Significance
Loss of retinoschisin (RS1)-mediated retinal cell–cell adhesion in the retina is the underlying cause of X-linked retinoschisis, leading to vision impairment in young males. Using cryo-electron microscopy, we show that RS1 forms paired back-to-back octameric rings. This molecular architecture provides a structural basis for understanding the disruptive effect of many disease-related mutants, because it localizes residues that are involved in the proper assembly of the oligomer. The back-to-back ring pairing is reminiscent of other junctional proteins, where multiple interactions between arrays of the same or similar proteins on two opposing membranes form a strong adhesion plaque.
Keywords: retinoschisin, X-linked retinoschisis, discoidin domain, cryo-electron microscopy, single particle analysis
Abstract
Retinoschisin (RS1) is involved in cell–cell junctions in the retina, but is unique among known cell-adhesion proteins in that it is a soluble secreted protein. Loss-of-function mutations in RS1 lead to early vision impairment in young males, called X-linked retinoschisis. The disease is characterized by separation of inner retinal layers and disruption of synaptic signaling. Using cryo-electron microscopy, we report the structure at 4.1 Å, revealing double octamer rings not observed before. Each subunit is composed of a discoidin domain and a small N-terminal (RS1) domain. The RS1 domains occupy the centers of the rings, but are not required for ring formation and are less clearly defined, suggesting mobility. We determined the structure of the discoidin rings, consistent with known intramolecular and intermolecular disulfides. The interfaces internal to and between rings feature residues implicated in X-linked retinoschisis, indicating the importance of correct assembly. Based on this structure, we propose that RS1 couples neighboring membranes together through octamer–octamer contacts, perhaps modulated by interactions with other membrane components.
The structural integrity and functioning of tissues are highly dependent on cell–cell junctions. These are typically composed of two similar opposing layers of interacting proteins embedded as oligomers or arrays in the cellular membranes. For example, paired arrays of cadherins are found in adherens junctions and desmosomes (1); of claudins and occludins in tight junctions (2, 3); and of connexin channels in gap junctions (4). The back-to-back architecture of these junctions is fundamental to their adhesive function.
The retina features another protein implicated in cell–cell adhesion, retinoschisin (RS1). RS1 is a 24-kDa protein expressed exclusively by photoreceptors and bipolar cells in the retina (5) and pineal gland (6). Loss-of-function mutants of RS1 lead to progressive visual impairment in a disease called X-linked retinoschisis (XLRS) (7–9). The clinical manifestation is a pathological separation of retinal layers, leading to structural splitting and cavity (or “schisis”) formation (10). Additionally, the bipolar cells show disorganization (11) and failure to maintain synaptic organization (9). Both XLRS patients and RS1 gene knockout mouse models (Rs1-KO) show similar phenotypes: schisis/splitting through retinal layers and synaptic transmission defects manifested as reduced b-wave amplitudes on the clinical electroretinogram (Fig. S1) (8, 12). RS1 gene therapy rescues the XLRS mouse phenotype (9, 13) and human RS1 gene therapy trials are underway (ClinicalTrials.gov: NCT02317887; NCT02416622).
Fig. S1.
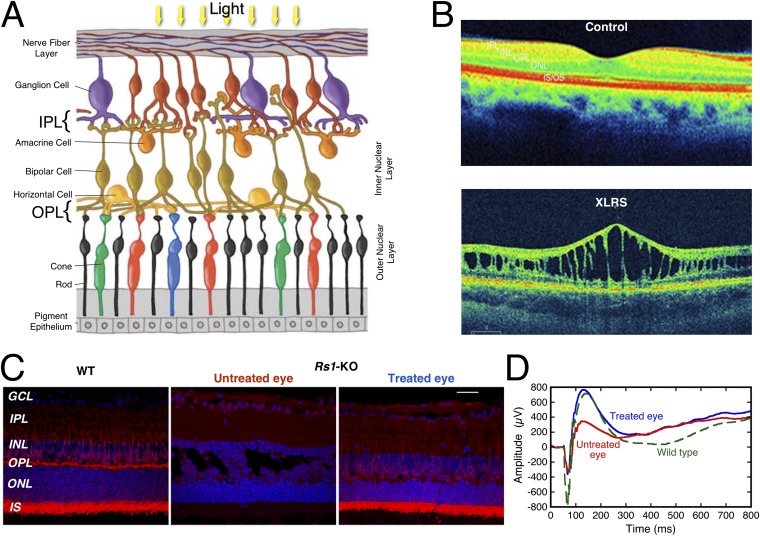
Disruption of the retina in XLRS and rescue by recombinant RS1. (A) The layered structure of the retina (adapted from ref. 52 with permission from the Scientific Research Society). (B) Spectral domain-optical coherence tomography (SD-OCT) images of the macular region of the retina from a normal control individual and from a patient with XLRS. (C) Expression of RS1 (red) in retinal sections for a wild-type mouse (Left), an untreated eye of a RS1 knockout mouse (Center), and the other eye treated with a viral vector coding for RS1-FLAG (Right). Blue indicates DAPI nuclear staining. (D) The electroretinograms (ERGs) for the treated (blue) and untreated (red) eyes of a knockout mouse show complete rescue by the recombinant RS1 compared with a wild-type eye (green). Visual perception begins when light reaching the retina activates the rhodopsin in rod and cone photoreceptors cells. The photoreceptors transduce the light energy into electrical impulses that travel to the brain via the optic nerve through the bipolar and ganglion cells. Rods function in dim light, whereas cones respond to bright light. Retinal interneurons, bipolar, horizontal, and amacrines integrate visual information for brightness, color contrast, and motion before transmittal to the ganglion cells. The outer plexiform layer (OPL) is a dense network of synapses where neural connections are established between axon terminals of rods/cones and dendrites of bipolar and horizontal cells. The inner plexiform layer (IPL) is where bipolar, amacrine, and ganglion cells interact and form synapses and the axons of ganglion cells converge to form the optic nerve. Integrity of retinal cell function and their organization are critical for visual function. XLRS, an early form of visual impairment in young males is caused by mutations in the X-linked RS1 gene. XLRS pathology is characterized by splitting of inner retinal layers and is mainly seen at the OPL and in the bipolar cell layer (inner nuclear layer). SD-OCT is a noninvasive diagnostic technique that uses light waves to take cross-section pictures of the retina at ∼20,000–40,000 scans per second and 3-μm resolution. With OCT, each of the retina’s distinctive layers can be seen allowing mapping and measuring their thickness. In contrast to the normal eye, an XLRS-affected eye shows prominent splitting of the inner nuclear layer (INL) and abnormally thin outer nuclear layer (ONL). [We thank C. A. Cukras (National Eye Institute, Bethesda, MD) for providing retina images]. The construct used in this paper is tagged at the C terminus with six histidines to aid in purification. This is expected to be active in vivo based on a related gene-therapy experiment done with a C-terminal FLAG tag. An AAV8 vector that contained the human RS1 cDNA with a FLAG tag (sequence: DYKDDDDK) at the C terminus driven by a CMV promoter was administered into the left eye of an Rs1-null mouse (Rs1-KO) by intravitreal injection at 40 d of age (P40). The contralateral right eye was injected with PBS. Retinal morphology and functions were analyzed 60 d postinjection (P100). Retinal sections were examined for RS1-FLAG expression by fluorescent microscopy, with expression most intense in the photoreceptor layer and in bipolar cells. The ERG of the PBS-injected control eye (red curve) shows a suppressed b-wave amplitude typical for Rs1-KO mice (∼672 µV), whereas the AAV-CMV-RS1-FLAG injected eye (blue curve) reverted to a normal b-wave amplitude similar to the wild type (green curve) (∼1120 µV). Therefore, the tagged RS1 protein augmented signaling across the rod-to-bipolar cell synapses that lie within the OPL. These results demonstrate: (i) that the injection of an adeno-associated virus (AAV)-based RS1-FLAG vector is able to transduce the retina and rescue the XLRS disease phenotype, and (ii) the FLAG tag does not influence the in vivo function of RS1 protein. GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; IS, inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer.
RS1 is secreted as a soluble disulfide bond-stabilized octamer (14, 15). Most of the monomer comprises a discoidin (DS) domain, a globular fold that is highly conserved over many proteins with diverse functions (16). Little progress has been made toward understanding the molecular mechanisms underlying the disease because of difficulty in producing the protein in sufficient quantities (5). In the absence of experimentally derived information, homology models of the RS1 monomer have been formulated based on existing X-ray structures of other DS domains (17–21). However, the arrangement of subunits in the putative octamer remained an open question. In addition, RS1 differs from the other junctional proteins listed above in that it has no obvious membrane-insertion elements.
In this study we were able to purify RS1 in sufficient quantity to support structural analysis by cryo-electron microscopy (cryo-EM). Here we present the structure of the mature RS1 as a 16-mer in a dihedral eightfold arrangement at a sufficient resolution in the DS domain (4.1 Å) to locate key elements, in particular the intramolecular and intermolecular disulfide bonds stabilizing the oligomer. The resolution for the RS1 domain is lower (∼9 Å), indicative of mobility, but suffices to position these domains relative to the DS rings. Consideration of the locations of some disease-related mutants indicates that their phenotypes are caused either by disrupting the DS fold or by affecting assembly of the oligomer. The symmetry of the double-ring shape favors a junctional model where the flat surfaces interact with apposing membranes.
Results
Recombinant RS1 Production.
The key to producing adequate quantities of RS1 was in appending an effective affinity tag. The mature RS1 cDNA (coding for residues 24–224) was cloned with a C-terminal 6His tag, expressed in insect cells, and purified by affinity chromatography as a secreted protein in the cell-culture medium. An N-terminal deletion mutant was also produced (truncated before residue 58). Affinity tags at both ends of the sequence have been shown to yield secreted oligomeric species: an myc tag inserted after the signal sequence (22) and a FLAG tag at the C terminus (23). The C-terminal FLAG tag was shown not to interfere with normal RS1 biological functioning (Fig. S1). The histidine-tagged protein used here (Fig. S2) is therefore not expected to behave differently from the wild-type. On a blue native gel, the protein ran as a single band at about 400 kDa (Fig. 1A), roughly twice the size expected for a single octamer.
Fig. S2.
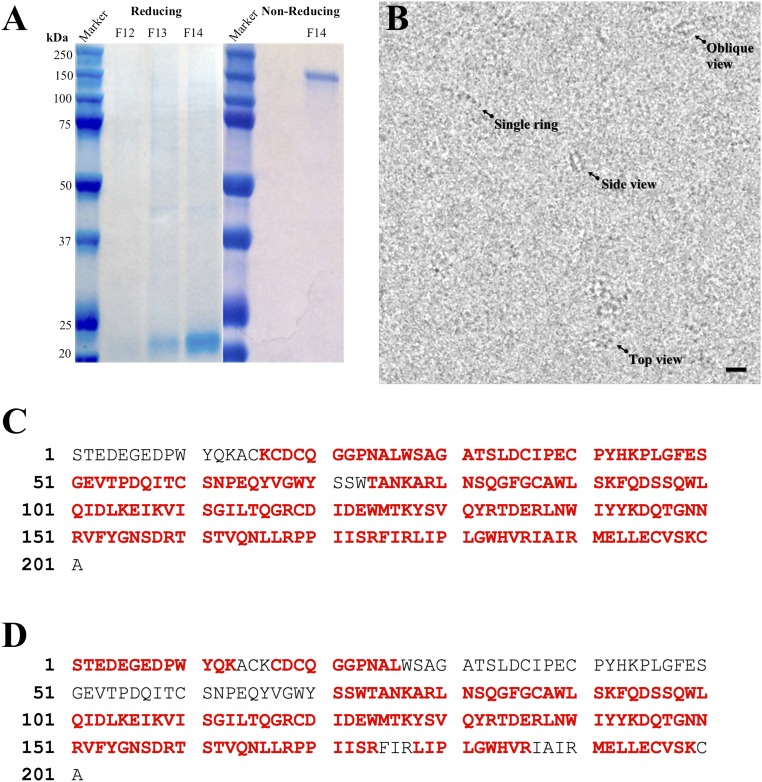
Identification of purified RS1-6His. (A) SDS/PAGE of purified RS1-6His in fractions eluted from the Mono Q column (F12–F14), showing the ∼23-kDa monomer in the reduced gel and the ∼180-kDa octamer in the nonreduced gel. (B) Field of RS1 particles embedded in vitrous ice, showing top, oblique, and side views of the double rings and few single rings. (moderately low-pass–filtered to improve visualization). (Scale bar, 100 Å.) (C) Coverage of the sequence (red: 90%) analyzed by LC-MS/MS of a chymotrypsin digest. (D) Coverage of the sequence (red: 72%) analyzed by LC-MS/MS of a trypsin digest. RS1-6His was expressed in baculovirus-infected Sf9 insect cells, and the protein secreted into the culture medium was purified on His Pur Cobalt resin followed by anion-exchange chromatography on a Mono Q column, as described in Methods. RS1-6His was eluted with a salt (NaCl) gradient in 20 mM Tris⋅HCl (pH 8.0) buffer and fractions run with β-mercaptoethanol (reducing) or without (nonreducing), stained with Coomassie blue. In the reducing gel, the major band at ∼23 kDa is the RS1 monomer, with minor bands corresponding to oligomers. Molecular weight markers are shown on the left side in each panel. The RS1-6His band was excised from a gel, reduced with DTT and alkylated with iodoacetamide. Half of the sample was digested in the gel with trypsin, and the other half with chymotrypsin. The digests were desalted using a Waters HLB µElution plate. The LC-MS/MS experiment was performed on an Orbitrap Elite Mass Spectrometer (Thermo Scientific) coupled to an Ultimate 3000 HPLC (Thermo-Dionex). Peptides were separated on an ES800 Easy-Spray column (75 μm × 15 cm, 3 μm C18 beads). The mobile phases were composed of 0.1% formic acid in 2% (vol/vol) acetonitrile (MPA) and 0.1% formic acid in 98% acetonitrile (MPB). The gradient began at 2% MPB rising to 27% MPB in 25 min at a flow rate of 300 nL/min. MS scans were acquired using an m/z range of 300–2,000, 60k resolution at m/z 400. MS/MS scans were acquired on the top five most abundant precursor ions using an isolation window of 1.9, CID fragmentation with decision tree method activated, an intensity threshold of 50,000 counts, and a dynamic exclusion of 9 sec Xcalibur RAW files were converted to peak list files in mgf format using Mascot Distiller (v2.5.1.0). The database search was performed using Mascot Daemon (2.4.0) against an in-house database, which contains the mature RS1 (Retinoschisin) protein sequence and the NCBI human database. The false discovery rate was less than 1%.
Fig. 1.
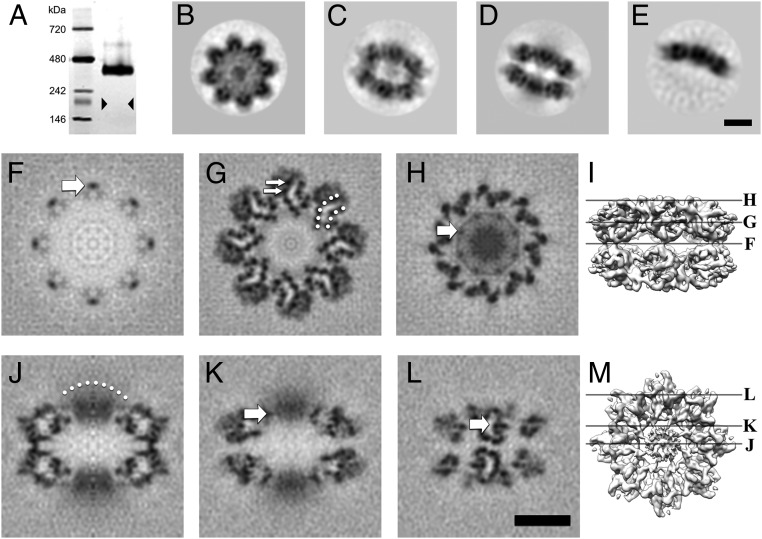
RS1 is predominantly a 16-mer arranged as back-to-back octamer rings. (A) A silver-stained blue native gel showing an oligomer of ∼400 kDa, much larger than an octamer (triangles indicate the expected band position). (B–E) Selected 2D class averages derived from micrographs of frozen-hydrated RS1 particles: (B) top view average; (C) oblique view average of double octamer rings; (D) side view average of double octamer rings; (E) side view average of single octamer rings. (F–M) Selected slices through the 3D reconstruction. (F) Central slice at the interface between the two octamer rings, the arrow indicating one of the narrow connections between the rings. (G) Slice 16.5 Å from the center, with the two β-sheets indicated by dotted arcs and two β-strands indicated by arrows. (H) Slice 33 Å from the center, the arrow indicates a node in the ring (the site of an intermolecular disulfide bond) around the diffuse density in the middle (8 RS1 domains). (I) Guide to the slice levels in F–H. (J) Central slice on a twofold axis, the dotted arc indicating a diffuse density composed of eight N-terminal RS1 domains. (K) Slice 10 Å from the center showing the node (arrow) corresponding to that in H. (L) Slice 48 Å from the center, showing the two curved β-sheets with low density in between (arrow). The latter is occupied by bulky side chains in the core of the domain not resolved in this map. (M) Guide to the slice levels in J–L. (Scale bars, 50 Å.)
RS1 Forms a Double-Ring Oligomer.
Cryo-electron micrographs of purified RS1 (Fig. S2B) show numerous double-line particles, some ring-like structures, and a few single-line particles. We interpret the double-line particles as side views of a two-layered structure, and the rings as top views. Two-dimensional classification and averaging indicated several classes; some examples are shown in Fig. 1 B–E. The top views have a typical eightfold appearance (Fig. 1B), consistent with single octamers or double octamers stacked in register. Fig. 1C shows an oblique view and Fig. 1D a side view, which is clearly a double-layered particle. Side views of a single-layered particle were a small minority (∼8%) (Fig. 1E). The 16-mer is consistent with the size observed in the blue native gel (16 × 24 kDa = 384 kDa) (Fig. 1A).
The N-Terminal RS1 Domain Is Not Required for Oligomerization.
To localize the RS1 domain and assess its influence on oligomerization, a mutant lacking the N-terminal 34 residues of the mature protein was produced and imaged by negative-stain electron microscopy. Micrographs of both the full-length wild-type (Fig. S3A) and the deletion mutant (Fig. S3B) show both top and side views, consistent with the double octameric ring structure. To examine the differences, the images were first subjected to 2D classification, yielding top- and side-view classes (Fig. S3 C and D). The prominent difference is the lower density in the middle of the mutant top view (Fig. S3D) compared with the wild-type (Fig. S3C). Radial profiles indicate that the density in this region of the mutant is at the same level as the background (Fig. S3E). Three-dimensional reconstructions further clarified the location of the RS1 domains (compare Fig. S3 F and G, blue arrows).
Fig. S3.
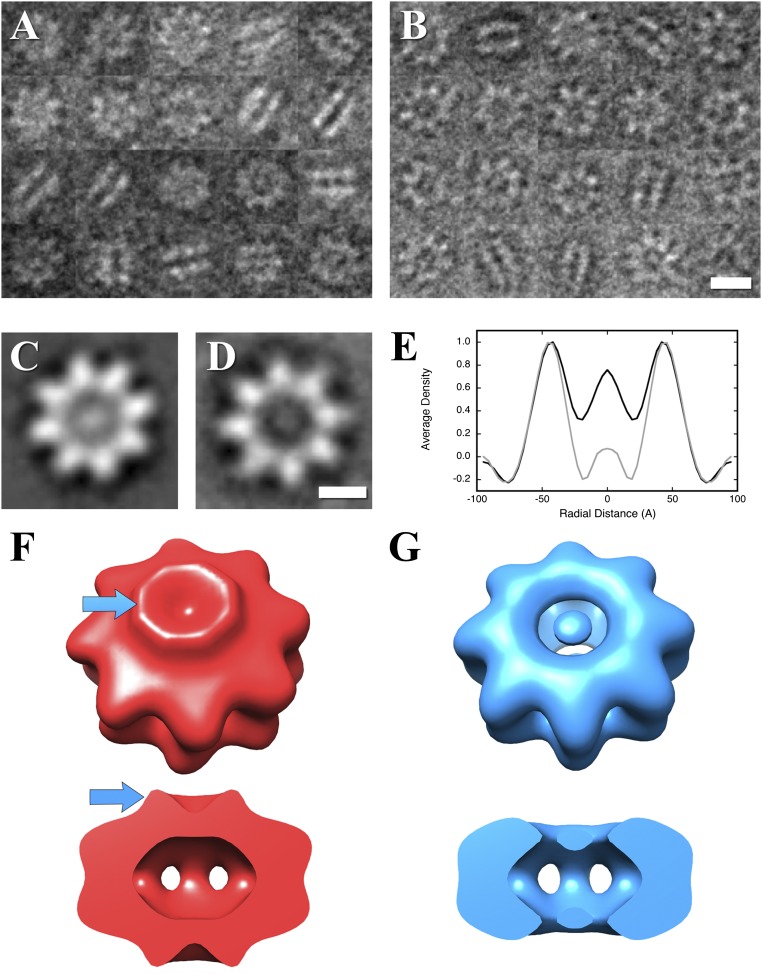
The RS1 domain does not affect oligomerization. (A) Negative-stain electron micrograph images of RS1 wild-type. (B) Negative-stain electron micrograph images of an N-terminal deletion mutant, RS1-∆N. (Scale bar, 100 Å.) (C) Two-dimensional class average of wild-type top views, showing a central density. (D) Two-dimensional class average of mutant top views without a central density. (Scale bar, 50 Å.) (E) Radial average profiles of the wild-type and mutant top views show that the density in the center of the mutant protein (radius 0–20 Å) is similar to the background (radius 70–95 Å). Side views in A and B show both wild-type and mutant to be double-ring structures for the great majority of the particles. (F) Reconstruction of the wild-type particle showing the RS1 domains as a ridge (blue arrows). (G) Reconstruction of the N-terminal deletion mutant, RS1-∆N, showing the absence of the RS1 domains.
High-Resolution Map of RS1.
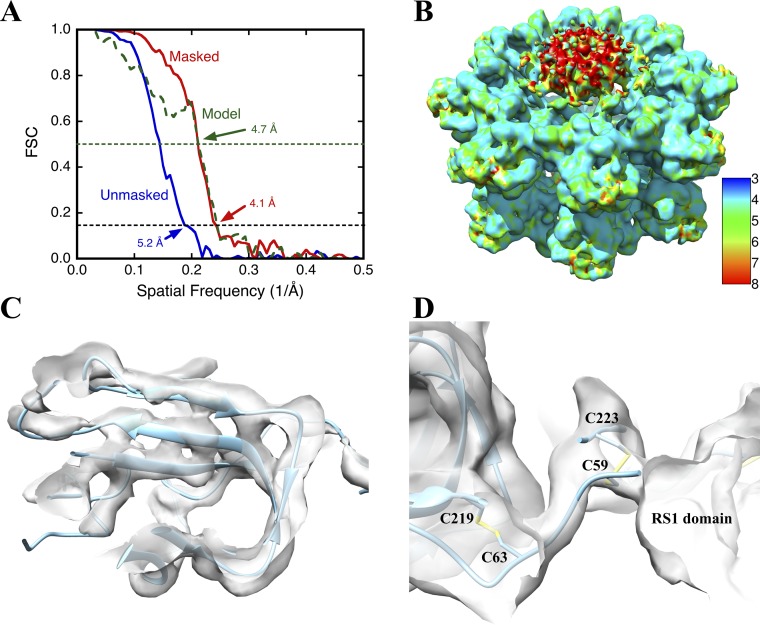
The cryo-EM data were used to determine a 3D structure with imposed dihedral eightfold symmetry (D8), yielding a global resolution of 5.2 Å (Fig. S4A). The two octameric rings are composed of detailed globular subunits. Masking the map to include only these structures yielded a resolution of 4.1 Å (Fig. S4A), also reflected in local resolution analysis (Fig. S4B). Sections through the map are shown in Fig. 1 F–M. The connections between the octameric rings are narrow (Fig. 1 F and J), although the quality of the map indicates that the avidity of the eight interactions is sufficient for stable association of the two rings. Each ring is composed of eight subunits structurally similar to canonical DS domains, with clearly distinguishable β-strands arranged in two β-sheets (Fig. 1G and Fig. S4C). A relatively diffuse region of density protrudes from the center of each ring (Fig. 1 H–K). This region, which has appreciably lower resolution than the DS rings (Fig. S4B), accounts for the RS1 domains. The diffuse nature of this density and its persistence through the eightfold axis suggests that the RS1 domains are not strictly symmetry-related, precluding resolving high detail. Fig. 1H shows a ring of density between the DS and RS1 domains, with nodes interpreted as sites for the intermolecular disulfide bonds formed by C59 and C223 (arrows in Fig. 1 H and K). Within each DS domain, the two β-sheets are evidently separated by a cavity (Fig. 1G).
Fig. S4.
Quality of the RS1 map. (A) The resolution of the RS1 reconstruction was estimated by Fourier shell correlation (FSC) between two half-maps unmasked (blue) and masked (red), using the 0.143 cut-off (black dashed line). Also shown is the FSC curve (green) between the full map (masked) and density generated from the fitted model, using a cut-off of 0.5 (green dashed line) to estimate the agreement. The mask was generated with a soft edge and covered only the DS domains. (B) Local resolution analysis of the RS1 map. The scale indicates the color-mapping of resolution values of 3 Å to 8 Å. The N-terminal RS1 domain in the center of the ring shows lower resolution (∼9 Å, red parts), consistent with a partially ordered structure. The rest of the structure is around 4 Å, sufficient to resolve β-strands. (C) Illustration of map detail. The three strands of one β-sheet are clearly resolved (separation distance of 4.7 Å; isosurface at 3.5σ). (D) The intrasubunit disulfide bond (C63–C219) is readily formed within the DS domain fold. The intersubunit disulfide bond (C59–C223) fits into a density node between subunits.
Homology Modeling and Defining Disulfide Bonds.
The 16 subunit densities in the map are unambiguously recognizable as having the DS fold. The RS1 DS domain has been modeled several times with confidence based on the high conservation of the fold (18–20). We used this approach in conjunction with the cryo-EM map, starting from a computationally derived model in the SWISS-MODEL repository (swissmodel.expasy.org) (24).
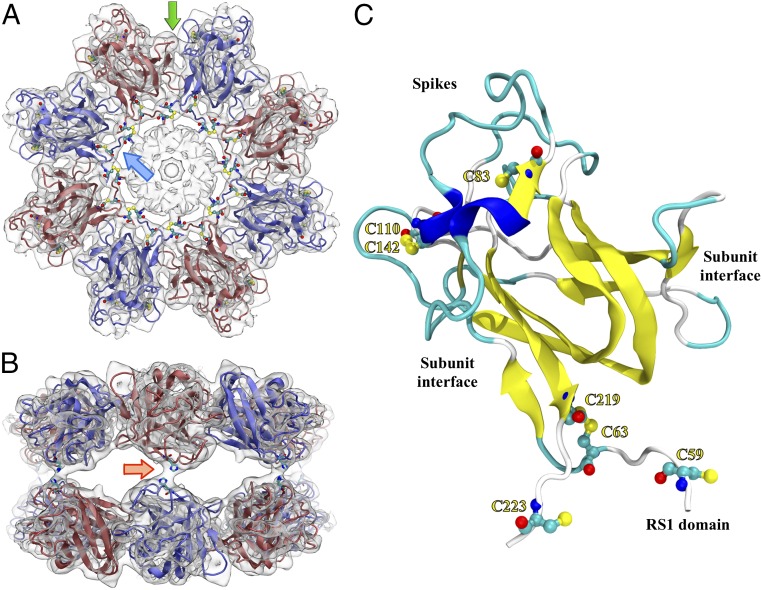
Of particular importance to the modeling was the contribution of the disulfide bonds to the DS fold and oligomeric assembly (14, 17). The highly conserved internal disulfide, C63–C219, and the C110–C142 bond were readily compatible with the model (a distance of ∼2 Å between the sulfur atoms). The N-terminal chain was extended to residue 58 and built into the ring density shown in Fig. 2C. This positioned C59 close enough to C223 of the neighboring subunit to form an intermolecular disulfide bond (at the node indicated in Fig. 2C; also see Fig. S4D). The N-terminal part of the polypeptide chain could not be traced further into the central density. Sixteen copies of this subunit model (residues 58–224) were built into the cryo-EM map. Explicit disulfides were generated for C59–C223 (between subunits), C63–C219, and C110–C142. This satisfies the constraints as given in Wu et al. (14).
Fig. 2.
Top (A) and side (B) views of the RS1 double octamer with one subunit model highlighted (C). (A) The blue arrow points to a rod-like density connecting neighboring subunits between the DS and RS1 domains. All cysteine residues are indicated, including the intramolecular disulfides, C63–C219 and C110–C142. C59 and C223 form disulfide bonds with neighboring subunits. The green arrow points to a subunit-subunit interface further detailed in Fig. 3. (B) The RS1 side view shows the connections between the octameric rings (one indicated by the red arrow). H207 is located in this contact site and is potentially involved in the interaction. (C) The monomer ribbon is colored to indicate β-strands (yellow), helices (blue), and coil (cyan) secondary structure.
The 16 DS-domain structure was refined by “flexible fitting” (25). Briefly, the process involved rebuilding parts of the structure to fit better into the experimental density, and refining it using molecular dynamics with the map as an added constraint. The resulting structure exhibits the core of the DS domain: two β-sheets packed against each other. The C59–C223 disulfides are arranged in a ring in each octamer between the RS1 and DS domains (blue arrow in Fig. 2). The two octameric rings are held together by connecting densities located on the dihedral twofold axes (red arrow in Fig. 2). The model was converted to a density map, and the agreement between the experimental and computed density maps assessed to be ∼4.7 Å (Fig. S4A).
Discussion
RS1 is unique among known cell-adhesion proteins in that it is secreted from retinal cells and has no evident membrane-insertions. Its importance is underscored by the dramatic structural disruption of the retina caused by the many mutations that lead to XLRS. An octameric state of RS1 was first described by Wu and Molday (17) and is taken to be one of the characteristics, together with secretion, of a functional protein. It is, however, hard to envisage how a single octameric ring would form a junction between opposing cellular membranes. Here we show that the assembled state of the mature wild-type protein is, in fact, a double octamer. In this context, the native gel analysis showed a complete absence of single octamers (Fig. 1A), suggesting that the few single-ring particles we observe by EM (Fig. 1E) are products of dissociation during EM grid preparation. As discussed below, the back-to-back double rings of RS1 present a structure with the symmetry needed to function as a homomeric junctional protein.
The Ring–Ring Interface.
The connections between the two rings appear tenuous (Fig. 1F), but suffice for a stable complex. The residues in the contact regions are 147–148 and 206–210. Of note, mutants in H207 and R209 still form octamers and are secreted (Table 1) (26). Several XLRS-related mutants have been described for R209 (27–29). These basic residues may be sensitive to their electrostatic environment, which would explain why the major species observed in sedimentation analysis is a single octamer (14).
Table 1.
The interfaces in the RS1 double octamer mapped to disease-related mutants and whether they are secreted as octamers
The Intermolecular Disulfides.
Both inter- and intramolecular disulfide bonds stabilize the octameric ring (14, 17). The internal C110–C142 bond was detected in previous homology models and is thought to stabilize the spikes protruding from the DS fold (17). Similarly, the emergence of the N- and C-termini on the same side of the DS domain places them close enough together to form the C63–C219 bond. At this point the RS1 map shows a rod-like density connecting neighboring subunits (blue arrow in Fig. 2). This is interpreted as the extension of the N terminus from C63, and is compatible with the formation of an intermolecular bond, C59–C223. Mutational studies indicated that both C63–C219 and C59–C223 bonds are involved in octamer formation, as mutants are secreted as mono- and dimers at lower levels (14).
The Interface Between Subunits in a Ring.
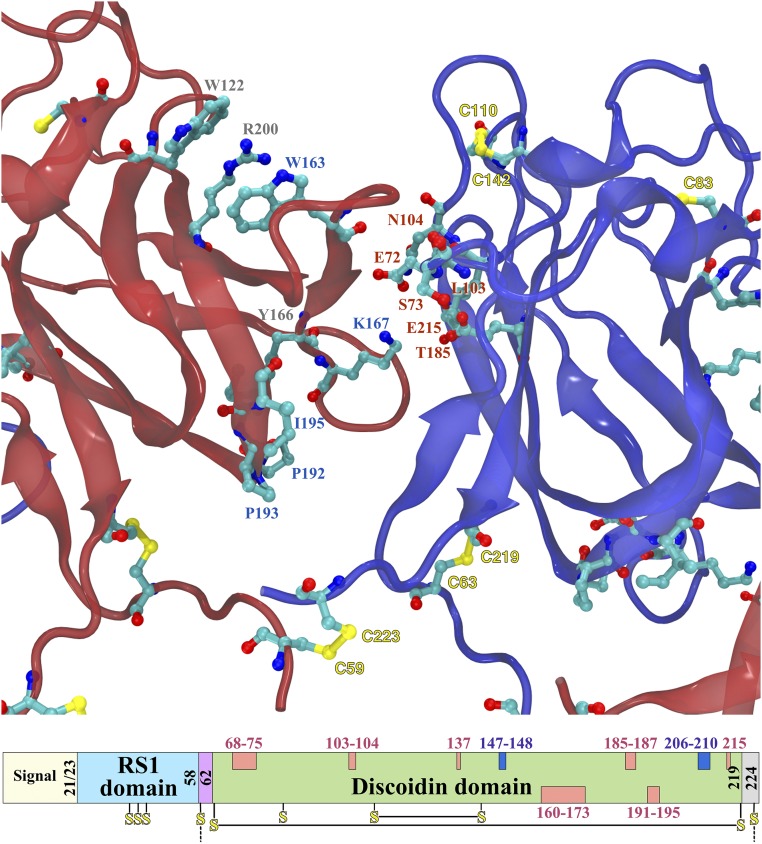
The structure determined for the DS domains is of sufficient quality that the positions of most residues may be assigned with confidence (Figs. 2 and 3). To identify residues involved in the intersubunit interface in the ring, the accessible surface of the monomer was compared with that of the octamer. Two loops appear to be buried in the interface on one side, each between pairs of β-strands (left side in Fig. 3). The first is a long stretch from 158 to 172 that is in close contact with the neighboring subunit. The second is a shorter sequence, 191–195, that is close enough to the neighboring subunit to exclude solvent, but may not be in direct contact. On the other side, several motifs contribute to the interface (right side in Fig. 3). It starts with a short strand-loop from 67 to 74, then residues 103 and 104 on a loop, and finally a surface of the β-sheet formed by 137, 139, 183–187, and 215.
Fig. 3.
XLRS-related mutations in the subunit interface and the corresponding positions in the sequence. The interface between adjacent subunits within an octameric ring (as seen from center) is composed of a long loop on the left subunit (red) and mainly the β-sheet face on the right subunit (blue) composed of residues 137, 139, 183–187, and 215. The interface loop (160–173) on the left are likely stabilized by the “sandwich” W122-R200-W163, and the interaction of Y166 with the 192–195 loop. A potential salt bridge can be formed between K167 and either E72 or E215. In the domain scheme at the bottom, the sequence locations of the subunit interfaces in the DS domain are shown in pink (those at the top are for the right side and those at the bottom for the left side). Residues potentially involved in the octamer ring connections are shown in dark blue.
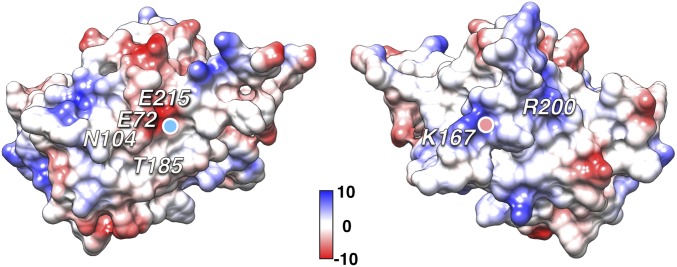
Mutations in many of the residues in the intersubunit interface are associated with XLRS. The side of the interface with the two loops feature debilitating mutations W163C, Y166H, K167E, P192S, P193L/S, I194N, and I195V (Fig. 3) (30, 31). The importance of W163 has been noted before in that it forms part of stacked “sandwich” with residues R200 and W122, likely stabilizing the long loop (158–172), coupling it to the core of the DS domain (20). The opposing side of the interface similarly includes several residues with XLRS-associated mutations: E72K, S73P, G74V, L103F, N104K, T185K, and E215V (Fig. 3). Many of the mutants involve a change in charge in the interface, and the interacting coulombic surfaces show charge complementarity (Fig. 4). Of note is K167, which fits into a pocket lined by E72 and E215 on the neighboring subunit, probably forming a salt bridge.
Fig. 4.
Charge complementarity of the two interacting RS1 subunit surfaces. The Coulombic surfaces of two subunits are shown with their N and C termini facing each other. Charge-altering disease-linked mutants in the interface residues include E72K, N104K, T185K, E215K (Left) and K167E (Right). The lysine 167 fits into the negatively charged pocket formed by the glutamates 72 and 215 (the light blue dot on the left surface indicates where K167 fits, and the pink dot on the right surface indicates where E72 fits). Arginine 200 (Right surface) does not interact with a neighboring subunit, but rather stabilizes interface residues on the ridge to the left of it. Scale in kcal/mol.e−. [Calculated in UCSF Chimera with default settings for Coulombic surfaces (48).]
The RS1 Domain.
The RS1 domain is not required to produce a double octameric ring (Fig. S3). These domains are less well resolved in our map (Fig. 1), and it may not have a unique structure. The three cysteines (C38, C40, and C42) have been implicated in oligomerization (14), but no mutants leading to disease have been reported. The density in the center of each ring represents eight RS1 domains, giving a total of 24 cysteines in close proximity. It is possible that these can cross-link in multiple ways, giving rise to a combinatorially complicated and nonunique structure. If so, the function of the protein may not rely on a specific structure for the RS1 domain. To our knowledge, no missense mutations linked to XLRS have been identified in the RS1 domain, whereas other mutations tend to result in downstream premature termination and a missing or nonfunctional protein. Thus, the role of this domain remains obscure.
The “Spikes.”
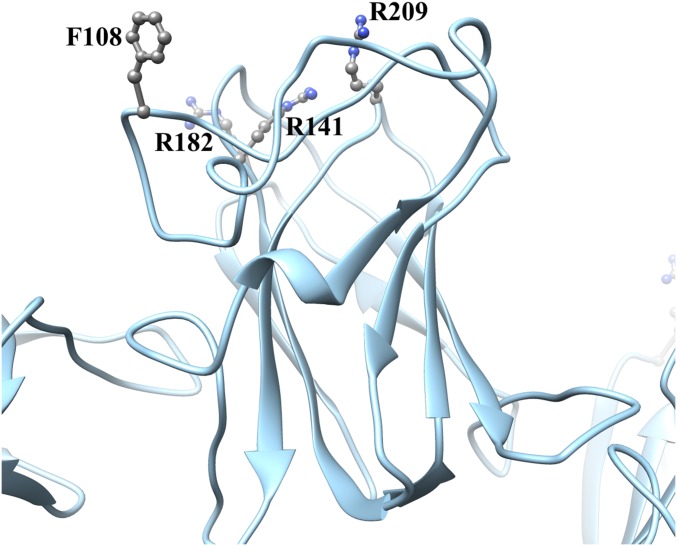
Opposite the N and C termini and pointing outwards from the octameric rings are loops that vary among DS domain proteins. Macedo-Ribeiro et al. (32) described the corresponding loops of the C2 domain of human coagulation factor V as three “spikes.” These spikes are commonly associated with the function of DS domain proteins, in particular ligand binding (16). Disease-linked mutations yielding secreted oligomeric RS1 (22, 26) occur mostly in this region (F108C, R209H, R141A/G/H/S, R182C) (Fig. S5). The most prominent loop has at its tip residues 87–93 (corresponding to spike 1), including the bulky Y89 and W92 associated with XLRS and implicated in binding to phospholipids (33). Several DS domain proteins bind to membranes, and in several cases it was proposed that hydrophobic residues in the spikes embed in the membrane [C2 domains of factor V (32), factor VIII (34), and lactadherin (35)]. A similar mechanism for the interaction of RS1 with a membrane was proposed (20). However, the double-ring structure of RS1 does not fit this interpretation. An alternative mechanism would involve the carbohydrate moieties of proteins and lipids binding laterally to the spikes, with the flat surface of the rings facing the membrane planes. Such lateral interactions may also include other participants, such as the Na/K ATPase (36), and with R141 implicated in binding of galactose (37) and an L-type voltage-gated calcium channel (38). However, a structural basis for such a mechanism still needs to be established.
Fig. S5.
Four XLRS-linked mutants located in the RS1 spikes. Mutants in these residues are in general secreted, with the exception of R141C/V/Q/E/K (26).
Requirements for the Functional State of RS1.
RS1 is produced as a disulfide-linked octamer in the endoplasmic reticulum (15). Many of the XLRS-related mutants do not manage to exit from the endoplasmic reticulum, because of defects in synthesis, folding, or assembly. The intersubunit interfaces, as well as the stabilizing disulfides, play key roles in the proper constitution of the octamer that is then secreted. The most similar other junctional protein is connexin, where the monomers are packed into arrays of hexamers in each of the opposing membranes, with sixfold interactions between each pair to form a channel (4). We envisage RS1 octamers forming the two halves of a junctional complex, packed in an array to increase the avidity of cell–cell interaction.
Immunoelectron microscopy located an abundance of RS1 between closely packed photoreceptor inner segments, where the separation between membranes is on the order of 100–200 Å (33). The connections between cells can be very complex, as manifested in the many proteins involved in tight junctions (2). Similarly, RS1 does not function in isolation, with other possible participants being charged lipids (33) and glycoproteins (36, 38, 39). In our scheme, the protruding loops (spikes) then interact laterally with these other actors, forming a densely packed assembly of proteins sandwiched between the membranes from opposing photoreceptor or bipolar cells. This places the RS1 domains closest to the membranes in a suitable orientation for interaction with surface charges or receptors. Future studies will need to focus on the molecular determinants of the structural and functional organization of RS1 in retinal membranes.
Methods
Cloning, Expression, and Purification.
Two versions of RS1 cDNA were cloned into bacmid vectors with N-terminal honey bee mellitin leader sequences (to ensure secretion), and C-terminal hexahistidine (6His) tags: (i) mature RS1 (residues 24–224 + 6His) and (ii) an N-terminal deletion mutant (residues 58–224 + 6His). The proteins were expressed in Sf9 cells and purified from the cell culture medium using a cobalt-agarose column, followed by a clean-up with a Mono Q column. The RS1-6His band (Fig. S2A) was excised from an SDS/PAGE gel and identified by LC-MS/MS (Fig. S2 C and D). The samples were finally concentrated by dialysis and spin column in preparation for microscopy (see SI Methods for details).
Blue Native Gel Electrophoresis.
A pooled and dialyzed fraction of RS1 from the Mono Q column was loaded onto a Native PAGE 3–12% Novex Bis-Tris gel and run in a XCell SureLock system (Invitrogen) in NativePAGE sample buffer with G-250 sample additive. The gel was stained with silver (Silver Express Silver Staining Kit, Invitrogen) and scanned with a LI-COR Odyssey Infrared Imaging System (Model 9120, LI-COR, Biosciences).
Negative-Stain Electron Microscopy.
Negatively stained samples were prepared by incubating a drop of purified RS1-6His or Ndel-RS1-6His solution (∼0.2 mg/mL) on a glow-discharged, carbon-coated, copper grid for 1–2 min, blotting the excess liquid off and staining with a 2% (wt/vol) uranyl acetate solution for 30 s. Micrographs were recorded on a Tecnai 12 electron microscope (FEI) equipped with a 2 K × 2 K energy-filtered CCD camera (Gatan) at 34,000× nominal magnification (79,000× at the camera, pixel size 3.8 Å). The EMAN2 software package (40) was used to pick particles and generate 2D class averages.
Cryo-EM.
RS1-6His samples (∼0.2 mg/mL) were applied to the carbon side of plasma-cleaned 2.0-µm hole, 2.0-µm space C-flat holey carbon grids (Protochips), and blotted from the copper side (i.e., backside). These were vitrified in a LEICA EM GP plunge freezing robot (Leica) at room temperature and 90% humidity. Dose-fractionated images were acquired using SerialEM (41) on a Polara F30 electron microscope (FEI) equipped with a Gatan K2 Summit direct electron detector camera (Gatan) using counting mode at 39,000× magnification, corresponding to a calibrated 1.03 Å per pixel scale. The dose rate was ∼10 e− per pixel per second for 3 s to give a total dose of ∼30 e−/Å2.
Image Processing.
The dose-fractionated images were aligned and summed using software provided by Li et al. (42). The EMAN2 software package (40) was used for picking 11,346 particles (224 × 224 pixels). Two-dimensional class averages were calculated using RELION v1.3 (43). An initial model was generated from the 2D top- and side-view class averages using EMAN2, and low-pass–filtered to 60 Å. Particles that were assigned to 2D classes showing only single rings or diffuse averages were excluded from further processing, bringing the final particle count to 9,096 (80%). On the basis of the double-ring structure in the remaining 2D class averages, D8 symmetry was imposed during 3D refinement iterations with RELION, also used for postprocessing the map (i.e., masking and filtering). Programs included in the RELION package were used for determining CTF parameters, CTFFIND 3 (44), and local resolution, ResMap (45). The overall resolution was determined by the gold standard FSC curve (46). Bsoft (47) was used as a toolbox for performing intermediary operations during the overall processing and reconstruction. The map was deposited in the EM Data Bank (www.ebi.ac.uk/pdbe/emdb, EMD-6425).
Homology Modeling.
A homology model of the RS1 monomer downloaded from the SWISS-MODEL website (swissmodel.expasy.org) (24) was fitted manually into the DS domain density of the 3D map using University of California, San Francisco (UCSF) Chimera (48). The N terminus was extended from L68 to D58 within a rod-like density extending from the DS domain to the neighboring subunit. The C terminus was also extended, from C219 to A224, positioning C223 close to C59 of a neighboring subunit. All 16 monomers were positioned within the density and the intramolecular (C110–C142 and C63–C219) and intermolecular (C59–C223) disulfides were connected. The full 16-mer model was then subjected to interactive MDFF using NAMD2 (25) and manual-modeling using COOT (49) and FoldIt (fold.it/portal/) (50). This process was iterated until it converged to a good fit into the electron density and satisfied most geometric constraints, as assessed using COOT (49) and MolProbity (51). The final MolProbity score is 2.26, better than the estimated resolution of the map at 4.1 Å. The final structure was deposited in the Protein Data Bank (www.rcsb.org/pdb, 3JD6).
SI Methods
Cloning of RS1.
Regions from a pTOPO-RS1 vector (12) were amplified using PCR (Verti Thermal Cycler, Applied Biosystems/Life Technologies) with a primer coding for a hexahistidine tag (6His) on the C terminus. Two versions of RS1 were generated: (i) mature RS1 (residues 24–224 + 6His) and (ii) an N-terminal deletion mutant (residues 58–224 + 6His). An adapter primer coding for the honey bee mellitin (HBM) leader sequence was added partway through the PCR amplification to ensure secretion. The PCR products were recombined into the Gateway donor vector, pDonr253, using the Gateway BP recombination reaction. The sequences of the resulting Entry clones (11109-E01 HBM-RS1aa24-224–6xHis; 11110-E02 HBM-RS1aa57-224–6xHis) were verified throughout the entire cloned region. These were subcloned by Gateway LR recombination into pDest-8 to create clone 11109-X01 (RS1-6His) and 11110-X-02 (Ndel-RS1-6His) for insect cell expression. The expression clone was transformed into Escherichia coli DH10Bac, and plated on selective media containing gentamycin, kanamycin, tetracycline, IPTG, and X-gal, as per the manufacturer’s protocols. White colonies were selected from these plates, and bacmid DNA was generated by alkaline lysis plasmid preparation and verified by PCR amplification across the bacmid junctions.
Expression and Purification of RS1.
Bacmid DNA was transfected into 1 × 107 Sf9 insect cells using polyethyleneimine, and baculovirus supernatant was harvested after incubation at 27 °C for 72 h. Supernatant (1 mL) was transferred to 50 mL High Five cells (1 × 107 cells/mL) and grown at 21 °C for 72 h. All purification steps were carried out at 4 °C, and purity was assessed using a 10% SDS/PAGE gel. The culture medium was extensively dialyzed against equilibration buffer (20 mM sodium phosphate pH 7.4, 0.5 M NaCl, and protease inhibitor mixture) and sedimented at 10,000 × g to remove the cellular debris. About 200 mL was loaded on a 10-mL cobalt-agarose column (HisPur Cobalt Resin, Thermo Scientific), washed with 10 volumes of buffer A (20 mM sodium phosphate pH 7.4, 0.5 M NaCl, 20 mM imidazole), and eluted with buffer B (buffer A but with 200 mM imidazole). Fractions with RS1-6His were pooled, dialyzed against buffer C (20 mM Tris⋅HCl, pH 8.0). About 15 mL was loaded on a Mono Q 5/50 GL column on an Äkta chromatography system (GE Healthcare Life Sciences), equilibrated with buffer C and eluted with a gradient of 0–1 M NaCl. Fractions were pooled, dialyzed against 20 mM Tris⋅HCl, 150 mM NaCl (pH 7.5), and stored at –80 °C. The RS1-6His band (Fig. S2A) was excised from an SDS/PAGE gel and identified by LC-MS/MS (Fig. S2 C and D). For electron microscopy, samples were concentrated by dialysis (Slide-A-Lyzer, 10K MWCO, Thermo Scientific) against dry silica (Spectra/Gel; Spectrum Labs), followed by centrifugation through a Centricon spin column (MWCO 100K, EMD Millipore).
Acknowledgments
We thank Dr. Emilios Dimitriadis (National Institute of Biomedical Imaging and Bioengineering) for assistance in assessing the initial protein preparations; Dr. Dennis Winkler (National Institute of Arthritis and Musculoskeletal and Skin Diseases) for assistance in installing and using SerialEM; and Dr. R. E. Anderson (University of Oklahoma) for helpful discussions. This work was supported by the Intramural Research Programs of the National Eye Institute, National Institute on Deafness and Other Communication Disorders, and National Institute of Arthritis and Musculoskeletal and Skin Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The cryo-EM map and the fitted structure reported in this paper have been deposited in the EMBL-EBI Protein Data Bank, www.ebi.ac.uk/pdbe/emdb (accession no. EMD-6425) and the Protein Data Bank, www.rcsb.org/pdb (accession no. PDB-3JD6), respectively.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519048113/-/DCSupplemental.
References
- 1.Al-Amoudi A, et al. The three-dimensional molecular structure of the desmosomal plaque. Proc Natl Acad Sci USA. 2011;108(16):6480–6485. doi: 10.1073/pnas.1019469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuse M. Molecular basis of the core structure of tight junctions. Cold Spring Harb Perspect Biol. 2010;2(1):a002907. doi: 10.1101/cshperspect.a002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haseloff RF, Dithmer S, Winkler L, Wolburg H, Blasig IE. Transmembrane proteins of the tight junctions at the blood-brain barrier: Structural and functional aspects. Semin Cell Dev Biol. 2015;38:16–25. doi: 10.1016/j.semcdb.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Sosinsky GE, Nicholson BJ. Structural organization of gap junction channels. Biochim Biophys Acta. 2005;1711(2):99–125. doi: 10.1016/j.bbamem.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Molday RS. Focus on molecules: Retinoschisin (RS1) Exp Eye Res. 2007;84(2):227–228. doi: 10.1016/j.exer.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Takada Y, et al. Retinoschisin expression and localization in rodent and human pineal and consequences of mouse RS1 gene knockout. Mol Vis. 2006;12:1108–1116. [PubMed] [Google Scholar]
- 7.Sauer CG, et al. Positional cloning of the gene associated with X-linked juvenile retinoschisis. Nat Genet. 1997;17(2):164–170. doi: 10.1038/ng1097-164. [DOI] [PubMed] [Google Scholar]
- 8.Weber BH, et al. Inactivation of the murine X-linked juvenile retinoschisis gene, Rs1h, suggests a role of retinoschisin in retinal cell layer organization and synaptic structure. Proc Natl Acad Sci USA. 2002;99(9):6222–6227. doi: 10.1073/pnas.092528599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou J, et al. Synaptic pathology and therapeutic repair in adult retinoschisis mouse by AAV-RS1 transfer. J Clin Invest. 2015;125(7):2891–2903. doi: 10.1172/JCI81380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tantri A, et al. X-linked retinoschisis: A clinical and molecular genetic review. Surv Ophthalmol. 2004;49(2):214–230. doi: 10.1016/j.survophthal.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Molday RS, Kellner U, Weber BH. X-linked juvenile retinoschisis: Clinical diagnosis, genetic analysis, and molecular mechanisms. Prog Retin Eye Res. 2012;31(3):195–212. doi: 10.1016/j.preteyeres.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng Y, et al. RS-1 gene delivery to an adult Rs1h knockout mouse model restores ERG b-wave with reversal of the electronegative waveform of X-linked retinoschisis. Invest Ophthalmol Vis Sci. 2004;45(9):3279–3285. doi: 10.1167/iovs.04-0576. [DOI] [PubMed] [Google Scholar]
- 13.Byrne LC, et al. Retinoschisin gene therapy in photoreceptors, Müller glia or all retinal cells in the Rs1h-/- mouse. Gene Ther. 2014;21(6):585–592. doi: 10.1038/gt.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu WW, Wong JP, Kast J, Molday RS. RS1, a discoidin domain-containing retinal cell adhesion protein associated with X-linked retinoschisis, exists as a novel disulfide-linked octamer. J Biol Chem. 2005;280(11):10721–10730. doi: 10.1074/jbc.M413117200. [DOI] [PubMed] [Google Scholar]
- 15.Gleghorn LJ, Trump D, Bulleid NJ. Wild-type and missense mutants of retinoschisin co-assemble resulting in either intracellular retention or incorrect assembly of the functionally active octamer. Biochem J. 2009;425(1):275–283. doi: 10.1042/BJ20091179. [DOI] [PubMed] [Google Scholar]
- 16.Kiedzierska A, Smietana K, Czepczynska H, Otlewski J. Structural similarities and functional diversity of eukaryotic discoidin-like domains. Biochim Biophys Acta. 2007;1774(9):1069–1078. doi: 10.1016/j.bbapap.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Wu WW, Molday RS. Defective discoidin domain structure, subunit assembly, and endoplasmic reticulum processing of retinoschisin are primary mechanisms responsible for X-linked retinoschisis. J Biol Chem. 2003;278(30):28139–28146. doi: 10.1074/jbc.M302464200. [DOI] [PubMed] [Google Scholar]
- 18.Sergeev YV, et al. Molecular modeling of retinoschisin with functional analysis of pathogenic mutations from human X-linked retinoschisis. Hum Mol Genet. 2010;19(7):1302–1313. doi: 10.1093/hmg/ddq006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sergeev YV, et al. Molecular modeling indicates distinct classes of missense variants with mild and severe XLRS phenotypes. Hum Mol Genet. 2013;22(23):4756–4767. doi: 10.1093/hmg/ddt329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraternali F, Cavallo L, Musco G. Effects of pathological mutations on the stability of a conserved amino acid triad in retinoschisin. FEBS Lett. 2003;544(1-3):21–26. doi: 10.1016/s0014-5793(03)00433-2. [DOI] [PubMed] [Google Scholar]
- 21.Wu JW, Liu HL. In silico investigation of the disease-associated retinoschisin C110Y and C219G mutants. J Biomol Struct Dyn. 2012;29(5):937–959. doi: 10.1080/07391102.2012.10507420. [DOI] [PubMed] [Google Scholar]
- 22.Dyka FM, Molday RS. Coexpression and interaction of wild-type and missense RS1 mutants associated with X-linked retinoschisis: Its relevance to gene therapy. Invest Ophthalmol Vis Sci. 2007;48(6):2491–2497. doi: 10.1167/iovs.06-1465. [DOI] [PubMed] [Google Scholar]
- 23.Vijayasarathy C, et al. Molecular mechanisms leading to null-protein product from retinoschisin (RS1) signal-sequence mutants in X-linked retinoschisis (XLRS) disease. Hum Mutat. 2010;31(11):1251–1260. doi: 10.1002/humu.21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37(Database issue):D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16(5):673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T, et al. Molecular pathology of X linked retinoschisis: Mutations interfere with retinoschisin secretion and oligomerisation. Br J Ophthalmol. 2006;90(1):81–86. doi: 10.1136/bjo.2005.078048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi T, et al. Four Japanese male patients with juvenile retinoschisis: Only three have mutations in the RS1 gene. Am J Ophthalmol. 2004;138(5):788–798. doi: 10.1016/j.ajo.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 28.Ma X, Li X, Wang L. Novel XLRS1 gene mutations cause X-linked juvenile retinoschisis in Chinese families. Jpn J Ophthalmol. 2008;52(1):48–51. doi: 10.1007/s10384-007-0488-4. [DOI] [PubMed] [Google Scholar]
- 29.Skorczyk A, Krawczyński MR. Four novel RS1 gene mutations in Polish patients with X-linked juvenile retinoschisis. Mol Vis. 2012;18:3004–3012. [PMC free article] [PubMed] [Google Scholar]
- 30.Hiriyanna KT, et al. Novel mutations in XLRS1 causing retinoschisis, including first evidence of putative leader sequence change. Hum Mutat. 1999;14(5):423–427. doi: 10.1002/(SICI)1098-1004(199911)14:5<423::AID-HUMU8>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 31.Chen J, et al. Novel mutations of the RS1 gene in a cohort of Chinese families with X-linked retinoschisis. Mol Vis. 2014;20:132–139. [PMC free article] [PubMed] [Google Scholar]
- 32.Macedo-Ribeiro S, et al. Crystal structures of the membrane-binding C2 domain of human coagulation factor V. Nature. 1999;402(6760):434–439. doi: 10.1038/46594. [DOI] [PubMed] [Google Scholar]
- 33.Vijayasarathy C, Takada Y, Zeng Y, Bush RA, Sieving PA. Retinoschisin is a peripheral membrane protein with affinity for anionic phospholipids and affected by divalent cations. Invest Ophthalmol Vis Sci. 2007;48(3):991–1000. doi: 10.1167/iovs.06-0915. [DOI] [PubMed] [Google Scholar]
- 34.Pratt KP, et al. Structure of the C2 domain of human factor VIII at 1.5 A resolution. Nature. 1999;402(6760):439–442. doi: 10.1038/46601. [DOI] [PubMed] [Google Scholar]
- 35.Lin L, Huai Q, Huang M, Furie B, Furie BC. Crystal structure of the bovine lactadherin C2 domain, a membrane binding motif, shows similarity to the C2 domains of factor V and factor VIII. J Mol Biol. 2007;371(3):717–724. doi: 10.1016/j.jmb.2007.05.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molday LL, Wu WW, Molday RS. Retinoschisin (RS1), the protein encoded by the X-linked retinoschisis gene, is anchored to the surface of retinal photoreceptor and bipolar cells through its interactions with a Na/K ATPase-SARM1 complex. J Biol Chem. 2007;282(45):32792–32801. doi: 10.1074/jbc.M706321200. [DOI] [PubMed] [Google Scholar]
- 37.Dyka FM, et al. Characterization and purification of the discoidin domain-containing protein retinoschisin and its interaction with galactose. Biochemistry. 2008;47(35):9098–9106. doi: 10.1021/bi800938g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi L, Jian K, Ko ML, Trump D, Ko GY. Retinoschisin, a new binding partner for L-type voltage-gated calcium channels in the retina. J Biol Chem. 2009;284(6):3966–3975. doi: 10.1074/jbc.M806333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedrich U, et al. The Na/K-ATPase is obligatory for membrane anchorage of retinoschisin, the protein involved in the pathogenesis of X-linked juvenile retinoschisis. Hum Mol Genet. 2011;20(6):1132–1142. doi: 10.1093/hmg/ddq557. [DOI] [PubMed] [Google Scholar]
- 40.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 45.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11(1):63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9(9):853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157(1):3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper S, et al. Predicting protein structures with a multiplayer online game. Nature. 2010;466(7307):756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kolb H. How the retina works. Am Scientist. 2003;91(1):28–35. [Google Scholar]