp of Klf2fl/fl control mice (Fig. 6A). In contrast, significantly more KLF2-deficient NK cells were present in the T-cell–rich area of the white pulp. These KLF2-deficient NKp46+ cells stained negatively for TCRβ, thus confirming that they were NK cells and not NKT cells. Within the spleen, IL-15 is typically presented by myeloid cells and VCAM-1+ stromal cells (36) located in the red pulp. Consistent with a lack of mature NK cells in Vav-cre; Klf2fl/fl mice, KLF2-deficient NK cells were preferentially located in IL-15–depleted areas of the white and red pulp (Fig. 6B and Fig. S3). To verify that differences in localization were directly attributable to KLF2-regulated NK cell migration and not maturation-associated homing patterns (5), we examined how similarly differentiated KLF2-replete and KLF2-deficient NK cells trafficked in vivo. In this instance, KLF2-deficient NK cells were harvested from tamoxifen-treated T2-cre; Klf2fl/fl mice before alterations in subset frequencies (Fig. 6C, i). Following cotransfer into wild-type animals, we found that KLF2+ NK cells primarily homed to the red pulp and associated with F4/80+ myeloid cells (Fig. 6C, ii), a cell type that transpresents IL-15 (10). In contrast, a significant number of KLF2-deficient NK cells migrated to the white pulp, as defined by MOMA-1 staining (Fig. 6C, iii), and were distal to myeloid cells. Therefore, we conclude that KLF2 directs NK cell trafficking under noninflammatory conditions, the implication being that this migration pattern supports late-stage NK cell survival.
Fig. 6.
KLF2 promotes NK cell migration toward IL-15–rich niches. (A) Immunohistochemistry of Klf2fl/fl and Vav-cre; Klf2fl/fl splenic serial sections costained for NKp46/MOMA-1 (Top) and NKp46/TCRβ (Bottom). Enlarged images that identify NK cells (arrows) and NKT cells (*) are shown at Right. Associated bar graph shows the average number of NKp46+ NK cells within 10 similarly sized MOMA-1–encased sections per mouse. n = 4 mice per cohort. (B) Klf2fl/fl (Top) and Vav-Cre; Klf2fl/fl (Bottom) splenic sections costained for NKp46 and IL-15. Middle and Right show enlarged areas of IL-15–rich and IL-15–poor splenic sections, respectively. NKp46+ cells (arrow) and IL-15+ tissue (#) are identified. Associated bar graph shows the average frequency of NK cells (five high-powered fields per mouse) identified in IL-15–rich and –poor niches. n = 3–4 mice per cohort. (C) Dynamic migration of mature NK cell populations within the spleen. Splenic NK cells were isolated (MACS-sorted) from tamoxifen-treated (5 d) Klf2fl/fl versus T2-cre; Klf2fl/fl mice then labeled with red or green cell tracker dyes, respectively. These cells were subsequently cotransferred into a wild-type recipient and splenic localization was assessed 24 h later. This experiment was repeated three times. (i) NK cell subset frequencies (contour plots) and degree of Klf2 excision (RT-PCR) within isolated NK cell populations before transfer. (ii) Immunohistochemistry of KLF2-sufficient (red) versus KLF2-deficient (green) NK cells in relation to F4/80+ myeloid cells. Transferred NK cell numbers were quantified from 10 individual low-power field images. (iii) Immunohistochemistry of cotransferred NK cell populations in relation to the white pulp, as outlined with MOMA-1 antibody. Average number of transferred cells per field was calculated from 25 individual images. **P < 0.01; ***P < 0.001.
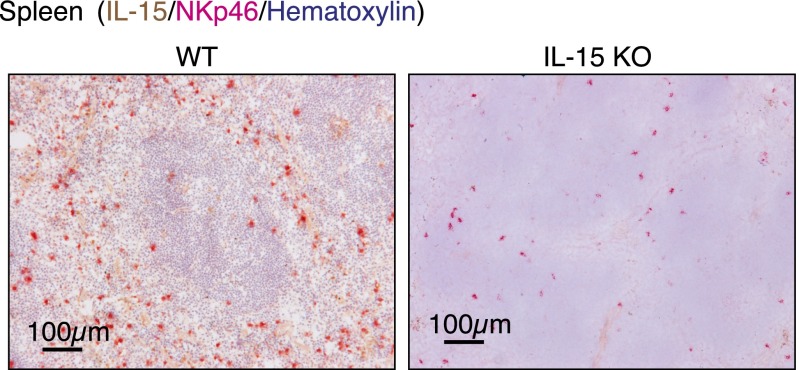
Fig. S3.
Spleens harvested from wild-type (WT) and IL-15–deficient (IL-15 KO) mice were costained for NKp46 (pink), hematoxylin (purple), and IL-15 (brown) to verify IL-15 antibody specificity.
KLF2 Regulates Mature NK Cell Homing Receptor Expression and Survival.
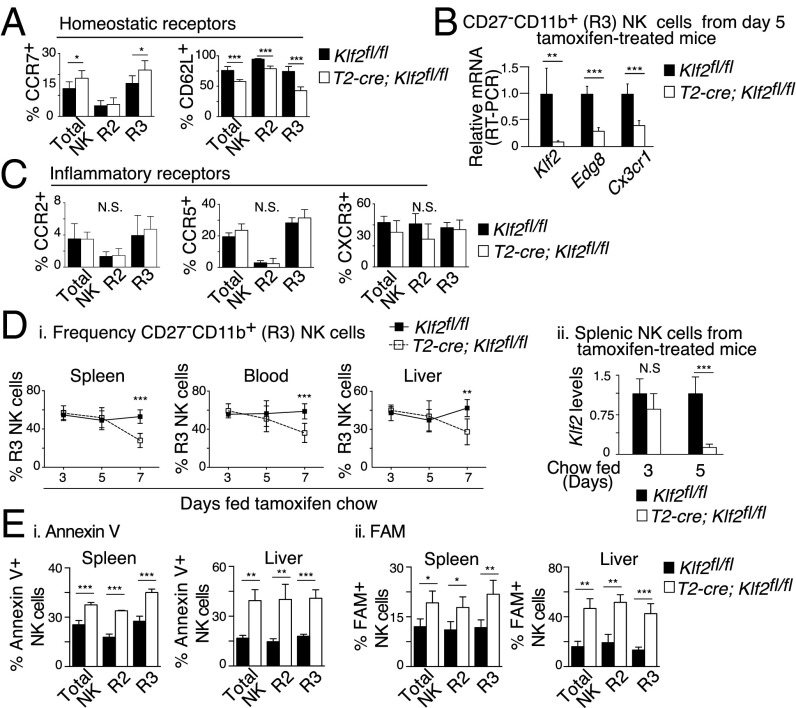
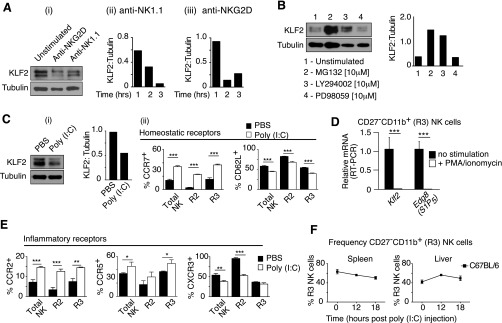
Mature NK cells circulate throughout the vasculature and quickly respond to inflammatory chemokine gradients without prior activation. As such, NK cells need to constitutively express a hybrid array of homing receptors that recognize ligands present under steady-state (e.g., CD62L, CX3CR1, S1P5) and inflammatory conditions (e.g., CCR2, CCR5, CXCR3). To determine whether any of these receptors were modified in response to Klf2 excision, mature NK cell populations harvested from tamoxifen-treated T2-cre; Klf2fl/fl mice were examined by flow cytometry (Fig. 7A) or by RT-PCR when antibodies were not available (Fig. 7B). Compared with mature NK cells from tamoxifen-treated littermate controls, KLF2-deficient cells expressed significantly less CD62L, Cx3cr1, and Edg8. Surprisingly, CCR7 was up-regulated following Klf2 excision, which suggested that a chemokine receptor typically associated with naïve T-cell trafficking was actively repressed by KLF2 in mature NK cells. Previous studies have noted that NK cells increase CCR7 expression and decrease CD62L surface levels following NK cell activation (37) or PI3K activity (16), respectively, and because KLF2 is degraded in a PI3K-associated manner following T-cell activation (19, 20), we were curious whether a similar process existed within the NK cell compartment. Indeed, stimulation of NK cells via activating receptors (NK1.1, NKG2D) promoted KLF2 proteolysis (Fig. S4A). Moreover, NK cells cultured in the presence of a PI3K-inhibitor impaired KLF2 degradation (Fig. S4B), the implication being that this signaling pathway regulates KLF2 levels. To assess how closely Klf2 excision replicated homing receptor regulation following NK cell activation, wild-type NK cells were stimulated with the toll-like receptor 3 agonist, polyinosinic-polycytidylic acid [poly(I:C)], then examined for differential receptor expression. Similar to KLF2-deficient cells, poly(I:C)-treated NK cells altered their surface levels of CCR7 and CD62L (Fig. S4C) and (PMA/ionomycin)-activated CD27−CD11b+ NK cells expressed less Edg8 mRNA (Fig. S4D). Of note, elevated CCR7 expression is consistent with observed NK cell entry into splenic white pulp following poly(I:C) challenge (38). Poly(I:C)-activated NK cells also altered their surface expression of CCR2, CCR5, and CXCR3—homing receptors that respond to inflammatory cytokines (Fig. S4E). In contrast, Klf2 excision did not appreciably affect the expression of these inflammatory homing receptors (Fig. 7C). From these studies, we conclude that KLF2 regulates expression of homing receptors that respond to homeostatic ligands; however, this transcription factor does not impact inflammatory chemokine receptors within the NK cell compartment.
Fig. 7.
KLF2 supports mature NK cell survival by regulating expression of homeostatic homing receptors. (A–C) Klf2fl/fl and T2-cre; Klf2fl/fl mice were placed on tamoxifen-infused chow for 5 d, then analyzed for expression of homing receptors that respond to constitutively expressed ligands (A and B) or inflammatory chemokines (C). (A) Surface expression of CCR7 and CD62L on CD122+Lin− NK cells (R2 = CD27+CD11b+, R3 = CD27−CD11b+), as determined by flow cytometry. This experiment was repeated three times with three mice per cohort. (B) Relative mRNA expression of Klf2, Edg8 (S1P5), and Cx3cr1 in FACS-sorted CD27−CD11b+ NK cells. This experiment was repeated twice. (C) Surface expression of CCR2, CCR5, and CXCR3 on CD122+Lin− NK cells. This experiment was repeated two to three times by using a minimum of three mice per group. (D, i) Frequency of mature (CD122+Lin−CD27−CD11b+) NK cells in the spleen, blood, and liver of Klf2fl/fl versus T2-cre; Klf2fl/fl mice placed on tamoxifen-infused chow for the indicated time points, as determined by flow cytometry. n = 3–11 mice per time point. (ii) Klf2 excision within MACS-sorted splenic NK cells was assessed at days 3 and 5 by RT-PCR. (E) KLF2 is necessary for mature NK cell survival under noninflammatory conditions. Frequency of Annexin V+ (i) and FAM+ (caspase active) (ii) NK cells isolated from Klf2fl/fl versus T2-cre; Klf2fl/fl mice placed on tamoxifen chow (d9). Mature NK cells (CD49b+Lin−) were defined as CD27+CD11b+ (R2) or CD27−CD11b+ (R3). This experiment was repeated three times by using three mice per cohort. *P < 0.05; **P < 0.01; ***P < 0.001; N.S., not significant.
Fig. S4.
Activated NK cells degrade KLF2 and alter homeostatic and inflammatory homing receptors. (A) Engagement of activation receptors promotes KLF2 degradation. Representative KLF2 immunoblot at 3 h (i) and densitometry plots of KLF2 relative to tubulin following stimulation of MACS-sorted NK cells with NK1.1 (i) or NKG2D (iii) antibody. This experiment was repeated twice. (B) KLF2 immunoblot of MACS-sorted NK cells cultured for 4 h ± MG132 (proteasome inhibitor), LY294002 (PI3K inhibitor), or PD98059 (MEK1 inhibitor). This experiment was repeated twice. (C) In vivo activation of NK cells promotes KLF2 degradation and altered expression of homeostatic homing receptors. (i) KLF2 immunoblot and densitometry of splenic NK cells harvested from control (PBS) versus poly(I:C)-treated mice 16 h after injection. This experiment was repeated twice. (ii) Flow cytometric analysis of splenocytes harvested from wild-type mice treated with PBS (black) versus poly(I:C) (white). Sixteen hours after injection, CD49b+Lin− NK cell subsets (R2, CD27+CD11b+; R3, CD27−CD11b+) were examined for surface expression of homing receptors that recognize constitutively expressed ligands. n = 3 mice per group, repeated twice. (D) Relative mRNA expression of Klf2 and Edg8 (S1P5) following (PMA + ionomycin) stimulation of FACS-sorted CD27−CD11b+ NK cells (4 h) harvested from wild-type mice. This experiment was performed once in triplicate. (E) Same as C(ii), except cytometric analysis focused on homing receptors that recognize chemokines associated with inflammation. (F) Frequency of mature (CD27−CD11b+) NK cells in the spleen and liver of wild-type mice over time following poly(I:C)-treatment. n = 3 mice per time point. *P < 0.05; **P < 0.01; ***P < 0.001.
With regard to how this alteration in homing receptors impacted the NK cell compartment, we found that mature NK cell numbers decreased over time when Klf2 was excised under noninflammatory conditions (Fig. 7D). Unlike poly(I:C) treatment that causes splenic NK cell displacement into the liver (ref. 39; Fig. S4F), Klf2 excision did not cause an accumulation of mature NK cells in any observed tissues. Instead, analysis of NK cells harvested from tamoxifen-treated Klf2fl/fl versus T2-cre; Klf2fl/fl littermates revealed that this reduction was due to increased apoptosis of mature NK cells (Fig. 7E). Importantly, KLF2-deficient NK cell apoptosis (d7 onwards) occurred after alterations in homing receptors (d5; Fig. 7 A–C) and aberrant migration (d5; Fig. 6C). From these results, we conclude that KLF2 controls NK cell migration via regulation of homeostatic homing receptors, the alteration of which does not support NK cell survival under steady-state conditions.
Discussion
Adoptive transfer of allogeneic NK cells is a promising cancer therapy (40, 41); however, to maximize its potential in the clinic, it is important to devise new ways of increasing NK cell numbers and prolonging effector functions in vivo. Results from this study indicate that low levels of KLF2 limit antigen-independent NK cell proliferation in all tissues and removal of this factor expands the proliferative burst associated with CD27+CD11b− NK cells. These findings are consistent with previous reports demonstrating that the PI3K-PDK1-mTOR signaling pathway promotes NK cell cycling (9, 17, 18, 42, 43), because we and others have shown that signaling receptors that activate the PI3K pathway suppress KLF2 expression in T cells (19, 20) and B cells (12). Surprisingly, Foxo1, which is negatively regulated by PI3K signaling and directly promotes Klf2 transcription in T cells (21, 22), does not appear to link PI3K-mediated activating events with KLF2 expression in NK cells. This disconnect between transcription factors is evidenced by an inverse expression pattern (in contrast to KLF2, Foxo1 expression decreases from stage 1→stage 3) and increased frequencies of CD27−CD11b+ NK cells in Foxo1 gene-targeted animals (23). This heretofore association between these two molecules raises the question as to what factors directly promote Klf2 transcription in NK cells. Addressing this issue may provide a therapeutic means of increasing NK cell numbers by suppressing KLF2 expression in a lineage-specific manner.
In addition to controlling NK cell cycling, KLF2 is necessary for late-stage NK cell survival under steady-state conditions. More specifically, KLF2 regulates homeostatic homing receptors so that these cells gain access to transpresented IL-15. In contrast, activated NK cells degrade KLF2 and alter their homing receptor expression patterns accordingly. Both quiescent and activated NK cells rely on IL-15 for survival (44–46), which implies that activated NK cells have access to IL-15 that is not available to KLF2-deficient NK cells under noninflammatory conditions. We propose that during an innate immune response that activates NK cells, myeloid cells that are capable of IL-15 transpresentation are likewise activated and recruited to inflammatory sites, thereby maintaining NK cell effector functions. In the absence of this IL-15–dependent support system, NK cell effector activity is quickly terminated. Such an event might occur at the conclusion of a productive NK cell immune response when myeloid cells contract. Likewise, inappropriate NK cell activation (e.g., an immune response directed against self via NKG2D) is predicted to starve these cells of IL-15 and maintain tolerance. This model may explain why mice and humans are devoid of NK cell-initiated autoimmunity, despite the fact that NK cells recognize self-ligands (e.g., stress molecules exhibited by tumor cells) (47) and do not require additional cells to become activated (48). Therefore, linking activated NK cell survival to colocalized IL-15 transpresentation may constitute a fundamental mechanism to ensure self-tolerance. Conversely, cancers may co-opt this tolerance mechanism to evade NK cell-mediated tumor surveillance. If this model of IL-15 dependence proves to be the case, then recruiting IL-15 transpresenting cells to environments that elicit NK cell activation may prevent NK cell exhaustion and restore antitumor immunity.
Materials and Methods
Mice.
Vav-cre; Klf2fl/fl and T2-cre; Klf2fl/fl mice were generated as described (15, 19); LysM-cre and B6.SJL (CD45.1+) mice were purchased from Jackson Laboratories. IL-15−/− mice were purchased from Taconic. Mice were housed in pathogen-free conditions in accordance with the Institutional Animal Care and Use Committee at Vanderbilt University.
Flow Cytometry.
Standard flow cytometric procedures were used to acquire data on a 5-laser LSRII (BD Biosciences); analysis was performed by using FlowJo (TreeStar) software.
Bone Marrow Chimeras.
Irradiated (2 × 500 cGy) Klf2fl/fl (CD45.2+) mice were reconstituted with 106 bone marrow cells (1:1 CD45.1+ versus Vav-cre; Klf2fl/fl CD45.2+), then analyzed 8 wk later by FACS.
Immunohistochemistry.
Cryosections were stained with the following antibodies: polyclonal goat NKp46 (R&D Systems), biotin-TCRβ (BD Biosciences), biotin-MOMA1 (Cedarlane), biotin-F4/80 (Tonbo), and biotin-IL-15 (R&D Systems). The anti-NKp46 antibody was visualized with secondary biotinylated F(ab')2 Frag donkey anti-goat IgG (H+L) (Jackson Immunoresearch). Nuclei were counterstained by using Meyer’s hematoxylin solution (Sigma). Images were acquired by using Nikon AZ 100 (Nikon) and NIS-Elements (Nikon) software. Additional staining information is provided in SI Materials and Methods.
NK Cell Apoptosis.
Klf2fl/fl and T2-cre; Klf2fl/fl littermates were placed on tamoxifen chow for 5 d, killed 4 d later, and NK cell populations were examined by flow cytometry. Annexin V surface expression (BD Biosciences) and caspase cleavage (Vibrant FAM kit; Life Technologies) were used as readouts for apoptosis based on manufacturers’ instructions.
Statistical Analysis.
Data were analyzed by using a two-tailed Student t test and displayed as the mean ± SD: *P < 0.05, **P < 0.01, ***P < 0.001 unless otherwise indicated.
SI Materials and Methods
Cell Isolation.
Single-cell suspensions were made from spleen, liver, lung, bone marrow, mesenteric lymph node, and blood for flow cytometry analysis. All organs isolated were mechanically dissociated and put through a 70-μm cell strainer. Single-cell suspensions in complete media were resuspended in 40% (vol/vol) Percoll and centrifuged for 12 min at 680 × g at room temperature to pellet the leukocyte fraction. Blood was collected in PBS supplemented with 2 mM EDTA via cardiac puncture. Lungs were minced with a razor blade and digested with collagenase IV (1 mg/mL; Worthington) in HBSS (Gibco), supplemented with 5 mM CaCl2 for 1 h at 37 °C. Following digestion, lungs were passed through a 70-μm cell strainer. Bone marrow was isolated from a single femur flushed with a 25-gauge needle. Erythrocytes were lysed by using ammonium-chloride-potassium lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA). Cells were counted by using a hemocytometer and trypan blue to exclude dead cells.
Antibody Staining.
Anti-Ly49A (A1), anti-Ly49C/I (5E6), anti-Ly49D (4E5), anti-Ly49G2 (4D11), anti-NK1.1 (PK136), anti-CD3ε (145-2C11), anti-CD8 (53-6.7) anti-CD11b (M1/70), anti-CD49a (Ha31/8), anti–Gr-1 (RB6-8C5), and anti-TCRβ (H57-597) were purchased from BD Biosciences. Anti-EOMES (Dan11mag), anti–Ki-67 (SolA15), anti-KLRG1 (2F1), anti-Ly49H (3D10), anti-NKG2D (A10), anti-CD19 (1D3), anti-CD27 (LG3A.10), anti-CD49b (DX5), anti-CD127 (A7R34), and anti-CD132 (TUGm2) were purchased from eBioscience. Anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD107a (1D4B), anti-CCR5 (HM-CCR5), anti-CCR7 (4B12), and anti-CXCR3 (CXCR3-173) were purchased from Biolegend. Anti-NKp46 (AF2225) and anti–IL-15 (AF447-SP) were purchased from R&D Systems. Anti-CD62L (MEL-14) and anti-F4/80 were purchased from Tonbo Biosciences. Anti-phosophoS6 was purchased from Cell Signaling Technologies. Anti-CCR2 (E68) was purchased from Abcam. Anti-granzyme B (FGB12) and chicken anti-rabbit IgG (H+L) that was used to recognize purified rabbit anti-CCR2 were purchased from Life Technologies. Purified goat anti-NKp46 was recognized by using donkey anti-goat IgG (H+L) (Jackson Immunoresearch; 705-606-147) in 2% (vol/vol) donkey sera to avoid bovine cross-reactivity. Intracellular and surface staining was performed by using standard procedures except CCR5, CCR7, and CXCR3 stains, which were done at 37 °C for 40 min in the presence of 0.05% sodium azide to enhance staining, then fixed with 4% (wt/vol) paraformaldehyde to prevent receptor internalization.
Cell Culture and MACS Sorting.
For functional assays, cells were cultured in Iscove's Modified Dulbecco's Medium supplemented with 10% (vol/vol) FBS (Gibco), 1× antibiotic/antimycotic (Gibco), 5 μM β-mercaptoethanol, and 1× nonessential amino acids (Sigma). IL-2, IL-12, and IL-15 were purchased from Peprotech. IL-18 was purchased from MBL. MACS-sorted NK cells were isolated by using the NK cell Isolation KitII (Miltenyi Biotech).
Ex Vivo Functional Assays.
NK cell degranulation and granzyme B expression were measured by flow cytometry following 2 h of priming cells with plate-bound anti-NK1.1 (10 μg/mL) or PMA/ionomycin (50 ng/mL + 500 ng/mL) and 4-h incubation of Golgi Plug/Stop (BD Biosciences)-treated cells to accumulate signal. NK cell cytotoxicity assays were performed by using IL-2–primed splenocytes (20 ng/mL, 24 h) that were cocultured with 2 × 104 YAC-1 target cells for 4 h at 37 °C. Cytotoxicity was assessed by using a CytoTox Non-Radioactive Cytotoxicity Assay kit (Promega). Proliferation following ex vivo Klf2 excision was examined by coculturing MACS-sorted NK cells (Miltenyi Biotec) harvested from Klf2fl/fl versus T2-cre; Klf2fl/fl littermates (1:1 ratio, 5 × 105/mL) in Complete media supplemented with IL-2 (20 ng/mL) and 4-OHT (400 nM) for 5 d. Klf2 excision was >95% as determined by RT-PCR. To examine ex vivo NK cell differentiation, CD11b-depleted bone marrow from Klf2fl/fl versus Vav-cre; Klf2fl/fl mice was cocultured on stromal support (CD45.1+ bone marrow) in media supplemented with IL-15 (20 ng/mL), IL-12 (20 ng/mL), and IL-18 (50 ng/mL) for 3–4 d, then analyzed by flow cytometry.
In Vivo Functional Assays.
NK cell cytotoxicity was measured by coinjecting 4 × 105 RMA and 6 × 105 RMA-S cells [labeled with carboxytetramethylrhodamine (CTMR) and carboxyfluorescein succinimidyl ester (CFSE) (Molecular Probes), respectively] into mice i.p., performing a peritoneal lavage 48 h later, then analyzing the ratio of target (RMA-S) to control (RMA) cells by flow cytometry. NK cell proliferation was assessed following i.p. injection of BrdU (1 mg/mL, every 12 h for 4.5 d) according to manufacturer’s instructions (BrdU Flow Kit; BD Biosciences). Alternatively, Ki-67 staining (eBioscience) was used to detect cell cycling.
Immunohistochemistry.
Before cryosectioning, spleens were embedded and snap frozen in OCT (Sakura) and stored at −80 °C. Five-micrometer serial cryosections were fixed in 100% acetone for 10 min, air dried for 10 min, rehydrated in TBS, and blocked in 10% (vol/vol) donkey sera with 50% (vol/vol) avidin solution for 30 min, followed by 10% (vol/vol) donkey sera with 50% (vol/vol) biotin solution for 30 min using the Avidin/Biotin Blocking kit (Vector Laboratories) to block endogenous avidin and biotin before staining. For NKp46/TCRβ and NKp46/MOMA-1 costaining, sections were incubated with titrated amounts of polyclonal goat anti-NKp46 and biotinylated anti-TCRβ or biotinylated anti-MOMA1 (Cedarlane) overnight at 4 °C or 2 h at room temperature. MOMA-1 and TCR-β biotinylated antibodies were visualized by using the VECTASTAIN ABC-AP (STANDARD) and VECTOR Red Alkaline Phosphatase (AP) substrate kit (Vector Labs) supplemented with levamisole (Vector Labs) to block endogenous phosphatase activity. Sections were then incubated with 1.8% (vol/vol) hydrogen peroxide and 0.2% sodium azide to quench endogenous peroxidases for 1 h, and reblocked by using the Avidin/Biotin Blocking kit to block biotinylated sites of the first antibody. The anti-NKp46 antibody was visualized by incubating with secondary biotinylated F(ab')2 Frag donkey anti-goat IgG (H+L), then VECTASTAIN Elite ABC Kit (Standard), followed by development using the DAB Peroxidase (HRP) Substrate Kit. Nuclei were counterstained by using Meyer’s hematoxylin solution (Sigma). Sections were dried overnight and cleared with xylene for 30 min before permanent mounting with Permount (Fisher). For dual staining for IL-15 and NKp46, sections were first stained for NKp46 by using alkaline phosphatase/Vector Red, then serially incubated with biotinylated anti-IL-15 and HRP/DAB as described. Specificity was determined by using similar titrations of biotinylated isotype controls for monoclonal antibodies, secondary antibody staining alone for polyclonal antibodies on sections stained in tandem, or IL-15 gene-deficient (KO) sections when appropriate.
Homing Receptor Regulation.
Wild-type mice were challenged i.p. with poly(I:C)-LMW (200 μg; InvivoGen) and killed 16 h later. Alternatively, Klf2fl/fl and T2-cre; Klf2fl/fl mice were placed on tamoxifen-infused chow (250 mg/kg; Harlan Teklad) for 5 d. In both cases, surface expression of homing receptors was assessed by flow cytometry in a subset-specific manner or transcription levels of homing receptors present in CD27−CD11b+ FACS-sorted NK cells was determined by RT-PCR.
Dynamic Migration Assay.
MACS-sorted NK cells were harvested from Klf2fl/fl and T2-cre; Klf2fl/fl littermates placed on tamoxifen-infused chow for 5 d. Before cell sorting, Klf2fl/fl and T2-cre; Klf2fl/fl splenocytes were labeled with cell tracker CMRA [10 μM] and CMFDA [10 μM] (Molecular Probes), respectively, for 40 min at 4 °C. Equal numbers of labeled NK cells (6 × 106) were injected (i.v.) into a wild-type recipient. The recipient spleen was harvested 24 h after transfer and fixed in PBS containing 4% (wt/vol) paraformaldehyde and 30% (wt/vol) sucrose before cryoembedding to preserve cell tracker dyes before immunohistochemical analysis. Eight-micrometer cryosections were then avidin/biotin/normal serum-blocked as described above before being incubated with biotinylated anti–MOMA-1 and anti-F4/80 antibodies. Biotinylated antibodies were visualized with streptavidin BV421 (BD Biosciences), and slides were mounted in fluorescence mounting media (DAKO). Fluorescent images were overlayed by using ImageJ software.
NK Cell Stimulation and Western Blots.
NK cells were stimulated with plate-bound NKG2D (25 μg/mL) or NK1.1 (25 μg/mL) antibody for select time points at 37 °C. These plates were supplemented with plate-bound fibronectin (10 μg/mL) and low-dose IL-15 (0.5 ng/mL) to improve cell viability. Alternatively, NK cells were labeled with biotinylated anti-NK1.1 (10 μg/mL) or anti-NKG2D (10 μg/mL) at 4 °C before cross-linking with streptavidin PE (10 μg/mL) at 37 °C for 1–3 h. To inhibit proteasome, PI3K, or MEK1 activity, NK cells were cultured with MG132 (10 μM), LY294002 (10 μM), or PD98059 (10 μM), respectively. Following stimulation, NK cells were washed with PBS and lysed in PBS-RIPA buffer (1.37 M NaCl, 27 mM KCl, 100 mM Na2HPO4, 18 mM KH2PO4, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, 2 mM EDTA, 2 mM EGTA) with protease inhibitor mixture 2 (Sigma) for 15 min at 4 °C before pelleting debris. Lysates were resolved on a 10% SDS/PAGE and probed by using anti-mouse KLF2 (Millipore) and anti–α-tubulin (Santa Cruz Biotechnology). Densitometry was measured by using ImageJ software.
Real-Time PCR Primer Sequences.
RNA was extracted by using the RNeasy Micro Kit (Qiagen), and cDNA was generated by using the iScript cDNA Synthesis Kit (Bio-Rad). Real-time reactions were performed by the iQ Sybr Green Supermix (Bio-Rad) and iCycler eQ Real-Time PCR Detection System (Bio-Rad). Data were expressed as 2^[(CT KLF2-deficient experimental gene – CT KLF2-deficient Gapdh) – (CT control experimental gene – CT control Gapdh)].
Klf2-F/R: GCGGCAAGACCTACACCAAGAG, CTTTCGGTAGTGGCGGGTAAGC
Gapdh-F/R: GCCTTCCGTGTTCCTACC, GCCTGCTTCACCACCTTC
Cx3cr1-F/R: CGACATTGTGGCCTTTGGAACCAT, AGATGTCAGTGATGCTCTTGGGCT
Edg8-F/R: CTTGCTATTACTGGATGTCGC, GTTGGAGGAGTCTTGGTTGC
Acknowledgments
We thank Dr. James Carlyle and Dr. Luc Van Kaer for discussions and critical reading of the manuscript. W.R. was supported by Immunobiology of Blood and Vascular Systems Training Grant T32HL069765.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521491113/-/DCSupplemental.
References
- 1.Bachanova V, Miller JS. NK cells in therapy of cancer. Crit Rev Oncog. 2014;19(1-2):133–141. doi: 10.1615/critrevoncog.2014011091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orange JS. Natural killer cell deficiency. J Allergy Clin Immunol. 2013;132(3):515–525, quiz 526. doi: 10.1016/j.jaci.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knorr DA, Bachanova V, Verneris MR, Miller JS. Clinical utility of natural killer cells in cancer therapy and transplantation. Semin Immunol. 2014;26(2):161–172. doi: 10.1016/j.smim.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller JS, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood. 2005;105(8):3051–3057. doi: 10.1182/blood-2004-07-2974. [DOI] [PubMed] [Google Scholar]
- 5.Walzer T, Vivier E. G-protein-coupled receptors in control of natural killer cell migration. Trends Immunol. 2011;32(10):486–492. doi: 10.1016/j.it.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Grégoire C, et al. The trafficking of natural killer cells. Immunol Rev. 2007;220:169–182. doi: 10.1111/j.1600-065X.2007.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiossone L, et al. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113(22):5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- 8.Kim S, et al. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol. 2002;3(6):523–528. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- 9.Yang M, et al. PDK1 orchestrates early NK cell development through induction of E4BP4 expression and maintenance of IL-15 responsiveness. J Exp Med. 2015;212(2):253–265. doi: 10.1084/jem.20141703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mortier E, et al. Macrophage- and dendritic-cell-derived interleukin-15 receptor alpha supports homeostasis of distinct CD8+ T cell subsets. Immunity. 2009;31(5):811–822. doi: 10.1016/j.immuni.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Hart GT, Wang X, Hogquist KA, Jameson SC. Krüppel-like factor 2 (KLF2) regulates B-cell reactivity, subset differentiation, and trafficking molecule expression. Proc Natl Acad Sci USA. 2011;108(2):716–721. doi: 10.1073/pnas.1013168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoek KL, et al. Follicular B cell trafficking within the spleen actively restricts humoral immune responses. Immunity. 2010;33(2):254–265. doi: 10.1016/j.immuni.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkelmann R, et al. B cell homeostasis and plasma cell homing controlled by Krüppel-like factor 2. Proc Natl Acad Sci USA. 2011;108(2):710–715. doi: 10.1073/pnas.1012858108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442(7100):299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 15.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9(3):292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 16.Leong JW, et al. PTEN regulates natural killer cell trafficking in vivo. Proc Natl Acad Sci USA. 2015;112(7):E700–E709. doi: 10.1073/pnas.1413886112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marçais A, et al. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat Immunol. 2014;15(8):749–757. doi: 10.1038/ni.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nandagopal N, Ali AK, Komal AK, Lee SH. The critical role of IL-15-PI3K-mTOR pathway in natural killer cell effector functions. Front Immunol. 2014;5:187. doi: 10.3389/fimmu.2014.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pabbisetty SK, et al. KLF2 is a rate-limiting transcription factor that can be targeted to enhance regulatory T-cell production. Proc Natl Acad Sci USA. 2014;111(26):9579–9584. doi: 10.1073/pnas.1323493111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinclair LV, et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat Immunol. 2008;9(5):513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabre S, et al. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008;181(5):2980–2989. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- 22.Kerdiles YM, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10(2):176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng Y, et al. Transcription factor Foxo1 is a negative regulator of natural killer cell maturation and function. Immunity. 2015;42(3):457–470. doi: 10.1016/j.immuni.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinette ML, et al. Immunological Genome Consortium Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16(3):306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sojka DK, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper MA, et al. In vivo evidence for a dependence on interleukin 15 for survival of natural killer cells. Blood. 2002;100(10):3633–3638. doi: 10.1182/blood-2001-12-0293. [DOI] [PubMed] [Google Scholar]
- 27.Jamieson AM, Isnard P, Dorfman JR, Coles MC, Raulet DH. Turnover and proliferation of NK cells in steady state and lymphopenic conditions. J Immunol. 2004;172(2):864–870. doi: 10.4049/jimmunol.172.2.864. [DOI] [PubMed] [Google Scholar]
- 28.Koka R, et al. Interleukin (IL)-15R[alpha]-deficient natural killer cells survive in normal but not IL-15R[alpha]-deficient mice. J Exp Med. 2003;197(8):977–984. doi: 10.1084/jem.20021836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burkett PR, et al. Coordinate expression and trans presentation of interleukin (IL)-15Ralpha and IL-15 supports natural killer cell and memory CD8+ T cell homeostasis. J Exp Med. 2004;200(7):825–834. doi: 10.1084/jem.20041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawamura T, Koka R, Ma A, Kumar V. Differential roles for IL-15R alpha-chain in NK cell development and Ly-49 induction. J Immunol. 2003;171(10):5085–5090. doi: 10.4049/jimmunol.171.10.5085. [DOI] [PubMed] [Google Scholar]
- 31.Schluns KS, et al. Distinct cell types control lymphoid subset development by means of IL-15 and IL-15 receptor alpha expression. Proc Natl Acad Sci USA. 2004;101(15):5616–5621. doi: 10.1073/pnas.0307442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soderquest K, et al. Monocytes control natural killer cell differentiation to effector phenotypes. Blood. 2011;117(17):4511–4518. doi: 10.1182/blood-2010-10-312264. [DOI] [PubMed] [Google Scholar]
- 33.Kuo CT, Veselits ML, Leiden JM. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277(5334):1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 34.Buckley AF, Kuo CT, Leiden JM. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc-dependent pathway. Nat Immunol. 2001;2(8):698–704. doi: 10.1038/90633. [DOI] [PubMed] [Google Scholar]
- 35.Ranson T, et al. IL-15 is an essential mediator of peripheral NK-cell homeostasis. Blood. 2003;101(12):4887–4893. doi: 10.1182/blood-2002-11-3392. [DOI] [PubMed] [Google Scholar]
- 36.Cui G, et al. Characterization of the IL-15 niche in primary and secondary lymphoid organs in vivo. Proc Natl Acad Sci USA. 2014;111(5):1915–1920. doi: 10.1073/pnas.1318281111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson MJ, Williams BT, Christopherson K, 2nd, Brahmi Z, Hromas R. Regulation of human natural killer cell migration and proliferation by the exodus subfamily of CC chemokines. Cell Immunol. 2000;199(1):8–14. doi: 10.1006/cimm.1999.1601. [DOI] [PubMed] [Google Scholar]
- 38.Grégoire C, et al. Intrasplenic trafficking of natural killer cells is redirected by chemokines upon inflammation. Eur J Immunol. 2008;38(8):2076–2084. doi: 10.1002/eji.200838550. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Xu J, Zhang W, Wei H, Tian Z. TLR3 ligand-induced accumulation of activated splenic natural killer cells into liver. Cell Mol Immunol. 2005;2(6):449–453. [PubMed] [Google Scholar]
- 40.Geller MA, Miller JS. Use of allogeneic NK cells for cancer immunotherapy. Immunotherapy. 2011;3(12):1445–1459. doi: 10.2217/imt.11.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leung W. Infusions of allogeneic natural killer cells as cancer therapy. Clin Cancer Res. 2014;20(13):3390–3400. doi: 10.1158/1078-0432.CCR-13-1766. [DOI] [PubMed] [Google Scholar]
- 42.Banh C, Miah SM, Kerr WG, Brossay L. Mouse natural killer cell development and maturation are differentially regulated by SHIP-1. Blood. 2012;120(23):4583–4590. doi: 10.1182/blood-2012-04-425009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo H, Samarakoon A, Vanhaesebroeck B, Malarkannan S. The p110 delta of PI3K plays a critical role in NK cell terminal maturation and cytokine/chemokine generation. J Exp Med. 2008;205(10):2419–2435. doi: 10.1084/jem.20072327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huntington ND, et al. Interleukin 15-mediated survival of natural killer cells is determined by interactions among Bim, Noxa and Mcl-1. Nat Immunol. 2007;8(8):856–863. doi: 10.1038/ni1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wittnebel S, et al. Membrane-bound interleukin (IL)-15 on renal tumor cells rescues natural killer cells from IL-2 starvation-induced apoptosis. Cancer Res. 2007;67(12):5594–5599. doi: 10.1158/0008-5472.CAN-06-4406. [DOI] [PubMed] [Google Scholar]
- 46.Pillet AH, Bugault F, Thèze J, Chakrabarti LA, Rose T. A programmed switch from IL-15- to IL-2-dependent activation in human NK cells. J Immunol. 2009;182(10):6267–6277. doi: 10.4049/jimmunol.0801933. [DOI] [PubMed] [Google Scholar]
- 47.Raulet DH, Guerra N. Oncogenic stress sensed by the immune system: Role of natural killer cell receptors. Nat Rev Immunol. 2009;9(8):568–580. doi: 10.1038/nri2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan CJ, Smyth MJ, Martinet L. Molecular mechanisms of natural killer cell activation in response to cellular stress. Cell Death Differ. 2014;21(1):5–14. doi: 10.1038/cdd.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]